Abstract
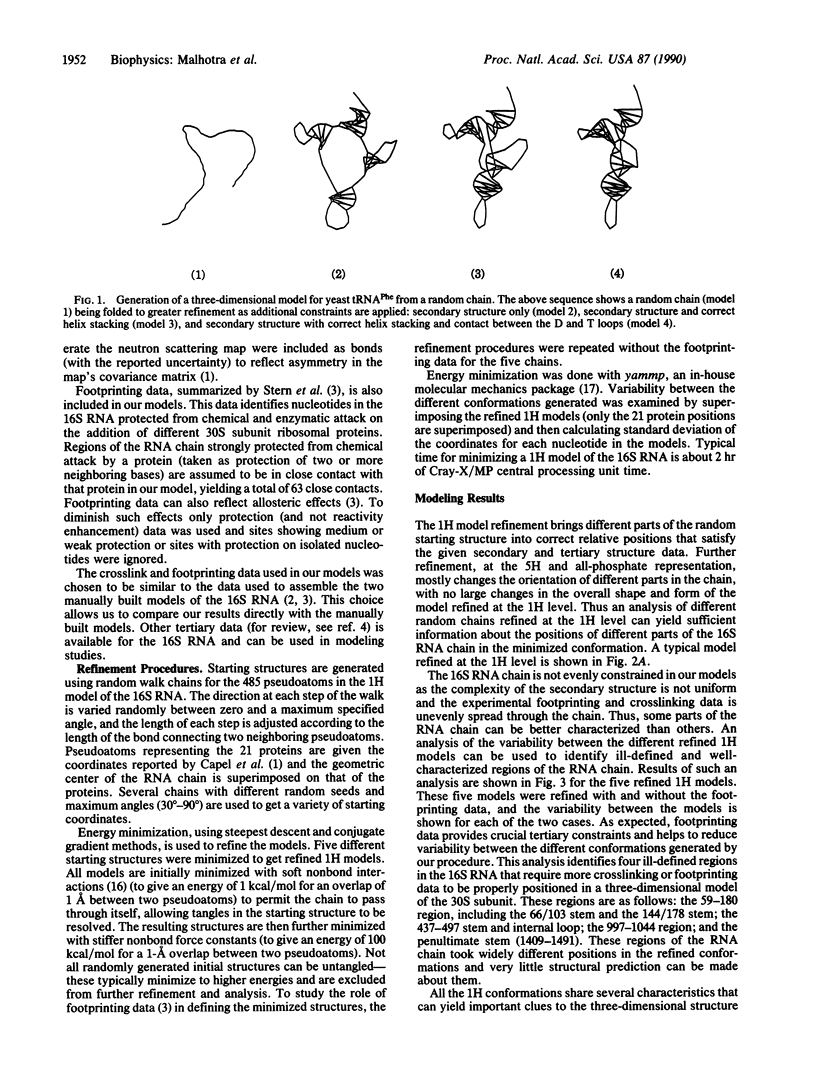
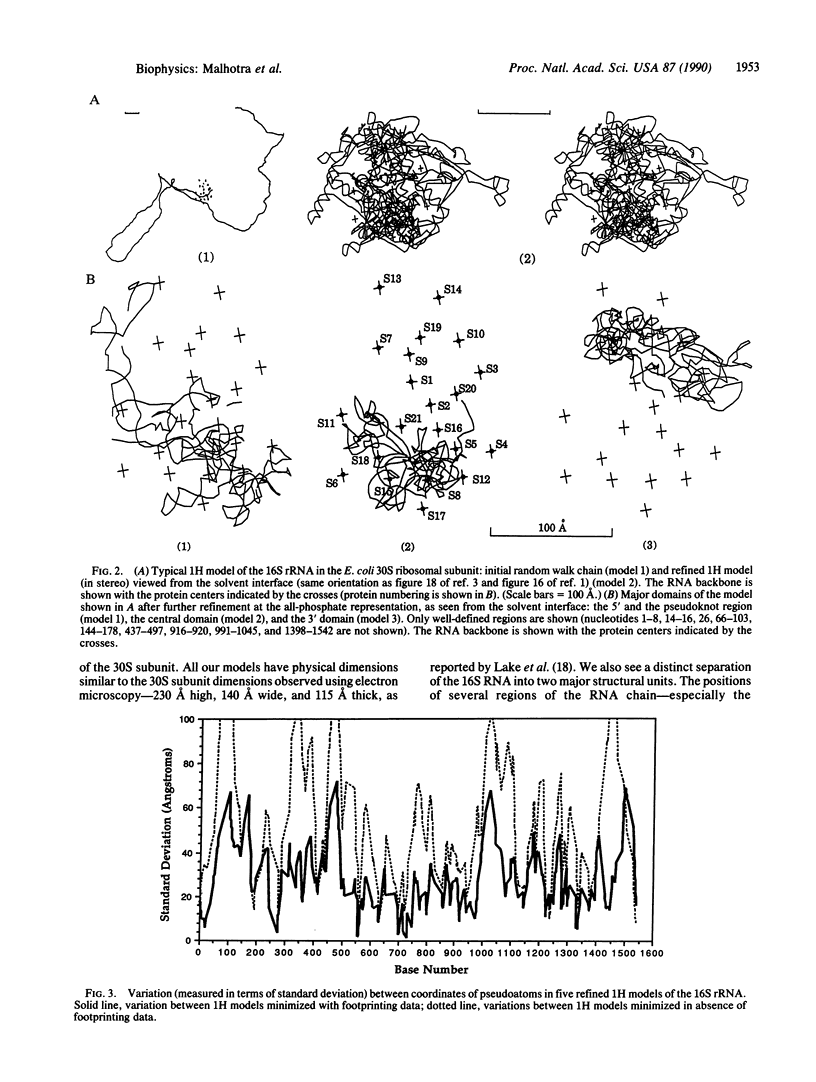
We introduce a computer-assisted procedure for folding large RNA chains into three-dimensional conformations consistent with their secondary structure and other known experimental constraints. The RNA chain is modeled using pseudoatoms at different levels of detail--from a single pseudoatom per helix to a single pseudoatom for each nucleotide. A stepwise procedure is used, starting with a simple representation of the macromolecule that is refined and then extrapolated into higher resolution for further refinement. The procedure is capable of folding different random-walk chains by using energy minimization, allowing generation of a range of conformations consistent with given experimental data. We use this procedure to generate several possible conformations of the 16S RNA in the 30S ribosomal subunit of Escherichia coli by using secondary structure and the neutron-scattering map of the 21 proteins in the small subunit. The RNA chain is modeled using a single pseudoatom per helix. RNA-RNA and RNA-protein crosslinks, reported in current literature, are included in our model. Footprinting data for different ribosomal proteins in the 16S RNA are also used. Several conformations of the 16S RNA are generated and compared to predict gross structural features of the 30S subunit as well as to identify regions of the 16S RNA that cannot be well-defined with current experimental data.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brimacombe R., Atmadja J., Stiege W., Schüler D. A detailed model of the three-dimensional structure of Escherichia coli 16 S ribosomal RNA in situ in the 30 S subunit. J Mol Biol. 1988 Jan 5;199(1):115–136. doi: 10.1016/0022-2836(88)90383-x. [DOI] [PubMed] [Google Scholar]
