Abstract
The motility of cilia and flagella is powered by dynein ATPases associated with outer doublet microtubules. However, a flagellar kinesin-like protein that may function as a motor associates with the central pair complex. We determined that Chlamydomonas reinhardtii central pair kinesin Klp1 is a phosphoprotein and, like conventional kinesins, binds to microtubules in vitro in the presence of adenosine 5′-[β,γ-imido]triphosphate, but not ATP. To characterize the function of Klp1, we generated RNA interference expression constructs that reduce in vivo flagellar Klp1 levels. Klp1 knockdown cells have flagella that either beat very slowly or are paralyzed. EM image averages show disruption of two structures associated with the C2 central pair microtubule, C2b and C2c. Greatest density is lost from part of projection C2c, which is in a position to interact with doublet-associated radial spokes. Klp1 therefore retains properties of a motor protein and is essential for normal flagellar motility. We hypothesize that Klp1 acts as a conformational switch to signal spoke-dependent control of dynein activity.
Keywords: Chlamydomonas, cilia, motility, radial spoke, RNA interference
Cilia and flagella have long been used as model systems for studies of microtubule-based motility by dynein motors. However, kinesins have also been found in these organelles. The best-studied kinesins in cilia and flagella are motors essential for intraflagellar transport (IFT), whose absence has been linked to defects in axoneme assembly in both motile and sensory cilia (1). In addition, two kinesin-like proteins have been localized to the central pair microtubule complex in Chlamydomonas flagella. One of these has only been identified as an antigen associated with the C1 microtubule through recognition by a pan-kinesin antibody (2, 3). The other, Klp1, was identified as the product of a gene up-regulated after deflagellation and characterized through analysis of cDNA clones (4). Antibodies directed against unique tail sequences of Klp1 detect a C2-associated 85-kDa protein. This protein, predicted to share general structural properties with other kinesins, has an N-terminal motor domain and a C-terminal tail that includes regions of predicted α-helical coiled coil structure. The Klp1 protein may therefore function as a motor protein and contribute in some fashion to the forces of ciliary and flagellar motion. Because Klp1 groups in the kinesin-9 family of kinesin-like proteins (5), all of whose members are exclusively expressed in tissues with motile cilia or flagella (testis, brain, lung) or in flagellated microorganisms (Giardia, Leishmania, Chlamydomonas) (6), the role of Klp1 in ciliary motility is likely to be evolutionarily conserved.
The central pair complex is an important regulator of flagellar dynein activity in 9 + 2 organelles. Although flagella in some organisms naturally lack central microtubules, those 9 + 0 (7) or 6 + 0 (8) organelles display a limited, helical motility pattern. Absence of the central pair from normally 9 + 2 organelles results in a complete lack of motility, as exemplified by several Chlamydomonas central pair assembly mutations (9, 10). Central pair microtubules are decorated by a tight array of associated proteins that does not appear to leave any unoccupied surface for progressive movement of a kinesin (11), suggesting that central pair kinesins do not function as typical motors. In Chlamydomonas, as in many other organisms, the central pair apparatus rotates one turn relative to the cage of outer doublets during propagation of each bend. One potential function of a central pair kinesin could therefore be the generation of a rotation force through transient interactions with surrounding radial spokes. However, we recently discovered that central pair rotation is a passive response to bend propagation and does not require interactions between radial spoke heads and the central apparatus (12). This finding suggests that Klp1 functions in another capacity, perhaps as a conformational switch.
To test the general properties of Klp1, we extracted the native protein from axonemes and determined that it resembles typical kinesins in its ability to bind to microtubules in a nucleotide-dependent fashion. As a direct test of Klp1 function, we transformed Chlamydomonas with constitutively expressed RNA interference (RNAi) constructs to reduce Klp1 protein levels, and obtained strains that assemble 20% or less of wild-type flagellar Klp1. Cells that retain 20% of normal Klp1 swim, but at a reduced velocity due to a lower beat frequency. Under enriched growth conditions, these cells have paralyzed flagella. EM image averages of knockdown axonemes identify Klp1 as a specific projection on the C2 central pair microtubule that may interact directly with radial spokes and regulate beat frequency.
Materials and Methods
Chlamydomonas reinhardtii strains 137c (CC 124) and arg2 (CC 1930) were obtained from Elizabeth Harris at the Chlamydomonas Genetics Center, Duke University (Durham, NC). All genetic manipulations followed standard procedures (13).
