Abstract
It is well established that lipid metabolism is drastically altered during tumor development and response to therapy. Choline kinase alpha (ChoKα) is a key mediator of these changes, as it represents the first committed step in the Kennedy pathway of phosphatidylcholine biosynthesis and ChoKα expression is upregulated in many human cancers. ChoKα activity is associated with drug resistant, metastatic, and malignant phenotypes, and represents a robust biomarker and therapeutic target in cancer. Effective ChoKα inhibitors have been developed and have recently entered clinical trials. ChoKα's clinical relevance was, until recently, attributed solely to its production of second messenger intermediates of phospholipid synthesis. The recent discovery of a non-catalytic scaffolding function of ChoKα may link growth receptor signaling to lipid biogenesis and requires a reinterpretation of the design and validation of ChoKα inhibitors. Advances in positron emission tomography, magnetic resonance spectroscopy, and optical imaging methods now allow for a comprehensive understanding of ChoKα expression and activity in vivo. We will review the current understanding of ChoKα metabolism, its role in tumor biology and the development and validation of targeted therapies and companion diagnostics for this important regulatory enzyme. This comes at a critical time as ChoKα-targeting programs receive more clinical interest.
Keywords: Choline kinase, Lipid metabolism, Tumor metabolism, Inhibitors
1. Introduction
Changes in lipid metabolism during tumorigenesis and tumor development have long been recognized, but the molecular mechanisms underlying these alterations are still being unraveled [1]. Among their vital roles, lipids serve as a major energy store, an essential component of cellular membranes, and a source of bioactive signaling molecules. The expansion of cancerous cells requires transcriptional promotion of lipid synthesis pathways, expression of fatty acid transporters, and conversion of biologically inert lipids into second messengers. Exposure of cancer cells to stress signals (e.g. acidic microenvironments, hypoxia, immune attack, and cytotoxic pharmacologic agents) often stimulates rapid production of lipid droplets as a survival mechanism that leads to therapy resistance [2–7]. Because of the dependence on lipid synthesis pathways for both growth and survival, many commonly mutated oncogenes code for proteins that regulate lipid synthesis [8]. Many prevalent oncogenes directly or indirectly control lipid composition by altering transcription or activity of lipid metabolic enzymes, however there are few lipid enzymes whose overexpression is sufficient to drive malignant transformation. A notable exception is choline kinase alpha (ChoKα; E.C. 2.7.1.32), which suggests choline is a rate-limiting substrate for lipid-mediated proliferation.
2. Choline metabolism
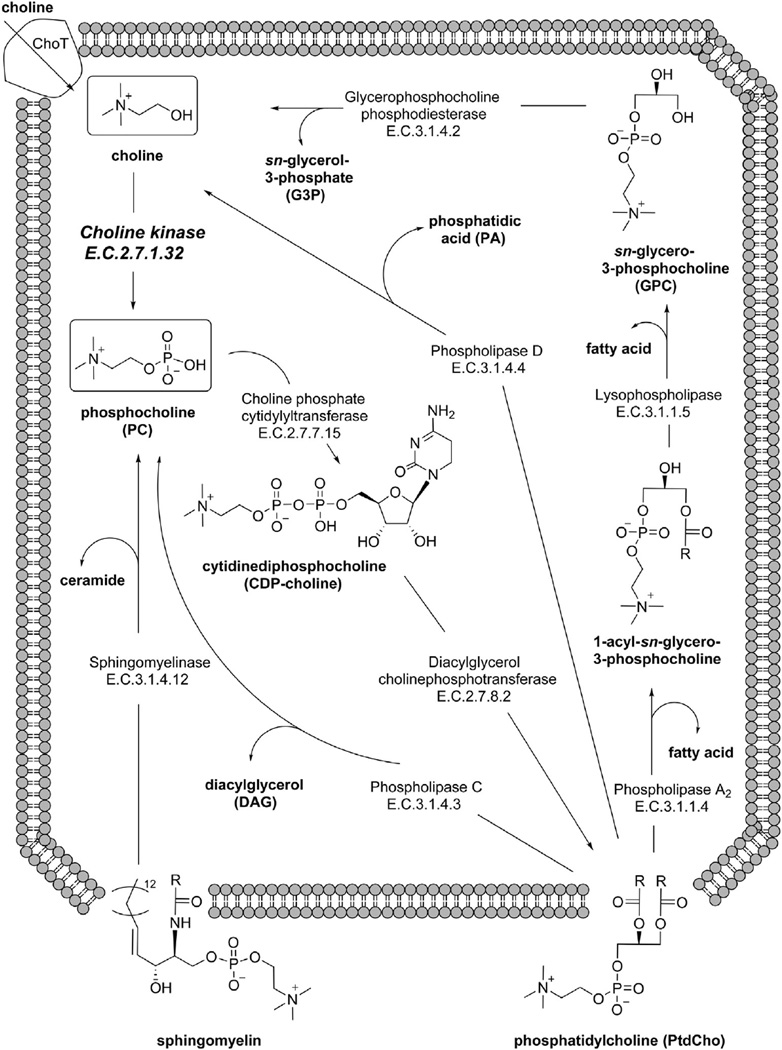
ChoKα catalyzes the phosphorylation of free choline to form phosphocholine (PC). PC is an important precursor to phosphatidylcholine (PtdCho), the predominant phospholipid in mammalian cell membranes (Fig. 1). Free choline is transported across the cell membrane by a number of transporters (ChoT), which can be classified into four major groups: high-affinity choline transporters (CHTs), choline transporter-like proteins (CTLs), organic cation transporters (OCTs), and organic cation/carnitine transporters (OCTNs). Each transporter group is composed of multiple genes and splice variants [9]. Members of each class have been found at elevated levels in tumors, although the expression of each transporter seems to differ between cancers [10]. CTL1 for example has been reported in colon cancer [11], lung cancer [12], and leukemia [13]. In a study of breast cancer cells, CHT1 and OCT2 were found to be the primary choline transporters [14]. The role of each choline transporter in each tumor type is still poorly understood, due to the complexity of the isoforms present in each transporter class.
Fig. 1.
Choline kinase plays a central role in phospholipid metabolism. PtdCho is synthesized in most cells via the Kennedy pathway, where choline (top left) is phosphorylated by choline kinase. DAG is then added via a CTP-mediated two-step mechanism. PtdCho can be metabolized at a variety of cleavage sites by the phospholipase enzymes. The products of both the PtdCho catabolic and anabolic steps have important signaling functions, which can be dysregulated in diseased tissue.
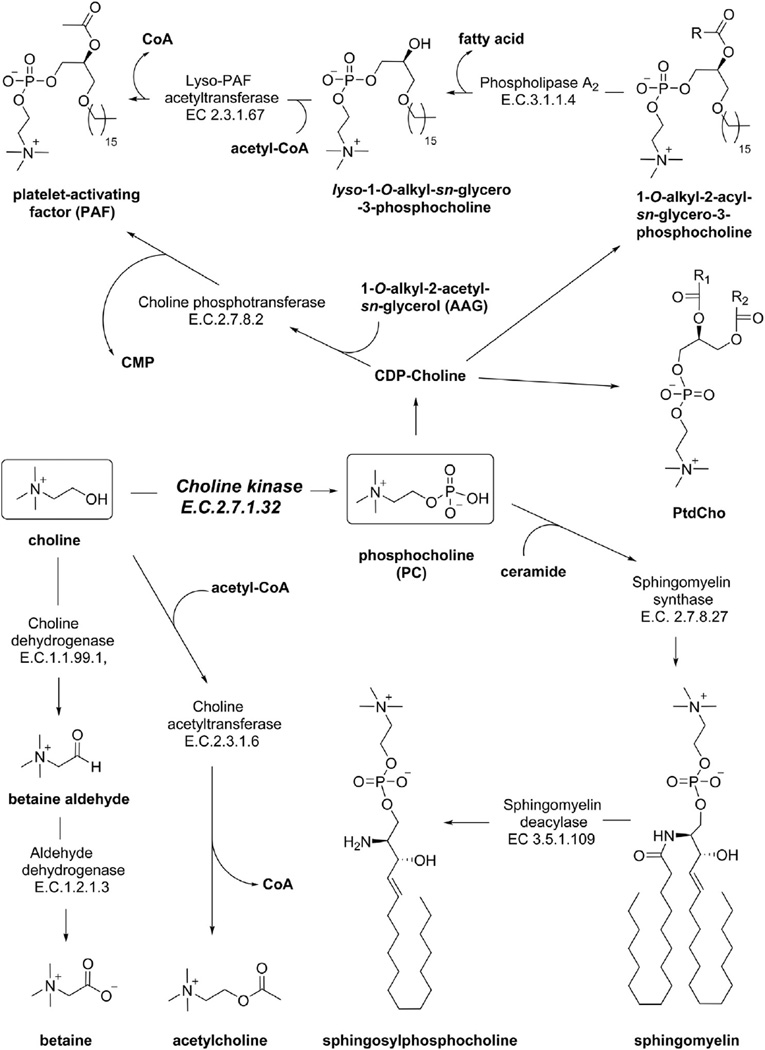
Upon entering the cell, choline can be incorporated into a variety of important cellular components, including acetylcholine, betaine, platelet activating factor (PAF), sphingomyelin, or PtdCho (Fig. 2). In healthy individuals, the CHT family is primarily expressed in neurons, which take up choline and synthesize the neurotransmitter acetylcholine in a one-step pathway catalyzed by choline acetyltransferase [15]. Choline can also be oxidized in the mitochondria at the hydroxyl position to form the common osmolyte betaine. Betaine:homocysteine S-methyltransferase transfers a methyl group from betaine to homocysteine, replenishing the methionine pool needed for the synthesis of the crucial methyl donor S-adenosylmethionine (SAM) [16]. Among other functions, SAM can be used by phosphatidylethanolamine N-methyltransferase (PEMT) to methylate phosphatidylethanolamine (PtdEtn) for PtdCho synthesis during choline deficiency [17]. In humans PEMT is almost exclusively expressed in the liver [18]. The majority of cells in the human body use choline as a head group for cell membrane lipids. The Kennedy pathway leads to formation of the diacylglycerophospholipid, PtdCho. PtdCho can then be converted to the sphingolipid sphingomyelin (SM) via transfer of PC to a ceramide by the enzyme sphingomyelin synthase [19]. Deacylation of sphingomyelin by sphingomyelin deacylase yields sphingosylphosphocholine [20].
Fig. 2.
The metabolic roles of choline. Choline (left box) can be oxidized to betaine, acetylated to acetylcholine, or phosphorylated to PC (right box). PC can be acetylated and serve as the head-group in many glycerophospholipids (e.g. PAF, PtdCho) or sphingolipids (e.g. sphingomyelin). The fate of choline and PC varies depending of tissue type and cell status.
2.1. Pathways of choline phospholipid metabolism
Anabolic production of PC occurs during the Kennedy pathway (Fig. 1) for PtdCho biosynthesis [21]. Free choline in the cytosolic space is rapidly phosphorylated to PC by the enzyme choline kinase (ChoK) [22]. PC conversion to CDP-choline is the rate-limiting step catalyzed by the enzyme choline phosphate cytidylyltransferase (CCT) [23]. CCT is inactive in soluble form but recruited to the nuclear membrane in a highly regulated fashion [10]. Cytidine monophosphate is then replaced with 1,2-sn-diacylglycerol (DAG) by the enzyme diacylglycerol cholinephosphotransferase (CPT) in the final step of PtdCho synthesis [24].
PC formation can also occur by a number of catabolic paths, which are mediated by a class of enzymes known as the phospholipases. PC can be cleaved directly from PtdCho by PtdCho-specific phospholipase C (PC-PLC), releasing a DAG second messenger that can go on to activate protein kinase C. While the activation of PC-PLC at the plasma membrane and the inhibition of its activity by the compound D609 have been studied, the gene(s) encoding its protein(s) have yet to be identified [25,26]. PC can also be derived by phospholipases using indirect methods. These methods include conversion of PtdCho to free choline and phosphatidic acid (PA) by phospholipase D (PLD), and the coordinated activity of phospholipase A2 (PLA2), lysophospholipase (LPL) and glycerophospholipase:phosphodiesterase (GPC:PDE) to render free choline. These enzymes have been found to be differentially expressed in some but not all cancers, and much work is still required to fully elucidate their involvement in PC production [27,28]. A number of mitogenic second messengers are released as byproducts of PtdCho cleavage (Fig. 1), including DAG, lysophosphatidic acid, PA, arachidonic acid, and PC itself [29–32].
2.2. Phosphocholine signaling
Generation of PC is required for the induction of DNA synthesis by a number of growth factors including PDGF, FGF, EGF and phorbol esters [33]. Inhibiting PC production prior to PDGF or FGF addition eliminates activation of the Raf-1 and MAP kinase signaling cascades responsible for relaying the mitogenic growth signal [34]. Microinjection of PC itself is sufficient to induce a mitogenic response [33], although a separate study demonstrated that PC added to cellular media was not taken up by cells but still exhibited a pro-mitogenic response [35]. These results suggest that extracellular ATP detection by the purinergic P2 receptors induces release of mitogenically active levels of PC from fibroblasts. PC acts synergistically with ATP, insulin, and/or sphingosine-1-phosphate to stimulate phosphatidylinositol-3′-kinase (PI3K) and pp70s6k, which regulate progression from the G1 to the S phase of the cell cycle [35]. PC levels have been found to increase substantially following oncogene transfection, loss of tumor suppressor function, and immortalization [36,37]. The full mechanistic relationship between PC levels and DNA synthesis has yet to be understood.
