Abstract
The three-dimensional structure of the complex formed between the cytosolic chaperonin CCT (chaperonin containing TCP-1) and phosducin (Pdc)-like protein (PhLP), a regulator of CCT activity, has been solved by cryoelectron microscopy. Binding of PhLP to CCT occurs through only one of the chaperonin rings, and the protein does not occupy the central folding cavity but rather sits above it through interactions with two regions on opposite sides of the ring. This causes the apical domains of the CCT subunits to close in, thus excluding access to the folding cavity. The atomic model of PhLP generated from several atomic structures of the homologous Pdc fits very well with the mass of the complex attributable to PhLP and predicts the involvement of several sequences of PhLP in CCT binding. Binding experiments performed with PhLP/Pdc chimeric proteins, taking advantage of the fact that Pdc does not interact with CCT, confirm that both the N- and C-terminal domains of PhLP are involved in CCT binding and that several regions suggested by the docking experiment are indeed critical in the interaction with the cytosolic chaperonin.
Keywords: molecular chaperones, PhLP, protein folding, electron microscopy
Molecular chaperones are a large class of proteins that assist other proteins in attaining their active conformation. Among them, chaperonins are a ubiquitous family of chaperones that have a common toroidal structure formed by the oligomerization of 60-kDa proteins. The toroid is made of two rings placed back-to-back with each ring enclosing a cavity where folding occurs (1). Chaperonins have been classically divided in two groups depending on whether they are found in eubacteria and in the endosymbiotic organelles (group I) (2) or in archaea and the cytosol of eukarya (group II) (3). The monomers of every chaperonin known share a very similar three-domain structure (4, 5): an equatorial domain that contains the nucleotide binding site and most of the interaction sites between the subunits of the same ring and of the opposite ring; an apical domain where the substrate binding site is located; and an intermediate domain that transmits to the apical domain the signals generated in the equatorial domain upon nucleotide binding. Chaperonins act on unfolded substrates by using a general mechanism that involves the recognition of the unfolded polypeptide by hydrophobic residues at the entrance of the chaperonin cavity, followed by folding of the polypeptide upon closure of the cavity induced by the binding of ATP and a cochaperonin (6).
A more specific mechanism seems to operate for the group II eukaryotic cytosolic chaperonin CCT (chaperonin containing TCP-1), whose toroidal structure is made up of two rings composed of eight different but homologous proteins (7). The work carried out with the major CCT substrates, actin and tubulin, has shown that the recognition mechanism operates through defined CCT subunits and specific domains of the substrates, which have already acquired a large degree of native-like conformation before interacting with CCT. The conformational changes undergone by CCT upon nucleotide binding would be used to actively fold the two cytoskeletal proteins (6) or to generate a structure apt to form a stable complex with other proteins (8).
In contrast, a novel role for CCT other than folding or complex formation seems to be behind its interaction with phosducin (Pdc)-like protein (PhLP). PhLP and its homologue Pdc are involved in the regulation of cell signaling through their interaction with the G protein βγ subunit complex (Gβγ). Binding of PhLP or Pdc prevents Gβγ from interacting with the Gα subunit or downstream effectors (9–13). Unlike protein folding substrates, PhLP has been shown to interact with CCT in its native form and to inhibit the chaperonin actin folding activity (14), suggesting that PhLP may be a regulator of CCT activity or conversely that CCT could control the availability of PhLP during G protein signaling (14). To gain further insight into the interaction between CCT and PhLP, we have carried out electron microscopy and biochemical analysis of the CCT:PhLP complex. The three-dimensional reconstruction of CCT:PhLP obtained by cryoelectron microscopy together with a docking analysis carried out with an atomic model of PhLP and with CCT binding experiments performed with various PhLP mutants has led to the determination of the regions of PhLP and the subunits of CCT involved in the formation of the CCT:PhLP complex, and to a hypothesis of the role of the CCT:PhLP interaction.
Materials and Methods
Protein Preparation. CCT was purified from soluble extracts of bovine testis as described in ref. 15. Gβ1γ1 was purified from bovine retina and recombinant rat PhLP, and the PhLP/Pdc chimeric proteins were expressed and purified from Escherichia coli as described in ref. 16. The CCT:PhLP complexes were formed by incubating CCT and PhLP in a 1:10 molar ratio for 30 min at 25°C. In the case of the CCT:PhLP:antibody immunocomplexes, preformed CCT:PhLP complexes were incubated with anti-CCTδ 8g monoclonal antibody (5:1 antibody:complex molar ratio) during 15 min at 25°C.
