Abstract
Aquaporins mediate rapid selective water transport across biological membranes. Elucidation of their precise physiological roles promises important insight into cellular and organismal osmoregulation. The genome of the yeast Saccharomyces cerevisiae encodes two similar but differentially regulated aquaporins. Here, we show that expression of AQY1 is stimulated during sporulation and that the Aqy1 protein is detectable exclusively in spore membranes. When spores are rapidly frozen, those that lack Aqy1 survive better, providing for a functional test of active spore water channels. Under ambient conditions, lack of Aqy1 reduces spore fitness. Because this reduction is independent from germination conditions, Aqy1 may be important during spore formation rather than subsequent maintenance or germination. Indeed, it seems that Aqy1 is degraded after spores have been formed and during germination. Taken together, Aqy1 is developmentally controlled and may play a role in spore maturation, probably by allowing water outflow. Taken together, we demonstrate a functional role of an aquaporin in gametogenesis, as well as in the formation of durable structures such as spores, a role that may have wider biological and medical implications.
Keywords: aquaporins, gametogenesis, gene expression, water transport, development
Aquaporins mediate the transport of water across biological membranes with high velocity and specificity (1). Members of this ancient protein family occur in all organisms, and each species of higher animals and plants expresses numerous different aquaporins (2, 3). This result suggests that aquaporins fulfill different fundamental physiological roles at the cellular or organismal level. For instance, mammalian AQP1 and AQP2 play critical roles in water resorption in the kidney (4), and AQP4 is a crucial aquaporin of the brain-blood barrier (5). Bacterial glycerol facilitators are needed for uptake of glycerol as a nutrient (6), and a yeast aquaglyceroporin controls the intracellular level of the osmolyte glycerol in osmoregulation (7). However, for the majority of the aquaporins, the precise physiological roles remain to be established, and their elucidation promises amazing insight into the mechanisms with which cells and organisms control their relationship to water.
The yeast Saccharomyces cerevisiae is a widely used and powerful model system in molecular cell biology. Its genome encodes two aquaporins and two aquaglyceroporins (8). The aquaporins Aqy1 and Aqy2 are 88% identical (excluding the short C terminus) (9–11). However, these proteins seem to be expressed under different conditions: whereas expression of the AQY2 gene and its product Aqy2 have been observed only in proliferating cells, AQY1 is expressed in resting cells and in particular during sporulation (12, 13). Different phenotypes for vegetative cells lacking these aquaporins have been reported, such as a growth advantage upon repeated cycles of high and low osmolarity (9, 11), as well as an impaired tolerance to repeated cycles of rapid freezing and thawing (14).
Yeast sporulation is a complex developmental process tightly coupled to meiosis. It resembles in many respects gametogenesis in higher organisms (12, 15). Sporulation results in formation of an ascus with four haploid spores. Spores are highly resistant to harsh environmental conditions, and they are surrounded by a specialized wall, the ascospore wall. Spores display a reduced water content and low metabolic activity (15). Sporulation is triggered by nitrogen starvation on poor carbon sources such as acetate. Spore germination is stimulated in the presence of a rich carbon source. The process of meiosis and sporulation has been studied by time course global gene expression analysis (12, 16) as well as different genetic screens, including large-scale analysis of knockout mutants (17, 18).
Here we show that Aqy1 is a spore-specific aquaporin. The protein seems to be poorly or not at all expressed in vegetative cells, but it becomes abundantly expressed late during spore formation. Mutants lacking AQY1 show reduced spore fitness.
Materials and Methods
Growth Condition. Yeast cells were grown in YPD (yeast extract/peptone/dextrose) medium or in YNB (yeast nitrogen base) lacking histidine (19). G418 resistance was tested on YPD containing 200 mg/liter geneticin. For sporulation, cells were pregrown in YPD to OD610 nm of 0.6 or 1.0, harvested by centrifugation, washed, resuspended in 1% KAc, and incubated at 25°C. For germination, an aliquot of the sporulation culture was harvested by centrifugation, and spores were prepared (see below) and spread on YPD plates.
Strain Constructions. Yeast strains used in this study were isogenic to SK1 (12) (MATα or MATa or MATα/MATa ho::hisG ura3 lys2 leu2::hisG trp1ΔFA his3), which was kindly provided by L. Huang (University of Massachusetts, Boston). For some initial experiments, we used strains isogenic to Σ1278b provided by G. R. Fink (20). AQY1 was deleted in SK1 by PCR with YDp-H (21) as template for the HIS3 gene and primers P1 and P2 (Table 1). Correct gene deletion was confirmed by PCR on genomic DNA by using primers P3/P4 and P5/P4. The gene for the green fluorescent protein (GFP) was integrated at the C terminus of AQY1. For this purpose, the yEGFP3-KanMX cassette from pUG30 was amplified by using primers P6/P7 and transformed into yeast. Correct integration was confirmed by PCR by using primers P3/P8 and by sequencing on both strands. Wild-type SK1 was converted to HIS3 by using the HIS3 gene amplified from strain YSH4 with primers P12/P13. The PCR product was transformed into SK1, and transformants were selected on His– plates and checked by tetrad analysis.