3. Choline kinase
The primary mediator of PC levels in cancer cells is believed to be the enzyme ChoK, which is responsible for phosphorylation of free choline upon cell entry or release by a catabolic process. Enhancement of intracellular PC by Ras transformation is a ChoK-dependent process, as ChoK inhibitors have been demonstrated to halt these alterations [38]. The growth factors that rely on PC production to exert their mitogenic effects also require ChoK; ChoK inhibition in primary human mammary epithelial cells blocks EGF, insulin, and hydrocortisone-induced DNA synthesis [39]. It can therefore be inferred that the production of PC as a mitogenic messenger is regulated by ChoK and cannot be salvaged in most cases by a compensatory catabolic mechanism.
There are three different isoforms of ChoK in mammals coded by two different genes. CHKA encodes the protein ChoKα, which exists in two splicing variants. ChoKα variant 2 includes an 18 amino acid insertion between Met150 and Gly151 [40]. While the importance of this splicing difference is not certain, some evidence indicates it may impact subcellular distribution [41]. The CHKB gene encodes the protein ChoKβ, which has approximately 60% sequence homology to the ChoKα isoforms [42]. ChoK is only functional in homodimer or heterodimer form, and the distribution of each isoform varies widely between tissues. While the testis and liver exhibit relatively high ChoKα expression, the heart and liver have the highest ChoKβ protein [43]. Each isozyme is capable of phosphorylating choline, as well as the structurally similar ethanolamine. Only ChoKα has been found to have a link to tumor transformation [44], but defects in ChoKβ can lead to muscular dystrophy [45]. While the ChoKα protein is more selective for the choline substrate, ChoKβ has higher affinity for ethanolamine [44]. CHKA but not CHKB deletion is embryonically lethal [46], further implicating the ChoKα variants as the crucial contributors to PtdCho biosynthesis. ChoK α/α complexes have the highest ChoK activity (higher Vmax and lower Km), β/β complexes have low activity, and α/β dimer complexes have intermediate catalytic activity [40,44].
3.1. ChoK mechanism of action
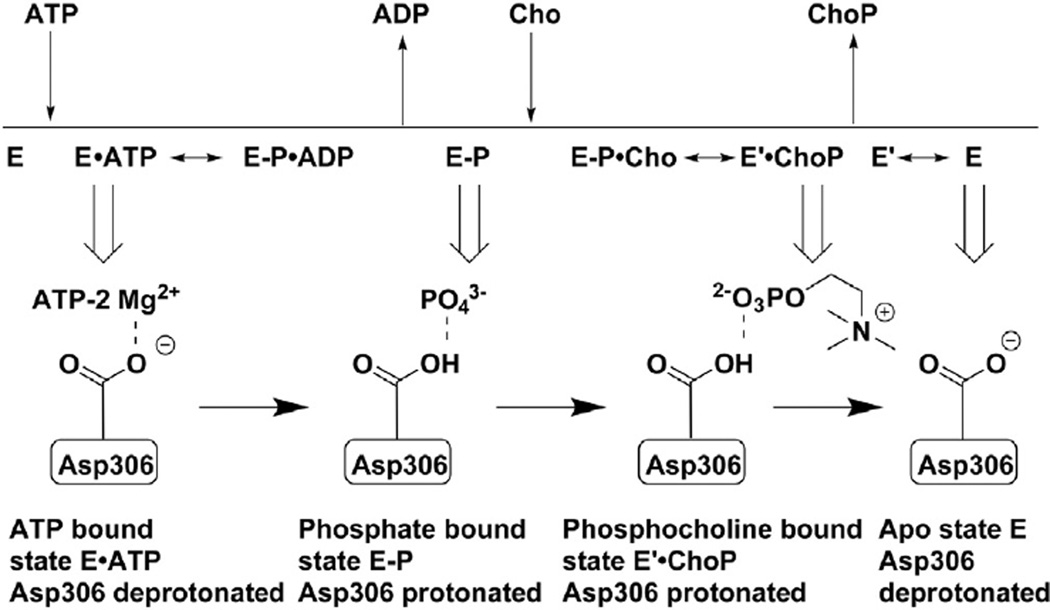
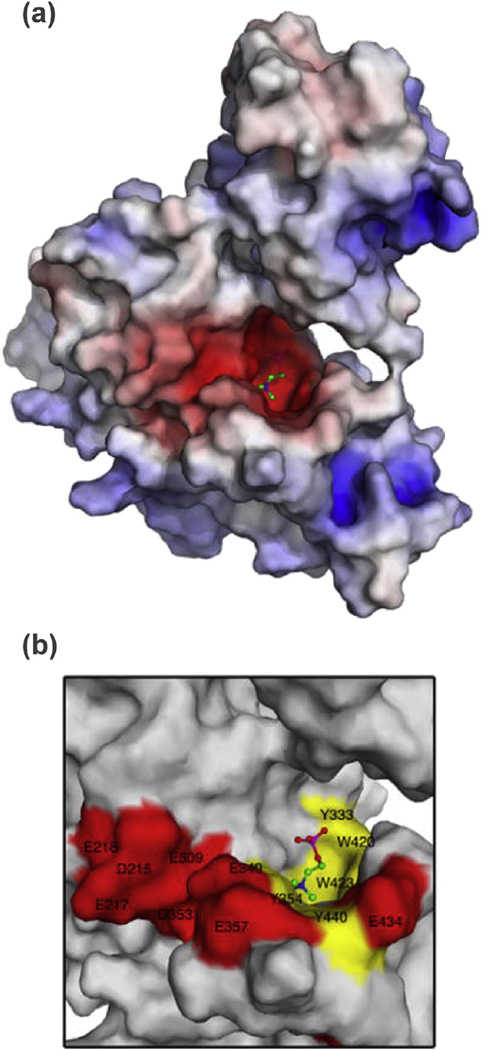
ChoK is similar to other kinases in that its N- and C-terminal lobes come together to form the ATP-binding site, but it is classified as an “atypical kinase” because there is a flexible ATP-binding loop in place of the glycine-rich P-loop found in typical eukaryotic kinases [47,48]. It has been known since its discovery that the conversion of the phosphate group from ATP to choline requires magnesium ions for reaction coordination [22]. It was only recently reported by Hudson et al. [49] that ChoK works by an iso double-displacement mechanism, meaning that no ternary choline-ATP-ChoK complex is formed during catalysis (Fig. 3). Instead, ATP transfers a phosphate group directly to the enzyme at residue Asp306, which upon protonation causes ADP to leave [49]. The protonated, phosphate-bound Asp306 favors the binding of choline, which causes a conformational shift in ChoK that deprotonates Asp306 and releases PC in a process that then makes the original ChoK conformation more favorable. This ping-pong mechanism was supported by the surprising finding that ChoK has ATPase activity even in the absence of choline [49]. The active site of ChoK is unlike the cleft-like structure of most protein kinases; the choline binding site exists in a tunnel-like pocket of hydrophobic residues with a rim of negatively-charged amino acids (Fig. 4) [50]. Reports generally agree on Km values for choline between 100 and 180 µM, Km values for ATP between 410 and 760 µM, and kcat values for the enzyme of 69–83 s−1 [49–51].
Fig. 3.
ChoKα phosphorylates choline via an unusual ping-pong mechanism. Unprotonated Asp306 at the active site of human ChoKα can accept a phosphate group by a Mg2+-coordinated reaction resulting in protonation of the amino acid and subsequent ejection of ADP. The phosphate-primed enzyme can accept choline, which induces a conformational shift in the enzyme resulting in PC exit. The deprotonated ChoKα enzyme reverts to its original conformation and is again ready for ATP binding. Adapted with permission from Hudson et al., 2013 [49].
Fig. 4.
ChoKα active site. (a) A surface representation of ChoKα with PC bound at the active site was colored corresponding to the electrostatic potential of the regional amino acid residues (red = −15 kT/e; blue = 15 kT/e). (b) A closer view of the choline-binding site was color-coded with hydrophobic regions in yellow and negatively charged regions in red. This view demonstrates the hydrophobic pocket and anionic rim of this atypical kinase active site. Figure adapted with permission from Malito et al. [50].
3.2. Enzyme induction
Heightened ChoK activity was first reported in rat hepatocytes following treatment of carcinogenic polyaromatic hydrocarbons; reversal of this phenomenon by addition of transcription inhibitors (cyclohexamide or actinomycin D) suggests the increase in activity was due to an increase in ChoK protein expression [52]. ChoKα, but not ChoKβ, expression can also be induced in liver cells by carbon tetrachloride (CCl4) [43]. This hepatotoxin was later found to cause upregulation of the oncogenic transcription factor c-jun that binds to an activator protein 1 (AP1) element site in the promoter region of CHKA [53]. Hypoxia response elements (HREs) in the CHKA promoter region have linked HIF1α activity to ChoKα expression [54,55]. Inhibitors of the mitochondrial electron transport chain in neuroblastoma cells can increase ChoKα expression and phospholipase activity, leading to elevated choline metabolites [56]. Additional reports have shown that the oncogene c-Myc can increase ChoKα expression ultimately leading to higher levels of PC [57,58]. ChoKα activation can be mediated by the oncogenic small GTPase RhoA and its effectors [59]. Ras mutation in NIH3T3 fibroblasts was shown to increase ChoK activity via Ral-GDS and PI3K downstream pathways [60]. Treatment of human prostate and colon carcinoma cells with the PI3K inhibitor PI-103 reduced ChoKα and PC levels, indicating PI3K is regulative of PtdCho biosynthesis by promoting CHKA expression [61]. In yeast, direct phosphorylation at Ser30 and Ser85 of choline kinase (CKI) by protein kinase A (PKA) increases PC production [62], although only rudimentary evidence in humans exists to suggest similar regulation of ChoK by PKA [63]. Phosphorylation of ChoKα at the Tyr197 and Tyr333 residues has been reported in human breast cancer cells, and these alterations increase the activity of ChoKα when in a complex with epidermal growth factor receptor (EGFR) and c-Src [41]. In human breast cancer cells, extracellular calcium has been used to stimulate ChoK activity and PC formation by a Rho-dependent pathway [64]. This is of interest because calcium, along with other growth factors, is released in large quantities at sites of metastases to bone [65,66].
3.3. Oncogenic role
The diversity of cancer-associated factors that can stimulate ChoK activity establishes the critical role this oncoprotein plays in tumorigenesis. Overexpression of ChoKα, but not ChoKβ, is sufficient to drive tumor transformation [44]. Amplified ChoKα expression was first reported in breast carcinomas, and these studies found elevated ChoK activity in 39% of patient-derived tumor specimens [67]. ChoKα deregulation has since been found in 47% of colon cancer, 56% of lung cancer, and 48% of prostate cancer tissues [68]. ChoKα over-activity has additionally been reported in ovary [69], endometrial [70], and pancreatic cancers [71]. A retrospective study of non-small-cell lung cancer patients who had received prior surgical tumor resection found that individuals with tumor ChoKα levels 1.91-times higher than healthy tissue had four-year survival rates of just 49%, compared to a survival rate of 71% in patients with ChoKα expression below this threshold [72]. ChoKα activity is clinically correlated with histological tumor grade in breast cancer, and is inversely proportional to estrogen receptor positivity in patients [67]. This is significant in that it implicates breast cancer patients that don't respond to the anti-estrogen tamoxifen may have resistance pathways mediated by ChoK. In murine models of bladder cancer, ChoKα overexpression is associated with larger, more metastatic tumors and contributes to a more aggressive carcinoma [73].
ChoKα expression and activity have been described as robust biomarkers for tumor aggressiveness in a growing number of cancers, with few exceptions. Although heat shock protein-90 (HSP-90) inhibitors promote the proteasomal degradation of tumor-driving oncoproteins, they have been reported to elevate PC levels in colon cancer and melanoma cells [74,75]. Subsequent gene expression profiling in 17AAG-treated breast cancer cells, however, found no change in ChoK expression and attributed the HSP-90-inhibitor effects to choline transporter and phospholipase induction [76]. HDAC inhibitors Vorinostat, LAQ-824, and belinostat provide another example running contrary to this conventional wisdom, in that they induce differentiation and apoptosis in cancer cells but cause a rise in PC and ChoK levels [77–80].