Molecular Biology. Hybridization probes were labeled with digoxygenin and detected by chemiluminescence (Roche Diagnostics) on Biomax ML film (Kodak). Wild-type genomic clones for the KLP1 gene were identified by hybridization of a bacterial artificial chromosome (BAC) library filter (Genome Systems, St. Louis) with the KLP1 cDNA (4), and BAC 12O10 was used as template to amplify the genomic fragment used to construct knockdown plasmid pGenC. The first three coding exons (900 bp) were amplified by using primers GCACATCGcAtATGGTGAAGCAAGCTG and AGCAggatcCTGTGGATTCACGGGCG, in which lowercase indicates bases that differ from Chlamydomonas, bold indicates the NdeI and BamHI sites used for cloning, and the underlined ATG is the translation initiation codon. The corresponding 290-bp region of the KLP1 cDNA (GenBank accession no. X78589) was amplified with primers CATCGAAttcGGTGAAGCAAGCTGT (EcoRI site in bold) and CACAGATCTGGCCGTACGCGAAGATA (BglII site in bold). The two amplification products were digested with BamHI and BglII, respectively, and ligated together, and the 1.2-kb product was digested with NdeI and EcoRI and used to replace the ble gene in pGenD-ble (14) to make pGenC. Glass bead-mediated transformation and selection of transformants used ARG plasmid pARG7.8, as described (15). Transformants containing integrated pGenC sequences were identified by PCR of genomic DNA using PsaD-specific primers AGGTGTTGCGCTCTTGACTCGTTGT and AGCTCTTCTCCATGGTACAGGCGG, which match nucleotides 761–785 and the complement of nucleotides 1435–1459 of PsaD (GenBank accession no. AF335592). Amplification with these primers generated a 0.7-kb fragment from the endogenous PsaD gene and a 0.5-kb fragment from introduced sequences.
Electron Microscopy. Axonemes prepared by standard methods were fixed and processed for electron microscopy as described (15). Negatives taken at an original magnification of ×50,000 were digitized on a flatbed scanner at a resolution of 1,200 dpi, and image contrast and gamma were adjusted in photoshop (Adobe Systems, Mountain View, CA). For image averaging, negatives containing axonemes with central pair complexes in approximate cross section were scanned at a pixel size 1.0 nm by using an AGFA Duoscan f40 scanner. Averages were computed by using methods described in detail (16, 17). Briefly, a model point was placed in the center of each central pair complex, and a program was run that extracted the images into a 120 × 120 pixel box. The individual central pair complexes were then aligned by applying a general linear transformation to solve for differences in translation, rotation, magnification, and stretch along a single axis, and the resulting aligned images were averaged. The densities in the images were normalized by using the doublet normalization procedure (16). The averages for wild-type and AC30 were then aligned to each other by marking fiducial points in the centers of 10 protofilaments and by using multiple linear regression to minimize the differences in the fiducial points between the images. Difference images were computed by using a program that subtracts one average image from another and uses standard deviation images to find the statistical significance of the difference at each pixel, as evaluated by a t statistic. The program then sets to zero all differences less significant than the specified level of significance (P = 0.0005). A total of 86 images were used for the wild-type average, and 71 images were used for the AC30 average.
Protein Biochemistry and Blotting. For NaCl extractions, axonemes were resuspended in HMDEK (30 mM Hepes/5 mM MgSO4/1 mM DTT/0.5 mM EGTA/25 mM potassium acetate/1 mM phenylmethylsulfonyl fluoride, pH 7.4) containing the indicated concentration of NaCl, left on ice for 30 min, and spun in a microfuge at top speed for 10 min at 4°C. To determine the sedimentation rate of Klp1, axonemes were extracted with HMDEK containing 0.6 M NaCl for 30 min on ice and centrifuged for 20 min at 4°C, and the supernatant was dialyzed against HMDEK and clarified by centrifugation. Proteins in the NaCl extract were separated by centrifugation on a 5–20% sucrose gradient (prepared with HMDEK) at 35,000 rpm (155,000 × g) for 16 h in a SW40 rotor. Gradients were collected from the bottom of the tube in 0.8-ml fractions. Sedimentation standards were thyroglobulin (19S) and catalase (11S) and were checked against the position of outer row dynein (21S) as an internal standard and the cpc1 complex (16S) run in a parallel gradient (18). Protein composition was determined by SDS/PAGE on 7% acrylamide gels stained with Coomassie blue. Dynein heavy chains (average Mr = 500,000) and Benchmark Protein Ladder (Invitrogen) were used as protein size standards. For immunoblotting, proteins were transferred to Immobilon-P membranes (Millipore) and probed with affinity-purified polyclonal anti-Klp1 (4) (obtained from M. Bernstein, Albert Einstein University, New York) at a 1:2,000 dilution; polyclonal affinity-purified anti-Fla10 (19) (obtained from J. L. Rosenbaum, Yale University, New Haven, CT) at a 1:5,000 dilution; and monoclonal anti-sea urchin kinesin SUK4 (20) (obtained from BabCo, Richmond, CA) at a 1:1,000 dilution. The antibodies against Klp1 and Fla10 were both raised against unique sequences in the nonconserved tail domains of these proteins, and their specificity has been previously documented. SUK4 recognizes several antigens in Chlamydomonas flagella that are retained in axonemes of central pair assembly mutants (2). Antibodies were detected with peroxidase-labeled goat-anti-rabbit or goat-anti-mouse (Bio-Rad) developed with enhanced chemiluminescence (ECL) reagents (Amersham Pharmacia). Proteins on blots were detected by staining with amido black.
