Abstract
Marked increased expression of cyclooxygenase 2 (COX-2), a prostaglandin-synthesizing enzyme that is pharmacologically inhibited by nonsteroid anti-inflammatory-type drugs, is a major early oncogenic event in the genesis of human colon neoplasia. We report that, in addition to inducing expression of COX-2, colon cancers further target the prostaglandin biogenesis pathway by ubiquitously abrogating expression of 15-hydroxyprostaglandin dehydrogenase (15-PGDH), a prostaglandin-degrading enzyme that physiologically antagonizes COX-2. We find that 15-PGDH transcript and protein are both highly expressed by normal colonic epithelia but are nearly undetectable in colon cancers. Using gene transfection to restore 15-PGDH expression in colon cancer cells strongly inhibits the ability of these cells to form tumors in immune-deficient mice and demonstrates 15-PGDH to have functional colon cancer tumor suppressor activity. In interrogating the mechanism for 15-PGDH expression loss in colon cancer, we determined that colonic 15-PGDH expression is directly controlled and strongly induced by activation of the TGF-β tumor suppressor pathway. These findings thus delineate an enzymatic pathway that induces colon cancer suppression, a pathway that is activated by TGF-β and mediated by 15-PGDH.
Keywords: colon, gastric
Up-regulation of cyclooxygenase 2 (COX-2, formally referred to as prostaglandin-endoperoxide synthase 2) expression is an early and key oncogenic event in human colon neoplasia, typifying 85% of colon cancers and 50% of colon adenomas (1). COX-2 is a cyclic endoperoxidase that catalyzes the rate-limiting step in prostaglandin biosynthesis (1). Nonsteroidal anti-inflammatory drugs (NSAIDs), which include indomethacin, sulindac, and celecoxib, are all COX-2 inhibitors (1), and NSAID agents have been shown to directly shrink the size of colon adenomas in some patients (2). In mice, genetic inactivation of COX-2 similarly blocks development of murine intestinal adenomas (3). However, the biochemical activity of COX-2 also has a normal physiologic antagonist in the prostaglandin-degrading enzyme 15-hydroxyprostaglandin dehydrogenase (15-PGDH), which catalyzes the inactivating NAD+-dependent conversion of the prostaglandin 15-OH to a 15-keto group (4). We were accordingly struck when, in a microarray-based comparison of colon cancers versus normal colon, 15-PGDH emerged as being among the genes most down-regulated in cancer. Moreover, parallel microarray-based studies also identified 15-PGDH as a gene showing among the strongest induction of expression in colon epithelial cells undergoing chronic treatment with TGF-β, a cytokine mediating a known colon cancer suppression pathway (5, 6). These observations suggested that colon cancer development may require two cooperating hits, one a dramatic up-regulation of expression of the COX-2 “oncogene,” and the other a dramatic down-regulation of an opposing and putative tumor suppressor gene, 15-PGDH. To explore this hypothesis, we embarked on studies to more completely characterize 15-PGDH expression in malignant versus normal colon cells, to determine the relationship between the TGF-β pathway and 15-PGDH expression, and to test for the hypothesized colon cancer suppressor activity of 15-PGDH.
Materials and Methods
Human Tissues. All colon tissues were collected under an Institutional Review Board-approved protocol at University Hospitals of Cleveland and underwent histologic review before use. Human tissue histology arrays with matched cancer and patient normal tissues representing gastric, breast, and lung cancers were purchased from Cybrdi (Gaithersburg, MD).
DNA Microarray Studies. RNA was isolated and cRNA generated as described (7). cRNA was hybridized to a custom Affymetrix GeneChip (Eos Hu03) designed by Eos Biotechnology (South San Francisco, CA) (8). A single Eos Hu03 GeneChip contains >59,000 probesets, which represent ≈45,000 mRNAs and EST clusters along with 6,200 ab initio predicted genes from the human genomic sequence not represented in the mRNA- and EST-expressed sequences at the time of chip design. Labeled cRNA was hybridized to the custom Affymetrix arrays by using standard protocols (Affymetrix, Santa Clara, CA), and raw image data were collected by using the Affymetrix expression array software. Data were normalized by using protocols and software developed at Eos Biotechnology (9). In brief, probe intensity values were background-subtracted and normalized to a γ distribution. An average intensity was calculated from these probe intensities by using a trimean (10).
Cell Culture. Vaco series colon cell lines were cultured as described (11, 12). FET was the kind gift of M. Brattain (Roswell Park Cancer Institute, Buffalo, NY) and was maintained in MEM (Invitrogen) containing 8% calf serum (HyClone).
Reagents and Antibodies. TGF-β1 was purchased from R & D Systems and was added to cell cultures at 10 ng/ml. A previously characterized polyclonal antiserum was raised in rabbits after injection of 15-PGDH protein purified from human placenta (13).