4. Choline kinase inhibition
There is growing clinical interest in the suppression of ChoKα activity to ameliorate the pro-growth, drug resistant, and invasive phenotype this enzyme imparts on cancers originating from a wide range of tissues. An siRNA screen of human kinases and phosphatases which are critical for cell survival found ChoKα knock-down to cause a two-fold increase in apoptosis [81]. The first study to explore RNAi knockdown of ChoKα in-depth found reduced proliferative markers and signs of differentiation in the highly-metastatic triple-negative MDA-MB-231 human breast cancer cell line [82]. Chua et al. performed an siRNA screen of more than 90 kinases to identify regulators of Akt, a critical molecular mediator in metabolism and cell survival pathways. In MDA-MB-468 breast cancer cells, both ChoKα and ChoKβ activities were found to promote phosphorylation of Akt at Ser473, activating Akt in a PI3K-dependent manner [83]. While ChoKα RNAi silencing prevents mitotic entry and induces apoptosis in HeLa cells, knockdown of ChoKβ in addition to ChoKα rescues this lethality implying that the ChoKα to ChoKβ ratio is key to regulating release from G0/G1 arrest [84]. The Lacal group used stable ChoKα knockdown by shRNA and found enhanced PARP cleavage in cervical, lung, and bladder cancer cells. This study also found ChoKα shRNA induces apoptosis in MDA-MB-231 cells and tumor xenografts, but is tolerated in untransformed human mammary epithelial cells [85]. Suppression of ChoKα expression also appears to sensitize cancer cells to chemotherapeutics [86, 87]. In 2009, a lentiviral approach to deliver ChoKα shRNA to MDA-MB-231 xenografts achieved 80% reduction in ChoKα protein and attenuated the Kennedy pathway [88]. An early study that sought to explain the observation that myo-inositol and choline addition to growth media reduces de novo PtdCho synthesis found this effect is due to down-regulation of ChoKα expression, rather than direct chemical inhibition of the protein [89].
4.1. Early inhibitors
In studying the interaction between ChoK and its natural substrates, propanol, butanol, and methyl-substituted derivatives of choline were described as the first ChoK inhibitors [90]. Wittenburg and Kornberg reported that ChoK activity could be stimulated by cysteine [22]. This early observation led Brostrom and Browning to study choline phosphorylation in the presence of the thiol group inhibitors and found ChoK inhibition by both N-ethylmaleimide and Ellman's reagent [91]. A preliminary study as far back as 1974 linked inhibition of ChoK to membrane morphological changes in cancer cells, and demonstrated anti-tumor activity using purinyl-6-histamine to inhibit choline phosphorylation [92]. An early review of agents known to modulate ChoK activity included many Kennedy pathway intermediates (e.g. ethanolamine, PC, CDP-choline) and choline derivatives (e.g. ethyl choline mustard, acetylcholine, betaine) [93]. A variety of muscarinic receptor antagonists (e.g. quinacrine), ATP analogs (5′-AMP, AMP-PNP), and ions capable of displacing Mg2+ have also been reported, although for many of these agents it is likely that this effect is non-specific and due to action on the choline transporters, widespread interaction with ATP-binding sites in kinases, or other metal-dependent enzymes [94]. Finally, ChoK is most catalytically efficient at alkaline pH (8.0–9.5) and can be effectually inactivated in acidic environments [22]. Although low pH is a common characteristic of tumor microenvironments [95], the intracellular pH is maintained in an optimal range (7.0–7.2) for ChoK activity.
4.2. Hemicholinium-3
Since its discovery as a ChoK inhibitor [96], the symmetric dicationic choline-mimetic hemicholinium-3 (HC-3, Fig. 5) has been a prototype for ChoK inhibitors [97]. Prior to its study as a ChoK inhibitor, HC-3 was used as a research tool for its ability to inhibit acetylcholine synthesis in the nervous system [98]. First discovered in 1954 [99], HC-3 was shown to disrupt ChoK activity in the brain by blocking sodium-dependent high-affinity choline uptake [100,101] and diminishing acetylcholine synthesis [102,103]. While HC-3 is capable of reducing PC levels and inhibiting growth factor-induced DNA synthesis in vitro [34], administering HC-3 at doses necessary for in vivo ChoK inhibition causes substantial off-target action on the neuronal high affinity choline transporters as well as acetylcholinesterase [104]. HC-3 toxicity results in a “curare-like” death due to its disruption of cholinergic neurotransmission [105]. The first exploration of HC-3 derivatives took place in the late-1980s and found that hydrophobic spacer length was critical in establishing the distance between quaternary ammonium groups [106–109]. A fourteen-atom spacing between the cationic head-groups yielded the most potent modulators of acetylcholine content in the caudate nucleus, one of the structures that makes up the basal ganglia and controls motor function [107]. Most tertiary ammonium derivatives are inactive, however the choline-like oxazinium group can be replaced with 4-methylpiperidine, suggesting that a hydroxyl group is not essential for choline uptake inhibition [106,109].
Fig. 5.
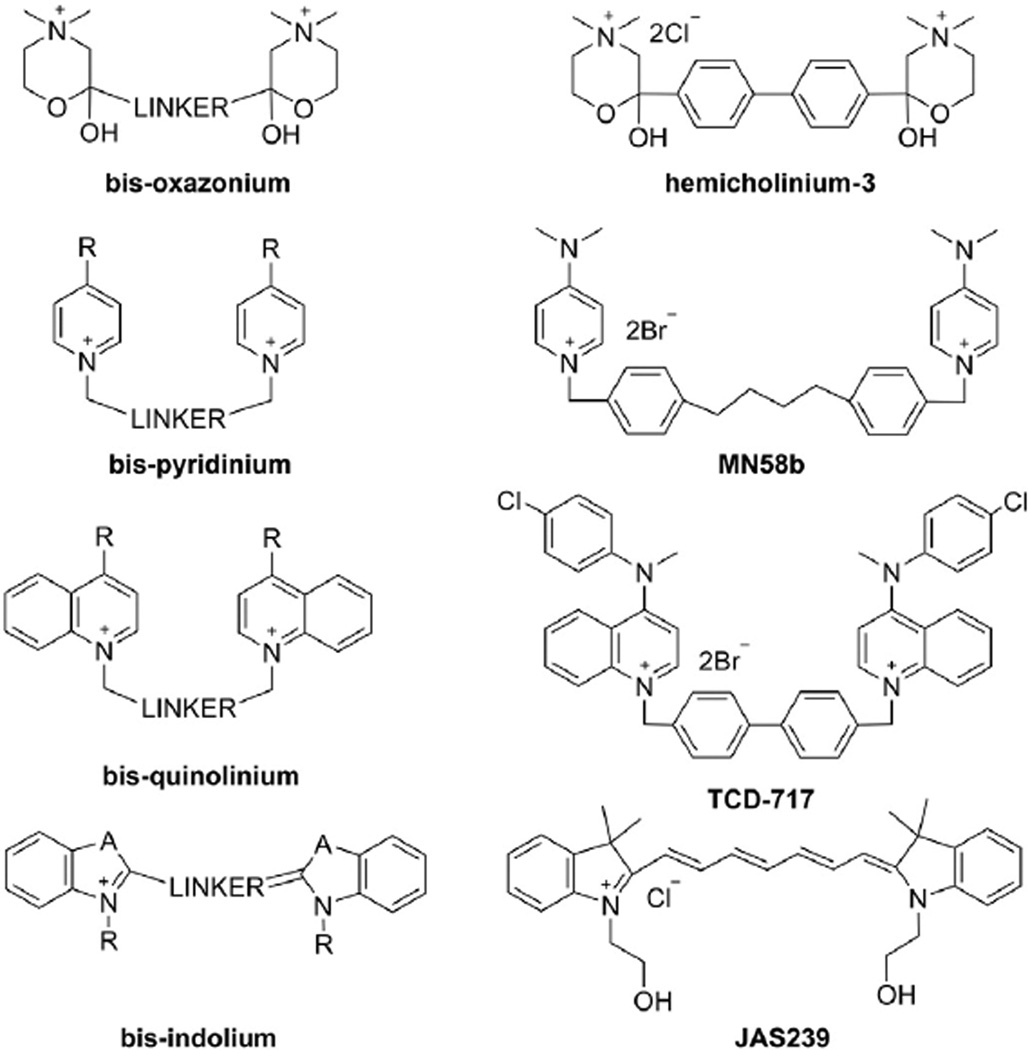
Symmetric ChoKα inhibitors. The general structure of each symmetric ChoKα inhibitor class (left column) is presented with a representative compound (right column) from that class. Hemicholinium-3 is the prototypical choline mimetic, but lacks specificity for ChoKα. MN58b and TCD-717 are highly selective analogs of hemicholinium-3 with promising antiproliferative properties. JAS239 is a near infrared fluorescent choline mimetic developed as an optical imaging contrast agent for cancer applications.
4.3. Bis-pyridiniums
The IC50 of HC-3 against purified ChoK is 500 µM, so as new links between cancer and choline phosphorylation were discovered in the 1990s, it became clear that more selective and potent ChoK inhibitors were needed. Based upon prior HC-3 derivatives, new ChoK inhibitors were developed in the lab of Juan Carlos Lacal. This initial class of bispyridinium compounds improved the potency against pure ChoK and the growth inhibitory concentration in cancer cells by several orders of magnitude [110]. Quantitative structure–activity relationships (QSAR) demonstrated that electron-donating groups substituted at the C4-position of the pyridinium head-groups (Fig. 5) give the strongest ex vivo ChoK inhibition, the –NMe2 moiety is one of the most electron-donating substituents known [111]. The most promising of these compounds, MN58b (Fig. 5), is effective against a variety of cancer cell lines, significantly retards xenograft growth [112,113], and most importantly, is more specific for ChoKα than HC-3 [114]. Improved activity was found when the 4,4′-biphenyl linker of HC-3 was replaced by a 1,2-ethylene(bisbenzyl) fragment [115]. Although tri-pyridinium structures exhibit superior potency against purified ChoKα, the additional charge on these molecules makes them impenetrable to the cell membrane and relatively weak antiproliferative agents [116]. Likewise, attempts to make these structures more rigid to reduce rotational bonds and conform to “drug-likeness” principles ultimately led to IC50s as low as 30 nM in purified recombinant ChoKα, but these bis-pyridinium cyclophane compounds are ineffective at inhibiting growth of HT29 colorectal adenocarcinoma cells [117]. A rigid biphenyl linker did yield the most potent compounds, and when paired with a 4-chloro-N-methylanilino substituent on the pyridinium moiety, gave rise to a number of promising agents [118]. Extensive molecular modeling of these compounds with the crystal structures of ChoKα have helped to highlight the importance of the linker moiety, distance between quaternary ammoniums, delocalization of positive charge by electron-donating groups, and steric hindrance [119].
4.4. Bis-quinoliniums
The first attempt to make HC-3 analogous ChoKα inhibitors found pyridine derivatives to be the most potent, however insights gleaned from the QSAR studies of these bis-pyridiniums suggest lipophilicity could be a major factor disconnecting ex vivo ChoK inhibition from in vitro antiproliferative efficacy [93]. Additionally, with a better understanding of steric and electrostatic interactions at the active site, it became attractive to substitute the pyridine moiety with a variety of substituted quinolinium or isoquinolinium fractions to improve the cell permeability [120]. The bis-quinoliniums (Fig. 5) have been demonstrated to have improved antiproliferative properties compared to the bis-pyridinium structures. This is due to overall greater hydrophobicity, although in both classes the same 3,3′-biphenyl linker and electron-donating substitutions have yielded the most potent ChoKα inhibitors [120]. These studies found that linker length has less of an effect on the antiproliferative properties than it does on ChoKα inhibition, and it was postulated that other mechanisms of cell death besides those induced by the interaction with ChoKα may exist for these compounds [121]. Nonetheless, there have been excellent ChoKα inhibitor candidates derived from this class, which have sub-micromolar IC50s and GI50s [122]. One clinical candidate, known as RSM-932 A or TCD-717 (Fig. 5) has recently completed Phase I clinical trials for advanced solid tumor treatment (NCT01215864) [123].
4.5. Non-symmetric inhibitors
The importance of inhibitor symmetry was questioned even at the early stages of this field. However, when half-inhibitor fragments were found to be poor inhibitors and highly toxic, it was proposed that the dimeric form of ChoKα must allow each di-cationic inhibitor head-group to interact with a unique catalytic site [93]. With the crystal structures of inhibitors docked at the ChoKα active site having since been elucidated, the distance between catalytic units is now known to be too great for these inhibitors to span between bound ChoKα dimers, although the possibility that the unbound cationic head-group can bind to the choline-binding site of a free ChoKα monomer or to the ATP-binding site on the same protein was never excluded [51].