Table 1. Oligonucleotides used.
| Oligonucleotide | Sequence | Effect |
|---|---|---|
| P1 | GGTGCTGTCTGTCAATACGGCACATAAAGTAACATGTAATTAACTATAACCAGTGAATTCCCGGGGATCC | Replacement AQY1::HIS3 |
| P2 | CGAGTATTATAACATTAAGTGCTAGTGAGCGAGAAATAAAGAAAAGGAGGTGACCATGATTACGCCAAGC | Replacement AQY1::HIS3 |
| P3 | GAACGATACCGACAAGCAAC | Confirming deletion |
| P4 | ATAAACTGGGCACACCAAG | Confirming deletion |
| P5 | GAGAGTGCGTTCAAGGCTCTTGCG | Confirming HIS presence |
| P6 | TCGCTCACTAGCACTTAATGTTATAATATTCGGCAAAAACTCTAAAGGTGAAGAATTATTCACTG | Insertion GFP |
| P7 | AGGGATATTAAAAACACTAATTACCTCAGTAGTATGGATGGCATAGGCCACTAGTGGATCTG | Insertion GFP |
| P8 | GGCGTGAATGTAAGCGTGACAT | Confirming GFP |
| P9 | ACATTAAGTGCTAGTGAGCG | Confirming insertion |
| P10 | CTATCAACTTTCGATGGTAGG | Probe 18S |
| P11 | TATGGTTAAGACTACGACGGT | Probe 18S |
| P12 | TTTGAACACGGCATTAGTCAG | Amplification of HIS3 |
| P13 | CTCGTTCAGAATGACACGTATAG | Amplification of HIS3 |
Underlined residues match plasmid sequence.
Northern Blot Analysis. Total RNA was isolated, separated by electrophoresis, and hybridized as described (21). Probes were prepared by PCR from genomic DNA by using primers P3/P9 (AQY1) and P10/P11 (18S) and labeled with 17 pmol [α-32P]dCTP, 3,000 Ci/mmol (1 Ci = 37 GBq). Signals were detected by using a Bio-Rad FX PhosphorImager. The 18S ribosomal RNA served to normalize transcript levels.
Protein Extraction and Subcellular Fractionation. Cells were chilled in ice-cold water, harvested by centrifugation, and washed in cell wash buffer (10 mM Tris·HCl, pH 7.5/0.5 mM sucrose/2.5 mM EDTA). Proteins were extracted, and plasma membranes were prepared as described (22). For fractionation of cell membranes, protein pellets were resuspended in lysis buffer (0.8 M sorbitol/10 mM Mops, pH 7.2/2mMNa4EDTA/mixture of protease inhibitors), broken with glass beads in a FastPrep (9 cycles of 20 s at full speed), and cellular debris was removed by two rounds of centrifugations (5 min at 1,600 × g). Protein extracts were fractionated on a 12–60% sucrose step gradient as described by Egner et al. (23).
Western Blot Analysis. Ten micrograms of total protein was incubated at 65°C for 10 min, loaded on a 12.5% SDS-polyacrylamide gel, and blotted on poly(vinylidene difluoride) (PVDF) membranes (Hybond-P, Amersham Pharmacia). The anti-GFP antibody was used at a 1/2,000 dilution (mixture of two mouse monoclonal anti-GFP antisera, Roche). Sucrose gradient fractions were denaturated at 40°C for 20 min and separated as above. Primary antibodies were rabbit anti-Pma1 (plasma membrane proton ATPase) antisera, 1/2,000, as a marker for plasma membrane [kindly provided by P. Ljungdahl (Ludwig Institute for Cancer Research, Stockholm), originally from A. Chang (University of Michigan, Ann Arbor)]; and a mouse anti-Dpm1 (dolichol phosphate mannose synthase) antisera, 1/1,000, as a marker for the endoplasmic reticulum (from Molecular Probes). The secondary antibodies (Sigma) coupled to horseradish peroxidase were applied in 1/5,000, 1/2,000 and 1/2,000 dilutions. Signals were visualized with the ECL plus substrate kit (Amersham Pharmacia) and the FUJIFILM LAS-1000 camera.