Generation of PhLP/Pdc Chimeras. The cDNA for wild-type rat PhLP and Pdc with a 3′ c-myc epitope tag were previously constructed in the pET15b vector (14). The PhLP/Pdc chimeras were made by PCR amplification of two PhLP cDNA fragments from this vector. The fragments were divided at an endonuclease restriction site within the Pdc sequence to be inserted or the PhLP sequence near the replacement point. If the restriction site was within the Pdc insert, fragments were amplified with primers complimentary to the sequence of PhLP at the replacement point with overhangs containing the Pdc sequence including the restriction site. If there was no restriction site within the Pdc insert, then a primer containing complementary nucleotides of PhLP next to the replacement point, the entire Pdc sequence to be inserted, and the additional PhLP sequence up to the restriction site was used. The other primer was complementary to PhLP sequence including the restriction site. Each fragment was then amplified by pairing these primers with either the T7 forward or reverse primers from pET15b flanking the PhLP cDNA. The fragments were cut at the restriction site, gel-purified, and ligated. The full-length chimeras were then PCR-amplified by using the T7 forward and reverse primers and inserted into the pET15b vector by using the NcoRI and BamHI restriction sites. For the P193R chimera, the single amino acid substitution was made by using the QuikChange protocol (Stratagene). All constructs were confirmed by DNA sequence analysis.
Binding of PhLP/Pdc Chimeras to CCT. Binding of the PhLP/Pdc chimeras to CCT was measured by coimmunoprecipitation and immunoblotting. Purified PhLP/Pdc chimeric proteins (250 nM) were added to 10% rabbit reticulocyte lysate in PBS with 0.5 mM PMSF and 0.5% Igepal CA-630 detergent in a 100-μl total volume and incubated for 15 min at 4°C. PhLP/Pdc complexes were immunoprecipitated by using an antibody to the C-terminal c-myc tag fused to each chimera and immunoblotted with an antibody to CCTα or Gβγ as described in ref. 14. Intensities of the CCTα bands from the PhLP/Pdc chimera were expressed as a percentage of the CCTα band intensity from the wild-type PhLP immunoprecipitates.
Electron Microscopy. For cryoelectron microscopy, 5-μl aliquots of a solution containing CCT:PhLP complexes were applied to glow-discharged holey carbon grids for 1 min, blotted for 5 sec, and frozen rapidly in liquid ethane at –180°C. Images were recorded at 20° tilt under minimum dose conditions in a FEI G2 FEG electron microscope equipped with a Gatan cold stage operated at 200 kV and recorded on Kodak SO-163 film at ×62,000 nominal magnification and between 1.5 and 2.5 μm underfocus. For electron microscopy of negatively stained samples, 5-μl aliquots were applied to glow-discharged carbon grids for 1 min and then stained for 1 min with 2% uranyl acetate. Images were recorded at 0° tilt in a JEOL 1200EX-II electron microscope operated at 100 kV and recorded at ×60,000 nominal magnification.
Image Processing, Two-Dimensional Averaging, and Three-Dimensional Reconstruction. Micrographs were digitized in a Zeiss SCAI scanner with a sampling window corresponding to 3.5 Å per pixel for negatively stained samples and 3.2 Å per pixel for vitrified samples. For two-dimensional classification and averaging, top and side views of CCT particles were selected, aligned by using a free-pattern algorithm, and classified by using self-organizing maps as described in ref. 15 to separate the PhLP-bound CCT particles from those free of PhLP.
The three-dimensional reconstruction of the CCT:PhLP complex was generated from randomly oriented particles whose orientation was determined by using the angular refinement algorithms provided by spider (17). The volumes were generated by using the back-projection method (20). No symmetrization was applied to any of the volumes obtained during the iterative procedure. The final resolution was estimated with the 0.5 criterion for the Fourier shell correlation coefficient between two independent reconstructions by using bsoft (18). Visualization of the volumes was carried out by using amira (http://amira.zib.de).
Modeling of PhLP and Docking of the CCT:PhLP Complex. The atomic model of PhLP was generated by homology modeling techniques using the sequences and atomic structures of four Pdc proteins (PDB ID codes 2TRC, 1AOR, 1B9Y, and 1B9X) with the dali comparison algorithm (19) at the swiss-model server facilities (20) (http://swissmodel.expasy.org//SWISS-MODEL.html). The atomic model of PhLP was then fitted manually into the three-dimensional reconstruction of the CCT:PhLP complex by using o (21).
Results and Discussion
The Formation of the CCT:PhLP Complex. To confirm the reported interaction of PhLP with CCT and to visualize the CCT:PhLP complexes, purified CCT was incubated in the absence or presence of a 10 molar excess of purified PhLP, and the samples were stained as described in Materials and Methods. Two typical views were observed under the electron microscope: the most common top view revealing the octameric nature of the CCT rings, and the less frequent side view showing the two-ring structure of the chaperonin. The latter view turned out to be the most informative in detecting the absence (Fig. 1A) or the presence of PhLP bound to the chaperonin oligomer (Fig. 1B), which seems to occur outside the folding cavity. PhLP protrudes from the apical region of the chaperonin in a manner similar to the interaction between CCT and its cochaperone prefoldin (PFD) (15). However, unlike what happens with PFD, the side views of the CCT:PhLP complex indicate that the interaction between PhLP and CCT occurs with only one of the chaperonin rings, regardless of the amount of PhLP added to the CCT solution, confirming the 1:1 stoichiometry for the CCT:PhLP complex described in ref. 14.
