Abstract
Epstein–Barr virus (EBV) infects human B lymphocytes and epithelial cells. We have compared the requirements for EBV glycoprotein-induced cell fusion between Chinese hamster ovary effecter cells and human B lymphoblasts or epithelial cells by using a virus-free cell fusion assay. EBV-encoded gB, gH, gL, and gp42 glycoproteins were required for efficient B cell fusion, whereas EBV gB, gH, and gL glycoproteins were required for Chinese hamster ovary effecter cell fusion with epithelial cell lines (AGS and SCC68) or the human embryonic kidney cell line 293-P. Fusion with human embryonic kidney 293-P cells was greater than fusion observed with B cells, indicative of an important role for cell contact. An antibody directed against the gH and gL complex inhibited epithelial cell fusion. Increased surface expression of gB alone as a result of truncations or point mutants in the carboxyl-terminal tail allowed gB-mediated fusion with epithelial cells, albeit at a lower level than with coexpression of gB, gH, and gL. Overall, gB appears to be the critical component for EBV glycoprotein-mediated cell fusion.
Keywords: viral entry, herpesvirus
The entry process of human herpesviruses entails a two-step process: binding followed by fusion. Specific herpesvirus-encoded glycoproteins and cell-surface receptors are important for both events (1–3). Entry may occur by free virus infecting a cell or by cell–cell spread (2). In either case, fusion is a prerequisite for infection. Despite herpesvirus genomes encoding many viral glycoproteins, the glycoproteins implicated in entry and fusion are limited and conserved (1). Most herpesviruses require the conserved glycoproteins gH, gL, and gB, as well as an additional, family-specific viral glycoprotein, such as gD for α-herpesviruses or gp42 for the γ-herpesvirus Epstein–Barr virus (EBV), for efficient entry and fusion (1). These nonconserved glycoproteins result in cell specificity by recognizing distinct cellular receptors. For example, EBV glycoprotein 42 binds to HLA class II on B cells, an interaction essential for the infection of B cells (4, 5). Additionally, EBV gp350/220 binds to the B cell-surface receptor CD21/CR2, and this binding mediates the initial interaction of EBV with B cells and is thought to tether the virus to promote the subsequent binding of gp42 to HLA class II (6, 7). The gp42–class II interaction is then proposed to trigger membrane fusion, a reaction mediated by gH (EBV gp85), gL (EBV gp25), and gB (EBV gp110) (8). The exact role each of these glycoproteins plays in fusion is unclear.
EBV can enter cells by means of two routes, depending on the host cell type. In primary human lymphocytes, EBV is endocytosed before fusion, whereas, in B cells in culture, the virion envelope directly fuses with the plasma membrane (9, 10). Similarly to B cells in culture, fusion of EBV with epithelial cells occurs by direct fusion of the virion envelope with the plasma membrane. Despite B lymphocytes being the primary site for latency, the tropism of EBV for epithelial cells is supported by the presence of EBV in nasopharyngeal carcinoma, gastric carcinoma, and oral hairy leukoplakia (11). Several other observations point to an alternative mechanism of entry for EBV into epithelial cells. Because most epithelial cells do not express class II molecules or CD21/CR2, the viral proteins gp42 and gp350 likely play little, if any, role in the entry of EBV into the epithelium. Furthermore, EBV infection of epithelial cells occurs more efficiently by cell–cell contact, suggesting that entry in these cells may require different glycoprotein-receptor combinations (12, 13).
Recently, a model was described in which the glycoproteins present in the EBV envelope switch, depending on the cell type in which EBV replicates (14). This model is compatible with earlier studies showing that the EBV virion envelope contains two different glycoprotein complexes: a tripartite complex of gp42, gH, and gL and a bipartite complex of gH and gL (ref. 15 and reviewed in ref. 16). In the most recent studies, a high level of gp42 was found in virions propagated in epithelial cells, resulting in virions that infect epithelial cells poorly. Virus produced by B cells express low levels of gp42 because of the sequestration of gp42 by HLA class II. Therefore, these viruses are deficient in B lymphocyte infection and, consequently, efficiently infect epithelial cells. Other results have indicated an important role for gH and gL in the infection of epithelial cells. Antibodies directed against the gH/gL complex neutralize EBV infection of SVKCR2 and AGS cells (14, 15). A soluble form of gL fused to the constant region fragment of IgG bound better to human embryonic kidney (HEK)-293, AGS, and NU-GC-3 cell lines when compared with the B cell lines, Raji and BJAB (17). Recently, soluble gH/gL was found to bind to both AGS and SVKCR2 cell lines, but not EBV-negative Akata cells, suggesting a specific receptor for gH/gL (gHgLR) is present on epithelial cells (18). Despite the conservation of gH and gL, the only binding partner of this complex identified thus far is the binding of human herpesvirus (HHV)-6 to CD46 (19), but this does not appear to be functionally significant.