Random Spores Analysis. Spores were prepared (19), incubated twice for 2 h at 30°C with 1,000 units of lyticase (Sigma) and 0.1% Tween 100, and checked microscopically. Spores were counted in a hemocytometer, and 100 spores were streaked per plate. For slow freezing experiments, aliquots were stored for 2 h in a –20°C freezer, thawed for 10 min in a 30°C water bath, and plated. For rapid freezing, aliquots were kept for 30 min in a –20°C ethanol bath or for 20 min in liquid nitrogen. Three aliquots were plated for each experiment, and experiments were repeated several times.
Microscopy. Phase-contrast and fluorescence microscopy was performed with a Leica DM-RXA microscope with epifluorescence illumination coupled to a Leica computer system for image integration (leica fw4000 program). DAPI staining was performed as described (24). All experiments were done at least in triplicate.
Results
Aqy1 Is Conserved in Yeasts. Recently, genome sequences from seven Saccharomyces species have been determined (25, 26). All contain AQY1 homologs in positions syntenic to AQY1 from S. cerevisiae. Although Aqy1 from most laboratory strains is inactivated by mutations in two critical residues, V121M and P255T (9), Aqy1 from all newly sequenced Saccharomyces species has the conserved V121 and P255. The same is true for aquaporins from five additional, more distantly related yeasts. Because all these proteins contain the sequence elements known to be important for aquaporin function, it seems that yeasts generally possess a potentially active Aqy1 and that laboratory S. cerevisiae strains are an exception.
The C terminus of S. cerevisiae Aqy1 showed strain-dependent polymorphism (9, 10). It seems that Aqy1 from all seven sequenced Saccharomyces species has a short C terminus. Hence, the longer version of the C terminus found in active Aqy1 from S. cerevisiae strain Σ1278 may be due to a mutation. The extended C terminus in Aqy1 Σ1278 caused reduced expression of the protein in Xenopus oocytes (10). Taken together, it seems that genuine yeast Aqy1 has the conserved V121 and P255 and a variable, commonly short C terminus.
Expression of AQY1 Is Stimulated in Sporulating Cells. Global expression analysis of the yeast sporulation gene expression program indicated that AQY1 expression is stimulated in sporulating diploid cells within the “Early II Phase” (12, 16) or “Expression Cluster 4” (16). We tested expression of AQY1 and AQY2 in a diploid Σ1278 strain shifted to sporulation medium. Expression of AQY2 remained undetectable throughout the experiment. AQY1 was expressed at a low level during the first 6 h and then increased gradually to reach a maximum ≈13 h after the shift (not shown). Hence, expression of AQY1 is indeed stimulated in sporulating cells.
Strain SK1 sporulates faster, synchronously, and with high efficiency and therefore is commonly used for studies of meiosis and sporulation. PCR amplification and diagnostic restriction analysis (10), as well as sequence analysis, revealed that SK1 contains the same (functional) AQY1 allele as Σ1278. We grew SK1 in complete YPD medium to an OD610 nm of 0.6 or 1.0 and shifted cells to KAc sporulation medium. During the first 6 h, AQY1 was expressed at a low level (Fig. 1A and data not shown). Approximately 8 h after the shift, the mRNA level for AQY1 increased strongly. This time point coincided with the appearance of asci.
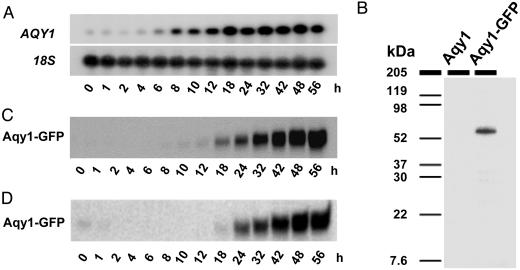
Fig. 1.
Aqy1 is expressed in sporulating cells. (A) Northern blot analysis. Cells of a diploid wild-type SK1 were shifted to KAc sporulation medium, and samples were taken at different time points. 18S RNA served as loading controls. (B) Western blot analysis using an anti-GFP antibody shows expression of a 59-kDa Aqy1-GFP fusion protein in spores. (C) Time-course Western blot analysis of Aqy1-GFP using an anti-GFP antibody from the same culture as in A. The appearance of asci was monitored microscopically and coincided with that of Aqy1-GFP. (D) Western blot analysis of Aqy1-GFP in a batch culture growing in YPD. SK1 cells start sporulating when glucose is exhausted. The appearance of asci was monitored microscopically and coincided with that of Aqy1-GFP.