For microtubule binding assays, 0.5 M NaCl extracts were dialyzed against HMDEK, and extracted axonemes were resuspended in HMDEK to the same final volume as the extract (final extracted axoneme concentration of 3 mg/ml protein). Bovine brain tubulin (Cytoskeleton, Denver) was assembled in vitro in 0.1 M Pipes (pH 6.9), 2 mM MgSO4, 1 mM EGTA, 1 mM GTP, sedimented in an airfuge at 100,000 × g for 5 min, and resuspended in HMDEK containing 20 μM taxol to a final concentration of 1 mg/ml protein. For binding assays, 20 μl each of extract and microtubules (or extracted axonemes) and 10 μl of nucleotide or buffer control were mixed, incubated at room temperature for 30 min, and spun at 100,000 × g for 5 min. Final concentrations of nucleotides were 4 mM (adenosine 5′-[β,γ-imido]triphosphate, AMP-PNP) or 10 mM (ATP). Pellets were resuspended in 60 μlof1× sample buffer, and supernatants were mixed with 10 μl of 6× sample buffer. For ATP release assays, pellets from AMP-PNP samples were resuspended in HMDEK containing taxol and ATP and incubated at room temperature for 30 min before processing as above. For immunoblotting, proteins were separated on 7% gels, transferred to Immobilon-P membranes (Millipore), and probed with affinity-purified rabbit anti-Klp1. To remove phosphates from Klp1 protein, 1/100 vol of a 10 units/μl stock of calf intestinal phosphatase (Promega) was added to a high-salt extract and incubated for 1 h at 37°C, followed by addition of sample buffer for SDS/PAGE.
Results
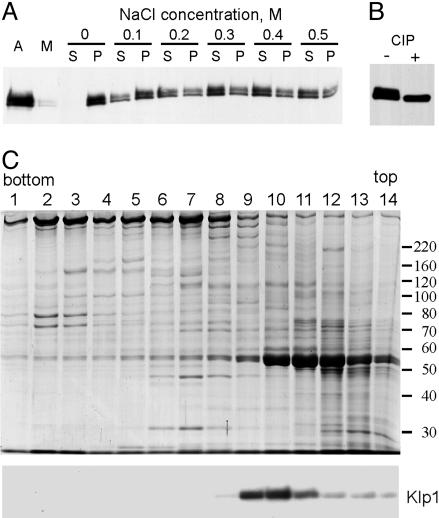
Klp1 Biochemistry. To characterize properties of Klp1, we first identified conditions that could remove Klp1 from axonemal central pair microtubules. Bernstein et al. (4) previously showed that Klp1 remained in the axonemal fraction during detergent treatment of whole flagella and was not removed from axonemes by 5 mM ATP. As illustrated in Fig. 1A, increasing concentrations of NaCl were effective at extracting Klp1 from axonemes, with most of the Klp1 extracted at 0.5 M NaCl. The presence or absence of 1 mM ATP had no effect on this extraction pattern (data not shown). The anti-Klp1 antibody detects a doublet on Western blots of axonemes (Fig. 1 A) as noted previously (4). We observed that both bands were extracted at similar salt concentrations, but that the relative amount of these two bands differed between different axonemal preparations, which suggested that these two bands represent posttranslationally modified versions of the same protein. To see whether these electrophoretic variants are related by phosphorylation, extracts were treated with phosphatase before preparation for SDS/PAGE. After phosphatase treatment, only the lower band was detected (Fig. 1B), indicating that the upper band is a phosphorylated form.
Fig. 1.
Klp1 is an axonemal phosphoprotein. (A) Western blot of axonemes (A), the membrane/matrix fraction (M), and pellets (P) or supernatants (S) from extraction of axonemes with the indicated concentrations of NaCl. Klp1 is not removed by detergent but becomes mostly soluble after treatment with 0.5 M NaCl. (B) Western blot of a 0.5 M NaCl extract before (–) and after (+) treatment with calf intestinal phosphatase. (C) Coomassie-stained gel (Upper) and anti-Klp1 Western blot (Lower) of fractions from sucrose gradient centrifugation of an axonemal 0.5 M NaCl extract. Klp1 sediments in fractions 9–10 at 8S.