One of the first indications for an essential role of gB in fusion resulted from studies with herpes simplex virus (HSV)-1 performed in the 1970s with a temperature-sensitive mutant of gB that was able to attach to cells but failed to enter without the addition of polyethylene glycol at the nonpermissive temperature (20). More recent studies of recombinant viruses with specific deletions in gB has shown that gB is essential for the production of infectious virus (21, 22). Finally, gB coexpressed with other viral glycoproteins is required for cell–cell fusion in cell-based fusion assays for pseudorabies virus, HSV-1, HSV-2, HHV-8, and EBV (8, 23–27).
gB is one of the most conserved proteins within the herpesvirus family and consists of a large amino-terminal ectodomain, several amphipathic helices just before the membrane spanning domain, and a C-terminal tail domain (28). The gB C terminus is one of the longest of the herpesvirus-encoded glycoproteins and contains important cellular-sorting signals and regulates virally induced membrane fusion (28). The role of the gB tail in regulating herpesvirus-induced membrane fusion is most apparent because many mutations that modulate fusion are located in the gB C terminus (23, 24, 29–38). For EBV, the C terminus also contains domains that are important for cell fusion and the cellular localization of gB (8, 21). Four arginine residues in the C terminus (836–839) are required for the localization of gB primarily to the endoplasmic reticulum (ER)/nuclear membrane (39). Deletion or point mutations in the C terminus of gB cause gB to be expressed more abundantly on the plasma membrane of transfected cells (21, 39).
There is no direct evidence for which herpesvirus-encoded proteins function as a fusogen. Evidence suggests that multiple herpesvirus glycoproteins participate in the fusion process. Both gH and gL from Varicella–Zoster virus and HHV-8 mediate cell fusion when tested in a cell-based fusion assay (27, 40). A mutant gB of HHV-8 also has some fusion potential when expressed at a higher level on the cell surface, although it functions less well than gH and gL (27). Varicella–Zoster virus gB alone does not mediate fusion, but polykaryocytes indicating fusion are observed by coexpression of gB and gE (41). For other herpesviruses (HSV-1, HSV-2, or pseudorabies virus), cell-based fusion assays require the expression of multiple viral proteins (23–26).
To further delineate the requirements for EBV-induced cell fusion, we have used a cell-based fusion assay. This assay allows for the importance of individual glycoproteins in the effector cells to be assessed, as well as the fusion competence of various target cell lines to be tested. We demonstrate that gB, gH, and gL are sufficient for optimal fusion of epithelial cells. In addition, epithelial cell fusion was mediated with gB mutants with enhanced cell-surface expression independent of other viral proteins, which suggests that gB is the major fusogenic protein for EBV and indicates that the possible existence of a epithelial receptor-specific for EBV. Finally, the studies described offer a model system to further understand the interactions of gB with other EBV-encoded glycoproteins, EBV cellular receptors in fusion and viral entry, and, by extension, other members of the herpesvirus family in general.
Materials and Methods
Cells, Plasmids, and Antibodies. All cells were cultured in media containing 10% serum and penicillin/streptomycin. Chinese hamster ovary (CHO)-K1, SCC68, and AGS cells were grown in Ham's F12 medium. Daudi cells were grown in RPMI medium 1640. Daudi cells were stably selected with G418 to express T7 RNA polymerase (42). PEAK cells are a HEK-293 derivative selected for high transfection frequency and are designated HEK-293-P (Edge Biosystems, Gaithersburg, MD). HEK-293-P, Vero, and HeLa cells were passaged in DMEM. The plasmids used in this study were as published in ref. 8. EGFP-N1 was purchased from Clontech.
Monoclonal antibodies E1D1 and F-2–1 were the gift from L. Hutt-Fletcher (Louisiana State University Health Sciences Center, Shreveport) and recognize the gH/gL complex and gp42, respectively. A large-scale preparation of the E1D1 antibody was made at the Northwestern University Monoclonal Antibody Facility. L2 was purchased from Chemicon and is directed against gB. Murine-specific biotinylated anti-IgG conjugate was purchased from Sigma. The streptavidin-horseradish peroxidase conjugate and the Ig isotype-matched control IgG2a antibody were purchased from Amersham Pharmacia.
Transfection. CHO-K1, AGS, HeLa, Vero, and HEK-293-P cell lines were transfected by using Lipofectamine 2000 (GIBCO/BRL). CHO-K1 cells were seeded in 12-well plates 1 day before transfection, and samples were done in triplicate. HEK-293-P, AGS, HeLa, and Vero cells were plated in 10-cm2 dishes 24 h before transfection. For Lipofectamine transfection, 70% confluent cells were placed in Opti-MEM (GIBCO/BRL) and incubated with DNA in lipid micelles for 12 h. CHO-K1 cells were transfected with 0.16 μg of pFgp350, pFgH, pFgL, pFgB, and gB mutants, 0.26 μg of pT7ENCLuc and 0.6 μg of pFgp42. The amount of DNA per transfection sample was kept constant by the addition of empty vector DNA in experiments in which specific glycoproteins were subtracted. Cells in 10-cm2 dishes were transfected with 10 μg of pCAGT7. SCC68 cells were electroporated with 40 μg of pCAGT7 by using a GenePulser (Bio-Rad). A total of 1 × 107 cells were electroporated at 0.24 kV and with 960-μF capacitance.
Fusion Assay. Effector CHO-K1 cells and the various target cell lines were transfected with plasmids encoding the glycoproteins as stated above. After 12 h, the effector cells and target cells were washed with PBS and detached by using Versene. After the cells were detached, the cells were counted with a Beckman Coulter Z1 particle counter and the two cell populations were mixed in equal amounts (0.2 × 106 cells per sample) and plated into a 24-well plate. Twenty-four hours later, the cells were lysed, and luciferase was quantitated by using the Promega Reporter Assay system. Substrate and samples were read on a Perkin–Elmer Victor2 multilabel counter. For the antibody inhibitory experiments, antibody was added when the two cell populations were mixed. As an alternative method to detect fusion, CHO-K1 cells were transfected with the plasmids encoding EBV glycoproteins, and HEK-293-P cells were transfected with EGFP-N1. Similar to the fusion assay above, after transfection for 12 h, the cells were mixed and incubated for 24 h. Cells were washed, fixed in 2% formaldehyde, and stained with 300 nM DAPI according to the manufacturer's protocol (Molecular Probes). The cells were visualized with a DMIRE2 inverted microscope (Leica, Deerfield, IL). Photos were taken with a capture controller (Hamamatsu, Middlesex, NJ) and openlab 3.1.7 software.