To correlate gene with protein expression, we fused AQY1 to the coding sequence of GFP. This fusion is functional because cells expressing Aqy1-GFP behaved like wild type in phenotypic tests reported below. The fusion protein was detected at the expected size of 59 kDa (32 kDa for Aqy1 plus 27 kDa for GFP; Fig. 1B). We could not detect Aqy1-GFP in vegetative cells, in accordance with previous findings using an anti-Aqy1 antibody (27). Aqy1-GFP was also undetectable during the first 10 h after shift to sporulation medium, but, once asci were visible, the protein rapidly accumulated (Fig. 1C).
We also tested Aqy1-GFP expression in an SK1 culture growing on YPD. In contrast to most strains, SK1 sporulates even in YPD medium when proliferation ceases. Aqy1-GFP was undetectable until cells ceased proliferation. Expression of the fusion protein again coincided with the appearance of asci (Fig. 1D). The weak signal at time 0 h is due to some asci transferred from the preculture.
Aqy1 Is Expressed in Spores. When sporulation was monitored microscopically, Aqy1-GFP was not detectable in vegetative or sporulating cells. Aqy1-GFP was visible only in asci that had clearly distinguishable spores (Fig. 2A). Moreover, only two of the four spores in each tetrad showed clear Aqy1-GFP expression. Note that the AQY1/AQY1-GFP diploid is heterozygous for Aqy1-GFP. DAPI staining confirmed that all four spores had developed normally and contained a nucleus (Fig. 2B). Hence, expression of Aqy1-GFP seems to occur after the spore genomes were physically separated from each other.
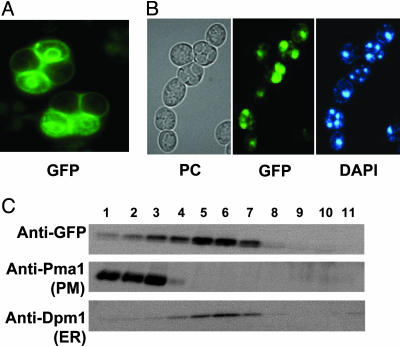
Fig. 2.
Aqy1-GFP localizes to spore membranes and is expressed in only two spores of a heterozygous diploid. (A) Detection of the GFP signal in living asci. (B) Comparison of asci in phase contrast (PC), detection of GFP and nuclear DNA (DAPI). (C) Western blot analysis of sucrose gradient fractions. Pma1 (plasma membrane proton ATPase) serves as marker for the plasma membrane (PM) and Dpm1 (dolichol phosphate mannose synthase) for the endoplasmic reticulum (ER). Top fraction 11 is at 12% sucrose, and bottom fraction 1 is at 60% sucrose.
Aqy1-GFP decorated the spore surface as well as an intracellular ring. DAPI staining detecting nuclear DNA revealed that the ring surrounds the nucleus (Fig. 2B) and hence likely represents the endoplasmic reticulum (ER). To confirm ER and plasma membrane localization of Aqy1-GFP, we performed membrane fractionation and Western blot analysis with marker proteins for ER (Dpm1, dolichol phosphate mannose synthase) and the plasma membrane (Pma1, plasma membrane proton ATPase). Indeed, it seems that Aqy1-GFP is localized both to the ER and to the plasma membrane (Fig. 2C). A similar localization was previously observed for yeast Aqy2 (11) (unpublished data).
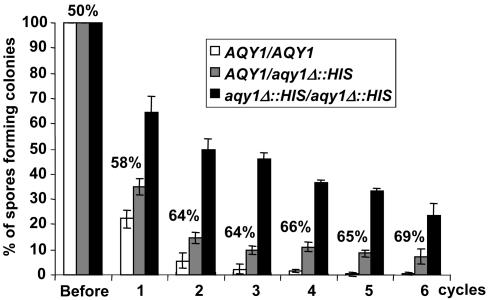
AQY1 Deletion Confers Resistance to Rapid Freezing. We prepared young spores from the homozygous wild type (AQY1/AQY1), the homozygous deletion mutant (aqy1Δ::HIS3/aqy1Δ::HIS3) and the heterozygous diploids (AQY1/aqy1Δ::HIS3), exposed those to a range of different harsh treatments, and subsequently tested their ability to give rise to colonies of vegetative cells. When spores were rapidly frozen in liquid nitrogen (Fig. 3), or lyophilized (not shown), deletion of AQY1 conferred a dramatic increase in survival. Although almost no spores from wild-type diploid were detected after four rounds of freezing/thawing, ≈25% of the spores of the homozygous aqy1Δ::HIS3/aqy1Δ::HIS3 mutant remained competent to form colonies even after six rounds. This effect is not due to the presence of the HIS3 gene in the aqy1Δ::HIS3 mutant because two isogenic AQY1 wild-type cells, one being his3– and the other HIS3+, showed the same poor spore viability at rapid freezing (data not shown). Heterozygous AQY1/aqy1Δ::HIS3 attained an intermediate level, which was, however, consistently lower than that expected from the mathematical average of wild-type and homozygous aqy1Δ::his3 diploids. Also, the relative proportion of aqy1Δ::HIS3 spores able to form colonies in heterozygous diploids was lower than expected. This result may be due to the lower spore fitness of aqy1Δ::HIS3 spores (see further). This test provides for a clear phenotype in aqy1Δ spores and hence a suitable assay system.