The architecture of flagellar axonemes requires that axonemal proteins assemble into large complexes. Because the C2 central pair microtubule is disassembled by 0.5 M NaCl (21), we reasoned that Klp1 might remain associated with other proteins after extraction. To find out, axonemal extracts containing Klp1 were centrifuged on a sucrose gradient. Under the conditions tested, Klp1 sedimented at 8S (Fig. 1C), similar to the sedimentation rates reported for conventional dimeric kinesins (22, 23), and not as part of a larger complex.
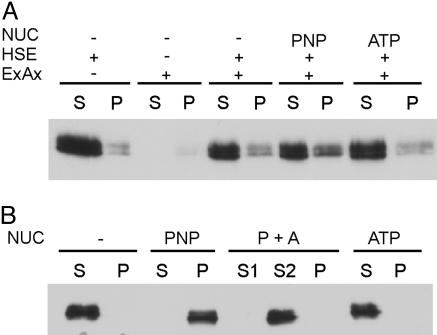
Conventional kinesins can interact directly with microtubules in a nucleotide-dependent fashion. To see whether the extracted Klp1 protein would rebind to extracted axonemal microtubules, salt concentration of the extract was reduced by dialysis and the extract was mixed with resuspended salt-extracted axonemes in a 1:1 ratio in the absence of nucleotide or in the presence of 10 mM ATP or 4 mM AMP-PNP. The amount of Klp1 that sedimented with the extracted axonemes in the absence of nucleotide or in the presence of ATP did not differ from the amount of Klp1 that sedimented in controls that lacked axonemes (Fig. 2A). When AMP-PNP was present, a small additional amount of Klp1 sedimented. Therefore, extracted axonemal doublet microtubules appear to support binding of a small amount of Klp1. When extracts containing Klp1 were mixed with reassembled bovine brain tubulin, Klp1 associated with the microtubules in the presence of AMP-PNP but not in the presence of ATP or in the absence of nucleotide. In addition, when the AMP-PNP pellet was resuspended in the presence of ATP and respun, Klp1 was released into the supernatant solution (Fig. 2B). Thus, Klp1 interaction with reassembled brain microtubules is modulated by nucleotides similarly to conventional kinesins.
Fig. 2.
Microtubule binding properties of Klp1. (A) Western blots of supernatants (S) and pellets (P) after incubation of dialyzed NaCl extract (HSE) with extracted axonemes (ExAx). Most of the Klp1 remains in the supernatant even in the presence of AMP-PNP (PNP). (B) Binding of Klp1 to reassembled brain microtubules. Klp1 binds in the presence of AMP-PNP (PNP), but not ATP, and is released from the AMP-PNP pellet by subsequent ATP treatment (P+A).
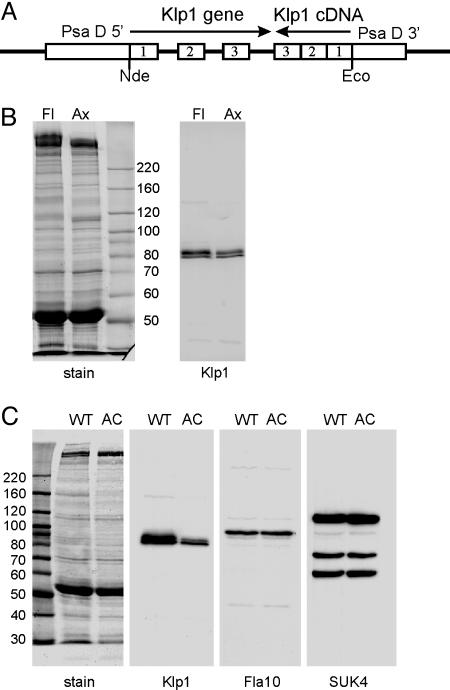
Klp1 Knockdown by RNAi. Because no known motility mutants map to the C. reinhardtii KLP1 locus, and tests of existing insertional mutant libraries failed to identify a KLP1 mutant strain, we turned to RNAi to knock down the amount of Klp1 protein and determine its contribution to flagellar motility. Based on previous reports of successful RNAi-mediated inhibition of gene expression in Chlamydomonas, we constructed chimeric intron-containing expression plasmid pGenC by using Klp1 genomic and cDNA sequences (Fig. 3A) as detailed in Materials and Methods. The insert in this plasmid spans the first three Klp1 introns joined with a cDNA fragment that spans the same coding region in reverse orientation, placed between 5′ and 3′ elements of the constitutively expressed PsaD gene. The expected expression product of pGenC is a mRNA that, after splicing, should form a 290-bp hairpin RNA. Plasmid DNA was cotransformed along with an ARG plasmid into arg2 strain cc 48, and arg+ transformants were screened by PCR for retention of pGenC sequences. When pGenC-positive transformants were grown in liquid, approximately half (14 of 31) swam at reduced velocity. Whole-cell Western blots confirmed that the amount of Klp1 protein was reduced in all of the slow-swimming strains and remained at wild-type levels in all of the strains swimming at wild-type velocities, indicating that the reduction in swimming velocity was related to a reduction in Klp1 protein (data not shown). Because IFT kinesins are required for flagellar assembly, we used sequences in construction of pGenC that are not 100% conserved between Klp1 and Fla10 in any 21-base window. However, some silencing affects or reductions