Cell ELISA. CHO-K1 cells used for the fusion assay as described above were also used to detect surface expression of the glycoproteins. When the cells were mixed together for the fusion assay, 4 × 104 CHO-K1 cells per well were plated into 96-well plates. After 24 h, the cells were washed and incubated with primary antibody directed against the glycoprotein or glycoprotein complex for half an hour. Subsequently, the cells were washed and fixed with 0.2% glutaraldehyde and 2% formaldehyde. The fixed cells were washed and the secondary biotin-conjugated anti-mouse IgG antibody and tertiary streptavidin-horseradish peroxidase antibody were added sequentially. Finally, peroxidase substrate was added and the plate was read on a Perkin–Elmer Victor2 multilabel counter.
Results
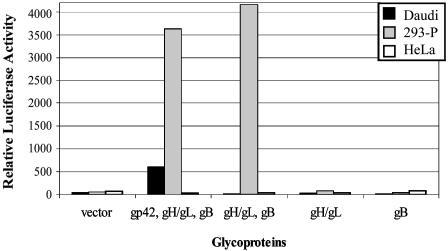
gB, gH, and gL Mediate Efficient Fusion with Some Human Epithelial Cell Lines. The purpose of this study was to investigate the requirements for EBV-induced cell fusion with epithelial cells and to examine whether differences exist between B cells and epithelial cells. For B cells, gp350 is not required for EBV-induced cell fusion, but gp42, gB, gH, and gL are necessary and sufficient (8). gp350 is dispensable for entry given that a virus lacking gp350 still infects numerous lymphoid and epithelial cell lines (43). To examine the requirements for epithelial cell fusion, CHO-K1 cells were transfected with different combinations of viral glycoproteins along with a plasmid containing the T7 promoter upstream of a luciferase reporter. These effector cells were then mixed with a variety of different target cells to assess the requirements for fusion for different cell types. The target cells were transfected with a plasmid encoding the T7 RNA polymerase so that, upon fusion of the two cell types, luciferase is expressed and can be measured to indicate the extent of fusion. As previously published (8), target Daudi B cells require the expression of gp42 with gB, gH, and gL in the effector cells for cell fusion to occur (Fig. 1). In contrast, gB, gH, and gL alone were required to induce cell fusion with the HEK-293-P cell line [a derivative of HEK-293 cells selected for high transfection efficiency (PEAK cells, Edge Biosystems)]. gH/gL or gB alone did not mediate fusion in any of the cell types tested. Expression of gp350 on the effector cell slightly increased fusion efficiency for both Daudi and HEK-293-P cells (data not shown). Expression of gp42 slightly inhibited fusion with HEK-293-P cells, although the decrease in luciferase activity was not significant (data not shown). Interestingly, fusion levels with HEK-293-P cells were increased in relation to Daudi B cells (Fig. 1). This result suggests a role of cell contact in the enhancement of fusion because HEK-293-P cells in this assay are attached to the same surface as the CHO-K1 cells and are intrinsically in close contact whereas the Daudi cells grow in suspension and fusion is therefore dependent on contact and adhesion. Previous studies have indicated that infection of some epithelial cell lines occurs more efficiently by means of cell–cell contact (12, 13).
Fig. 1.
gp42 is not necessary for EBV-induced epithelial cell fusion. Various combinations of EBV glycoproteins were transfected into CHO-K1 cells as indicated. The cells were mixed 1:1 with either Daudi, HEK-293-P, or HeLa cells, and, 24 h later, the cells were harvested and relative luciferase activity was measured. These data depict one representative experiment of a total of five experiments.
We next asked whether the glycoproteins needed for fusion with HEK-293-P cells were similar with other epithelial cell lines. Because of the association of EBV with gastric carcinoma, the AGS cell line was tested. AGS is a human stomach adenocarcinoma cell line susceptible to EBV infection (44). The human cervix epithelial adenocarcinoma cell line, HeLa, was chosen because of previous discrepancies regarding susceptibility of these cells to EBV infection (12, 45). An additional target, SCC68, a human oral carcinoma cell line, was tested because of the association of EBV with nasopharyngeal carcinoma (46). AGS and SCC68 cells were efficient target cells when the effector CHO-K1 cells expressed gB, gH, and gL, similar to HEK-293-P cells (data not shown). Vero (African green monkey kidney cell line), HeLa and CHO-K1 cell lines were not functional targets for fusion (Fig. 1 and data not shown), which was not because of insufficient T7 RNA polymerase expression in target Vero, HeLa, or CHO-K1 cells, because cotransfection of an EGFP reporter plasmid and/or the luciferase driven by the T7 promoter plasmid produced high levels of expression of luciferase or EGFP (data not shown). These results indicate that close cell–cell contact is not all that is required for epithelial cell infection, because fusion was not observed for Vero, HeLa, or CHO-K1 cells, despite the cells growing in the same monolayer as the CHO-K1 cells expressing the EBV glycoproteins. Effector cell fusion with target epithelial cells occurred in the absence of gp42, indicating as previously shown that gp42 and the interaction with class II is not essential for epithelial cell entry (15, 47). Overall, these data demonstrate that neither gp42 nor gp350 are necessary for epithelial fusion and that gB, gH, and gL are sufficient for fusion to HEK-293-P, AGS, and SCC68 cells.