15-PGDH Immunohistochemistry. Briefly, 5-μM-thick formalin-fixed paraffin-embedded tissue sections were baked at 60°C for 75 min, deparaffinized, and rehydrated. Antigen retrieval was performed by steaming (Black and Decker Flavor Scenter, Handy Steamer HS800, Black and Decker, Hampstead, MD) at 96°C for 5 min in 10 mM citrate buffer (pH 6.0), plus a cool-down period of 20 min. Reduction of peroxidases was accomplished by incubating in 3% H2O2 in water for 30 min at room temperature. Avidin–biotin blocking was performed for 15 min each, followed by nonspecific protein blocking (Serum-Free Protein Block, Dako, Carpenteria, CA) performed for 60 min. Primary antibody was diluted in 1% BSA (Boehringer Mannheim) and incubated overnight at 4°C in humidified chambers. The slides were washed thoroughly, and Protein Block was added again for 30 min. LSAB+ anti-rabbit kit (Dako) was used for development, applying the secondary antibody and horseradish peroxidase-conjugated streptavidin per the manufacturer's instructions. Finally, diaminobenzidine (Dako) was added to the slides for 10 min. All washes were done with TBS (50 mM Tris·HCl/150 mM NaCl, pH 7.6) diluted in deionized water. The sections were then counterstained by using Harris modified hematoxylin stain (Fisher Scientific) for 1 min.
Production of Recombinant 15-PGDH. Recombinant 15-PGDH (rPGDH) protein was encoded from the pBad-Topo vector (Invitrogen) expressed in one-shot bacteria (Invitrogen) and induced by l-arabinose (Sigma). rPGDH was purified with the B-Per 6× His-Fusion Protein Purification Kit (Pierce). Bacteria transformed with an empty pBad vector were put through a parallel purification process to yield a bacterial control lysate. Blocking of anti-15-PGDH sera was performed by adding 2 mg of rPGDH protein or an equal amount of bacterial control lysate to antibody followed by rotation at room temperature for 2 h.
Northern Hybridization. Ten micrograms of total RNA was separated on a 1% formaldehyde agarose gel, transferred to Nytran SuPerCharge (Schleicher & Schuell), and hybridized in Express-Hyb Buffer (Clontech) to a 15-PGDH-coding cDNA probe labeled with 32P-dATP (Strip-EZ DNA labeling kit; Ambion, Austin, TX). The blot was exposed to a phosphor screen and visualized by using a STORM optical scanner (Molecular Dynamics).
Real-Time PCR. cDNA was reverse transcribed with AMV reverse transcriptase (Roche Applied Sciences). Detection of mature spliced 15-PGDH by real-time PCR used PCR amplification primers 5′-TGCT TCA A AGCATGGCATAG-3′ and 5′-AACAAAGCCTGGACAAATGG-3′, respectively, located in 15-PGDH exons 5 and 6, and a fluorogenic hybridization probe 5′FAM-CTCAGCAGCGTTGGCTGCTAATCTTA-BHQ-3′ detected in an Icycler optical module (Bio-Rad). A 25-μl reaction mix contained 300 nM primer and 250 nM probe in 1× Supermix (Bio-Rad). Thermal cycling was initiated at 95°C for 3 min, followed by 50 cycles of 95°C for 15 sec and 60°C for 1 min. β-2-Microglobulin (B2M) was amplified and detected by using human B2M TaqMan predeveloped assay reagents (Applied Biosystems), with PCR initiated at 95°C for 10 min, followed by 50 cycles of 95°C for 15 sec and 60°C for 1 min. The level of mature 15-PGDH RNA was determined as the ratio of 15-PGDH/B2M = 2 exp (CTB2M–CTPGDH).
For detection of the nascent unspliced 15-PGDH RNA, nuclear RNA was prepared by using a PARIS kit (Ambion), treated with RQ1 DNase (Promega) per the manufacturer's protocol, and reverse transcribed. A product specific for the unspliced 15-PGDH transcript was amplified with primers 5′-TTTATTGTTTGTCCGTCTATTTCGTGAGC-3′ and 5′-ACACCTGCTTCAAGATTCCAATCCAC-3′, located, respectively, in intron 1 and exon 2. A product specific for the unspliced GAPDH transcript was amplified with primers 5′-AAAGGGCCCTGACAACTCTT-3′ and 5′-GGTGGTCCAGGGGTCTTACT-3′ (14) located in the introns flanking GAPDH exon 9. PCR amplifications were performed by using IQ SYBR Green Supermix (Bio-Rad), according to the manufacturer's protocol. Thermal cycling was initiated at 95°C for 5 min, followed by 50 cycles of 95°C for 15 sec, 63.8°C for 30 sec, and 72°C for 30 sec. No amplified product was detected from negative control RNA samples to which reverse transcriptase was not added.