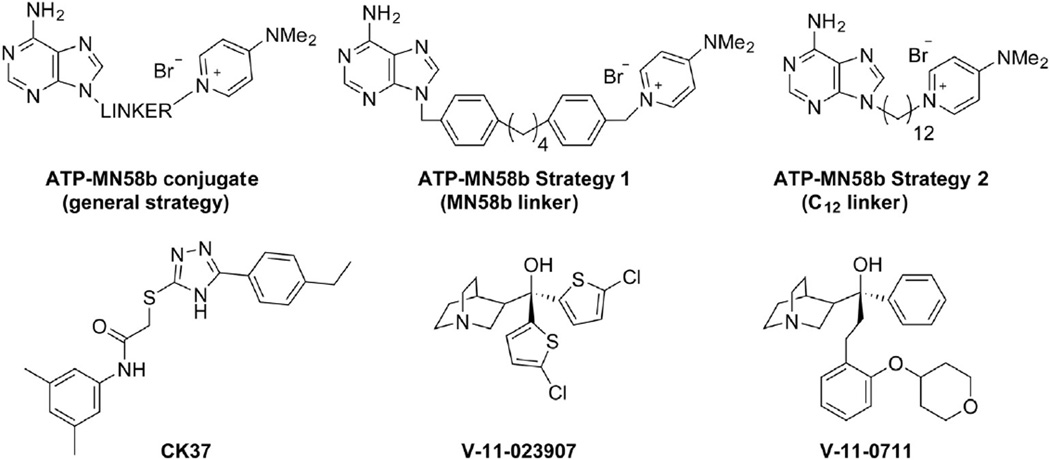
Two separate efforts have been made to design asymmetric ChoKα inhibitors which link pyridinium head-groups with ATP-like structures (Fig. 6) [124,125]. Schiaffino-Ortega et al. were unable to identify ATP-competitive inhibitors in their panel of ChoKα inhibitors [125] (Fig. 6), but one of these structures (ATP-MN58b Strategy 1) was used to demonstrate that inhibition of one ChoKα monomer can reduce inhibitor binding at the active site of the coupled monomer [126]. This negative co-operativity between ChoKα dimers provides greater insight into the interaction between ChoKα dimers and the potential for asymmetric conformations to alter ligand binding. Trousil et al. reported greater potency for symmetric MN58b derivatives than in rationally designed dual choline/ATP mimetic compounds exemplified in Fig. 6 by the ATP-MN58b Strategy 2 structure, but concluded that compounds with higher-affinity for the adenosine-binding site were needed to outcompete ATP at physiological concentrations [124]. With each of these strategies, little correlation was found between the potency of ChoKα inhibition and the antiproliferative effect.
Fig. 6.
Asymmetric ChoKα inhibitors. A rational design approach to improve potency involved the addition of adenine moieties to MN58b (ATP-MN58b conjugate) in an effort to bind the choline and ATP binding sites simultaneously. Sahún-Roncero et al. [126] used an MN58b linker group in a mixed ATP-MN58b inhibitor (Strategy 1), while Trousil et al. [124] independently found a 12-carbon linkage (ATP-MN58b Strategy 2) was capable of inhibition in vitro. Next generation ChoKα inhibitors were developed using computational (CK37) and compound library (V-11-023,907 and V-11-0711) screening approaches.
Use of the ChoKα structure for in silico screening led to the discovery of N-(3,5-dimethylphenyl)-2-[[5-(4-ethylphenyl)-1H-1,2,4-triazol-3-yl]sulfanyl]acetamide (referred to as CK37; Fig. 6) [127]. Modeling of the active site for structure-directed discovery at Vertex Pharmaceuticals has yielded the highly potent and kinase-selective ChoKα inhibitors V-11-023907 and V-11-0711 (Fig. 6) [49,128]. These have proven to be useful tools for studying ChoKα mechanism and how it relates to tumorigenesis.
5. Cancer cell death by ChoKα inhibition
There are several pharmacodynamic markers of ChoKα inhibition, which have been identified using a combination of RNAi and small molecule chemical inhibitors. The earliest studies with bis-pyridinium ChoKα inhibitors found they are tolerated by untransformed NIH-3T3 cells and only cytotoxic following oncogenic (Ras, Raf, Src, or Mos) overexpression [110]. It was presumed at this time that ChoKα's involvement in cell cycle signaling is based solely on its ability to regulate production of the mitogenic second messenger, PC, and that the anticancer effect of ChoKα inhibition was primarily due to attenuation of this growth signal and depletion of substrates required for PtdCho biosynthesis [93]. Indeed, depletion of PC, tCho, and ChoK activity has been measured using both ChoKα-targeted RNAi and small molecule approaches in both in vitro and in vivo settings [82,112–114]. While this is a rational conclusion, it does not explain why normal cells are resistant to apoptosis under the same conditions.
5.1. Cancer cell sensitivity
In 2004, it was discovered in primary lymphocytes that the ChoKα inhibitor MN58b causes dephosphorylation of the cell cycle checkpoint retinoblastoma protein (pRb), driving these cells into a reversible quiescent state until drug removal [129]. Due to defunct cell cycle regulation in lymphoma cells, the inability to dephosphorylate pRb causes Jurkat cells to continue proliferating. The cells compensate for inhibited de novo PC production by cleaving the PC head-group from the abundant membrane sphingolipid, sphingomyelin [129]. The accumulation of ceramides as a byproduct leads to tumor-specific apoptosis, as confirmed by cleavage of PARP due to caspase-9 activation [129]. Caspase-3 activation can be detected prior to a loss in mitochondrial potential and cytochrome-c release, suggesting apoptosis by a non-intrinsic pathway [130]. MN58b has no effect on other enzymes involved in the PtdCho anabolic/catabolic cycles, interference in the MAPK pathway, or DNA-intercalation [131]. Although MN58b has no measurable effect on MAPK signaling, an independent study found that siRNA knockdown of ChoKα does inhibit the MAPK cascade by reducing phosphatidic acid (PA) levels crucial for Ras anchorage and activation [132]. To support this hypothesis, this group developed a ChoKα inhibitor known as CK37 and found disrupted actin cytoskeleton and membrane ruffling in addition to suppressed signaling pathways downstream of Ras [127]. The only known relationship between ChoKα and the plasma membrane is recruitment by EGFR in a c-Src dependent manner; whether this occurs specifically at focal adhesion sites has yet to be explored [41]. This remains an important area of interest, as recent studies suggest that a scaffolding role, rather than or in addition to PC production, may be responsible for ChoKα's role in cancer [128].
5.2. Antiproliferative effect of ChoK inhibitors
The classic understanding of the effects of ChoKα inhibition began to unravel when it was discovered that V-11-0711 dramatically decreases PC pools, but causes a panel of cancer cell lines to enter a quiescent, rather than an apoptotic state [128]. This work found that reduction of ChoKα protein, not PC levels, is the crucial mechanism of cell death [128], making it unclear why some but not all ChoKα inhibitors elicit an antitumor response. The discovery that MN58b and TCD-717, but not ChoKα siRNA, elicit an endoplasmic reticulum (ER) stress response finally provided a clue to explain this discrepancy [133]. MN58b and TCD-717 cause a distinct activation of the unfolded protein response in cancer cells by elevating protein chaperones and the pro-apoptotic transcription factor CHOP [133]. Inhibitor-induced apoptosis in cancer cells can be marginally reversed by addition of ChoKα siRNA, suggesting the conformational changes in ChoKα elicited by these inhibitors cause the inhibited enzyme to be recognized as a misfolded protein. This is non-lethal in normal cells due to a more moderate ER-stress response triggered by the pro-survival transcription factor ATF4 and its downstream network of genes responsible for ER repair [133]. It appears that among the molecules capable of blocking the choline-binding site, there is a smaller subset with antiproliferative activity. This therapeutic response is dependent on an inhibitor's ability to trap ChoKα in an inactive position, prompting cancer cells toward apoptosis by provoking an overly sensitive unfolded protein response. The difference between cytotoxic (e.g. bis-pyridinium, bis-quinolinium) and cytostatic (e.g. V-11-0711) ChoKα inhibitors may be the length of the molecule and the ability to extend from the choline-binding site to one of the adjacent clefts formed by residues which open or close depending on enzyme conformation [134]. The possibility of undetected off-target binding, however, remains a concern until these discrepancies are resolved.
6. Methods for ChoKα inhibitor validation
The difficulty in confirming ChoKα specificity is due to the contributions from other phospholipid and choline-handling enzymes that can confound the interpretation of changes in PC levels. Most notably, drugs with known effects on phospholipase activity, such as the COX-II inhibitor indomethacin, have been studied for oncological use [135]. It is known that mitochondrial complex inhibitors are capable of enhancing activity through the Kennedy pathway [56], and screening of quaternary ammonium-containing structures for ChoK inhibition may be confounded by mitochondrial interactions. A reliable panel of pharmacodynamic biomarkers of ChoKα inhibition has not been established, due to conflicting cellular responses to ChoKα targeted RNAi and small molecule inhibitors.
It is yet unknown whether inactivated ChoKα protein is more crucial than PC depletion to these inhibitors' antiproliferative activities, although there is increasing evidence which supports this hypothesis [128,133]. It is also not known at this time the extent of interaction of existing ChoKα inhibitors, such as MN58b or TCD717, with choline transporters. The choline transporters and ChoKα have a reciprocal relationship: intracellular choline phosphorylation by ChoKα drives the concentration gradient required for choline transport [131], and choline transport controls the substrate availability for ChoKα catalysis [14]. Because the choline transporters are at the cell surface, it is possible that their preferential exposure to polar, less cell-permeant, bis-cationic agents leads to false-positives during ChoKα inhibitor screening. This would explain some of the in vivo toxicities that have been reported for less potent ChoKα inhibitors [114]. Strategies to detect ChoKα activity in its native state are required to validate these therapies. Unfortunately, choline tracers are dependent on choline transporters and cannot distinguish choline transport inhibitors from choline phosphorylation inhibitors [136].
6.1. Magnetic resonance spectroscopy
Magnetic resonance spectroscopy (MRS) provides a profile of endogenous metabolites located within an MRI-selected voxel [137] and a number of biomarkers unique to malignant tissue have been reported [138,139]. Increases in phosphomonoesters (PME) in 31P spectra and total choline-containing metabolites (tCho) in 1H spectra [28,140,141] have been described in a range of cancers, including breast, ovarian, prostate, lung, bone, colon, brain, hepatic, and lymphocytic tumors. The tCho peak is a composite resonance in in vivo 1H MR spectra consisting of the sum of free choline (3.20 ppm), PC (3.22 ppm), and glycerophosphocholine (GPC, 3.23 ppm). The nine chemically equivalent protons in the choline N-methyl groups yield a strong singlet resonance at 3.2 ppm that is detectable by 1H MRS (Fig. 7a). Tumor tissue extracts studied using high resolution MRS have revealed that the major contributor to the tCho signal is PC [138]. PME resonances, on the other hand, contain contributions from PC as well as phosphoethanolamine.
Fig. 7.
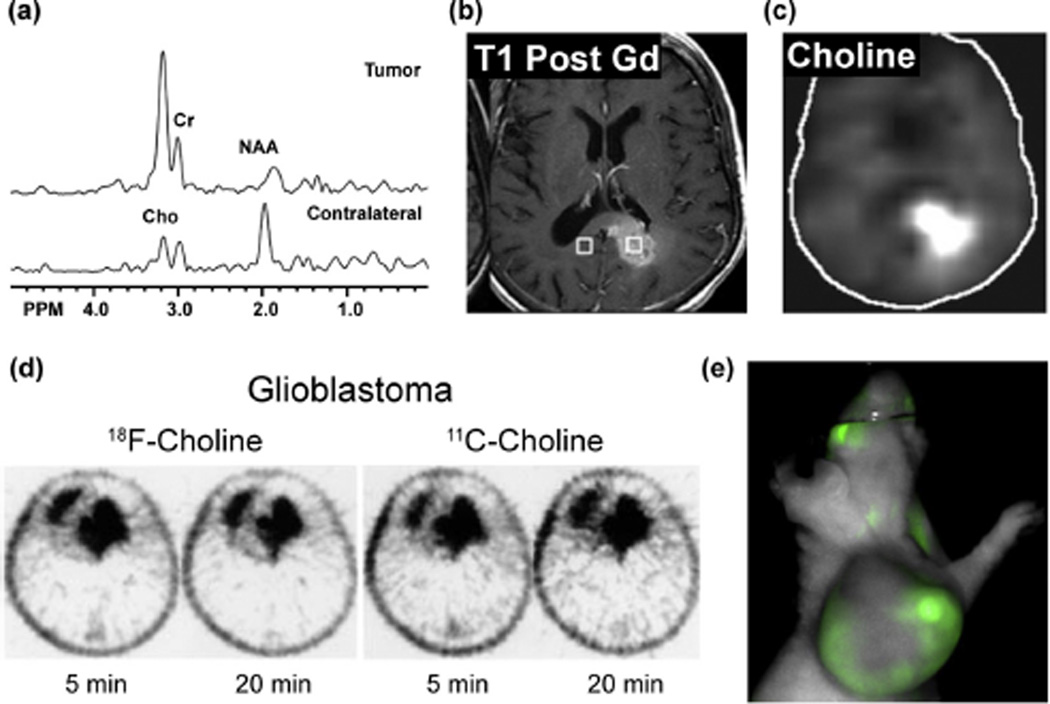
Choline metabolite accumulation as a biomarker in molecular imaging modalities. (a) Single voxel MR spectra acquired for a glioblastoma multiforme tumor or contralateral normal tissue show elevated tCho and diminished N-acetylaspartate (NAA) in the cancer tissue. (b) Gadolinium-enhanced T1-weighted MRI image shows location of the glioblastoma multiforme tumor in the corpus callosum and contralateral normal region, with spectral regions demarcated by the right and left voxels respectively. (c) Chemical shift imaging at 3.2 ppm gives a map of choline distribution (Cho) and demonstrates elevated tCho in the tumor region. Figure adapted with permission from Horská and Barker, 2010 [150]. (d) PET scans of a patient with Grade IV glioblastoma acquired 5 and 20 min after administration of either 18F- or 11C-choline (right) demonstrates comparable tumor avidity between the two tracers (Figure adapted with permission from Hara, et al., 2003 [161]). (e) An athymic nude mouse with a 9 L glioma xenograft was injected i.v. with 20 nmol JAS239, and NIR optical images were acquired after 4 h using a LiCOR Pearl (Ex 710 nm, Em 800 nm), revealing significant tumor accumulation (unpublished results).