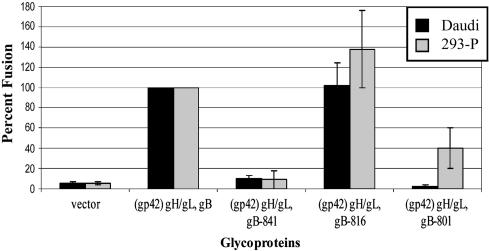
The Cytoplasmic Tail of gB Has Different Functional Domains When Fusion to B Cells Is Compared with Fusion to Epithelial Cells. Because their transfection efficiencies, HEK-293-P cells were used in the remaining experiments. Similar to other herpesvirus gB (29, 35, 38), we have shown that the gB cytoplasmic tail domain is an important regulator of EBV-induced membrane fusion (8). Previously, we created truncation mutants at three amino acids: 841 (gB-841); 816 (gB-816), which removed the ER retention sequence; and 801 (gB-801), which deleted the final 46 aa of gB, including the ER retention sequence. The gB-816 and gB-801 constructs are both expressed on the cell surface at higher levels than wild-type because of loss of the ER retention sequence (21).
By using the minimal glycoprotein requirements for both B cell and epithelial cell fusion, the domains of the cytoplasmic tail were tested in the cell–cell fusion assay. Truncation mutants (gB-801, gB-816, and gB-841) were expressed with gH and gL for epithelial cells and with gp42, gH, and gL for B cells. The percentage of fusion was calculated for each of the mutants by setting the minimal requirements (B cells: gp42, gB, gH, and gL; epithelial cells: gB, gH, and gL) at 100%. This analysis also allowed for the comparison of multiple experiments as the relative luciferase activity differed between experiments because of transfection rates. The overall transfection frequency of each gB construct was similar between cells (data not shown). The gB-816 truncation mutant increased fusion for B cells and epithelial cells, suggesting that either the increase in cell-surface expression of gB is important or that there is a negative regulatory domain between amino acids 816–857. Fusion was greater to the HEK-293-P cells than Daudi cells with this mutant. Interestingly, expression of gB-801 resulted in reduced fusion levels with HEK-293-P cells, but no fusion for B cells (Fig. 2). Both the gB-816 and gB-801 mutants were expressed more abundantly on the plasma membrane, suggesting that the levels of surface-expressed or -exposed gB may be important for fusion, particularly for epithelial cells, because the gB-801 mutant has a significant difference in fusion levels between the two cell types. Very low levels of fusion [somewhat lower than previously observed (8)] were observed for the gB-841 mutant for both HEK-293-P and Daudi cells (Fig. 2).
Fig. 2.
A mutant form of gB mediates epithelial cell fusion. gB truncation constructs were substituted for gB in the cell fusion assay. gp42 was included in the transfection mix in CHO-K1 cells overlaid with Daudi cells. Luciferase activity was normalized to wild-type transfection, gp42, gB, and gH/gL for Daudi and gB and gH/gL for HEK-293-P cells. The data shown are averages of four independent experiments. The vertical lines indicate the standard deviations.
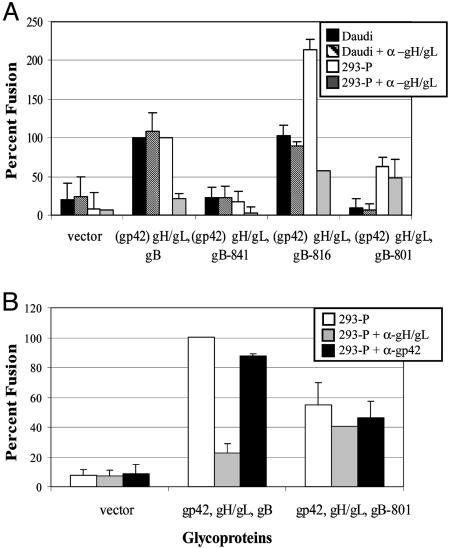
Antibodies to gH/gL Have No Effect on Fusion with the gB-801 Mutant in Contrast to Wild-Type gB and the gB-816 Mutant. To further investigate the requirements for EBV-induced cell fusion, we took advantage of antibodies that have differential effects, depending on the cell type that EBV infects. The gp42 monoclonal antibody, F-2-1, inhibits B cell entry but not epithelial cell entry, whereas the gH/gL monoclonal antibody E1D1 inhibits only epithelial cell entry (15, 47–49). These monoclonal antibodies were added when the two cell populations were mixed. As expected, the addition of the gH/gL antibody did not inhibit EBV-induced B cell fusion with wild-type gB or when any of the gB truncation mutants were expressed (Fig. 3A). In the HEK-293-P cells, the gH/gL antibody inhibited wild-type gB, gH, and gL fusion as well as the minimal fusion observed with the gB-841 mutant (Fig. 3A). Interestingly, although there was a dramatic reduction of fusion in the gB-816 mutant by the addition of the gH/gL antibody, the resulting level of fusion was similar to the level of fusion observed for the gB-801 mutant with or without the gH/gL antibody, suggesting that the two gB mutants with higher cell-surface expression may mediate fusion independent of gH and gL. Finally, to confirm the absence of a role of gp42 in epithelial cell fusion, CHO-K1 cells were transfected with gp42, gB, gH, and gL and fusion was tested. As would be expected from earlier studies indicating an absence of a role of gp42 in epithelial fusion (14, 15), the gp42 antibody had no effect on fusion with HEK-293-P cells. In contrast, as would also be expected from earlier studies (47) and the results presented in Fig. 3A, the gH/gL antibody inhibited EBV-induced cell fusion with only wild-type gB (Fig. 3B) and not with the gB-801 mutant, which again supports of fusion being mediated by the gB-801 mutant independent of gH and gL.