in translation efficiency occur even when the interfering RNA and mRNA retain mismatches (24). In addition, the Chlamydomonas IFT kinesin is a heterodimer of Fla10 and an uncharacterized kinesin II family member whose mRNA could share regions of identity with Klp1 mRNA. To test the possibility that an IFT kinesin was affected by pGenC expression, we measured flagellar length in slow-swimming pGenC transformants (11.1 ± 0.8 μm; n = 20) but found no significant change from the average length of flagella in untransformed cells (11.7 ± 0.9 μm; n = 20). Therefore, the reduction in swimming velocity associated with pGenC expression is the result of an altered flagellar motility pattern rather than a reduction in flagellar length. The primary motility defect identified in strains with reduced levels of Klp1 was a reduced beat frequency. Each transformant displayed a range of swimming velocities and beat frequencies within a clonal population, but maximal rates were reduced from wild type, with the most severely affected strains reduced to a maximum beat frequency of ≈35 Hz (compared with wild-type frequency of 60 Hz).
Fig. 3.
Axonemes of knockdown strains have reduced levels of Klp1. (A) Plasmid used for RNAi-mediated knockdown of Klp1. Portions of the Klp1 gene and cDNA were joined in opposite orientations and inserted into an expression plasmid containing the promoter and untranslated regions from the Chlamydomonas PsaD gene. Transcription and splicing should generate a double-stranded hairpin RNA. (B) SDS/PAGE of knockdown strain AC30 flagella (Fl) and axonemes (Ax). Gels were stained with Coomassie blue (stain) or blotted and probed with anti-Klp1 (Klp1). The blot shows that the residual Klp1 in knockdown strains is tightly associated with the axoneme. (C) Blots of wild-type (WT) and AC30 (AC) axonemes were stained with amido black (stain) or probed with anti-Klp1, anti-Fla10, or pan-kinesin antibody SUK4. Klp1 levels are reduced by 80% in AC30 axonemes, whereas other axonemal kinesin levels are unchanged. The sizes (in kDa) of molecular mass standards are indicated next to each stained panel.
None of the strains tested showed a complete absence of Klp1 in whole cell samples, suggesting that some Klp1 protein might still be present in flagella. Western blots of flagellar protein samples were used to identify strains with the lowest levels of Klp1 expression. Results indicated that most transformants retained ≈20% of the wild-type level of flagellar Klp1, and that the remaining Klp1 was associated with axonemal microtubules (Fig. 3B).
Quantification of flagellar Klp1 showed that, of the 14 strains tested, all had Klp1 levels <50% of wild type, but none had levels <18.5%. One of several strains with the lowest flagellar Klp1 levels (AC30) was selected for further analysis. Immunoblots of AC30 axonemes confirmed that Klp1 levels are reduced, whereas other flagellar kinesin-like proteins, including IFT kinesin Fla10 and proteins recognized by anti-sea urchin kinesin antibody SUK4, remain at wild-type levels (Fig. 3C). Average flagellar beat frequency of the most rapidly swimming AC30 cells was 36 ± 8Hz(n = 25), but some cells within the population had much lower frequencies, and ≈5% of the cells were non-motile and had full-length flagella that occasionally twitched but failed to propagate bends. These paralyzed flagella frequently formed strong principal or reverse bends near their base so that the two flagella crossed, but these curves spontaneously straightened rather than propagating. When AC30 cells were transferred to acetate-enriched growth media, all cells became paralyzed. A test of medium components revealed that paralysis was induced by a >50 mM increase in ionic strength and did not depend on the nature of the solute. A backcross of AC30 to wild-type cells produced 2:2 segregation of the pGenC insertion, which cosegregated with reduced motility and reduced levels of Klp1. Knockdown strains that resulted from this cross showed strain-dependent motility and Klp1 levels.