PME and tCho accumulation is one way that cancer cells are able to provide the metabolic precursors necessary for proliferation, and has been linked to overall poorer prognosis in numerous cancers [142]. Similarly, early drops in PMEs and tCho are predictive of responsiveness to both chemo- and radiotherapies [28,143–145]. The MR technique can be translated to study human or animal tumors, perfused cell models, biopsy tissue, and cell extracts, with the capacity to resolve individual contributors to the PME and tCho peaks improving in that order [146]. Comparison of spectra from tumor-derived cells versus their normal counterparts has revealed elevated PC levels in breast [14,36], glioma [147], ovarian [148], and prostate [149] cells. Because it only measures the total steady-state pool of PC, MRS is unable to detect the relative contribution of the anabolic and catabolic pathways by which PC is generated.
MRS-based studies can detect steady-state metabolite levels, but transporters and enzymes from many pathways can affect these pools. The Phase-I clinical trial for TCD-717 in advanced solid tumors had an efficacy outcome measure based upon Response Evaluation Criteria in Solid Tumors (RECIST), but two years into the trial was amended to include in vivo MRS-based measurements of tCho to validate patient response (ClinicalTrials.gov Identifier: NCT01215864). Measurement of tCho provides a more specific metric to monitor ChoK inhibition than RECIST criteria, but necrosis, hypoxia, and other local environmental stresses can alter tCho levels and obfuscate the interpretation of these results [150]. Due to low baseline lipid levels in the brain, MRI/MRS has found its greatest value in imaging brain tumors (Fig. 7a–c). In vivo tCho measurement by MRS was recently shown to be feasible for validation of the ChoKα inhibitor MN58b in rat models of glioma [113].
6.2. Positron emission tomography
Positron emission tomography (PET) is a sensitive imaging method that allows in vivo distribution of exogenous tracers to be observed. Due to high glucose uptake in the brain, 11C-choline was developed as an alternative to the widely used oncologic PET agent 18F-fluorodeoxyglucose for identifying brain tumors [151]. Both 11C-choline and its analog 18F-fluorocholine have had particular utility in detecting brain (Fig. 7d) and prostate tumors, as 18F-fluorodeoxyglucose detection in these tissues is difficult due to uptake in surrounding tissues [152,153]. The shorter radioactive half-life of 11C-choline (t1/2 = 20 min) compared to 18F-fluorocholine (t1/2 = 110 min) limits its routine use to facilities in close proximity to a cyclotron [154], however its biodistribution and phospholipid incorporation more closely matches that of endogenous choline [155]. Choline-analog PET tracers appear most promising for restaging prostate tumors after therapy [156], but there are still mixed opinions on the clinical utility due to the high false-positive rate in benign hyperplastic tissues [157]. It is not known how the co-dependence on choline transport and phosphorylation affects the likelihood of false positives using these choline analogs.
Although PET tracers that selectively bind to ChoKα have yet to be developed, the choline transport inhibitor HC-3 has been radiolabeled in an effort to identify regions of high choline uptake [158]. Zheng et al. used systemically-delivered 11C- or 18F-labeled HC-3 to image CHT1 transporters in 9 L glioma rats and found a tumor/muscle ratio as high as 5, however poor tumor delivery was observed relative to the high renal and liver accumulation [158]. To our knowledge, this was the first effort to label a choline-like inhibitor for imaging purposes and represents an interesting strategy to validate ChoKα inhibitors were a ChoKα specific probe to be identified.
6.3. Optical imaging
A major challenge has been that MRS and PET imaging methods cannot distinguish ChoKα expression and inhibition from the complex activities of the poorly understood choline transporters. The search for ChoKα-specific inhibitors has provided medicinal chemistry insight into pharmacophore features that can be adopted into molecular imaging probes with improved selectivity for ChoKα over other choline pathway enzymes and transporters [121]. An optical imaging (OI) approach was recently developed as a ChoKα-specific imaging strategy, by incorporating choline mimetic moieties into a fluorescent scaffold [159]. The carbocyanine derivative JAS239 was reported to inhibit ChoKα and diminish proliferation of triple negative breast cancer cells, without impairing ChoT. This work reveals a novel approach to validate ChoKα inhibitors and exclude neurotoxic ChoT inhibitors like HC-3. Accumulation of JAS239 in glioma xenografts with elevated ChoK activity has been imaged in vivo using near infrared fluorescence OI (Fig. 7e). Minimal light scattering and tissue absorbance at near infrared wavelengths [160] permits JAS239 detection through several millimeters of tissue, and makes ChoKα-targeted OI probes a promising cancer staging technique moving forward.
7. Conclusion
Cell survival depends on tight regulation of lipid metabolism to maintain homeostasis, however these processes are often highjacked by cancer cells to gain competitive proliferative, metastatic, and drug resistant phenotypes. ChoKα represents a critical node in phospholipid synthesis and second messenger production. Constitutive activation of this enzyme can be an oncogenic driving force. Aberrant choline metabolism is an established biomarker of tumor malignancy, and has gained attention as a diagnostic indicator and therapeutic target. Molecular imaging applications using endogenous or labeled choline for tumor contrast are already in the clinic, however the measurements made by these methods are not specific to ChoKα and have yet to be used routinely during disease monitoring. New specific ChoKα inhibitors have enhanced our understanding of this kinase's mechanism, but have also raised many new questions about ChoKα's role as a cell signaling mediator and provider of biosynthetic precursors. It is critical that these tool compounds be adapted as molecular imaging probes to allow a more complete interpretation of the choline-based MRS and PET techniques used clinically in brain and prostate cancer patients. In vivo tracers based on specific inhibitors such as TCD-717, CK-37, and V-11-023907 will benefit oncologic imaging by helping to distinguish choline accumulation from ChoKα over-expression. As PC production may not be the driving force of ChoKα's tumor-driving activity, in vivo ChoKα-targeted companion diagnostics such as JAS239 will be critical to validate future clinical ChoKα inhibition therapies.
Acknowledgments
This work was supported by the NIH R01 CA129176, R01 EB018645 (E.J. Delikatny), T32 GM8076 (S.P. Arlauckas), F31 CA180328 (S.P. Arlauckas) and DoD Breast Cancer Concept Award BC076631 (E.J. Delikatny).
Footnotes
Conflict of interest
The authors disclose no potential conflicts of interest.
References
- 1.Currie E, Schulze A, Zechner R, Walther TC, Farese RVJ. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delikatny EJ, Cooper WA, Brammah S, Sathasivam N, Rideout DC. Nuclear magnetic resonance-visible lipids induced by cationic lipophilic chemotherapeutic agents are accompanied by increased lipid droplet formation and damaged mitochondria. Cancer Res. 2002;62:1394–1400. [PubMed] [Google Scholar]
- 3.Milkevitch M, Shim H, Pilatus U, Pickup S, Wehrle JP, Samid D, et al. Increases in NMR-visible lipid and glycerophosphocholine during phenylbutyrate-induced apoptosis in human prostate cancer cells. Biochim Biophys Acta Mol Cell Biol Lipids. 2005;1734:1–12. doi: 10.1016/j.bbalip.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Schmitz JE, Kettunen MI, Hu D-E, Brindle KM. 1H MRS-visible lipids accumulate during apoptosis of lymphoma cells in vitro and in vivo. Magn Reson Med. 2005;54:43–50. doi: 10.1002/mrm.20529. [DOI] [PubMed] [Google Scholar]
- 5.Milkevitch M, Beardsley NJ, Delikatny EJ. Phenylbutyrate induces apoptosis and lipid accumulations via a peroxisome proliferator-activated receptor gamma-dependent pathway. NMR Biomed. 2010;23:473–479. doi: 10.1002/nbm.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delikatny EJ, Chawla S, Leung DJ, Poptani H. MR-visible lipids and the tumor microenvironment. NMR Biomed. 2011;24:592–611. doi: 10.1002/nbm.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boren J, Brindle KM. Apoptosis-induced mitochondrial dysfunction causes cytoplasmic lipid droplet formation. Cell Death Differ. 2012;19:1561–1570. doi: 10.1038/cdd.2012.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 9.Michel V, Yuan Z, Ramsubir S, Bakovic M. Choline transport for phospholipid synthesis. Exp Biol Med (Maywood) 2006;231:490–504. doi: 10.1177/153537020623100503. [DOI] [PubMed] [Google Scholar]
- 10.Glunde K, Bhujwalla ZM, Ronen SM. Choline metabolism in malignant transformation. Nat Rev Cancer. 2011;11:835–848. doi: 10.1038/nrc3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kouji H, Inazu M, Yamada T, Tajima H, Aoki T, Matsumiya T. Molecular and functional characterization of choline transporter in human colon carcinoma HT-29 cells. Arch Biochem Biophys. 2009;483:90–98. doi: 10.1016/j.abb.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Wang T, Li J, Chen F, Zhao Y, He X, Wan D, et al. Choline transporters in human lung adenocarcinoma: expression and functional implications. Acta Biochim Biophys Sin Shanghai. 2007;39:668–674. doi: 10.1111/j.1745-7270.2007.00323.x. [DOI] [PubMed] [Google Scholar]
- 13.Fullerton MD, Wagner L, Yuan Z, Bakovic M. Impaired trafficking of choline transporter-like protein-1 at plasma membrane and inhibition of choline transport in THP-1 monocyte-derived macrophages. AJP: Cell Physiol. 2006;290:C1230–C1238. doi: 10.1152/ajpcell.00255.2005. [DOI] [PubMed] [Google Scholar]
- 14.Eliyahu G, Kreizman T, Degani H. Phosphocholine as a biomarker of breast cancer: molecular and biochemical studies. Int J Cancer. 2007;120:1721–1730. doi: 10.1002/ijc.22293. [DOI] [PubMed] [Google Scholar]
- 15.Wu D, Hersh LB. Choline acetyltransferase: celebrating its fiftieth year. J Neurochem. 1994;62:1653–1663. doi: 10.1046/j.1471-4159.1994.62051653.x. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez B, Pajares MA, Martinez-Ripoll M, Blundell TL, Sanz-Aparicio J. Crystal structure of rat liver betaine homocysteine S-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding. J Mol Biol. 2004;338:771–782. doi: 10.1016/j.jmb.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Cui Z, Vance DE. Expression of phosphatidylethanolamine N-methyltransferase-2 is markedly enhanced in long term choline-deficient rats. J Biol Chem. 1996;271:2839–2843. doi: 10.1074/jbc.271.5.2839. [DOI] [PubMed] [Google Scholar]
- 18.Vance DE. Physiological roles of phosphatidylethanolamine N-methyltransferase. Biochim Biophys Acta. 1831;2013:626–632. doi: 10.1016/j.bbalip.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 19.Ullman MD, Radin NS. The enzymatic formation of sphingomyelin from ceramide and lecithin in mouse liver. J Biol Chem. 1974;249:1506–1512. [PubMed] [Google Scholar]
- 20.Berger A, Rosenthal D, Spiegel S. Sphingosylphosphocholine, a signaling molecule which accumulates in Niemann–Pick disease type A, stimulates DNA-binding activity of the transcription activator protein AP-1. Proc Natl Acad Sci. 1995;92:5885–5889. doi: 10.1073/pnas.92.13.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss SB, Smith SW, Kennedy EP. Net synthesis of lecithin in an isolated enzyme system. Nature. 1956;178:594–595. doi: 10.1038/178594a0. [DOI] [PubMed] [Google Scholar]
- 22.Wittenberg J, Kornberg A. Choline phosphokinase. J Biol Chem. 1953;202:431–444. [PubMed] [Google Scholar]