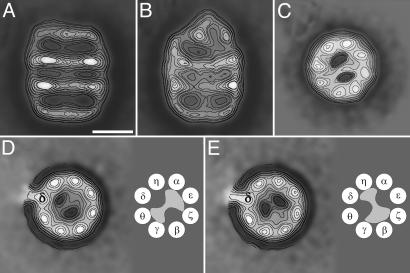
Fig. 1.
Two-dimensional average images of negatively stained CCT:PhLP complex. (A) Average image of side views obtained from 243 CCT particles of apo-CCT. (B) Average image obtained from 286 side views of CCT:PhLP complexes. (C) Average image obtained from 4,225 top views of CCT:PhLP complexes. (D and E) Average images of the two types of top views of CCT:PhLP:8g (anti-CCTδ) immunocomplexes (average of 324 and 626 particles). The subunit labeled by the antibody is marked with “δ.” (Scale bar, 100 Å.) A schematic model of the each mode of PhLP binding, with the topology of the CCT subunits according to ref. 24, accompanies each average image.
CCT Subunits Involved in PhLP Binding. The side view of the CCT:PhLP complex depicted in Fig. 1B also suggests the interaction of PhLP with regions on opposite sides of the CCT cavity. This orientation is confirmed by the average top view image of the same complex (Fig. 1C), which shows that an asymmetric mass traverses the chaperonin cavity and interacts with two CCT subunits on one side of the cavity and three CCT subunits in the other side. This interaction is geometry-dependent, similar to what has already been described for actin (22) and tubulin (23). To determine whether the interaction is also subunit-specific we made use of a monoclonal antibody reacting against the CCTδ subunit (8g) (22). Aliquots of the immunocomplexes were negatively stained (to contrast only one of the CCT rings), and 950 top views were processed. After the classification procedures, two main populations were obtained with PhLP present in the CCT cavity whose average images are represented in Fig. 1 D and E, respectively. Both images reproduce a similar mass crossing the CCT cavity. The specificity of the monoclonal antibody and the known topology of the CCT ring (24) allowed determination of the CCT subunits involved in PhLP binding. The average image shown in Fig. 1D represents 65% of the CCT:PhLP complexes and points to an interaction of PhLP with CCTγ/θ on one side of the CCT cavity and CCTα/ε/ξ on the other side. In the average image representing the remaining 35% of the CCT:PhLP complexes (Fig. 1E), PhLP seems to interact with CCTδ/η on one side of the cavity and CCTξ/β/γ on the other side. The structural basis for these two different modes of interaction and their physiologically relevance remains to be determined. Nevertheless, in either structure PhLP binding occludes the CCT cavity, possibly explaining why PhLP competes with other substrates for their interaction with CCT and therefore regulates the chaperonin folding activity (14).
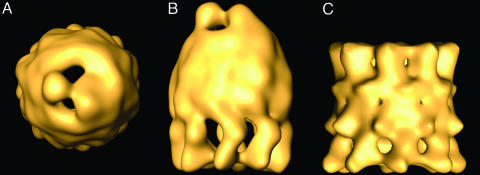
Three-Dimensional Structure of the CCT:PhLP Complex. To further characterize the interaction between PhLP and the cytosolic chaperonin, a three-dimensional reconstruction of the CCT:PhLP complex was carried out by cryoelectron microscopy and image processing. After image classification, a homogeneous population of 2,625 particles was obtained and used to generate a three-dimensional reconstruction of the CCT:PhLP complex (Fig. 2 A and B). The reconstruction reveals an asymmetric, bullet-shaped structure as was observed in the two-dimensional average image of the side view of the same complex (Fig. 1B). Compared with the three-dimensional reconstruction of apo-CCT (Fig. 2C), the CCT:PhLP complex shows important differences, especially in the PhLP-bound CCT ring. One difference has to do with the mass clearly attributed to PhLP that sits at the entrance of the cavity and protrudes from it. In contrast to the interaction of CCT with actin (22) tubulin (23), or its cochaperone, PFD (15), no part of the PhLP mass penetrates into the folding cavity but simply interacts with two opposite sides of the top apical region. The level of resolution of the CCT:PhLP complex (26 Å) allows visualization of the PhLP mass as a two-domain structure connected by a small linker. The two domains are clearly asymmetric, the small one interacting with two CCT subunits and the large one with three subunits (Fig. 2 A). Another difference is a PhLP-induced movement of the apical domains of the CCT subunits, reducing the diameter of the entrance of the folding cavity from ≈80 Å to ≈55 Å and leaving the entrance almost occluded by the presence of PhLP (Fig. 2 A). This finding confirms the flexibility of the apical domains, which are capable of undergoing large conformational changes within the functional cycle and of accommodating substrates of different sizes (25). These large conformational changes of the apical domains induced by PhLP suggest a high-affinity interaction, consistent with the 190 nM Kd reported in ref. 14. The high binding affinity appears to derive from a concerted action of the two PhLP domains and all eight CCT subunits, probably involving multiple contacts. Finally, the reconstruction also confirms the binding of PhLP to only one of the CCT rings (14) and strongly suggests that the movement of the apical domains in the PhLP-bound ring transmits an allosteric signal through the equatorial domains so that no PhLP molecule is able to bind to the opposite ring.