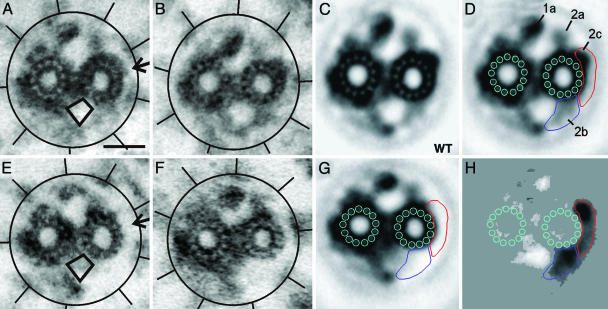
Localization of Klp1 on the C2 Microtubule. Previous immunolocalization showed that Klp1 is associated with the C2 central pair microtubule but could not further determine which C2 structures might contain Klp1. To begin such an analysis, thin-section electron micrographs of isolated, demembranated flagellar axonemes from wild type and AC30 were compared. The only structure that appeared altered in klp1 knockdown axonemes was a density associated with the C2 microtubule, projection 2c (Fig. 4 A and B, arrow). A portion of the 2c density was missing in 85% of AC30 axonemes (n = 77) vs. 10% of wild-type axonemes (n = 86). In many AC30 axonemal images, the 2b density was also reduced or displaced. Lack of a typical 2b density was noted in 58% of AC30 axonemes vs. 17% of wild-type axonemes. The 2b density often appeared displaced into a region that normally lacked density (rhomboid outline in Fig. 4 A and E). For more detailed analysis of structural changes associated with knockdown of Klp1, cross-section images of axonemes were digitized and used to generate image averages (Fig. 4 C and G). A density difference map (Fig. 4H) shows that projection 2b and part of projection 2c are the only structures whose densities are markedly reduced in Klp1 knockdown axonemes. Although 2c was considered to represent a single entity in our previous structural analyses (11, 15), the retention of part of the 2c density at wild-type levels in AC30 axonemes indicates that 2c is a composite of two separable structures. The light regions in Fig. 4H highlight regions of increased density in the AC30 average. These include most markedly two regions of shifted density due to repositioning of projections 1a and 2b. Repositioning of 1a results from a shift in the relative orientation of C1 and C2 microtubules, which can best be visualized by superpositioning the images (see Movie 1, which is published as supporting information on the PNAS web site). Repositioning of projection 2b density appears related to destabilization of its attachment to the C2 microtubule. Fig. 5 summarizes the wild-type structures that are most affected in the knockdown strain and that we believe correspond to Klp1. A stereoanaglyph image that exposes relevant surfaces of the central pair shows the 16-nm periodicity of all structures in this region (25). Structures tentatively identified as Klp1 appear continuous with the rest of the 2c density in this wild-type axoneme. In the diagram (Fig. 5 Inset), regions shaded red correspond to density that is lost in knockdown strains but is not affected by destabilization of density 2b in cpc1 mutant axonemes (15).
Fig. 4.
Localization of Klp1 by electron microscopy. The central pair complex and surrounding radial spokes of wild-type axonemes (A and B) and AC30 axonemes (E and F) show the loss of one density (arrow in A) and the displacement of density into the region indicated by a rhomboid. Circles and lines highlight the location of radial spokes in each image. Normalized image averages of wild-type (C) and AC30 (G) central pair complexes provide greater detail but blur the displaced images of density C2b. The wild-type average from C is repeated in D with the addition of density labels and outlines that aid in alignment of the images in C, G, and H. A difference image (H) shows density changes between wild-type and knockdown averages significant at the 0.005% level and reveals the shape of the regions of reduced density caused by loss of Klp1 (darker areas) as well as regions of increased density (lighter areas). Darker regions in H are outlined as areas related to density 2b (blue) and 2c (red), and the same outlines are shown in D and G. Increased density related to C1 projection 1a is caused by a rotational shift in the relative orientation of C1 and C2 microtubules (see Movie 1). (Scale bar, 20 nm.)
Fig. 5.