- Brimacombe R. The emerging three-dimensional structure and function of 16S ribosomal RNA. Biochemistry. 1988 Jun 14;27(12):4207–4214. doi: 10.1021/bi00412a001. [DOI] [PubMed] [Google Scholar]
- Capel M. S., Kjeldgaard M., Engelman D. M., Moore P. B. Positions of S2, S13, S16, S17, S19 and S21 in the 30 S ribosomal subunit of Escherichia coli. J Mol Biol. 1988 Mar 5;200(1):65–87. doi: 10.1016/0022-2836(88)90334-8. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hingerty B., Brown R. S., Jack A. Further refinement of the structure of yeast tRNAPhe. J Mol Biol. 1978 Sep 25;124(3):523–534. doi: 10.1016/0022-2836(78)90185-7. [DOI] [PubMed] [Google Scholar]
- Levitt M. A simplified representation of protein conformations for rapid simulation of protein folding. J Mol Biol. 1976 Jun 14;104(1):59–107. doi: 10.1016/0022-2836(76)90004-8. [DOI] [PubMed] [Google Scholar]
- Levitt M. Protein folding by restrained energy minimization and molecular dynamics. J Mol Biol. 1983 Nov 5;170(3):723–764. doi: 10.1016/s0022-2836(83)80129-6. [DOI] [PubMed] [Google Scholar]
- Levitt M., Warshel A. Computer simulation of protein folding. Nature. 1975 Feb 27;253(5494):694–698. doi: 10.1038/253694a0. [DOI] [PubMed] [Google Scholar]
- Nagano K., Harel M., Takezawa M. Prediction of three-dimensional structure of Escherichia coli ribosomal RNA. J Theor Biol. 1988 Sep 17;134(2):199–256. doi: 10.1016/s0022-5193(88)80202-9. [DOI] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Tan R. K., Harvey S. C. Molecular mechanics model of supercoiled DNA. J Mol Biol. 1989 Feb 5;205(3):573–591. doi: 10.1016/0022-2836(89)90227-1. [DOI] [PubMed] [Google Scholar]