Fig. 3.
gH/gL antibody inhibits EBV-induced epithelial cell fusion but not B cell fusion. CHO-K1 cells were transfected as described for Fig. 2. At the time of overlay, E1D1 (gH/gL) (A) or E1D1 and F-2-1 (gp42) (B) antibody was added. Data are averages of two independent experiments, with the standard deviations indicated by vertical lines.
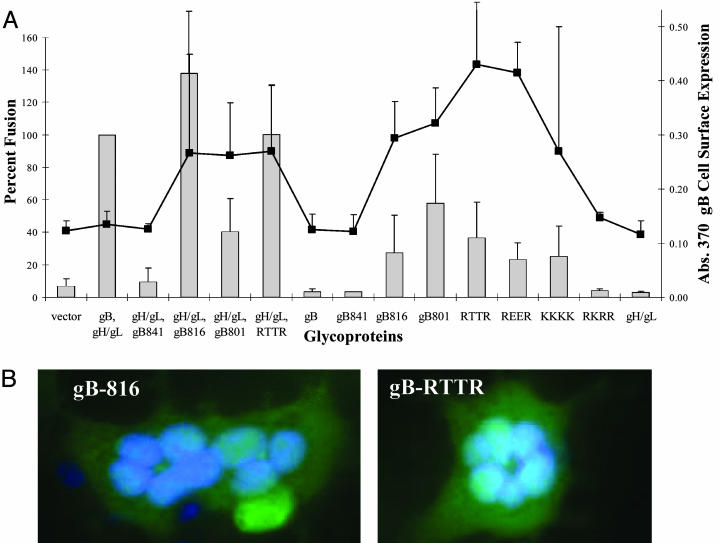
gB Surface Expression Is a Determinant of gH- and gL-Independent Cell Fusion. To further investigate the possibility that gB mutants may mediate fusion to epithelial cells independent of other EBV glycoproteins, we transfected the gB truncation mutants and gB mutants with point mutations in the ER-retention signal into CHO-K1 effector cells with and without gH and gL. Previous studies demonstrated that mutation of the two central arginine residues (RTTR and REER) or all of the arginine residues (KKKK) of the gB ER-retention signal results in an increase of expression of gB on the cell surface (39). Mutation of a single arginine (RKRR) resulted in a protein with localization similar to wild-type gB (39). After transfection, cell-surface expression of gB and fusion was measured (Fig. 4A). For the gB-801 mutant, fusion was similar with or without the inclusion of gH and gL (Fig. 4A). When gB was tested in the absence of gH and gL, fusion was only observed with the gB-816 and gB-801 mutants and mutants in the ER-retention signal with high levels of gB surface expression, which indicates that gB cell surface expression is a determinant of gH- and gL-independent cell fusion (Fig. 4A). For all of the gB mutants with the exception of the gB-801 mutant, fusion was lower or undetectable in the absence of gH and gL (Fig. 4A). We tested whether the increased expression of the gB mutants allowed fusion to HeLa cells previously shown to not mediate fusion, and none of the mutants mediated fusion to HeLa cells (data not shown). To confirm complete cell–cell fusion and not just the formation of a fusion pore, cell–cell fusion was also examined by means of microscopy. As shown in Fig. 4B with two of the gB mutants (gB-816 and RTTR), multinucleated cells were visible upon DAPI staining. Cells expressing gB, gH, and gL and the gB mutants with higher gB surface expression exhibited similar numbers of multinucleated cells. The gB-816-, gH-, and gL-transfected cells contained larger multinucleated cells, which is consistent with the luciferase data (data not shown).
Fig. 4.
Surface expression of gB mutants mediate fusion independent of gH/gL. Transfection of cells with various gB mutant constructs with gH and gL or alone were tested in the fusion assay. The target cells, HEK-293-P, were mixed 1:1 with the effector CHO-K1 cells, and some of the effector cells were transferred to a 96-well plate for cell ELISA to detect cell-surface expression. Twenty-four hours later, the cells from the fusion assay were harvested (shown in bars) and, alongside, a cell ELISA to detect cell-surface expression of the glycoproteins was performed by using the L2 antibody (Chemicon) (indicated by the line). Data are the average of three independent experiments, with standard deviations marked by vertical lines. (B) CHO-K1 cells were cotransfected with plasmids encoding EBV glycoproteins and EGFP-N1. Cells were fixed with 2% formaldehyde, stained with 1 μg/ml DAPI, and visualized with a Leica DMIRE2 microscope. Photos were taken with a Hamamatsu capture controller camera.