Fig. 3.
Spores of deletion mutants of AQY1 better survive rapid freezing in liquid nitrogen. Spores were prepared from AQY1/AQY1 wild type, AQY1/aqy1Δ::HIS3 heterozygous diploids, and aqy1Δ::HIS3/aqy1Δ::HIS3 homozygous mutants, frozen, and thawed for six cycles. Numbers indicate the proportion of aqy1Δ::HIS3 spores from AQY1/aqy1Δ::HIS3 heterozygous diploids that produced colonies. Numbers >50% indicate that such spores have a survival advantage.
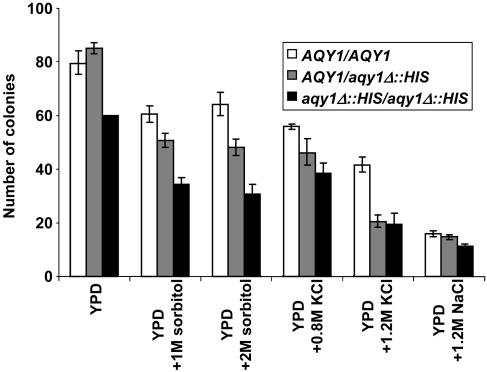
aqy1Δ::HIS3 Spores Show Reduced Fitness. To test whether Aqy1 performs functionally important roles, the three diploids were sporulated. We did not observe a difference between the three strains in their efficiency to produce asci. Spores were prepared, and equal numbers were plated on different media for germination. On average, the homozygous aqy1Δ::HIS3/aqy1Δ::HIS3 diploid produced 38% (between 20% and 50%) fewer viable spores, and heterozygous AQY1/aqy1Δ::HIS3 diploid produced on average 18% fewer viable spores than wild type (Fig. 4). A reduction was observed under standard as well as under different osmotic stress conditions, suggesting that reduced fitness is due to events that occur before germination. Consistent with this notion, we did not observe a significant difference between wild-type and aqy1Δ mutant spores with respect to the time point when colonies appeared on germination plates or in colony size. We also did not observe a significant difference in the lag phase of cultures when germination was allowed in liquid culture (not shown). Reduced fitness of aqy1Δ::HIS3 spores was also observed when spores underwent repeated cycles of slow freezing (not shown).
Fig. 4.
Deletion of AQY1 reduces spore fitness. Spores were prepared from AQY1/AQY1 wild type, AQY1/aqy1Δ::HIS3 heterozygous diploids, and aqy1Δ::HIS3/aqy1Δ::HIS3 homozygous mutants, and 100 spores per plate were allowed to germinate under normal and different osmotic stress conditions. The y axis represents the number of colonies per plate, and the standard deviation is based on three plates per sample and at least three independent experiments.
Fate of Aqy1p During Spore Maintenance and Germination. Because it seemed that Aqy1 plays a role during sporulation but probably not during germination, we followed Aqy1-GFP once spores had been formed. After ≈10 days in sporulation medium, it seemed that the ER and plasma membrane pool of Aqy1-GFP diminished and a bright spot appeared that grew with time (Fig. 5A). This is likely a degradation product. Hence, there seems to be turnover of Aqy1-GFP in spores. In Western blot analysis (data not shown), we observed that a band corresponding to free GFP (27 kDa) appeared 12–18 h after the Aqy1-GFP band became visible, indicating that Aqy1 turnover starts soon after sporulation has been completed.
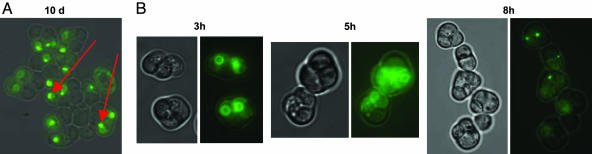
Fig. 5.