- 23.Kennedy EP, Weiss SB. The function of cytidine coenzymes in the biosynthesis of phospholipides. J Biol Chem. 1956;222:193–214. [PubMed] [Google Scholar]
- 24.Kennedy EP. Metabolism of lipides. Annu Rev Biochem. 1957;26:119–148. doi: 10.1146/annurev.bi.26.070157.001003. [DOI] [PubMed] [Google Scholar]
- 25.Ramoni C, Spadaro F, Barletta B, Dupuis ML, Podo F. Phosphatidylcholine-specific phospholipase C in mitogen-stimulated fibroblasts. Exp Cell Res. 2004;299:370–382. doi: 10.1016/j.yexcr.2004.05.037. [DOI] [PubMed] [Google Scholar]
- 26.Mawn TM, Popov AV, Beardsley NJ, Stefflova K, Milkevitch M, Zheng G, et al. In vivo detection of phospholipase C by enzyme-activated near-infrared probes. Bioconjug Chem. 2011;22:2434–2443. doi: 10.1021/bc200242v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Podo F. Tumour phospholipid metabolism. NMR Biomed. 1999;12:413–439. doi: 10.1002/(sici)1099-1492(199911)12:7<413::aid-nbm587>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 28.Podo F, Canevari S, Canese R, Pisanu ME, Ricci A, Iorio E. MR evaluation of response to targeted treatment in cancer cells. NMR Biomed. 2011;24:648–672. doi: 10.1002/nbm.1658. [DOI] [PubMed] [Google Scholar]
- 29.Agwu DE, McPhail LC, Chabot MC, Daniel LW, Wykle RL, McCall CE. Choline-linked phosphoglycerides. A source of phosphatidic acid and diglycerides in stimulated neutrophils. J Biol Chem. 1989;264:1405–1413. [PubMed] [Google Scholar]
- 30.Nishizuka Y. Intracellular signaling by hydrolysis of phospholipids and activation of protein kinase C. Science. 1992;258:607–614. doi: 10.1126/science.1411571. [DOI] [PubMed] [Google Scholar]
- 31.Moolenaar WH. Bioactive lysophospholipids and their G protein-coupled receptors. Exp Cell Res. 1999;253:230–238. doi: 10.1006/excr.1999.4702. [DOI] [PubMed] [Google Scholar]
- 32.Du J, Sun C, Hu Z, Yang Y, Zhu Y, Zheng D, et al. Lysophosphatidic acid induces MDA-MB-231 breast cancer cells migration through activation of PI3K/PAK1/ERK signaling. PLoS One. 2010;5:e15940. doi: 10.1371/journal.pone.0015940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cuadrado A, Carnero A, Dolfi F, Jiménez B, Lacal JC. Phosphorylcholine: a novel second messenger essential for mitogenic activity of growth factors. Oncogene. 1993;8:2959–2968. [PubMed] [Google Scholar]
- 34.Jiménez B, del Peso L, Montaner S, Esteve P, Lacal JC. Generation of phosphorylcholine as an essential event in the activation of Raf-1 and MAP-kinases in growth factors-induced mitogenic stimulation. J Cell Biochem. 1995;57:141–149. doi: 10.1002/jcb.240570114. [DOI] [PubMed] [Google Scholar]
- 35.Chung T, Crilly KS, Anderson WH, Mukherjee JJ. ATP-dependent choline phosphate-induced mitogenesis in fibroblasts involves activation of pp70 S6 kinase and phosphatidylinositol 3′-kinase through an extracellular site. J Biol Chem. 1997;272:3064–3072. doi: 10.1074/jbc.272.5.3064. [DOI] [PubMed] [Google Scholar]
- 36.Aboagye EO, Bhujwalla ZM. Malignant transformation alters membrane choline phospholipid metabolism of human mammary epithelial cells. Cancer Res. 1999;59:80–84. [PubMed] [Google Scholar]
- 37.Mori N, Delsite R, Natarajan K, Kulawiec M, Bhujwalla ZM, Singh KK. Loss of p53 function in colon cancer cells results in increased phosphocholine and total choline. Mol Imaging. 2004;3:319–323. doi: 10.1162/15353500200404121. [DOI] [PubMed] [Google Scholar]
- 38.Ramírez de Molina A, Rodríguez-González A, Penalva V, Lucas L, Lacal JC. Inhibition of ChoK is an efficient antitumor strategy for Harvey-, Kirsten-, and N-ras-transformed cells. Biochem Biophys Res Commun. 2001;285:873–879. doi: 10.1006/bbrc.2001.5250. [DOI] [PubMed] [Google Scholar]
- 39.Ramírez de Molina A, Báñez-Coronel M, Gutiérrez R, Rodríguez-González A, Olmeda D, Megías D, et al. Choline kinase activation is a critical requirement for the proliferation of primary human mammary epithelial cells and breast tumor progression. Cancer Res. 2004;64:6732–6739. doi: 10.1158/0008-5472.CAN-04-0489. [DOI] [PubMed] [Google Scholar]
- 40.Aoyama C, Liao H, Ishidate K. Structure and function of choline kinase isoforms in mammalian cells. Prog Lipid Res. 2004;43:266–281. doi: 10.1016/j.plipres.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Miyake T, Parsons SJ. Functional interactions between choline kinase α, epidermal growth factor receptor and c-Src in breast cancer cell proliferation. Oncogene. 2012;31:1431–1441. doi: 10.1038/onc.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aoyama C, Nakashima K, Ishidate K. Molecular cloning of mouse choline kinase and choline/ethanolamine kinase: their sequence comparison to the respective rat homologs. Biochim Biophys Acta. 1998;1393:179–185. doi: 10.1016/s0005-2760(98)00062-9. [DOI] [PubMed] [Google Scholar]
- 43.Aoyama C, Ohtani A, Ishidate K. Expression and characterization of the active molecular forms of choline/ethanolamine kinase-alpha and -beta in mouse tissues, including carbon tetrachloride-induced liver. Biochem J. 2002;363:777–784. doi: 10.1042/0264-6021:3630777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallego-Ortega D, Ramírez de Molina A, Ramos MA, Valdes-Mora F, Barderas MG, Sarmentero-Estrada J, et al. Differential role of human choline Kinase α and β enzymes in lipid metabolism: implications in cancer onset and treatment. PLoS One. 2009;4:e7819. doi: 10.1371/journal.pone.0007819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sher RB, Aoyama C, Huebsch KA, Ji S, Kerner J, Yang Y, et al. A rostrocaudal muscular dystrophy caused by a defect in choline kinase beta, the first enzyme in phosphatidylcholine biosynthesis. J Biol Chem. 2006;281:4938–4948. doi: 10.1074/jbc.M512578200. [DOI] [PubMed] [Google Scholar]
- 46.Wu G, Aoyama C, Young SG, Vance DE. Early embryonic lethality caused by disruption of the gene for choline kinase alpha, the first enzyme in phosphatidylcholine biosynthesis. J Biol Chem. 2008;283:1456–1462. doi: 10.1074/jbc.M708766200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peisach D, Gee P, Kent C, Xu Z. The crystal structure of choline kinase reveals a eukaryotic protein kinase fold. Structure. 2003;11:703–713. doi: 10.1016/s0969-2126(03)00094-7. [DOI] [PubMed] [Google Scholar]
- 48.Scheeff ED, Bourne PE. Structural evolution of the protein kinase-like superfamily. PLoS Comput Biol. 2005;1:e49. doi: 10.1371/journal.pcbi.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudson CS, Knegtel RM, Brown K, Charlton PA, Pollard JR. Kinetic and mechanistic characterisation of choline kinase-α. Biochim Biophys Acta Protein Proteomics. 2013;1834:1107–1116. doi: 10.1016/j.bbapap.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Malito E, Sekulic N, Too WCS, Konrad M, Lavie A. Elucidation of human choline kinase crystal structures in complex with the products ADP or phosphocholine. J Mol Biol. 2006;364:136–151. doi: 10.1016/j.jmb.2006.08.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hong BS, Allali-Hassani A, Tempel W, Finerty PJ, Mackenzie F, Dimov S, et al. Crystal structures of human choline kinase isoforms in complex with hemicholinium-3: single amino acid near the active site influences inhibitor sensitivity. J Biol Chem. 2010;285:16330–16340. doi: 10.1074/jbc.M109.039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishidate K, Tsuruoka M, Nakazawa Y. Induction of choline kinase by polycyclic aromatic hydrocarbon carcinogens in rat liver. Biochem Biophys Res Commun. 1980;96:946–952. doi: 10.1016/0006-291x(80)91446-1. [DOI] [PubMed] [Google Scholar]
- 53.Aoyama C, Ishidate K, Sugimoto H, Vance DE. Induction of choline kinase alpha by carbon tetrachloride (CCl4) occurs via increased binding of c-jun to an AP-1 element. Biochim Biophys Acta Mol Cell Biol Lipids. 2007;1771:1148–1155. doi: 10.1016/j.bbalip.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Glunde K, Shah T, Winnard PT, Raman V, Takagi T, Vesuna F, et al. Hypoxia regulates choline kinase expression through hypoxia-inducible factor-1 signaling in a human prostate cancer model. Cancer Res. 2008;68:172–180. doi: 10.1158/0008-5472.CAN-07-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bansal A, Harris RA, DeGrado TR. Choline phosphorylation and regulation of transcription of choline kinase in hypoxia. J Lipid Res. 2011;53:149–157. doi: 10.1194/jlr.M021030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baykal AT, Jain MR, Li H. Aberrant regulation of choline metabolism by mitochondrial electron transport system inhibition in neuroblastoma cells. Metabolomics. 2008;4:347–356. doi: 10.1007/s11306-008-0125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morrish F, Neretti N, Sedivy JM, Hockenbery DM. The oncogene c-Myc coordinates regulation of metabolic networks to enable rapid cell cycle entry. Cell Cycle. 2008;7:1054–1066. doi: 10.4161/cc.7.8.5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28:2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramírez de Molina A, Gallego-Ortega D, Sarmentero J, Báñez-Coronel M, Martín-Cantalejo Y, Lacal JC. Choline kinase is a novel oncogene that potentiates RhoA-induced carcinogenesis. Cancer Res. 2005;65:5647–5653. doi: 10.1158/0008-5472.CAN-04-4416. [DOI] [PubMed] [Google Scholar]
- 60.Ramírez de Molina A, Penalva V, Lucas L, Lacal JC. Regulation of choline kinase activity by Ras proteins involves Ral-GDS and PI3K. Oncogene. 2002;21:937–946. doi: 10.1038/sj.onc.1205144. [DOI] [PubMed] [Google Scholar]
- 61.Al-Saffar NMS, Jackson LE, Raynaud FI, Clarke PA, Ramirez de Molina A, Lacal JC, et al. The phosphoinositide 3-kinase inhibitor PI-103 downregulates choline kinase leading to phosphocholine and total choline decrease detected by magnetic resonance spectroscopy. Cancer Res. 2010;70:5507–5517. doi: 10.1158/0008-5472.CAN-09-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu Y, Sreenivas A, Ostrander DB, Carman GM. Phosphorylation of Saccharomyces cerevisiae choline kinase on Ser30 and Ser85 by protein kinase A regulates phosphatidylcholine synthesis by the CDP-choline pathway. J Biol Chem. 2002;277:34978–34986. doi: 10.1074/jbc.M205316200. [DOI] [PubMed] [Google Scholar]
- 63.Wieprecht M, Wieder T, Geilen CC. N-[2-bromocinnamyl (amino) ethyl]-5-isoquinolinesulphonamide (H-89) inhibits incorporation of choline into phosphatidylcholine via inhibition of choline kinase and has no effect on the phosphorylation of CTP:phosphocholine cytidylyltransferase. Biochem J. 1994;297:241–247. doi: 10.1042/bj2970241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang C, Hydo LM, Liu S, Miller RT. Activation of choline kinase by extracellular Ca2+ is Ca2+-sensing receptor, Gα12 and Rho-dependent in breast cancer cells. Cell Signal. 2009;21:1894–1900. doi: 10.1016/j.cellsig.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 65.Silver IA, Murrills RJ, Etherington DJ. Microelectrode studies on the acid microenvironment beneath adherent macrophages and osteoclasts. Exp Cell Res. 1988;175:266–276. doi: 10.1016/0014-4827(88)90191-7. [DOI] [PubMed] [Google Scholar]
- 66.Hoey RP, Sanderson C, Iddon J, Brady G, Bundred NJ, Anderson NG. The parathyroid hormone-related protein receptor is expressed in breast cancer bone metastases and promotes autocrine proliferation in breast carcinoma cells. Br J Cancer. 2003;88:567–573. doi: 10.1038/sj.bjc.6600757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramírez de Molina A, Gutierrez R, Ramos MA, Silva JM, Silva J. Increased choline kinase activity in human breast carcinomas: clinical evidence for a potential novel antitumor strategy. Oncogene. 2002;21:4317–4322. doi: 10.1038/sj.onc.1205556. [DOI] [PubMed] [Google Scholar]