Discussion
The fusion step of herpesvirus entry is not well understood but is critical for the infection of target cells with free virus and the transmission of virus from infected cells to uninfected cells by cell–cell contact. To directly examine fusion, we used a virus-free cell–cell fusion assay to study the requirements and ability of EBV glycoproteins to induce fusion. We determined that efficient fusion is induced with EBV gB, gH, and gL when CHO-K1 effector cells are cocultivated with the target cells HEK-293-P, AGS, and SCC68. We also found that a mutant form of gB was sufficient to induce fusion alone, indicating that gB may be the critical fusion protein of EBV. A mutant form of HHV-8 gB is also able to induce fusion independent of other viral glycoproteins (27). With the HHV-8 gB mutant, fusion levels were at most 12% of the fusion levels observed when HHV-8 gB, gH, and gL were tested, which is lower than the amount of fusion we observed with our EBV gB mutant. Interestingly, higher fusion levels, ≈25% when compared with gB, gH, and gL fusion levels, were observed with HHV-8 gH and gL alone. These results are consistent with studies showing that Varicella–Zoster virus gH and gL alone, but not gB alone, form polykaryocytes in culture (40, 41). Along with our studies, the Varicella–Zoster virus and HHV-8 studies highlight the complexity of herpesvirus-induced cell fusion and contrast the potential roles that each of these virally encoded proteins have acquired for fusion.
One significant finding of the current study is that specific regions of the EBV gB C terminus modulate fusion in the presence of gH and gL. Loss of the region between 841–857 greatly reduced fusion with B cells and epithelial cells. This result is somewhat surprising, because the two larger gB deletions (gB-816 and gB-801) gained function in fusion. The gB-816 mutant can mediate both B cell and epithelial fusion, whereas the gB-801 mutant only functions in epithelial fusion. The difference with the gB-801 mutant between the two cell types may be due to the mechanism EBV utilizes to induce fusion within these different cell types. Early reports on EBV entry showed EBV fuses by means of endocytosis with B cells and by direct cell fusion with epithelial cells (10). Perhaps the region from 801–816 functions in endocytosis in B cell fusion, and, therefore, this region is not necessary for direct virion–cell fusion, suggesting that the differences between the fusion mechanisms are related to gB function. The gB-816 mutant, when expressed with gH and gL, showed enhanced fusion for both epithelial and B cells. The increased surface expression of gB-816 mutant is not responsible for the enhanced fusion observed, because point mutations in the ER-retention signal with similar levels of surface expression did not result in enhanced fusion.
Several functions for the EBV gB C-terminal tail in fusion can be entertained. The gB tail may regulate fusion by binding a cellular or viral protein. Interestingly, the results with the gB-801 mutant suggest this may be true, because inclusion of gH and gL does not enhance fusion with this mutant when compared with the other forms of gB examined. gH and gL may directly enhance fusion by altering the conformation of gB, positioning gB in a metastable form favorable for receptor binding or in promoting membrane interaction. This conformational change may be mediated by interactions of the gH/gL complex with the gB tail. The gH/gL complex may also enhance fusion by directly binding to a cellular receptor, although a specific cellular receptor for EBV gH or gL has not been identified. Currently, there is no data to suggest the binding of a cellular protein to the EBV gB tail. Future structural and biochemical studies will need to be performed to investigate these possibilities.
Another means that the EBV gB tail may modulate fusion is by regulating the oligomerization or conformation of gB. Recent results with HSV-2 gB C-terminal truncation mutants argue against this hypothesis, because all of the mutants that functioned in fusion showed similar oligomerization and conformation properties (29). Interestingly, the largest gB truncation, which did not function in fusion, appeared to be conformation-defective, indicating that the HSV-2 gB tail may influence the HSV-2 gB conformation. More recent results with the cytoplasmic tail of the F fusion protein from the paramyxovirus simian virus 5 indicate that tail domains of viral fusion proteins can regulate the oligomerization, conformation, and subsequent fusion competence of viral fusion proteins (50). Therefore, additional studies of the oligomerization and conformation of the EBV gB truncation mutants are clearly warranted to determine whether the EBV gB tail domain regulates the oligomerization or conformation of gB.
EBV gB may not only function in fusion but may also bind to a receptor on epithelial cells to trigger fusion. No cellular receptors for EBV gB have been identified, and this possibility has not been well studied. Previous studies have shown that the amount of gB in the virion differs between EBV strains with some strains, such as B95-8 having low levels when compared with other EBV glycoproteins (45, 51). EBV gB is expressed primarily in the ER and nuclear membrane and not on the plasma membrane (51–53). The ER and nuclear membrane expression appears to be critical for a previously identified role of gB in virion morphogenesis from the infected cell nucleus (21). The amount of gB found in the virion is an important determinant of the infection of various cell types (45) and may be an important mechanism to regulate viral entry into different cell types. The low expression of gB in the virion may explain why epithelial cells in vitro and in vivo are less susceptible to infection if a cellular receptor for gB exists. Two independent reports showed that gH-negative virions retain a small amount of virus binding to the surface of gastric carcinoma cell lines (54, 55). In both of these studies, the amount of gB present in the virion was not examined. Furthermore, because other herpesvirus gB interact with cellular receptors, such as the interaction of HHV-8 and human cytomegalovirus gB with integrins (56–58), the possibility of EBV gB having a receptor warrants further investigation.