Fig. 2.
Three-dimensional reconstruction of the CCT:PhLP complex by cryoelectron microscopy. (A) Top view of the CCT:PhLP complex. (B) Side view of the same volume. (C) Side view of the three-dimensional reconstruction of apo-CCT (23).
Docking Analysis of PhLP into the Three-Dimensional Structure of the CCT:PhLP Complex. PhLP belongs to a family of widely expressed regulators of G protein signaling (26). Although no atomic structure is available for PhLP, there is a high degree of sequence homology between PhLP and Pdc (41% amino acid identity) (27), another member of the family for which several atomic structures are available (12, 28, 29). This similarity allowed us to generate an atomic model of PhLP by homology modeling techniques (see Materials and Methods). The atomic model (Fig. 3) lacked the first 50 residues of the rat PhLP sequence, which are not present in Pdc, and the last 24 residues not defined in the atomic structures of Pdc. The model naturally shows very similar structural features to the Pdc atomic structure (Fig. 3B): a unstructured N-terminal domain built up by three α-helices (H1–H3) and a more compact C-terminal domain showing a typical thioredoxin fold (12, 30), with a core formed by a five-stranded β-sheet (S1–S5) flanked by four α-helices (H4–H7). The two domains are linked by a flexible loop that connects H3 and S1.
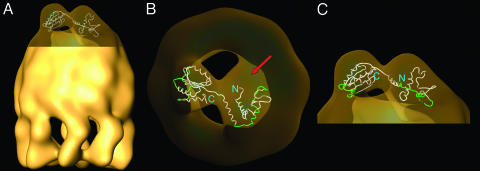
Fig. 3.
Docking of the atomic model of PhLP into the three-dimensional reconstruction of the CCT:PhLP complex. (A) Docking of the atomic model of PhLP into the CCT:PhLP volume. (B and C) Two enlarged views of the docking of the PhLP atomic model (drawn in tubes) into the CCT:PhLP complex (depicted in transparent fashion). The red arrow in B indicates a region of the PhLP mass that could be filled by the 50 residues of the N-terminal sequence of PhLP not present in the PhLP atomic model. The green regions in the atomic model of PhLP are those suggested by the docking analysis to be involved in CCT binding.
A docking analysis was carried out by fitting the atomic model of PhLP into the mass of the CCT:PhLP complex attributable to PhLP (Fig. 3). The fit is very good only when the C-terminal domain is assigned to the smaller, more compact of the two PhLP masses of the reconstructed volume. The N-terminal domain fits well into the larger mass, and although there is a portion of the mass that is not filled, this could be attributed to the 50 residues of the N-terminal domain not present in the atomic model (red arrow in Fig. 3B).
An analysis of the docking results suggests the involvement of several regions of PhLP in the binding of CCT (Fig. 3 B and C). In the N-terminal domain, a large stretch of amino acids runs parallel to the apical domains of the three CCT subunits that are in close proximity to PhLP (Fig. 3C) and suggests a possible binding interface (Fig. 4A). This region (K109–E135) encompasses part of the long H1–H2 loop, H2, the H2–H3 loop, and the N-terminal part of H3. In addition, the 50 N-terminal residues not present in the atomic model could potentially be involved in CCT binding through an interaction with the third CCT subunit (red arrow in Fig. 3B). In the C-terminal domain, three regions are likely candidates for interaction with the two CCT subunits (Fig. 3 B and C), the loops between S2 and H5 (E189–G194), H6 and S4 (G223–N231), and S5 and H7 (V249–D258). Additionally, part of the last 24 residues of the sequence, not present in the atomic model, might be placed in the bottom part of the PhLP mass and therefore could also be involved in CCT binding. In all, the electron microscopy shows clearly that both domains of PhLP are involved in CCT binding, and the docking of the atomic model of PhLP into the three-dimensional reconstruction of the CCT:PhLP complex points to several specific regions of both N- and C-terminal domains of PhLP as involved in CCT binding.
Fig. 4.