- 68.Ramírez de Molina A, Rodríguez-González A, Gutiérrez R, Martínez-Piñeiro L, Sánchez J, Bonilla F, et al. Overexpression of choline kinase is a frequent feature in human tumor-derived cell lines and in lung, prostate, and colorectal human cancers. Biochem Biophys Res Commun. 2002;296:580–583. doi: 10.1016/s0006-291x(02)00920-8. [DOI] [PubMed] [Google Scholar]
- 69.Iorio E, Ricci A, Bagnoli M, Pisanu ME, Castellano G, Di Vito M, et al. Activation of phosphatidylcholine cycle enzymes in human epithelial ovarian cancer cells. Cancer Res. 2010;70:2126–2135. doi: 10.1158/0008-5472.CAN-09-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trousil S, Lee P, Pinato DJ, Ellis JK, Dina R, Aboagye EO, et al. Alterations of choline phospholipid metabolism in endometrial cancer are caused by choline kinase alpha overexpression and a hyperactivated deacylation pathway. Cancer Res. 2014;74:6867–6877. doi: 10.1158/0008-5472.CAN-13-2409. [DOI] [PubMed] [Google Scholar]
- 71.Penet MF, Shah T, Bharti S, Krishnamachary B, Artemov D, Mironchik Y, et al. Metabolic imaging of pancreatic ductal adenocarcinoma detects altered choline metabolism. Clin Cancer Res. 2015;21:386–395. doi: 10.1158/1078-0432.CCR-14-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramírez de Molina A, Sarmentero-Estrada J, Belda-Iniesta C. Expression of choline kinase alpha to predict outcome in patients with early-stage non-small-cell lung cancer: a retrospective study. Lancet Oncol. 2007;8:889–897. doi: 10.1016/S1470-2045(07)70279-6. [DOI] [PubMed] [Google Scholar]
- 73.Hernando E, Sarmentero-Estrada J, Koppie T, Belda-Iniesta C, Ramírez de Molina V, Cejas P, et al. A critical role for choline kinase-alpha in the aggressiveness of bladder carcinomas. Oncogene. 2009;28:2425–2435. doi: 10.1038/onc.2009.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chung Y, Troy H, Banerji U, Jackson LE, Walton MI, Stubbs M, et al. Magnetic resonance spectroscopic pharmacodynamic markers of the heat shock protein 90 inhibitor 17-allylamino, 17-demethoxygeldanamycin (17AAG) in human colon cancer models. J Natl Cancer Inst. 2003;95:1624–1633. doi: 10.1093/jnci/djg084. [DOI] [PubMed] [Google Scholar]
- 75.Beloueche-Babari M, Arunan V, Jackson LE, Perusinghe N, Sharp SY, Workman P, et al. Modulation of melanoma cell phospholipid metabolism in response to heat shock protein 90 inhibition. Oncotarget. 2010;1:185–197. doi: 10.18632/oncotarget.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brandes AH, Ward CS, Ronen SM. 17-allyamino-17-demethoxygeldanamycin treatment results in a magnetic resonance spectroscopy-detectable elevation in choline-containing metabolites associated with increased expression of choline transporter SLC44A1 and phospholipase A2. Breast Cancer Res. 2010;12:R84. doi: 10.1186/bcr2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sankaranarayanapillai M, Tong WP, Maxwell DS, Pal A, Pang J, Bornmann WG, et al. Detection of histone deacetylase inhibition by noninvasive magnetic resonance spectroscopy. Mol Cancer Ther. 2006;5:1325–1334. doi: 10.1158/1535-7163.MCT-05-0494. [DOI] [PubMed] [Google Scholar]
- 78.Chung Y, Troy H, Kristeleit R, Aherne W, Jackson LE, Atadja P, et al. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of a novel histone deacetylase inhibitor, LAQ824, in human colon carcinoma cells and xenografts. Neoplasia. 2008;10:303–313. doi: 10.1593/neo.07834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beloueche-Babari M, Chung Y-L, Al-Saffar NMS, Falck-Miniotis M, Leach MO. Metabolic assessment of the action of targeted cancer therapeutics using magnetic resonance spectroscopy. Br J Cancer. 2010;102:1–7. doi: 10.1038/sj.bjc.6605457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ward CS, Eriksson P, Izquierdo-Garcia JL, Brandes AH, Ronen SM. HDAC inhibition induces increased choline uptake and elevated phosphocholine levels in MCF7 breast cancer cells. PLoS One. 2013;8:e62610. doi: 10.1371/journal.pone.0062610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MacKeigan JP, Murphy LO, Blenis J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat Cell Biol. 2005;7:591–600. doi: 10.1038/ncb1258. [DOI] [PubMed] [Google Scholar]
- 82.Glunde K, Raman V, Mori N, Bhujwalla ZM. RNA interference-mediated choline kinase suppression in breast cancer cells induces differentiation and reduces proliferation. Cancer Res. 2005;65:11034–11043. doi: 10.1158/0008-5472.CAN-05-1807. [DOI] [PubMed] [Google Scholar]
- 83.Chua BT, Gallego-Ortega D, Ramírez de Molina A, Ullrich A, Lacal JC, Downward J. Regulation of Akt(ser473) phosphorylation by choline kinase in breast carcinoma cells. Mol Cancer. 2009;8:131. doi: 10.1186/1476-4598-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gruber J, See Too WC, Wong MT, Lavie A, McSorley T, Konrad M. Balance of human choline kinase isoforms is critical for cell cycle regulation: implications for the development of choline kinase-targeted cancer therapy. FEBS J. 2012;279:1915–1928. doi: 10.1111/j.1742-4658.2012.08573.x. [DOI] [PubMed] [Google Scholar]
- 85.Báñez-Coronel M, Ramírez de Molina A, Rodríguez-González A, Sarmentero J, Ramos MA, García-Cabezas MA, et al. Choline kinase alpha depletion selectively kills tumoral cells. Curr Cancer Drug Targets. 2008;8:709–719. doi: 10.2174/156800908786733432. [DOI] [PubMed] [Google Scholar]
- 86.Mori N, Glunde K, Takagi T, Raman V, Bhujwalla ZM. Choline kinase down-regulation increases the effect of 5-fluorouracil in breast cancer cells. Cancer Res. 2007;67:11284–11290. doi: 10.1158/0008-5472.CAN-07-2728. [DOI] [PubMed] [Google Scholar]
- 87.Granata A, Nicoletti R, Tinaglia V, De Cecco L, Pisanu ME, Ricci A, et al. Choline kinase-alpha by regulating cell aggressiveness and drug sensitivity is a potential druggable target for ovarian cancer. Br J Cancer. 2014;110:330–340. doi: 10.1038/bjc.2013.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Krishnamachary B, Glunde K, Wildes F, Mori N, Takagi T, Raman V, et al. Noninvasive detection of lentiviral-mediated choline kinase targeting in a human breast cancer xenograft. Cancer Res. 2009;69:3464–3471. doi: 10.1158/0008-5472.CAN-08-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hosaka K, Murakami T, Kodaki T, Nikawa J, Yamashita S. Repression of choline kinase by inositol and choline in Saccharomyces cerevisiae. J Bacteriol. 1990;172:2005–2012. doi: 10.1128/jb.172.4.2005-2012.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hodgson E, Dauterman WC, Mehendale HM, Smith E, Khan MAQ. Dietary choline requirements, phospholipids and development in Phormia regina. Comp Biochem Physiol. 1969;16:343–359. [Google Scholar]
- 91.Brostrom MA, Browning ET. Choline kinase from brewers' yeast. Partial purification, properties, and kinetic mechanism. J Biol Chem. 1973;248:2364–2371. [PubMed] [Google Scholar]
- 92.Mayer D, Werner D. Inhibition of choline kinase by selectively cytotoxic purinyl-6-histamine. Biochem Pharmacol. 1974;23:1227–1230. doi: 10.1016/0006-2952(74)90300-1. [DOI] [PubMed] [Google Scholar]
- 93.Lacal JC. Choline kinase: a novel target for antitumor drugs. IDrugs. 2001;4:419–426. [PubMed] [Google Scholar]
- 94.Ishidate K. Phosphatidylcholine Metabolism. first. CRC Press; 1989. Choline transport and choline kinase; pp. 9–32. [Google Scholar]
- 95.Zhang X, Lin Y, Gillies RJ. Tumor pH and its measurement. J Nucl Med. 2010;51:1167–1170. doi: 10.2967/jnumed.109.068981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ansell GB, Spanner SG. The inhibition of brain choline kinase by hemicholinium-3. J Neurochem. 1974;22:1153–1155. doi: 10.1111/j.1471-4159.1974.tb04352.x. [DOI] [PubMed] [Google Scholar]
- 97.Hamza M, Lloveras J, Ribbes G, Soula G, Douste-Blazy L. An in vitro study of hemicholinium-3 on phospholipid metabolism of Krebs II ascites cells. Biochem Pharmacol. 1983;32:1893–1897. doi: 10.1016/0006-2952(83)90055-2. [DOI] [PubMed] [Google Scholar]
- 98.Gomez MV, Domino EF, Sellinger OZ. Effect of hemicholinium-3 on choline distribution in vivo in the canine caudate nucleus. Biochem Pharmacol. 1970;19:1753–1760. doi: 10.1016/0006-2952(70)90167-x. [DOI] [PubMed] [Google Scholar]
- 99.Long JP, Schueler FW. A new series of cholinesterase inhibitors. J Am Pharm Assoc. 1954;43:79–86. doi: 10.1002/jps.3030430204. [DOI] [PubMed] [Google Scholar]
- 100.Yamamura HI, Snyder SH. High affinity transport of choline into synaptosomes of rat brain. J Neurochem. 1973;21:1355–1374. doi: 10.1111/j.1471-4159.1973.tb06022.x. [DOI] [PubMed] [Google Scholar]
- 101.Guyenet P, Lefresne P, Rossier J, Beaujouan JC, Glowinski J. Inhibition by hemicholinium-3 of (14C)acetylcholine synthesis and (3H)choline high-affinity uptake in rat striatal synaptosomes. Mol Pharmacol. 1973;9:630–639. [PubMed] [Google Scholar]
- 102.Macintosh FC. Effect of HC-3 on acetylcholine turnover. Fed Proc. 1961;20:562–568. [PubMed] [Google Scholar]
- 103.Gardiner JE. The inhibition of acetylcholine synthesis in brain by a hemicholinium. Biochem J. 1961;81:297–303. doi: 10.1042/bj0810297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cannon JG. Structure–activity aspects of hemicholinium-3 (HC-3) and its analogs and congeners. Med Res Rev. 1994;14:505–531. doi: 10.1002/med.2610140503. [DOI] [PubMed] [Google Scholar]
- 105.Boobis AR, Gibson A, Stevenson RW. Ethanol protection against hemicholinium toxicity in mice. Biochem Pharmacol. 1975;24:485–488. doi: 10.1016/0006-2952(75)90134-3. [DOI] [PubMed] [Google Scholar]
- 106.Tedford CE, Reed D, Bhattacharyya B, Bhalla P, Cannon JG, Long JP. Evaluation of 4-methylpiperidine analogs of hemicholinium-3. Eur J Pharmacol. 1986;128:231–239. doi: 10.1016/0014-2999(86)90770-3. [DOI] [PubMed] [Google Scholar]
- 107.Bhattacharyya B, Sokoll MD, Cannon JG, Long JP. Pharmacologic evaluation and structure activity relationships of a series of hemicholinium-3 (HC-3) analogs. Arch Int Pharmacodyn Ther. 1987;288:136–146. [PubMed] [Google Scholar]
- 108.Cannon JG, Lee TM, Chang Y, Nyanda AM, Bhattacharyya B, Flynn JR, et al. Structure–activity relationship studies of hemicholinium (HC-3) congeners. Pharm Res. 1988;5:359–364. doi: 10.1023/a:1015955527100. [DOI] [PubMed] [Google Scholar]
- 109.Sheff KY, Tedford CE, Flynn JR, Yorek MA, Cannon JG, Long JP. Stereoisomers of quaternary and tertiary 4-methyl piperidine analogs of hemicholinium-3. J Pharmacol Exp Ther. 1988;247:640–644. [PubMed] [Google Scholar]
- 110.Hernández-Alcoceba R, Saniger L, Campos J, Núñez MC, Khaless F, Gallo MA, et al. Choline kinase inhibitors as a novel approach for antiproliferative drug design. Oncogene. 1997;15:2289–2301. doi: 10.1038/sj.onc.1201414. [DOI] [PubMed] [Google Scholar]
- 111.Campos J, del Carmen Núñez M, Rodríguez V, Entrena A, Hernández-Alcoceba R, Fernández F, et al. LUMO energy of model compounds of bispyridinium compounds as an index for the inhibition of choline kinase. Eur J Med Chem. 2001;36:215–225. doi: 10.1016/s0223-5234(01)01219-3. [DOI] [PubMed] [Google Scholar]
- 112.Al-Saffar NMS, Troy H, Ramírez de Molina A, Jackson LE, Madhu B, Griffiths JR, et al. Noninvasive magnetic resonance spectroscopic pharmacodynamic markers of the choline kinase inhibitor MN58b in human carcinoma models. Cancer Res. 2006;66:427–434. doi: 10.1158/0008-5472.CAN-05-1338. [DOI] [PubMed] [Google Scholar]
- 113.Kumar M, Arlauckas SP, Saksena S, Verma G, Ittyerah R, Pickup S, et al. Magnetic resonance spectroscopy for detection of choline kinase inhibition in the treatment of brain tumors. Mol Cancer Ther. 2015;14:899–908. doi: 10.1158/1535-7163.MCT-14-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hernández-Alcoceba R, Fernández F, Lacal JC. In vivo antitumor activity of choline kinase inhibitors: a novel target for anticancer drug discovery. Cancer Res. 1999;59:3112–3118. [PubMed] [Google Scholar]
- 115.Campos J, Núñez MC, Conejo-García A. QSAR-derived choline kinase inhibitors: how rational can antiproliferative drug design be? Curr Med Chem. 2003;10:1095–1112. doi: 10.2174/0929867033457539. [DOI] [PubMed] [Google Scholar]