Aqy1-GFP turnover after sporulation and in germinating cells. (A) Aqy1-GFP seems to be degraded during spore maintenance. Pictures were taken from asci that had been kept on sporulation medium for 10 days. (B) Aqy1-GFP rapidly disappears from membranes upon germination. Fresh asci were transferred to YPD medium to allow germination, and GFP was monitored for 8 h. Arrows indicate green spots likely to represent a deposit of free GFP.
We then monitored Aqy1-GFP in young spores shifted to YPD for germination. Within ≈8 h, when cells started to germinate, Aqy1-GFP disappeared from the plasma membrane and ER, and a bright spot of GFP deposit appeared (Fig. 5B). Taken together, it seems that Aqy1 is degraded during spore maintenance and in germinating cells.
Discussion
Aqy1 Is a Yeast Spore Aquaporin. Although the mRNA of AQY1 is detectable at a low level in slowly growing or resting yeast cells (this work and ref. 13), we could not detect the Aqy1-GFP protein in vegetative cells (this work and ref. 9). This finding raises the question of how previously reported phenotypes for an AQY1 deletion mutant [increased tolerance to osmotic cycles and reduced freeze tolerance (9, 14)] can be explained. Aqy1 might be produced at very low levels in vegetative cells (9), and possibly the difference between such low levels and complete absence accounts for the observed effects. However, massively increased Aqy1 expression during sporulation suggests a specific role under these conditions.
It has previously been reported that expression of AQY1 is stimulated in sporulating diploid cells within the early II phase (12) or expression cluster 4 (16). Other genes expressed during these stages encode functions in meiotic chromosome pairing and recombination. These events occur before the first meiotic division and hence well before separation of the haploid genomes (15). From our data, it seems that expression of Aqy1 coincided with the appearance of asci, indicating that Aqy1 rather classifies as a late protein within the meiosis and sporulation program. In particular, it seems that the haploid genome expressing AQY1-GFP cannot serve as source for significant levels of Aqy1-GFP in another spore of the same ascus. This finding suggests that Aqy1 is produced only after the nuclear lobes with the four haploid genomes have been separated from each other by cytoplasmic barriers that prevent passage of newly synthesized mRNA, i.e., after meiosis II (28). It should be noted, however, that aqy1Δ spores from heterozygous diploids behaved in functional tests intermediate to wild-type spores and aqy1Δ spores from homozygous aqy1Δ/aqy1Δ diploids. This finding indicates that small amounts of Aqy1 protein might be produced before complete spore separation and hence be present in aqy1Δ spores from heterozygous diploids.
These data also indicate some discrepancy between the gene and protein expression pattern: although the AQY1 mRNA is detectable in slowly growing or resting vegetative cells, the protein level seems to be increased only in spores. Hence, it is possible that Aqy1 expression is also controlled posttranscriptionally.
Possible Roles of Aqy1 in Spore Formation. After meiosis, the four haploid genomes are located within four nuclear lobes with one spindle pole body each. The spindle poles serve as starting point for formation of plaques, which differentiate to the prospore wall, eventually separating the four spores (15). The spore wall consists of two inner layers of primarily glucan, a third layer consisting almost completely of chitosan and an outer proteinaeous layer rich in dityrosine. The two outer layers confer the characteristic resistance of spores against environmental stress (29–32). Hence, the spore wall is quite distinct from that of vegetative cells and likely is much less flexible.
Spores accumulate large amounts of trehalose as well as other material, resulting in a relative dry weight about twice as large as that of vegetative cells. Hence, the relative water content of spores is lower than that of vegetative cells (33). The accumulated components generate an inward-directed force for water, likely leading to an increased turgor pressure counteracted by the spore wall. Aqy1 may be important to facilitate the efflux of water out of spores against its concentration gradient. The driving force for water efflux is likely turgor pressure mediated by the spore wall. According to this scenario, Aqy1 would serve as a valve to release excess pressure. Inability to release excess pressure at this stage of spore maturation may damage the spore surface and result in reduced spore viability, as we observed for the mutants lacking Aqy1. Apart from release of excess turgor pressure, a reduced spore water content may contribute to diminishing metabolic activity and establishment of spore dormancy.
Upon spore germination, water may be driven into spores passively, supported by hydrolysis of trehalose to glucose (34). Pressure is then probably diminished by the simultaneous degradation of the spore wall, releasing the new vegetative cell. How the newly developing cell controls volume and turgor and whether Aqy2 plays a role is an interesting question for future studies. The role of Aqy1, however, seems to be restricted to the stages of spore formation/maturation, because our data suggest that bulk Aqy1 is degraded during spore maintenance and germination, and we did not observe effects of deletion of AQY1 on spore germination. However, we cannot exclude at this point that some Aqy1 is present during germination and performs some function.