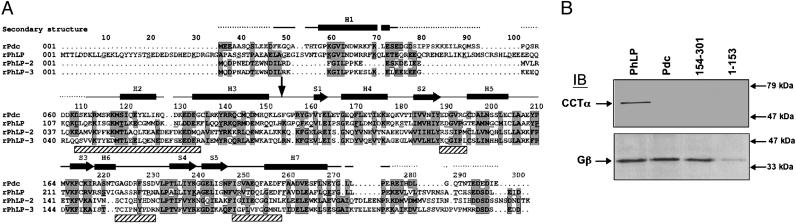
Both domains of PhLP participate in CCT binding. A sequence alignment of rat Pdc, PhLP1, PhLP2, and PhLP3 is shown in A. Conserved residues are indicated with gray boxes, and secondary structural elements for Pdc (12) are indicated above the sequence (H for helix and S for β-strand). Shaded boxes below the structural elements represent regions implicated in CCT binding by the docking analysis. A vertical arrow at residue 154 marks the loop between the N-and C-terminal domains. The PhLP/Pdc(154–301) chimera contains the N-terminal domain of PhLP and the C-terminal domain of Pdc and vice versa for the PhLP/Pdc(1–153) chimera. In B, the binding of these proteins to CCT or Gβγ was determined by immunoprecipitation of the PhLP chimeras and immunoblotting for CCTα and Gβ as described in Materials and Methods. Immunoblots show representative data from three separate experiments. Positions of molecular weight standards are shown on the right.
Biochemical Analysis of the CCT–PhLP Interaction. To assess the validity of this structural model of the PhLP–CCT interaction, the binding properties of a set of chimeric proteins were generated in which the PhLP sequences implicated in CCT binding by the docking analysis were replaced with the corresponding Pdc sequence. This mapping strategy takes advantage of the fact that although both proteins are homologous, only PhLP interacts with CCT (14). A set of two chimeras was generated in which the N-terminal (residues 1–153) and C-terminal (residues 154–301) domains of PhLP were switched with the corresponding region of Pdc (Fig. 4A). The two chimeras were then assayed for CCT and Gβγ binding by coimmunoprecipitation and immunoblotting, the latter serving as a control for the ability of the chimeras to maintain their functional activity and therefore their native conformation. The results in Fig. 4B show that neither chimera binds CCT yet both are able to bind Gβγ, indicating that both the N- and C-terminal domains of PhLP are required for CCT binding. The diminished Gβγ binding of PhLP/Pdc(1–153) is anticipated, given the fact that the N-terminal domain of PhLP contributes more to Gβγ binding than that of Pdc, and that the C-terminal domain of PhLP contributes less than the homologous region of Pdc (16). These results clearly confirm the structural data showing that contacts from both N- and C-terminal domains of PhLP are required for CCT binding.
The next step was to investigate in detail which specific regions of PhLP are involved in CCT binding, using the information provided by the docking analysis. Several PhLP/Pdc chimeric proteins were generated in both N- and C-terminal domains of PhLP and were also assayed for CCT and Gβγ binding (Figs. 5 and 6).
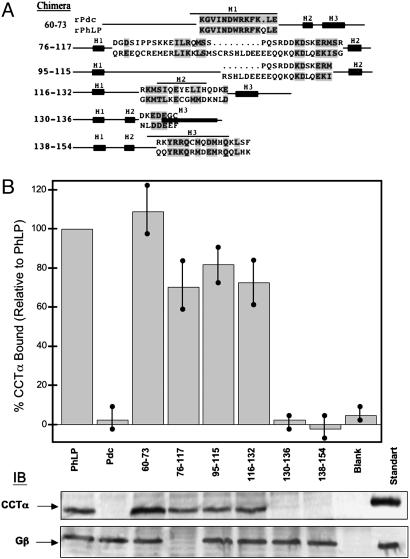
Fig. 5.
Binding of PhLP/Pdc chimeras within the N-terminal domain to CCT. Chimeras of PhLP within the N-terminal domain were made by inserting Pdc sequence as shown in A. The numbers indicate the residues of PhLP that were replaced with the corresponding Pdc residues and conserved residues within the replacements are located in gray boxes. Binding of these PhLP chimeras to CCT or Gβγ was measured as in Fig. 4. (B) Representative immunoblots for CCTα and Gβ, as well as a graphical representation of the CCTα binding data normalized to wild-type PhLP. Bars represent the mean ± standard error from seven separate experiments. No PhLP was added to the blank sample. The standard lanes contain 700 ng of purified CCT (90 ng of CCTα) or 25 ng of Gβγ (21 ng of Gβ).
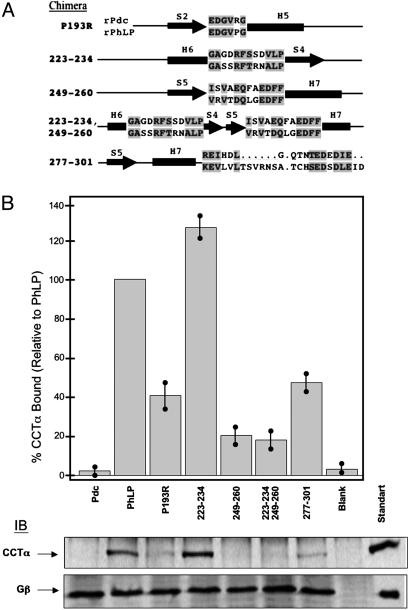
Fig. 6.