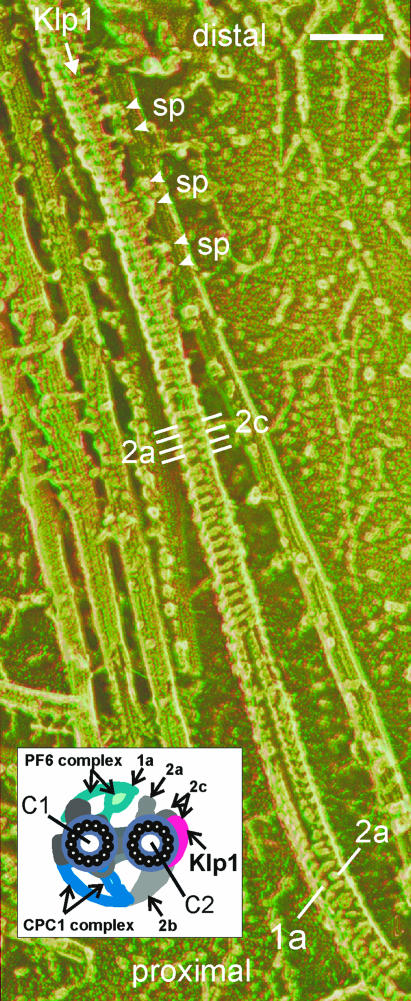
Summary of structures related to Klp1. Anaglyph stereo view of a disrupted wild-type Chlamydomonas axoneme prepared by the quick-freeze deep-etch method shows a central pair complex with a gradual left-handed helical twist (best viewed with red–green anaglyph glasses, green lens on the right), modified from Fig. 3 of Goodenough and Heuser (25). The apparent attachment of pairs of radial spokes (sp) to central pair projections in the Klp1 region may be an artifact of specimen preparation. [Image reproduced from with permission from ref. 25 (Copyright 1987, The Rockefeller University Press, New York)]. Inset summarizes densities seen in cross-section views, some of which are also labeled on the micrograph. (Scale bar, 100 nm.)
Discussion
Kinesins participate in many microtubule-related activities, including microtubule destabilization as well as plus- and minus-end-directed motility (26). Because of its distribution along the edge of a central pair microtubule that is heavily populated with microtubule-associated proteins, the Chlamydomonas Klp1 protein is not expected to function as either a traditional motor or a polymerization regulator. Klp1 retains microtubule interaction characteristics similar to conventional kinesins (tight binding in the presence of AMP-PNP, release by ATP) and yet is not extracted from axonemes by ATP. This kinesin-like protein is therefore likely to interact with the surface of the C2 central pair microtubule and change both conformation and microtubule affinity of its catalytic domain upon binding and hydrolysis of ATP but must have additional interactions that anchor it to a specific C2 protofilament. Conformational changes in Klp1 could alter the conformation of an associated protein or alter the affinity of Klp1 for an associated protein, thus acting as a regulatory switch. Both an AKAP (27) and a phosphatase (28) have been identified as central pair proteins, suggesting that changes in phosphorylation of central pair substrates could be important for motility regulation. Our results show that Klp1 is a phosphoprotein, which suggests that changes in phosphorylation of Klp1 could modulate its activity in response to physiological signals.
Reduced motility of Klp1 knockdown strains shows that the density missing from these flagella is important for normal dynein regulation. Our structural analysis shows that Klp1 is localized to a part of density 2c and along the edge of the C2 microtubule but is also essential for correct positioning of another central pair density, 2b. The 2b density is frequently missing from axonemes of central pair complex assembly mutant cpc1 (15), but other C2-associated densities, including the density that we identify as Klp1, are unaffected by loss of 2b in cpc1 axonemes. Therefore, Klp1 may be important for anchoring one end of 2b, but 2b is not needed to anchor Klp1. In addition, because most of the motility deficit of cpc1 flagella results from reductions in intraflagellar ATP concentrations and is restored in vitro by reactivation of cpc1 axonemes in 1 mM ATP (18), loss of 2b does not correlate with a major change in flagellar motility. Flagellar paralysis in Klp1 knockdown strains most likely results directly from loss of Klp1 function, although we cannot rule out the possibility that additional proteins may be localized in the density that is missing from knockdown axonemes and may play important roles in motility regulation.
Mutations in the pf6 gene likewise prevent assembly of one density associated with the central pair as seen in cross-sectional images, C1 microtubule projection 1a (Fig. 5 Inset), and are associated with an almost total lack of motility (10, 29). Flagella of pf6–1 cells occasionally beat with frequencies as high as 5 Hz. The PF6 gene product is a large protein of unknown function (29) that acts as a scaffold for several other novel proteins (10, ∥). Although central pair motility regulation involves interactions between central pair projections and radial spokes, none of the central pair and radial spoke proteins that directly participate has been identified. We hypothesize that Klp1 is a spoke-binding protein, or a switch that modulates the spoke interaction of a neighboring central pair protein. Identification of proteins that interact with Klp1 in situ will be an important step toward characterization of the Klp1-dependent dynein regulation pathway.
Supplementary Material
Acknowledgments
Masako Nakatsugawa, Judy Freshour, and Karly Judson provided technical assistance. This work was supported by National Institutes of Health Grant GM44228 (to D.R.M.). E.O. was supported by National Institutes of Health Biotechnology Resource Grant RR-00052 (to J. R. McIntosh).