Western Blot Analysis. Cell lysates prepared in RIPA buffer (Upstate Biotechnology, Lake Placid, NY) (50 mM Tris·HCL/1% Nonidet P-40/0.25% Na-deoxycholate/150 mM NaCl/1 mM EDTA/1 mM PMSF) supplemented with protease inhibitor mixture (Roche Applied Sciences) were separated on 12% SDS/PAGE Ready Gels (Pierce) (30 μg per lane) and transferred to Immobilon poly(vinylidene difluoride) membrane (Millipore). The blots were blocked with 5% milk, probed with anti-PGDH antibody at a 1:10,000 dilution and with anti-β-actin antibody at a 1:100,000 dilution, developed by using horseradish-peroxidase-conjugated secondary antibodies (Jackson ImmunoResearch), visualized by using an Enhanced Chemiluminescence Plus detection kit (Amersham Biosciences), following the manufacturer's instructions, and then scanned on a PhosphorImager (Molecular Dynamics).
Assay of 15-PGDH Enzyme Activity. Crude homogenates were prepared by sonicating the cells or tumor tissue in 1 ml of 50 mM Tris·HCl, pH 8.0, with 0.1 mM DTT, followed by centrifugation at 13,000 × g for 10 seconds in a tabletop microfuge. The clarified supernatants were then assayed for 15-PGDH activity, as described (15).
Ki67 Immunofluorescence. FET cells were fixed with cold methanol for 15 min at –20°C, blocked with 10% goat serum for 15 min, incubated with 1:200 diluted KI-S5 rabbit anti-Ki67 polyclonal antibody (Dako) at 4°C, followed by incubation with a 1:400 dilution of FITC-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch).
15-PGDH Expression Vectors. A wild-type 15-PGDH cDNA was amplified from normal colon cDNA by using PCR primers 5′-CAGCAGTGGCTGCACCATG-3′ and 5′-ATTTGTGCTTATTTTCAGCTATGGC-3′ and cloned into a pcDNA6 expression vector (Invitrogen) to yield pcDNA6-Wt-PGDH. In addition, two missense mutations (Y151L and K155E) were introduced by site-directed mutagenesis to generate pCDNA6-Mu-PGDH, encoding a control enzymatically inactive 15-PGDH. In addition, 6.3 kb of genomic DNA surrounding the PGDH promoter was subcloned into the PGL3 (Promega) luciferase reporter vector system to assay for elements conferring TGF-β regulation on the PGDH core promoter.
15-PGDH Stable Transfections. Vaco-400 colon cancer cells were transfected with pcDNA6-empty vector or expression vectors encoding wild-type (pcDNA6-Wt-PGDH) or mutant (pcDNA6-Mu-PGDH) 15-PGDH by using effectine (Qiagen, Valencia, CA) according to the manufacturer's protocol. Stable clones were selected by growth in 10 μg/ml blasticidin with individual colonies isolated by using cloning rings. Colonies arising from control empty vector transfections were pooled. In vitro growth curves of transfected cells were generated by plating 2,000 cells per well on 12-well plates and counting 3 wells from the plates every other day.
Tumorigenesis Assays. Cells from exponentially growing cultures were injected s.c. behind the anterior forelimb of 5- to 6-week-old BALB/c athymic mice at an inoculum of 5 × 106 cells per site. Tumors arising were measured externally at weekly intervals by using calipers with tumor volume determined as V = (L × W2) × 0.5, where L is the largest dimension, and W is the largest dimension perpendicular to L. Growth curves were plotted from determinations of the mean volume of xenograft tumors arising from 10 replicate injections.
Results
15-PGDH Is Expressed in Normal but Not Malignant Colonic Epithelium. 15-PGDH expression in normal and malignant colon epithelium was initially characterized by assay on GeneChip expression microarrays. Notably, in 38 colon cancer samples, median 15-PGDH expression was at an undetectable level and was at least 17-fold below the median expression in normal colon (Fig. 1A). In contrast, 15-PGDH was reproducibly well expressed in each of 21 samples of control normal colon epithelium (Fig. 1 A). Loss of 15-PGDH expression was similarly detected in early localized cancers and in advanced metastatic cases (respectively, Dukes' stages B, C, and D) (Fig. 1 A). Analysis of an independent set of 20 new colon cancers by using real-time PCR confirmed an average 10-fold loss of PGDH expression in cancers versus matched normal colon, with marked loss exhibited by 18 of the 20 tumors (Fig. 1B). Northern analysis also demonstrated a similar loss of PGDH transcript expression among colon cancers (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 1.
15-PGDH mRNA expression in normal and malignant colon. (A) 15-PGDH expression in normal colon and primary colon cancers of Dukes' stages B, C, and D, as measured on expression microarrays in average intensity (AI) units. Horizontal bars indicate median expression levels for normal colon versus all colon cancer samples. Twenty-five AI units represent null expression on the microarray. (B) Real-time PCR assay of 15-PGDH. Shown is the ratio of 15-PGDH expression in 20 colon cancers versus matched normal colon. Values represent averages of six replicates.