Binding of PhLP/Pdc chimeras within the C-terminal domain to CCT. Chimeras of PhLP within the C-terminal domain were made by inserting Pdc sequence in the loops between the predicted secondary structural elements as shown in A. The numbers indicate the residues of PhLP that were replaced with the corresponding Pdc residues and conserved residues within the replacements are located in gray boxes. Binding of these PhLP chimeras to CCT or Gβγ was measured as in Fig. 4. (B) Representative immunoblots for CCTα and Gβ, as well as a graphical representation of the CCTα binding data normalized to wild-type PhLP. Bars represent the mean ± standard error from six separate experiments. The lanes contain the same amounts of protein as in Fig. 5.
In the N-terminal domain, six PhLP/Pdc chimeric proteins were designed to cover most of the secondary structures elements of this domain (Fig. 5A): PhLP/Pdc(60–73), in which the putative H1 of PhLP had been switched to the corresponding Pdc sequence; PhLP/Pdc(76–117) and PhLP/Pdc(95–115), covering all or only the C-terminal half of the H1–H2 loop respectively; PhLP/Pdc(116–132), encompassing the last few residues of the H1–H2 loop, H2, and the H2–H3 loop; PhLP/Pdc(130–136), covering the H2–H3 loop and the three N-terminal residues of H3; and finally, PhLP/Pdc(138–154), encompassing H3. The CCT binding assay with the PhLP/Pdc(60–73) chimera showed no decrease with respect to wild-type PhLP (Fig. 5B), consistent with the docking analysis showing no interaction of H1 with CCT. Binding assays with chimeras PhLP/Pdc(76–117), PhLP/Pdc(95–115), and PhLP/Pdc(116–132) revealed a small decrease in the interaction with the chaperonin (≈20–30%), indicating that the H1–H2 loop, H2, and the N-terminal part of the H2–H3 loop individually make only minor contributions to chaperonin binding. The CCT binding assays with chimeras PhLP/Pdc(130–136) and PhLP/Pdc(138–154) showed a complete suppression of chaperonin binding. The combined information obtained from chimeras PhLP/Pdc(116–132), PhLP/Pdc(130–136), and PhLP/Pdc(138–154) points to H3 and the C-terminal part of the H2–H3 loop as critical for CCT binding (Fig. 5). According to the docking analysis, the H2–H3 loop and the N-terminal part of H3 make contact with CCT. In the PhLP/Pdc(130–136) chimera, three nonconservative changes, L131K, E135G, and F136G, abolish CCT binding (Fig. 5), suggesting that the stretch of negative charge D132DEE surrounded by hydrophobic residues is required for CCT binding. Furthermore, in the PhLP/Pdc(138–154) chimera, H3 residues Q138Q that are on the same side of H3 as E134E are replaced with R and K, respectively, increasing the positive charge in this face of H3 (Fig. 5). Interestingly, the negatively charged character of this region is conserved in other PhLP members like PhLP2 and PhLP3 (Fig. 4A), which are also believed to interact with CCT (31, 32). Indeed, replacement of the D132DEE stretch with alanines in human PhLP abolishes its CCT binding ability (data not shown). Thus, it appears that the negatively charged stretch in the H2–H3 loop and at the N terminus of H3 is critical for CCT binding.
In the C-terminal domain, five chimeras were generated based on the information extracted from the docking analysis (Fig. 6A). They were PhLP/Pdc(P193R), in which only a single mutation was necessary to generate the Pdc sequence for the S2–H5 loop; PhLP/Pdc(223–234), encompassing the H6–S4 loop; PhLP/Pdc(249–260), encompassing the S5–H7 loop; and PhLP/Pdc(223–234/249–260), a double-loop chimera switching both of these two later loops. A fifth chimera, PhLP/Pdc(277–301), covered the last 24 residues of the PhLP sequence, a region whose structure was not predicted by the homology modeling experiment but which could be potentially involved in chaperonin binding. The CCT binding assays (Fig. 6B) revealed a 60% decrease in chaperonin binding for PhLP/Pdc(P193R) compared with wild-type PhLP, suggesting that residue P193 is involved in the interaction with CCT, probably through the maintenance of a certain local conformation. Other binding assays showed a small 25% increase in binding with PhLP/Pdc(223–234) and a large 80% decrease in binding with chimera PhLP/Pdc(249–260). A similar 80% decrease was observed for the double-loop chimera PhLP/Pdc(223–234/249–260). These results indicate that the H6–S4 loop is not involved in CCT interaction, whereas the S5–H7 loop has an important role in chaperonin binding. In the PhLP/Pdc(249–260) chimera, there is only one nonconservative substitution (R250S; Fig. 6A), suggesting that the positive charge at R250 plays a role in CCT binding. Replacement of the last 24 residues of the C-terminal domain of PhLP with chimera PhLP/Pdc(277–301) generated a significant 50% decrease in CCT binding, suggesting that this region is also involved in the interaction with the chaperonin.