Considering our findings, it is interesting to speculate that the possible progenitor virus of EBV was able to enter epithelial cells by using gB, gH, and gL. The acquisition or evolution of gp350/220 and gp42 to bind CD21/CR2 and HLA Class II, respectively, and to trigger fusion mediated by gB, gH, and gL would have allowed EBV to move from the portal of entry to the targeting of B cells to provide a cell type to establish a latent infection. Recently, another EBV glycoprotein, BMRF2, was reported to bind to integrins on polarized oropharyngeal cells and appears to be important for the infection of polarized oropharyngeal cells (59). BMRF2 may be another protein acquired by the virus for infection of specific cell types. BMRF2 may explain the disparity between fusion not seen with HeLa cells in our fusion assay but infection of HeLa cells by viruses expressing abundant gB (59). Further studies are needed to explore the role of other EBV glycoproteins in fusion and to determine whether there is a specific cellular receptor for gB. Additionally, future studies should result in a better understanding of the viral and host factors required for the infection and persistence of EBV in the human host.
Acknowledgments
We thank Jasmina Omerović (Northwestern University, Chicago) for the stable T7 Daudi cells, Nanette Susmarski for cell line expertise, Boris Popov (Monoclonal Antibody Facility, Northwestern University Feinberg School of Medicine) for preparing antibody, Lindsey Hutt-Fletcher for providing E1D1 and F-2-1 antibodies, and the members of the Longnecker and Spear laboratories for help and support. This work was supported by Public Health Service Grants CA62234, CA73507, and CA93444 from the National Cancer Institute; Public Health Service Grant DE13127 from the National Institute of Dental and Craniofacial Research (to R.L.); and the Carcinogenesis Training Program through National Cancer Institute/National Institutes of Health Grant T32CA009560 (to M.P.M.). R.L is a Stohlman Scholar of the Leukemia and Lymphoma Society of America.
Author contributions: M.P.M. and R.L. designed research; M.P.M. performed research; M.P.M. contributed new reagents/analytic tools; M.P.M. and R.L. analyzed data; and M.P.M. and R.L. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HHV, human herpesvirus; HSV, herpes simplex virus; ER, endoplasmic reticulum; EBV, Epstein–Barr virus; HEK, human embryonic kidney; CHO, Chinese hamster ovary.
References
- 1.Spear, P. G. & Longnecker, R. (2003) J. Virol. 77, 10179–10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young, J. A. T. (2001) in Fields Virology, ed. Howley, P. M. (Lippincott, Philadelphia), Vol. 1, pp. 87–103. [Google Scholar]
- 3.Kieff, E. & Rickinson, A. B. (2001) in Fields Virology, ed. Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2511–2573. [Google Scholar]
- 4.Spriggs, M. K., Armitage, R. J., Comeau, M. R., Strockbine, L., Farrah, T., Macduff, B., Ulrich, D., Alderson, M. R., Mullberg, J. & Cohen, J. I. (1996) J. Virol. 70, 5557–5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, X. & Hutt-Fletcher, L. M. (1998) J. Virol. 72, 158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanner, J., Weis, J., Fearon, D., Whang, Y. & Kieff, E. (1987) Cell 50, 203–213. [DOI] [PubMed] [Google Scholar]
- 7.Fingeroth, J. D., Weis, J. J., Tedder, T. F., Strominger, J. L., Biro, P. A. & Fearon, D. T. (1984) Proc. Natl. Acad. Sci. USA 81, 4510–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haan, K. M., Lee, S. K. & Longnecker, R. (2001) Virology 290, 106–114. [DOI] [PubMed] [Google Scholar]
- 9.Miller, N. & Hutt-Fletcher, L. M. (1992) J. Virol. 66, 3409–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nemerow, G. R. & Cooper, N. R. (1984) Virology 132, 186–198. [DOI] [PubMed] [Google Scholar]
- 11.Rickinson, A. B. & Kieff, E. (2001) in Fields Virology, ed. Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2574–2627. [Google Scholar]
- 12.Imai, S., Nishikawa, J. & Takada, K. (1998) J. Virol. 72, 4371–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speck, P. & Longnecker, R. (2000) J. Natl. Cancer Inst. 92, 1849–1851. [DOI] [PubMed] [Google Scholar]
- 14.Borza, C. M. & Hutt-Fletcher, L. (2002) Nat. Med. 8, 594–599. [DOI] [PubMed] [Google Scholar]
- 15.Li, Q., Turk, S. M. & Hutt-Fletcher, L. M. (1995) J. Virol. 69, 3987–3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutt-Fletcher, L. M. & Lake, C. M. (2001) Curr. Top. Microbiol. Immunol. 258, 51–64. [DOI] [PubMed] [Google Scholar]
- 17.Maruo, S., Yang, L. & Takada, K. (2001) J. Gen. Virol. 82, 2373–2383. [DOI] [PubMed] [Google Scholar]
- 18.Borza, C. M., Morgan, A. J., Turk, S. M. & Hutt-Fletcher, L. M. (2004) J. Virol. 78, 5007–5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santoro, F., Greenstone, H. L., Insinga, A., Liszewski, M. K., Atkinson, J. P., Lusso, P. & Berger, E. A. (2003) J. Biol. Chem. 278, 25964–25969. [DOI] [PubMed] [Google Scholar]