To determine the origin of 15-PGDH expression in normal colon mucosa, we performed 15-PGDH immunohistochemistry (13). In multiple normal colon samples, 15-PGDH protein proved reproducibly expressed by epithelial cells located at the upper reaches and luminal surface of the colonic crypts, regions where colonocytes are nondividing and about to undergo apoptosis (Fig. 2 and Fig. 8, which is published as supporting information on the PNAS web site) (5). Specificity of staining was demonstrated by complete blocking of immunostaining by recombinant 15-PGDH protein (Fig. 8) and by anti-15-PGDH antibody detecting only a single correctly sized protein band on Western analysis of normal colon (Fig. 6). Staining for Ki-67 (Fig. 8) showed a clear demarcation between proliferating Ki-67 positive cells at the crypt base and 15-PGDH positive cells at the crypt surface.
Fig. 2.
Immunohistochemical assay of 15-PGDH in normal colon mucosa (N1–N4) versus matched colon cancers (T1–T4) from the same four patients. Normal mucosa was sampled at distances ranging from 5 to 17 cm distant from the tumors.
Fig. 6.
Tumor suppression of 15-PGDH reconstituted colon cancer cells. (A) 15-PGDH Western analysis and enzyme activity levels assayed in normal colon epithelium (colon-1 and colon-2) and in subclones of Vaco-400 transfected with wild-type 15-PGDH (S2-36, S3-2-32, S3-3-31, and S4-2-14) or with an enzymatically inactive mutant (Y151L and K155E) 15-PGDH (M3-2-71, M3-3 41, and M3-2-22). Additional control pools of Vaco-400 cells were transfected with an empty expression vector (EM pool-2 and EM pool-4). (B) Growth of xenograft tumors in athymic mice injected with: two Vaco-400 empty vector transfectant cell pools (black lines); three Vaco-400 mutant 15-PGDH transfectant subclones (green lines); and four Vaco-400 wild-type 15-PGDH transfectant subclones (red lines). Plots show mean outcomes from 10 injections with standard errors indicated by half bars rising above the graphed values.
In marked contrast to the findings in normal colon mucosa, dramatic loss of PGDH protein was evidenced in 16 of 17 colon cancer cases, in which cases immunostainable 15-PGDH was essentially absent (Fig. 2). In each of these cases, 15-PGDH expression was well detected in immunostaining of the patients' matched normal colonic mucosa (Fig. 2). Similar dramatic loss of 15-PGDH expression was also evidenced on immunohistochemical assay of human gastric cancers, with 10 of 13 gastric cancers showing absences of 15-PGDH protein expression, although robust 15-PGDH immunostaining was present in each patient's normal gastric mucosa. (Fig. 9, which is published as supporting information on the PNAS web site). In contrast, immunostaining of lung adenocarcinomas showed that the known expression of 15-PGDH by normal lung tissue was uniformly well maintained by these lung cancers (Fig. 10, which is published as supporting information on the PNAS web site). Thus, loss of 15-PGDH expression appears to be particularly characteristic to human gastrointestinal cancers.
Induction of Colonic 15-PGDH by TGF-β. To examine potential mechanisms for loss of 15-PGDH expression in colon cancers, we examined 15-PGDH expression among a panel of 22 colon cancer cell lines. On GeneChip microarrays, 15-PGDH expression in colon cancer cell lines showed the same average 17-fold reduction relative to normal colon mucosa as had been noted in colon cancer tumor tissues (data not shown), a finding that was further verified by real-time PCR (Table 1, which is published as supporting information on the PNAS web site). Nonetheless, a 15-PGDH transcript, expressed at a very low level, could be recovered by RT-PCR amplification from these colon cancer cell lines. On sequencing, these transcripts proved to be wild-type in all cases.
One clue to the mechanism for near absence of 15-PGDH expression in colon cancers came from parallel studies, in which we identified strong PGDH induction in nonmalignant colon cells treated with TGF-β, a cytokine mediating a tumor suppressor signaling pathway that colon cancers commonly inactivate, for example, by mutations in TGF-β RII receptors or downstream Smad signaling elements (5, 11, 16–19). Using expression microarrays, we initially observed a >12-fold induction of 15-PGDH in nontransformed Vaco-330 colon epithelial cells after chronic 72-hour exposure to TGF-β (Fig. 11, which is published as supporting information on the PNAS web site). Northern analysis confirmed that 15-PGDH transcripts, of previously described 3.4- and 2.0-kb sizes (20), were undetectable in control Vaco-330 cells but became well detected in cells treated with TGF-β for 48 or 72 h (Fig. 3A). To determine the generality of TGF-β regulation of 15-PGDH in colonocytes, we reactivated TGF-β signaling in a colon cancer cell line, Vaco-410, in which somatic mutation had inactivated the endogenous TGF-β RII (16). Whereas the Vaco-410 colon cancer cells lacked 15-PGDH expression, Vaco-410 cells transfected with a wild-type RII gene regained a strong induction of 15-PGDH upon TGF-β treatment (Fig. 3B). Finally, we demonstrated TGF-β-regulated 15-PGDH in a third model, FET, a nontumorigenic colon cancer cell line that has retained TGF-β responsiveness and that exhibits TGF-β autocrine activity (21). Real-time PCR-demonstrated FET cells express a basal level of 15-PGDH transcript, which further increased by 5- to 6-fold over baseline upon 24- or 48-h exposure to exogenous TGF-β (Fig. 3C). TGF-β induction of 15-PGDH mRNA was accompanied by similar increases in 15-PGDH protein (Fig. 3D) and by a similar 6-fold increase in 15-PGDH enzyme activity (Fig. 3E). Thus, TGF-β induction of 15-PGDH is reproducible across multiple colon cell line models and results in increased 15-PGDH transcript, protein, and enzymatic activity.