Author contributions: R.Y. and D.R.M. designed research; R.Y., S.G., and D.R.M. performed research; R.Y., E.O., and D.R.M. analyzed data; and D.R.M. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: IFT, intraflagellar transport; RNAi, RNA interference; AMP-PNP, adenosine 5′-[β,γ-imido]triphosphate.
Footnotes
Dymek, E. E., Wargo, M. J. & Smith, E. F. (2003) Mol. Biol. Cell 14, 433a (abstr.).
References
- 1.Pazour, G. J. & Rosenbaum, J. L. (2002) Trends Cell Biol. 12, 551–555. [DOI] [PubMed] [Google Scholar]
- 2.Johnson, K. A., Haas, M. A. & Rosenbaum, J. L. (1994) J. Cell Sci. 107, 1551–1556. [DOI] [PubMed] [Google Scholar]
- 3.Fox, L. A., Sawin, K. E. & Sale, W. S. (1994) J. Cell Sci. 107, 1545–1550. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, M., Beech, P. L., Katz, S. G. & Rosenbaum, J. L. (1994) J. Cell Biol. 125, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence, C. J., Dawe, R. K., Christie, K. R., Cleveland, D. W., Dawson, S. C., Endow, S. A., Goldstein, L. S., Goodson, H. V., Hirokawa, N., Howard, J., et al. (2004) J. Cell Biol. 167, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miki, H., Setou, M., Kaneshiro, K. & Hirokawa, N. (2001) Proc. Natl. Acad. Sci. USA 98, 7004–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbons, B. H., Gibbons, I. R. & Baccetti, B. (1983) J. Submicrosc. Cytol. 15, 15–20. [PubMed] [Google Scholar]
- 8.Schrevel, J. & Besse, C. (1975) J. Cell Biol. 66, 492–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witman, G. B., Plummer, J. & Sander, G. (1978) J. Cell Biol. 76, 729–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dutcher, S. K., Huang, B. & Luck, D. J. L. (1984) J. Cell Biol. 98, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchell, D. R. (2003) Cell Motil. Cytoskeleton 55, 188–199. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell, D. R. & Nakatsugawa, M. (2004) J. Cell Biol. 166, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, E. H. (1989) The Chlamydomonas Sourcebook (Academic, San Diego).
- 14.Fischer, N. & Rochaix, J.-D. (2001) Mol. Genet. Genomics 265, 888–894. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell, D. R. & Sale, W. S. (1999) J. Cell Biol. 144, 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mastronarde, D. N., O'Toole, E. T., McDonald, K. L., McIntosh, J. R. & Porter, M. E. (1992) J. Cell Biol. 118, 1145–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Toole, E., Mastronarde, D., McIntosh, J. R. & Porter, M. E. (1995) Methods Cell Biol. 47, 183–191. [DOI] [PubMed] [Google Scholar]
- 18.Zhang, H. & Mitchell, D. R. (2004) J. Cell Sci. 117, 4179–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozminski, K. G., Beech, P. L. & Rosenbaum, J. L. (1995) J. Cell Biol. 131, 1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingold, A. L., Cohn, S. A. & Scholey, J. M. (1988) J. Cell Biol. 107, 2657–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adams, G. M. W., Huang, B., Piperno, G. & Luck, D. J. L. (1981) J. Cell Biol. 91, 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bloom, G. S., Wagner, M. C., Pfister, K. K. & Brady, S. T. (1988) Biochemistry 27, 3409–3416. [DOI] [PubMed] [Google Scholar]
- 23.DeLuca, J. G., Newton, C. N., Himes, R. H., Jordan, M. A. & Wilson, L. (2001) J. Biol. Chem. 276, 28014–28021. [DOI] [PubMed] [Google Scholar]
- 24.Zeng, Y., Yi, R. & Cullen, B. R. (2003) Proc. Natl. Acad. Sci. USA 100, 9779–9784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodenough, U. W. & Heuser, J. E. (1985) J. Cell Biol. 100, 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldstein, L. S. & Philp, A. V. (1999) Annu. Rev. Cell Dev. Biol. 15, 141–183. [DOI] [PubMed] [Google Scholar]
- 27.Gaillard, A. R., Diener, D. R., Rosenbaum, J. L. & Sale, W. S. (2001) J. Cell Biol. 153, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, P. F., Fox, L., Colbran, R. J. & Sale, W. S. (2000) J. Cell Sci. 113, 91–102. [DOI] [PubMed] [Google Scholar]
- 29.Rupp, G., O'Toole, E. & Porter, M. E. (2001) Mol. Biol. Cell 12, 739–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.