- 20.Sarmiento, M., Haffey, M. & Spear, P. G. (1979) J. Virol. 29, 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S. K. & Longnecker, R. (1997) J. Virol. 71, 4092–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai, W. Z., Person, S., Warner, S. C., Zhou, J. H. & DeLuca, N. A. (1987) J. Virol. 61, 714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp, B. G., Nixdorf, R. & Mettenleiter, T. C. (2000) J. Virol. 74, 6760–6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muggeridge, M. I. (2000) J. Gen. Virol. 81, 2017–2027. [DOI] [PubMed] [Google Scholar]
- 25.Turner, A., Bruun, B., Minson, T. & Browne, H. (1998) J. Virol. 72, 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pertel, P. E., Fridberg, A., Parish, M. L. & Spear, P. G. (2001) Virology 279, 313–324. [DOI] [PubMed] [Google Scholar]
- 27.Pertel, P. E. (2002) J. Virol. 76, 4390–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pereira, L. (1994) Infect. Agents Dis. 3, 9–28. [PubMed] [Google Scholar]
- 29.Fan, Z., Grantham, M. L., Smith, M. S., Anderson, E. S., Cardelli, J. A. & Muggeridge, M. I. (2002) J. Virol. 76, 9271–9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bzik, D. J., Fox, B. A., DeLuca, N. A. & Person, S. (1984) Virology 137, 185–190. [DOI] [PubMed] [Google Scholar]
- 31.Gage, P. J., Levine, M. & Glorioso, J. C. (1993) J. Virol. 67, 2191–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cai, W. H., Gu, B. & Person, S. (1988) J. Virol. 62, 2596–2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DeLuca, N., Bzik, D. J., Bond, V. C., Person, S. & Snipes, W. (1982) Virology 122, 411–423. [DOI] [PubMed] [Google Scholar]
- 34.Nixdorf, R., Klupp, B. G. & Mettenleiter, T. C. (2001) J. Gen. Virol. 82, 215–226. [DOI] [PubMed] [Google Scholar]
- 35.Nixdorf, R., Klupp, B. G., Karger, A. & Mettenleiter, T. C. (2000) J. Virol. 74, 7137–7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nixdorf, R., Klupp, B. G. & Mettenleiter, T. C. (2001) J. Virol. 75, 11526–11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Favoreel, H. W., Van Minnebruggen, G., Nauwynck, H. J., Enquist, L. W. & Pensaert, M. B. (2002) J. Virol. 76, 6845–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foster, T. P., Melancon, J. M. & Kousoulas, K. G. (2001) Virology 287, 18–29. [DOI] [PubMed] [Google Scholar]
- 39.Lee, S. K. (1999) Virology 264, 350–358. [DOI] [PubMed] [Google Scholar]
- 40.Duus, K. M., Hatfield, C. & Grose, C. (1995) Virology 210, 429–440. [DOI] [PubMed] [Google Scholar]
- 41.Maresova, L., Pasieka, T. J. & Grose, C. (2001) J. Virol. 75, 9483–9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva, A. L., Omerovic, J., Jardetzky, T. S. & Longnecker, R. (2004) J. Virol. 78, 5946–5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janz, A., Oezel, M., Kurzeder, C., Mautner, J., Pich, D., Kost, M., Hammerschmidt, W. & Delecluse, H. J. (2000) J. Virol. 74, 10142–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshiyama, H., Imai, S., Shimizu, N. & Takada, K. (1997) J. Virol. 71, 5688–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuhierl, B., Feederle, R., Hammerschmidt, W. & Delecluse, H. J. (2002) Proc. Natl. Acad. Sci. USA 99, 15036–15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh, S., Munshi, H. G., Sen, R., Linz-McGillem, L. A., Goldman, R. D., Lorch, J., Green, K. J., Jones, J. C. & Stack, M. S. (2002) Cancer 95, 2524–2533. [DOI] [PubMed] [Google Scholar]
- 47.Wang, X., Kenyon, W. J., Li, Q., Mullberg, J. & Hutt-Fletcher, L. M. (1998) J. Virol. 72, 5552–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strnad, B. C., Schuster, T., Klein, R., Hopkins, R. F., III, Witmer, T., Neubauer, R. H. & Rabin, H. (1982) J. Virol. 41, 258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balachandran, N., Pittari, J. & Hutt-Fletcher, L. M. (1986) J. Virol. 60, 369–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waning, D. L., Russell, C. J., Jardetzky, T. S. & Lamb, R. A. (2004) Proc. Natl. Acad. Sci. USA 101, 9217–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gong, M. & Kieff, E. (1990) J. Virol. 64, 1507–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gong, M., Ooka, T., Matsuo, T. & Kieff, E. (1987) J. Virol. 61, 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Emini, E. A., Luka, J., Armstrong, M. E., Keller, P. M., Ellis, R. W. & Pearson, G. R. (1987) Virology 157, 552–555. [DOI] [PubMed] [Google Scholar]
- 54.Molesworth, S. J., Lake, C. M., Borza, C. M., Turk, S. M. & Hutt-Fletcher, L. M. (2000) J. Virol. 74, 6324–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oda, T., Imai, S., Chiba, S. & Takada, K. (2000) Virology 276, 52–58. [DOI] [PubMed] [Google Scholar]
- 56.Akula, S. M., Pramod, N. P., Wang, F. Z. & Chandran, B. (2002) Cell 108, 407–419. [DOI] [PubMed] [Google Scholar]
- 57.Feire, A. L., Koss, H. & Compton, T. (2004) Proc. Natl. Acad. Sci. USA 101, 15470–15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, F. Z., Akula, S. M., Sharma-Walia, N., Zeng, L. & Chandran, B. (2003) J. Virol. 77, 3131–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tugizov, S. M., Berline, J. W. & Palefsky, J. M. (2003) Nat. Med. 9, 307–314. [DOI] [PubMed] [Google Scholar]