These biochemical data are generally consistent with the structural model proposed from the docking analysis, confirming most of the suggested contacts and clearly showing that PhLP interacts with CCT through specific regions in both N- and C-terminal domains. In all, the CCT binding experiments shown here suggest that apart from the stringent binding site in the region encompassing part of the H2–H3 loop and H3, PhLP interacts with CCT through the concerted interaction of several regions of both N- and C-terminal domains, similar to what has been described for actin and tubulin (22, 23).
As mentioned earlier, the CCT:PhLP structure displays an interesting similarity to that of the CCT:PFD complex (15) in that PhLP sits above the CCT folding cavity, contacting the apical domains of the CCT subunits and occluding the entrance into the cavity. The function of the cochaperone PFD is to assist in the folding of actin and tubulin by binding to their nascent polypeptide chains and delivering them to CCT for folding into their native structures (15). Several lines of evidence suggest a similar role of PhLP with regard to Gβγ folding and/or assembly. First, genetic deletion of PhLP in Dictyostelium discoideum blocked G protein signaling and membrane localization of the Gβγ complex (33). According to these authors, PhLP could be involved in facilitating the correct folding of Gβ or its assembly into the Gβγ complex. Second, the need of chaperones for the correct folding of WD40 proteins like Gβ has been already demonstrated (34, 35). Interestingly, the interaction of some of these WD40 proteins with CCT has also been described, and a folding role of CCT has been demonstrated for some of them (36, 37). In fact, CCT seems to interact with at least 17% of the yeast WD40 proteins including Ste4, the yeast Gβ subunit (38). Third, a proteomic analysis of the protein complexes in yeast revealed an interaction between CCT, yeast PhLP2, and VID27, a Gβ protein homologue (31). Fourth, a recent genetic study reveals that the cochaperoning role of PhLP with respect to CCT could be extended to the folding of β-tubulin (32). In light of these data, it is tempting to suggest that PhLP acts as a cochaperone in concert with CCT to catalyze the folding of Gβ proteins and/or the assembly of the Gβγ complexes.
Acknowledgments
We thank Prof. K. R. Willison for the kind gift of his anti-CCTδ 8g monoclonal antibody. This work was supported by Ministry of Education and Science (Spain) Grants BMC2001-0950 (to J.M.V.), BFU2004-00232 (to J.M.V.), and BMC-00996 (to J.L.C.), National Science Foundation Grant MCB-0131361 (to B.M.W.) and National Institutes of Health Grant EY12287 (to B.M.W.).
Author contributions: J.M.-B., B.M.W., J.L.C., and J.M.V. designed research; J.M.-B., S.B., T.H., P.J.L., J.N.M., B.M.W., and J.M.V. performed research; J.M.-B., S.B., T.H., P.J.L., J.N.M., B.M.W., J.L.C., and J.M.V. analyzed data; and B.M.W. and J.M.V. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CCT, chaperonin containing TCP-1; Pdc, phosducin; PFD, prefoldin.
References
- 1.Ellis, R. J. (1996) in The Chaperonins, ed. Ellis, R. J. (Academic, San Diego), pp. 2–25.
- 2.Ellis, R. J. & Hartl, F. U. (1999) Curr. Opin. Struct. Biol. 9, 102–110. [DOI] [PubMed] [Google Scholar]
- 3.Gutsche, I., Essen, L. O. & Baumeister, W. (1999) J. Mol. Biol. 293, 295–312. [DOI] [PubMed] [Google Scholar]
- 4.Braig, K., Otwinowski, Z., Hegde, R., Boisvert, D. C., Joachimiak, A., Horwich, A. L. & Sigler, P. B. (1994) Nature 371, 578–586. [DOI] [PubMed] [Google Scholar]
- 5.Ditzel, L., Löwe, J., Stock, D., Stetter, K. O., Huber, H., Huber, R. & Steinbacher, S. (1998) Cell 93, 125–138. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Puertas, P., Martín-Benito, J., Carrascosa, J. L., Willison, K. R. & Valpuesta, J. M. (2004) J. Mol. Recognit. 17, 85–94. [DOI] [PubMed] [Google Scholar]
- 7.Willison, K. R. (1999) in Molecular Chaperones and Folding Catalysts, ed. Bukau, B. (Harwood Academic, Amsterdam), pp. 555–571.
- 8.Valpuesta, J. M., Carrascosa, J. L. & Willison, K. R. (2004) in Handbook of Protein Folding, eds. Buchner, J. & Kiefhaber, T. (Wiley, New York), in press.