Fig. 3.
TGF-β induction of 15-PGDH. (A) Northern analysis of 15-PGDH in TGF-β-treated (10 ng/ml) Vaco-330 colon adenoma cells at 48 and 72 h of incubation, with corresponding ethidium bromide staining of the 28S ribosomal subunit shown below. (B) Northern analysis of 15-PGDH in TGF-β-treated (10 ng/ml, 48 h) Vaco-410 colon cancers cells transfected with wild-type RII (V410/RII) or control empty vector (V410/Neo). (C) Real-time PCR of 15-PGDH induction by TGF-β (10 ng/ml) in FET colon cancer cells. (D) Western analysis of 15-PGDH protein induction by TGF-β (10 ng/ml) in FET colon cancer cells. (E) Induction of 15-PGDH enzyme activity (pmol/min per mg protein) in TGF-β-treated (10 ng/ml) FET cells. Values are averages of duplicate determinations. (F) Northern analysis of 15-PGDH induction by TGF-β (10 ng/ml, 18 h) in cycloheximide treated (CHX+) versus untreated (CHX–) FET cells.
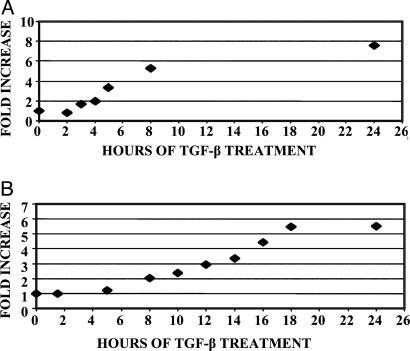
PGDH Is a Direct Target of the TGF-β Signaling Pathway. To determine the kinetics of 15-PGDH induction by TGF-β and to examine whether induction was at the transcriptional level, we used real-time PCR to measure levels of nascent unspliced nuclear 15-PGDH mRNA. This approach in part reflected technical difficulties with assay of 15-PGDH transcription by using nuclear run-ons. Unspliced nascent RNAs generally exhibit a short half-life, and hence alterations in their level provide surrogate measures of changes in gene transcription rates (22). Induction of unspliced 15-PGDH transcript could be demonstrated as early as 3 h after TGF-β addition to FET cells, with nascent transcript increased by 2-fold (Fig. 4A). Induction of unspliced 15-PGDH transcript increased to nearly 6-fold by 8 h of TGF-β treatment. This suggests a similar 6-fold TGF-β-induced increase of 15-PGDH transcription rates, which would fully account for the 6-fold TGF-β-induced increase that mature spliced 15-PGDH transcript attains at 18 h (Fig. 4B). The slower kinetics of accumulation of the mature 15-PGDH transcript is consistent with this transcript having relatively slow turnover, suggesting a message half-life of ≈5 h.
Fig. 4.
Kinetics of TGF-β induction of 15-PGDH. (A) Time course of TGF-β (10 ng/ml) induction of unspliced 15-PGDH precursor transcript in FET cells as measured by real-time PCR. Values are normalized to levels of the unspliced GAPDH precursor transcript and represent averages of duplicate experiments for which each real-time PCR measurement was performed in triplicate. Values at all duplicate time points agreed to within 17% of the mean. (B) Time course of TGF-β (10 ng/ml) induction of mature 15-PGDH transcript in FET cells, as measured by real-time PCR. Values are normalized to endogenous β-2 microglobulin and represent averages of duplicate experiments for which each real-time PCR measurement was performed in triplicate. Values at all duplicate experiments agreed to within 18% of the mean.
TGF-β induction of 15-PGDH transcript proved equally robust in cycloheximide-treated FET cells as in untreated controls (Fig. 3F). Thus 15-PGDH transcript is induced as a direct target of the TGF-β signaling pathway, without the need for intermediate protein synthesis. As described for other transcripts (23), cycloheximide induced a modest increase in 15-PGDH mRNA (Fig. 3F), which did not, however, interfere with our ability to assay the dominant TGF-β-induced increased expression. We attempted to identify a TGF-β response element by using the pGL3 luciferase reporter system to assay a region, extending 6 kb upstream of the transcription start site plus downstream through the first exon and first intron, for elements able to convey TGF-β up-regulation to the 15-PGDH core promoter. These studies were unrevealing, suggesting this putative element is likely located at some distance from the 15-PGDH core promoter.