Most commonly used laboratory strains do not possess active Aqy1 but show spore viability close to 100%. This result may be due to compensatory suppressor mutations that arose during “evolution” in the laboratory, where high spore viability is an important criterion for strain selection. Why the aquaporins are mutated in laboratory strains remains unclear. Rapid freezing of spores, which favors survival of mutants lacking Aqy1, does not seem to be a common procedure. Perhaps moderate expression of Aqy1 during vegetative growth generates a weak selective disadvantage under laboratory conditions.
Implications. It has been suggested that mammalian AQP7 and AQP8, which are expressed in testis and sperm, could play a role in facilitating water efflux in the cytoplasmic condensation that occurs in developing sperm (35). This physiological function could therefore be analogous to that of yeast Aqy1.
The involvement of Aqy1 in sporulation provides for interesting implications of aquaporins in the generation of spores and pollen. These structures have in common reduced relative water content, the ability to survive harsh conditions and remain viable for long periods. Involvement of aquaporins in spore fitness in Dictyostelium has been documented (36, 37). In plants, where aquaporins are particularly abundant, their involvement has not yet been reported in pollen development but rather in pollen germination upon contact with the stigma (38). It will also be of interest to investigate whether aquaporins are involved in spore formation of pathogenic bacteria, because they might serve as target for drugs controlling spreading and infection.
Acknowledgments
We thank members of the Hohmann group for critical reading of the manuscript and Peter Dahl for assistance with yeast strain constructions. This work was supported by the European Commission (contracts BIO4-CT98-0024, FMRX-CT97-0128, and QLK3-CT2000-52116). S.H. holds a research position from the Swedish Research Council, Vetenskapsrådet.
Author contributions: S.H. and F.S.-W. designed research; F.S.-W. and N.P. performed research; F.S.-W., N.P., and S.H. analyzed data; and S.H. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: YPD, yeast extract/peptone/dextrose; ER, endoplasmic reticulum.
References
- 1.Hohmann, S., Agre, P. & Nielsen, S. (2001) in Current Topics in Membranes, eds. Benos, D. J. & Simon, S. A. (Academic, San Diego), Vol. 51, pp. 390. [Google Scholar]
- 2.Borgnia, M., Nielsen, S., Engel, A. & Agre, P. (1999) Annu. Rev. Biochem. 68, 425–458. [DOI] [PubMed] [Google Scholar]
- 3.Johanson, U., Karlsson, M., Johansson, I., Gustavsson, S., Sjovall, S., Fraysse, L., Weig, A. R. & Kjellbom, P. (2001) Plant Physiol. 126, 1358–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen, S., Frokiaer, J., Marples, D., Kwon, T. H., Agre, P. & Knepper, M. A. (2002) Physiol. Rev. 82, 205–244. [DOI] [PubMed] [Google Scholar]
- 5.Papadopoulos, M. C., Krishna, S. & Verkman, A. S. (2002) Mt. Sinai J. Med. 69, 242–248. [PubMed] [Google Scholar]
- 6.Heller, K. B., Lin, E. C. & Wilson, T. H. (1980) J. Bacteriol. 144, 274–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hohmann, S. (2002) Microbiol. Mol. Biol. Rev. 66, 300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hohmann, S., Bill, R. M., Kayingo, I. & Prior, B. A. (2000) Trends Microbiol. 8, 33–38. [DOI] [PubMed] [Google Scholar]
- 9.Bonhivers, M., Carbrey, J. M., Gould, S. J. & Agre, P. (1998) J. Biol. Chem. 273, 27565–27572. [DOI] [PubMed] [Google Scholar]
- 10.Laizé, V., Tacnet, F., Ripoche, P. & Hohmann, S. (2000) Yeast 16, 897–903. [DOI] [PubMed] [Google Scholar]
- 11.Carbrey, J. M., Bonhivers, M., Boeke, J. D. & Agre, P. (2001) Proc. Natl. Acad. Sci. USA 98, 1000–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu, S., DeRisi, J., Eisen, M., Mulholland, J., Botstein, D., Brown, P. O. & Herskowitz, I. (1998) Science 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 13.Laizé, V., de Jesus Ferreira, M. C. & Hohmann, S. (2000) in Molecular Biology and Physiology of Water and Solute Transport, eds. Hohmann, S. & Nielsen, S. (Kluwer, New York), pp. 415–422.