- 116.Conejo-García A. Choline kinase inhibitory effect and antiproliferative activity of new 1,1′,1″-(benzene-1,3,5-triylmethylene)tris{4-[(disubstituted)amino]pyridinium} tribromides. Eur J Med Chem. 2003;38:109–116. doi: 10.1016/s0223-5234(02)00004-1. [DOI] [PubMed] [Google Scholar]
- 117.Conejo-García A, Campos JM, Sánchez-Martín RM, Gallo MA, Espinosa A. Bispyridinium cyclophanes: novel templates for human choline kinase inhibitors. J Med Chem. 2003;46:3754–3757. doi: 10.1021/jm030792i. [DOI] [PubMed] [Google Scholar]
- 118.Conejo-García A, Báñez-Coronel M, Sánchez-Martín RM, Rodríguez-González A, Ramos A, Ramírez de Molina A, et al. Influence of the linker in bispyridium compounds on the inhibition of human choline kinase. J Med Chem. 2004;47:5433–5440. doi: 10.1021/jm0496537. [DOI] [PubMed] [Google Scholar]
- 119.Janardhan S, Srivani P. 2D and 3D quantitative structure–activity relationship studies on a series of bis-pyridinium compounds as choline kinase inhibitors. QSAR Comb Sci. 2006;25:860–872. [Google Scholar]
- 120.Sánchez-Martín R, Campos JM, Conejo-García A, Cruz-López O, Báñez-Coronel M, Rodríguez-González A, et al. Symmetrical bis-quinolinium compounds: new human choline kinase inhibitors with antiproliferative activity against the HT-29 cell line. J Med Chem. 2005;48:3354–3363. doi: 10.1021/jm049061o. [DOI] [PubMed] [Google Scholar]
- 121.Campos JM, Sánchez-Martín RM, Conejo-García A, Entrena A, Gallo MA, Espinosa A. (Q)SAR studies to design new human choline kinase inhibitors as antiproliferative drugs. Curr Med Chem. 2006;13:1231–1248. doi: 10.2174/092986706776872961. [DOI] [PubMed] [Google Scholar]
- 122.Gómez Pérez V, McSorley T, See Too WC, Konrad M, Campos JM. Novel 4-amino bis-pyridinium and bis-quinolinium derivatives as choline kinase inhibitors with antiproliferative activity against the human breast cancer SKBR-3 cell line. Chem Med Chem. 2012;7:663–669. doi: 10.1002/cmdc.201100505. [DOI] [PubMed] [Google Scholar]
- 123.Lacal JC, Campos JM. Preclinical characterization of RSM-932A, a novel anticancer drug targeting the human choline kinase alpha, an enzyme involved in increased lipid metabolism of cancer cells. Mol Cancer Ther. 2015;14:31–39. doi: 10.1158/1535-7163.MCT-14-0531. [DOI] [PubMed] [Google Scholar]
- 124.Trousil S, Carroll L, Kalusa A, Aberg O, Kaliszczak M, Aboagye EO. Design of symmetrical and nonsymmetrical N,N-dimethylaminopyridine derivatives as highly potent choline kinase alpha inhibitors. Med Chem Commun. 2013;4:693. doi: 10.1039/C3MD00068K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schiaffino-Ortega S, López-Cara LC, Ríos-Marco P, Carrasco-Jimenez MP, Gallo MA, Espinosa A, et al. New non-symmetrical choline kinase inhibitors. Bioorg Med Chem. 2013:1–29. doi: 10.1016/j.bmc.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 126.Sahún-Roncero M, Rubio-Ruiz B, Saladino G, Conejo-García A, Espinosa A, Velázquez-Campoy A, et al. The mechanism of allosteric coupling in choline kinase α1 revealed by the action of a rationally designed inhibitor. Angew Chem Int Ed. 2013;52:4582–4586. doi: 10.1002/anie.201209660. [DOI] [PubMed] [Google Scholar]
- 127.Clem BF, Clem AL, Yalcin A, Goswami U, Arumugam S, Telang S, et al. A novel small molecule antagonist of choline kinase-α that simultaneously suppresses MAPK and PI3K/AKT signaling. Oncogene. 2011;30:3370–3380. doi: 10.1038/onc.2011.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Falcon SC, Hudson CS, Huang Y, Mortimore M, Golec JM, Charlton PA, et al. A non-catalytic role of choline kinase alpha is important in promoting cancer cell survival. Oncogenesis. 2013;2:e38–e34. doi: 10.1038/oncsis.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rodríguez-González A, Ramírez de Molina A, Fernández F, Lacal JC. Choline kinase inhibition induces the increase in ceramides resulting in a highly specific and selective cytotoxic antitumoral strategy as a potential mechanism of action. Oncogene. 2004;23:8247–8259. doi: 10.1038/sj.onc.1208045. [DOI] [PubMed] [Google Scholar]
- 130.Rodríguez-González A, Ramírez de Molina A, Báñez-Coronel M, Megías D, Lacal JC. Inhibition of choline kinase renders a highly selective cytotoxic effect in tumour cells through a mitochondrial independent mechanism. Int J Oncol. 2005;26:999–1008. [PubMed] [Google Scholar]
- 131.Rodríguez-González A, de Molina AR, Fernández F, Ramos MA, Núñez MDC, Campos J, et al. Inhibition of choline kinase as a specific cytotoxic strategy in oncogene-transformed cells. Oncogene. 2003;22:8803–8812. doi: 10.1038/sj.onc.1207062. [DOI] [PubMed] [Google Scholar]
- 132.Yalcin A, Clem B, Makoni S, Clem A, Nelson K, Thornburg J, et al. Selective inhibition of choline kinase simultaneously attenuates MAPK and PI3K/AKT signaling. Oncogene. 2010;29:139–149. doi: 10.1038/onc.2009.317. [DOI] [PubMed] [Google Scholar]
- 133.Sanchez-Lopez E, Zimmerman T, del Pulgar TG, Moyer MP, Sanjuan JCL, Cebrian A. Choline kinase inhibition induces exacerbated endoplasmic reticulum stress and triggers apoptosis via CHOP in cancer cells. 2013;4 doi: 10.1038/cddis.2013.453. e933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rubio-Ruiz B, Figuerola-Conchas A. Discovery of a new binding site on human choline kinase α1: design, synthesis, crystallographic studies, and biological evaluation of asymmetrical bispyridinium derivatives. J Med Chem. 2014;57:507–515. doi: 10.1021/jm401665x. [DOI] [PubMed] [Google Scholar]
- 135.Ackerstaff E, Gimi B, Artemov D, Bhujwalla ZM. Anti-inflammatory agent indomethacin reduces invasion and alters metabolism in a human breast cancer cell line. Neoplasia. 2007;9:222–235. doi: 10.1593/neo.06673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hara T, Bansal A, DeGrado TR. Choline transporter as a novel target for molecular imaging of cancer. Mol Imaging. 2006;5:498–509. [PubMed] [Google Scholar]
- 137.Glunde K, Artemov D, Penet MF, Jacobs MA, Bhujwalla ZM. Magnetic resonance spectroscopy in metabolic and molecular imaging and diagnosis of cancer. Chem Rev. 2010;110:3043–3059. doi: 10.1021/cr9004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Daly PF, Cohen JS. Magnetic resonance spectroscopy of tumors and potential in vivo clinical applications: a review. Cancer Res. 1989;49:770–779. [PubMed] [Google Scholar]
- 139.Gillies RJ, Morse DL. In vivo magnetic resonance spectroscopy in cancer. Annu Rev Biomed Eng. 2005;7:287–326. doi: 10.1146/annurev.bioeng.7.060804.100411. [DOI] [PubMed] [Google Scholar]
- 140.de Certaines JD, Larsen VA, Podo F. In vivo 31P MRS of experimental tumours. NMR Biomed. 1993;6:345–365. doi: 10.1002/nbm.1940060602. [DOI] [PubMed] [Google Scholar]
- 141.Glunde K, Serkova NJ. Therapeutic targets and biomarkers identified in cancer choline phospholipid metabolism. Pharmacogenomics. 2006;7:1109–1123. doi: 10.2217/14622416.7.7.1109. [DOI] [PubMed] [Google Scholar]
- 142.Negendank W. Studies of human tumors by MRS: a review. NMR Biomed. 1992;5:303–324. doi: 10.1002/nbm.1940050518. [DOI] [PubMed] [Google Scholar]
- 143.Delikatny EJ, Roman SK, Hancock R, Jeitner TM, Lander CM, Rideout DC, et al. Tetraphenylphosphonium chloride induced MR-visible lipid accumulation in a malignant human breast cell line. Int J Cancer. 1996;67:72–79. doi: 10.1002/(SICI)1097-0215(19960703)67:1<72::AID-IJC13>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 144.Shukla-Dave A, Poptani H, Loevner LA, Mancuso A, Serrai H, Rosenthal DI, et al. Prediction of treatment response of head and neck cancers with P-31 MR spectroscopy from pretreatment relative phosphomonoester levels. Acad Radiol. 2002;9:688–694. doi: 10.1016/s1076-6332(03)80314-8. [DOI] [PubMed] [Google Scholar]
- 145.Lee S-C, Poptani H, Pickup S, Jenkins WT, Kim S, Koch CJ, et al. Early detection of radiation therapy response in non-Hodgkin's lymphoma xenografts by in vivo 1H magnetic resonance spectroscopy and imaging. NMR Biomed. 2010;23:624–632. doi: 10.1002/nbm.1505. [DOI] [PubMed] [Google Scholar]
- 146.Ackerstaff E, Glunde K, Bhujwalla ZM. Choline phospholipid metabolism: a target in cancer cells? J Cell Biochem. 2003;90:525–533. doi: 10.1002/jcb.10659. [DOI] [PubMed] [Google Scholar]
- 147.Gillies RJ, Barry JA, Ross BD. In vitro and in vivo 13C and 31P NMR analyses of phosphocholine metabolism in rat glioma cells. Magn Reson Med. 1994;32:310–318. doi: 10.1002/mrm.1910320306. [DOI] [PubMed] [Google Scholar]
- 148.Iorio E, Mezzanzanica D, Alberti P, Spadaro F, Ramoni C, D'Ascenzo S, et al. Alterations of choline phospholipid metabolism in ovarian tumor progression. Cancer Res. 2005;65:9369–9376. doi: 10.1158/0008-5472.CAN-05-1146. [DOI] [PubMed] [Google Scholar]
- 149.Ackerstaff E, Pflug BR, Nelson JB, Bhujwalla ZM. Detection of increased choline compounds with proton nuclear magnetic resonance spectroscopy subsequent to malignant transformation of human prostatic epithelial cells. Cancer Res. 2001;61:3599–3603. [PubMed] [Google Scholar]
- 150.Horská A, Barker PB. Imaging of brain tumors: MR spectroscopy and metabolic imaging. Neuroimaging Clin N Am. 2010;20:293–310. doi: 10.1016/j.nic.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hara T, Kosaka N, Shinoura N, Kondo T. PET imaging of brain tumor with [methyl-11C]choline. J Nucl Med. 1997;38:842–847. [PubMed] [Google Scholar]
- 152.Beauregard J-M, Williams SG, DeGrado TR, Roselt P, Hicks RJ. Pilot comparison of F-fluorocholine and F-fluorodeoxyglucose PET/CT with conventional imaging in prostate cancer. J Med Imaging Radiat Oncol. 2010;54:325–332. doi: 10.1111/j.1754-9485.2010.02178.x. [DOI] [PubMed] [Google Scholar]
- 153.Contractor K, Challapalli A, Barwick T, Winkler M, Hellawell G, Hazell S, et al. Use of [11C]choline PET-CT as a noninvasive method for detecting pelvic lymph node status from prostate cancer and relationship with choline kinase expression. Clin Cancer Res. 2011;17:7673–7683. doi: 10.1158/1078-0432.CCR-11-2048. [DOI] [PubMed] [Google Scholar]
- 154.DeGrado TR, Baldwin SW, Wang S, Orr MD, Liao RP, Friedman HS, et al. Synthesis and evaluation of (18)F-labeled choline analogs as oncologic PET tracers. J Nucl Med. 2001;42:1805–1814. [PubMed] [Google Scholar]
- 155.Bansal A, Shuyan W, Hara T, Harris RA, DeGrado TR. Biodisposition and metabolism of [18F]fluorocholine in 9L glioma cells and 9L glioma-bearing Fisher rats. Eur J Nucl Med Mol Imaging. 2008;35:1192–1203. doi: 10.1007/s00259-008-0736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Vali R, Loidl W, Pirich C, Langesteger W, Beheshti M. Imaging of prostate cancer with PET/CT using (18)F-fluorocholine. Am J Nucl Med Mol Imaging. 2015;5:96–108. [PMC free article] [PubMed] [Google Scholar]
- 157.Umbehr MH, Muntener M, Hany T, Sulser T, Bachmann LM. The role of 11C-choline and 18F-fluorocholine positron emission tomography (PET) and PET/CT in prostate cancer: a systematic review and meta-analysis. Eur Urol. 2013;64:106–117. doi: 10.1016/j.eururo.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 158.Zheng Q-H, Gao M, Mock BH, Wang S, Hara T, Nazih R, et al. Synthesis and biodistribution of new radiolabeled high-affinity choline transporter inhibitors [11C]hemicholinium-3 and [18F]hemicholinium-3. Bioorg Med Chem. 2007;17:2220–2224. doi: 10.1016/j.bmcl.2007.01.105. [DOI] [PubMed] [Google Scholar]
- 159.Arlauckas SP, Popov AV, Delikatny EJ. Direct inhibition of choline kinase by a near-infrared fluorescent carbocyanine. Mol Cancer Ther. 2014;13:2149–2158. doi: 10.1158/1535-7163.MCT-14-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 161.Hara T, Kondo T, Hara T, Kosaka N. Use of 18F-choline and 11C-choline as contrast agents in positron emission tomography imaging-guided stereotactic biopsy sampling of gliomas. J Neurosurg. 2003;99:474–479. doi: 10.3171/jns.2003.99.3.0474. [DOI] [PubMed] [Google Scholar]