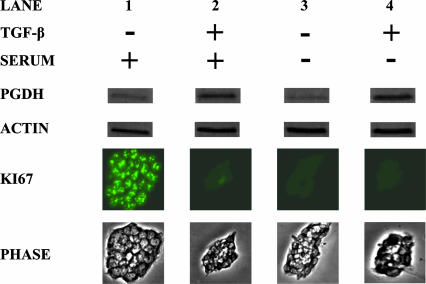
Finally, we examined whether 15-PGDH induction requires TGF-β signaling or might simply be a marker of nonproliferating colonic epithelial cells. Whereas 15-PGDH expression was well induced in FET cells growth arrested by TGF-β (Fig. 5, lane 2 versus lane 1), 15-PGDH was not induced in FET cell growth arrested by serum deprivation (Fig. 5, lane 3). Moreover, cell growth arrested by serum deprivation could robustly induce 15-PGDH upon further treatment with TGF-β (Fig. 5, lane 4).
Fig. 5.
15-PGDH induction by TGF-β but not by cell growth arrest. Shown is Western blot assay of 15-PGDH protein expression in FET cells treated with TGF-β (10 ng/ml) (TGF-β +) versus control (TGF-β –) and maintained in basal medium plus 8% serum (serum +) or minus the addition of (serum –). Proliferative status of the cells is determined by immunofluorescent assay for Ki67, and colony size was visualized by phase microscopy. Assays were performed on day 5 of cell culture.
Tumor Suppressor Activity of 15-PGDH. The enzymatic activity of 15-PGDH as a COX-2 antagonist, plus the observed loss of 15-PGDH expression in colon cancer, suggested that 15-PGDH might have tumor suppressor activity. To test this hypothesis, we used stable transfection to reconstitute 15-PGDH expression in Vaco-400 colon cancer cells, in which TGF-β signaling is inactivated due to somatic mutation of TGF-β RII (16). Cells were transfected with an expression vector encoding a wild-type 15-PGDH, with a control vector encoding an enzymatically inactive doubly mutant 15-PGDH (Y151L, K155E) (24), or with a control empty expression vector. Transfected clones expressing wild-type PGDH were reconstituted to levels of protein and enzyme activity that ranged from one-third to one-half that of normal colonic mucosa (Fig. 6A). Control mutant 15-PGDH transfected clones expressed 15-PGDH protein at levels comparable to wild-type transfectants. However, control clones bearing either mutant 15-PGDH or an empty expression vector displayed <1% of the 15-PGDH enzyme activity of normal colon epithelium (Fig. 6A). Although wild-type 15-PGDH transfected clones were reconstituted to levels below normal colon, these cells still proved to be markedly suppressed in their capacity to form tumors when injected into athymic mice (Fig. 6B). Whereas each control clone produced tumors in 100% of injected mice, two of the four wild-type 15-PGDH reconstituted clones showed essentially no tumor formation, and the remaining two clones showed marked suppression of tumor formation relative to controls (Fig. 6B and Fig. 12, which is published as supporting information on the PNAS web site). Moreover, the few xenograft tumors that did arise from 15-PGDH-transfected Vaco-400 cells had all lost 15-PGDH expression. Although wild-type 15-PGDH-reconstituted cells were markedly suppressed in ability to form tumors in athymic mice, they were in cell culture indistinguishable from control mutant 15-PGDH transfectants, both showing similar growth rates and attaining similar cell densities at confluence (Fig. 13, which is published as supporting information on the PNAS web site). The suppression by 15-PGDH of in vivo tumorigenic growth, but not of growth in cell culture, is consistent with suggestions from several models that the tumor-promoting effect of increased prostaglandin synthesis is principally mediated via increased tumor angiogenesis (25–28).
Discussion
Our observations principally identify 15-PGDH as a gene with tumor suppressor activity in human gastrointestinal malignancies. We find 15-PGDH is normally expressed in human gastrointestinal mucosa, and that this expression is lost by malignant colon and gastric epithelial cells. Restoration of 15-PGDH expression in malignant cells strongly suppresses the ability of these cells to grow as tumors in athymic mice. These findings, moreover, show that human gastrointestinal cancers multiply target the pathway regulating prostaglandin biogenesis and demonstrate not only increased expression of the COX-2 prostaglandin synthesizing “oncogene” but also marked loss of expression of the 15-PGDH prostaglandin degrading the “tumor suppressor” gene. The importance of this pathway was initially recognized by the finding of increased COX-2 expression in colon adenomas and colon cancers (1) and by the demonstration that genetic disruption of COX-2 rendered mice resistant to colon tumor induction by APC mutations (3). Moreover, pharmacological inhibition of COX-2 was shown to shrink precancerous colon adenomas in humans (2) and to suppress the ability of selected colon cancer cell lines to grow as tumors in athymic mice (29). Future studies will now be warranted to explore the potential that the “second hit” to the prostaglandin synthetic pathway provided by loss of the 15-PGDH activity could provide a mechanism for tumor progression or a mechanism for colon cancers becoming generally resistant to drugs that partially inhibit COX-2.