- 9.Bauer, P. H., Muller, S., Puzicha, M., Pippig, S., Obermaier, B., Helmreich, E. J. M. & Lohse, M. J. (1992) Nature 358, 73–76. [DOI] [PubMed] [Google Scholar]
- 10.Lee, R. H., Ting, T. D., Lieberman, B. S., Tobias, D. E., Lolley, R. N. & Ho, Y.K. (1992) J. Biol. Chem. 267, 25104–25112. [PubMed] [Google Scholar]
- 11.Hawes, B. E., Touhara, K., Kurose, H., Lefkowitz, R. J. & Inglese, J. (1994) J. Biol. Chem. 269, 29825–29830. [PubMed] [Google Scholar]
- 12.Gaudet, R., Bohm, A. & Sigler, P. B. (1996) Cell 87, 577–588. [DOI] [PubMed] [Google Scholar]
- 13.McLaughlin, J. N., Thulin, C. D., Bray, S. M., Martin, M. M., Elton, T. S. & Willardson, B. M. (2002) J. Biol. Chem. 277, 34885–34895. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin, J. N., Thulin, C. D., Hart, S. D., Resing, K. A., Ahn, N. G. & Willardson, B. M. (2002) Proc. Natl. Acad. Sci. USA 99, 7962–7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martín-Benito, J., Boskovic, J., Gómez-Puertas, P., Carrascosa, J. L., Simons, C., Lewis, S. A., Bartolini, F., Cowan, N. C. & Valpuesta, J. M. (2002) EMBO J. 21, 6377–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savage, J. R., McLaughlin, J. N., Skiba, N. P., Hamm, H. E. & Willardson, B. M. (2000) J. Biol. Chem. 275, 30399–30407. [DOI] [PubMed] [Google Scholar]
- 17.Frank, J. (1996) Three-dimensional Electron Microscopy of Macromolecular Assemblies (Academic, San Diego), pp. 182–246.
- 18.Heymann, J. B. (2001) J. Struct. Biol. 133, 156–169. [DOI] [PubMed] [Google Scholar]
- 19.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 20.Guex, N. & Peitsch, M. C. (1997) Electrophoresis 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- 21.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 22.Llorca, O., McCormack, E., Hynes, G., Grantham, J., Cordell, J., Carrascosa, J. L., Willison, K. R., Fernández, J. J. & Valpuesta, J. M. (1999) Nature 402, 693–696. [DOI] [PubMed] [Google Scholar]
- 23.Llorca, O., Martín-Benito, J., Ritco-Vonsovici, M., Grantham, J., Hynes, G., Willison, K. R., Carrascosa, J. L. & Valpuesta, J. M. (2000) EMBO J. 19, 5971–5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liou, A. K. & Willison, K. R. (1997) EMBO J. 16, 4311–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grantham, J., Llorca, O., Valpuesta, J. M. & Willison, K. R. (2000) J. Biol. Chem. 275, 4587–4591. [DOI] [PubMed] [Google Scholar]
- 26.Schroder, S. & Lohse, M. J. (2000) Naunyn-Schmiedeberg's Arch. Pharmacol. 362, 435–439. [DOI] [PubMed] [Google Scholar]
- 27.Miles, M. F., Barhite, S., Sganga, M. & Elliott, M. (1993) Proc. Natl. Acad. Sci. USA 90, 10831–10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loew, A., Ho, Y. K., Blundell, T. & Bax, B. (1998) Structure (London) 6, 1007–1019. [DOI] [PubMed] [Google Scholar]
- 29.Gaudet, R., Savage, J. R., McLaughlin, J. N., Willardson, B. M. & Sigler, P. B. (1999) Mol. Cell 3, 649–660. [DOI] [PubMed] [Google Scholar]
- 30.Martin, J. L. (1995) Structure (London) 3, 245–250. [DOI] [PubMed] [Google Scholar]
- 31.Aloy, P., Böttcher, B., Ceulemans, C., Leutwein, C., Mellwig, C., Fischer, S., Gavin, A. C., Bork, P., Superti-Furga, G., Serrano, L. & Russell, R. B. (2004) Science 303, 2026–2029. [DOI] [PubMed] [Google Scholar]
- 32.Lacefield, S. & Solomon, F. (2003) Genetics 165, 531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaauw, M., Knol, J. C., Kortholt, A., Roelofs, J., Ruchira, Postma, M., Visser, A. J. W. G. & Van Haastert, P. J. M. (2003) EMBO J. 22, 5047–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clapham, D. E. & Neer, E. J. (1993) Nature 365, 403–406. [DOI] [PubMed] [Google Scholar]
- 35.García-Higuera, I., Gaitatzes, C., Smith, T. F. & Neer, E. J. (1998) J. Biol. Chem. 273, 9041–9049. [DOI] [PubMed] [Google Scholar]
- 36.Siegers, K., Bölter, B., Schwarz, J. P., Böttcher, U. M. K., Guha, S. & Hartl, F. U. (2003) EMBO J. 22, 5230–5240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Camasses, A., Bodganova, A., Shevchenko, A. & Zachariae, W. (2003) Mol. Cell 12, 87–100. [DOI] [PubMed] [Google Scholar]
- 38.Valpuesta, J. M., Martín-Benito, J., Gómez-Puertas, P., Carrascosa, J. L. & Willison, K. R. (2002) FEBS Lett. 529, 11–16. [DOI] [PubMed] [Google Scholar]