- 14.Tanghe, A., Van Dijck, P., Dumortier, F., Teunissen, A., Hohmann, S. & Thevelein, J. M. (2002) Appl. Environ. Microbiol. 68, 5981–5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kupiec, M., Byers, B., Esposito, R. E. & Mitchell, A. P. (1997) in The Molecular and Cellular Biology of the Yeast Saccharomyces: Cell Cycle and Cell Biology, eds. Pringle, J. R., Broach, J. R. & Jones, E. W. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 880–1036.
- 16.Primig, M., Williams, R. M., Winzeler, E. A., Tevzadze, G. G., Conway, A. R., Hwang, S. Y., Davis, R. W. & Esposito, R. E. (2000) Nat. Genet. 26, 415–423. [DOI] [PubMed] [Google Scholar]
- 17.Rabitsch, K. P., Toth, A., Galova, M., Schleiffer, A., Schaffner, G., Aigner, E., Rupp, C., Penkner, A. M., Moreno-Borchart, A. C., Primig, M., et al. (2001) Curr. Biol. 11, 1001–1009. [DOI] [PubMed] [Google Scholar]
- 18.Deutschbauer, A. M., Williams, R. M., Chu, A. M. & Davis, R. W. (2002) Proc. Natl. Acad. Sci. USA 99, 15530–15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherman, F., Fink, G. R. & Hicks, J. B. (1983) Methods in Yeast Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Gimeno, C. J., Ljungdahl, P. O., Styles, C. A. & Fink, G. R. (1992) Cell 68, 1077–1090. [DOI] [PubMed] [Google Scholar]
- 21.Berben, G., Dumont, J., Gilliquet, V., Bolle, P.-A. & Hilger, F. (1991) Yeast 7, 475–477. [DOI] [PubMed] [Google Scholar]
- 22.Tamás, M. J., Luyten, K., Sutherland, F. C. W., Hernandez, A., Albertyn, J., Valadi, H., Li, H., Prior, B. A., Kilian, S. G., Ramos, J., et al. (1999) Mol. Microbiol. 31, 1087–1104. [DOI] [PubMed] [Google Scholar]
- 23.Egner, R., Mahe, Y., Pandjaitan, R. & Kuchler, K. (1995) Mol. Cell. Biol. 15, 5879–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gangloff, S., de Massy, B., Arthur, L., Rothstein, R. & Fabre, F. (1999) EMBO J. 18, 1701–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. (2003) Nature 423, 241–254. [DOI] [PubMed] [Google Scholar]
- 26.Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B. A. & Johnston, M. (2003) Science 301, 71–76. [DOI] [PubMed] [Google Scholar]
- 27.Meyrial, V., Laize, V., Gobin, R., Ripoche, P., Hohmann, S. & Tacnet, F. (2001) Eur. J. Biochem. 268, 334–343. [DOI] [PubMed] [Google Scholar]
- 28.Emanuel, J. R. & Magee, P. T. (1981) J. Bacteriol. 145, 1342–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briza, P., Bogengruber, E., Thur, A., Rutzler, M., Munsterkotter, M., Dawes, I. W. & Breitenbach, M. (2002) Yeast 19, 403–422. [DOI] [PubMed] [Google Scholar]
- 30.Briza, P., Winkler, G., Kalchhauser, H. & Breitenbach, M. (1986) J. Biol. Chem. 261, 4288–4294. [PubMed] [Google Scholar]
- 31.Briza, P., Ellinger, A., Winkler, G. & Breitenbach, M. (1988) J. Biol. Chem. 263, 11569–11574. [PubMed] [Google Scholar]
- 32.Christodoulidou, A., Briza, P., Ellinger, A. & Bouriotis, V. (1999) FEBS Lett. 460, 275–279. [DOI] [PubMed] [Google Scholar]
- 33.Roth, R. (1970) J. Bacteriol. 101, 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thevelein, J. M. (1984) Microbiol. Rev. 48, 42–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calamita, G., Mazzone, A., Bizzoca, A. & Svelto, M. (2001) Biochem. Biophys. Res. Commun. 288, 619–625. [DOI] [PubMed] [Google Scholar]
- 36.Mitra, B. N., Yoshino, R., Morio, T., Yokoyama, M., Maeda, M., Urushihara, H. & Tanaka, Y. (2000) Gene 251, 131–139. [DOI] [PubMed] [Google Scholar]
- 37.Flick, K. M., Shaulsky, G. & Loomis, W. F. (1997) Gene 195, 127–130. [DOI] [PubMed] [Google Scholar]
- 38.Ikeda, S., Nasrallah, J. B., Dixit, R., Preiss, S. & Nasrallah, M. E. (1997) Science 276, 1564–1566. [DOI] [PubMed] [Google Scholar]