The finding that TGF-β strongly induces colonic 15-PGDH demonstrates an unsuspected negative regulation of prostaglandin biogenesis by TGF-β. TGF-β mediates a potent tumor suppressor activity in gastrointestinal cancers, and these tumors commonly target TGF-β signaling for genetic inactivation by mutations in type II TGF-β receptors or in Smad signal transducing elements (5, 11, 16–19, 30). The potency of TGF-β-mediated tumor suppression likely reflects the induction by TGF-β of several complementary negative regulators of tumor growth, including p15, p21, and possibly DEC1, as well as TGF-β-mediated suppression of c-Myc (31–35). The findings of this study suggest that the TGF-β-mediated induction of 15-PGDH and crosstalk between the TGF-β and prostaglandin pathways may represent a significant additional effector of TGF-β-mediated suppression of cancer in the gastrointestinal tract, a tissue in which this tumor suppressor pathway is particularly robust. Indeed, the ubiquitous loss of 15-PGDH expression among colon cancers is a bioassay supporting that inactivation of the TGF-β signaling pathway is also ubiquitous among human colon cancers in vivo.
In sum, these findings delineate the presence in human gastrointestinal cells of a physiologic enzymatic tumor suppressor pathway that is activated by TGF-β and mediated by 15-PGDH.
Supplementary Material
Acknowledgments
We thank Hui Li and Stephen Fink for helpful discussion and Rachel Kassai for technical assistance. This work is supported by Public Health Service Grants UO1-CA82901 (to S.D.M), RO1-HL46296 (to H.-H.T.), and P30-CA43703 to the Case Western Reserve Comprehensive Cancer Center, by a grant from the State of Ohio Biomedical Research and Technology Transfer Commission, and by grants from the National Colon Cancer Research Alliance and Entertainment Industry Foundation (to S.D.M.) and the Kentucky Lung Cancer Research Program (to H-H.T.). S.D.M. is an Investigator of the Howard Hughes Medical Institute.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: 15-PGDH, 15-hydroxyprostaglandin dehydrogenase; COX-2, cyclooxygenase 2.
References
- 1.Gupta, R. A. & Dubois, R. N. (2001) Nat. Rev. Cancer 1, 11–21. [DOI] [PubMed] [Google Scholar]
- 2.Giardiello, F., Hamilton, S., Krush, S., Piatadosi, S., Hylind, L., Celano, P., Booker, S., Robinson, C. & Offerhaus, G. (1993) N. Engl. J. Med. 328, 1313–1316. [DOI] [PubMed] [Google Scholar]
- 3.Oshima, M., Dinchuk, J. E., Kargman, S. L., Oshima, H., Hancock, B., Kwong, E., Trzaskos, J. M., Evans, J. F. & Taketo, M. M. (1996) Cell 87, 803–809. [DOI] [PubMed] [Google Scholar]
- 4.Tai, H. H., Ensor, C. M., Tong, M., Zhou, H. & Yan, F. (2002) Prostaglandins Other Lipid Mediat. 68–69, 483–493. [DOI] [PubMed] [Google Scholar]
- 5.Markowitz, S., Dawson, D. M., Willis, J. & Willson, J. K. (2002) Cancer Cell. 1, 233–236. [DOI] [PubMed] [Google Scholar]
- 6.Grady, W. M. & Markowitz, S. D. (2002) Annu. Rev. Genomics Hum. Genet. 3, 101–128. [DOI] [PubMed] [Google Scholar]
- 7.Platzer, P., Upender, M. B., Wilson, K., Willis, J., Lutterbaugh, J., Nosrati, A., Willson, J. K., Mack, D., Ried, T. & Markowitz, S. (2002) Cancer Res. 62, 1134–1138. [PubMed] [Google Scholar]
- 8.Eaves, I. A., Wicker, L. S., Ghandour, G., Lyons, P. A., Peterson, L. B., Todd, J. A. & Glynne, R. J. (2002) Genome Res. 12, 232–243. [PubMed] [Google Scholar]
- 9.Ghandour, G. & Glynne, R. J. (2000) International Patent W00079645.
- 10.Tukey, J. W. (1977) Exploratory Data Analysis (Addison–Wesley, Reading, MA).
- 11.Markowitz, S., Wang, J., Myeroff, L., Parsons, R., Sun, L., Lutterbaugh, J., Fan, R., Zborowska, E., Kinzler, K., Vogelstein, B., et al. (1995) Science 268, 1336–1338. [DOI] [PubMed] [Google Scholar]
- 12.Willson, J., Bittner, G., Oberley, T., Meisner, G. & Weese, J. (1987) Cancer Res. 47, 2704–2713. [PubMed] [Google Scholar]
- 13.Zhou, H. & Tai, H. H. (1999) Biochem. Biophys. Res. Commun. 257, 414–417. [DOI] [PubMed] [Google Scholar]
- 14.Saha, S., Bardelli, A., Buckhaults, P., Velculescu, V. E., Rago, C., St Croix, B., Romans, K. E., Choti, M. A., Lengauer, C., Kinzler, K. W., et al. (2001) Science 294, 1343–1346. [DOI] [PubMed] [Google Scholar]
- 15.Tai, H. H. (1976) Biochemistry 15, 4586–4592. [DOI] [PubMed] [Google Scholar]
- 16.Grady, W., Myeroff, L., Swinler, S., Rajput, A., Thiagalingam, S., Lutterbaugh, J., Neumann, A., Brattain, M., Chang, J., Kim, S.-J., et al. (1999) Cancer Res. 59, 320–324. [PubMed] [Google Scholar]
- 17.Eppert, K., Scherer, S., Ozcelik, H., Pirone, R., Hoodless, P., Kim, H., Tsui, L.-C., Bapat, B., Gallinger, S., Andrulis, I., et al. (1996) Cell 86, 543–552. [DOI] [PubMed] [Google Scholar]
- 18.Riggins, G. J., Thiagalingam, S., Rozenblum, E., Weinstein, C., Kern, S., Hamilton, S., Willson, J. K. V., Markowitz, S. D., Kinzler, K. & Vogelstein, B. (1996) Nat. Genet. 13, 347–349. [DOI] [PubMed] [Google Scholar]
- 19.Thiagalingam, S., Lengauer, C., Leach, F., Schutte, M., Hahn, S., Overhauser, J., Willson, J. K. V., Markowitz, S., Hamilton, S., Kern, S., et al. (1996) Nat. Genet. 13, 343–346. [DOI] [PubMed] [Google Scholar]
- 20.Ensor, C. M., Yang, J. Y., Okita, R. T. & Tai, H. H. (1990) J. Biol. Chem. 265, 14888–14891. [PubMed] [Google Scholar]
- 21.Wu, S., Theodorescu, D., Kerbel, R., Willson, J., Mulder, K., Humphrey, L. & Brattain, M. (1992) J. Cell. Biol. 116, 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrotti, D., Cesi, V., Trotta, R., Guerzoni, C., Santilli, G., Campbell, K., Iervolino, A., Condorelli, F., Gambacorti-Passerini, C., Caligiuri, M. A., et al. (2002) Nat. Genet. 30, 48–58. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell, R. L., Henning-Chubb, C., Huberman, E. & Verma, I. M. (1986) Cell 45, 497–504. [DOI] [PubMed] [Google Scholar]
- 24.Ensor, C. M. & Tai, H. H. (1994) Biochim. Biophys. Acta 1208, 151–156. [DOI] [PubMed] [Google Scholar]
- 25.Tsujii, M., Kawano, S., Tsuji, S., Sawaoka, H., Hori, M. & DuBois, R. N. (1998) Cell 93, 705–716. [DOI] [PubMed] [Google Scholar]
- 26.Chang, S. H., Liu, C. H., Conway, R., Han, D. K., Nithipatikom, K., Trifan, O. C., Lane, T. F. & Hla, T. (2004) Proc. Natl. Acad. Sci. USA 101, 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, D. & DuBois, R. N. (2004) Proc. Natl. Acad. Sci. USA 101, 415–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonoshita, M., Takaku, K., Sasaki, N., Sugimoto, Y., Ushikubi, F., Narumiya, S., Oshima, M. & Taketo, M. M. (2001) Nat. Med. 7, 1048–1051. [DOI] [PubMed] [Google Scholar]
- 29.Sheng, H., Shao, J., Kirkland, S. C., Isakson, P., Coffey, R. J., Morrow, J., Beauchamp, R. D. & DuBois, R. N. (1997) J. Clin. Invest. 99, 2254–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park, J., Kim, S.-J., Bang, Y.-J., Park, J.-G., Kim, N. K., Roberts, A. & Sporn, M. (1994) Proc. Natl. Acad. Sci. USA 91, 8772–8776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derynck, R., Akhurst, R. J. & Balmain, A. (2001) Nat. Genet. 29, 117–129. [DOI] [PubMed] [Google Scholar]
- 32.Siegel, P. M. & Massague, J. (2003) Nat. Rev. Cancer 3, 807–821. [DOI] [PubMed] [Google Scholar]
- 33.Zawel, L., Yu, J., Torrance, C. J., Markowitz, S., Kinzler, K. W., Vogelstein, B. & Zhou, S. (2002) Proc. Natl. Acad. Sci. USA 99, 2848–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrow, M. & Moses, H. (1995) Cancer Res. 55, 1452–1457. [PubMed] [Google Scholar]
- 35.Wakefield, L. M. & Roberts, A. B. (2002) Curr. Opin. Genet. Dev. 12, 22–29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.