Abstract
OBJECTIVE
Insulin resistance (IR) is a hallmark of obesity and metabolic disease. Circulating extracellular RNAs (ex-RNAs), stable RNA molecules in plasma, may play a role in IR, though most studies on ex-RNAs in IR are small. We sought to characterize the relationship between ex-RNAs and metabolic phenotypes in a large community-based human cohort.
RESEARCH DESIGN AND METHODS
We measured circulating plasma ex-RNAs in 2,317 participants without diabetes in the Framingham Heart Study (FHS) Offspring Cohort at cycle 8 and defined associations between ex-RNAs and IR (measured by circulating insulin level). We measured association between candidate ex-RNAs and markers of adiposity. Sensitivity analyses included individuals with diabetes. In a separate cohort of 90 overweight/obese youth, we measured selected ex-RNAs and metabolites. Biology of candidate microRNAs was investigated in silico.
RESULTS
The mean age of FHS participants was 65.8 years (56% female), with average BMI 27.7 kg/m2; participants in the youth cohort had a mean age of 15.5 years (60% female), with mean BMI 33.8 kg/m2. In age-, sex-, and BMI-adjusted models across 391 ex-RNAs in FHS, 18 ex-RNAs were associated with IR (of which 16 were microRNAs). miR-122 was associated with IR and regional adiposity in adults and IR in children (independent of metabolites). Pathway analysis revealed metabolic regulatory roles for miR-122, including regulation of IR pathways (AMPK, target of rapamycin signaling, and mitogen-activated protein kinase).
CONCLUSIONS
These results provide translational evidence in support of an important role of ex-RNAs as novel circulating factors implicated in IR.
Introduction
Insulin resistance (IR) is a hallmark of human obesity and associated with the risk of developing diabetes and cardiovascular disease. IR can exist across the spectrum of BMI from lean (<25 kg/m2) to overweight/obese (>25 kg/m2). These findings indicate that a BMI-centric definition of obesity may not capture its underlying biology (1). Investigation of clinical and molecular markers that define architecture of IR has intensified, focusing on adipose tissue distribution and function, metabolite profiles, gut microbial diversity, and epigenetic and genetic variation. Recently, RNA located outside of cellular structures (extracellular RNAs [ex-RNAs]), circulating RNA molecules that are stable in plasma, have emerged as potential novel mediators in IR, potentially orchestrating control over networks of gene expression. Indeed, animal models suggest exquisite regulation of circulating ex-RNAs in the development and resolution of obesity and in metabolic cross talk between various organs involved in adipocyte dysfunction (2), suggesting their importance as clinical and functional biomarkers. As such, studies in small groups of patients with obesity and IR have identified candidate ex-RNAs associated with metabolic dysfunction (3–6), though there is absence of validation in large at-risk populations and against metabolic phenotypes (e.g., visceral and hepatic adiposity) known to impact cardiometabolic risk.
In this study, we investigate plasma-circulating ex-RNA abundance in two separate cohorts across the life span of human obesity—the 8th Framingham Heart Study (FHS) Offspring Cohort (adults) and the POOL study (obese/overweight youth)—to study ex-RNAs associated with IR. We further investigate the relationship between these ex-RNAs and several clinical hallmarks of metabolic dysfunction. We subsequently tested the association of two top candidate ex-RNAs discovered in FHS with IR in the POOL study and defined a relationship between miR-122 and IR independent of adverse metabolite profiles in youth.
Research Design and Methods
Framingham Heart Study
The Framingham Heart Study (FHS) is a community-based, prospective study of cardiovascular disease conducted in Framingham, MA, with serial examinations every 4–8 years and concomitant in-depth phenotyping of metabolic traits over multiple prior examinations. The study design has been published (7). Standard anthropometric indices were measured as reported (8). Diabetes was defined as fasting plasma glucose ≥126 mg/dL, hemoglobin A1c ≥6.5% (where measured), or treatment with either insulin or a hypoglycemic agent.
Blood collected in Framingham for the FHS Offspring Exam Cycle 8 (March 2005 to January 2008) was analyzed in this study. Venipuncture was performed on study participants in a supine position. Blood was collected into blood collection tubes with a liquid-buffered sodium citrate additive (0.105 mol), centrifuged, and plasma separated and frozen at −80°C within 90 min of collection. Insulin was measured by ELISA (Roche e411; Roche Diagnostics, Indianapolis, IN; intra-assay coefficient of variation [CV] 2.0%). HOMA of IR (HOMA-IR), a marker of IR, was calculated as the product of insulin (μIU/mL) and glucose (mmol/L) divided by 22.5 (9). Insulin (pmol/L) was converted to μIU/mL by multiplying by 0.144, and glucose (mg/dL) was converted to mmol/L by multiplying by 0.0555. Interleukin-6 (IL-6; ELISA, R&D Systems, Minneapolis, MN; intra-assay CV 3.7%) and soluble tumor necrosis factor receptor II (TNFRII; ELISA; R&D Systems) were measured at the time of FHS Offspring Exam Cycle 8. Total adiponectin from FHS Offspring Exam Cycle 7 (1998–2001) was included, measured by ELISA (R&D Systems) as described (10). For ex-RNA analysis, an aliquot of 170 µL plasma samples was transferred to our laboratory in March 2014 and stored at −80°C for analysis. A subset of participants underwent abdominal computed tomographic (CT) imaging (June 2002 to April 2005) for quantification of visceral and subcutaneous adipose tissue volume and fat attenuation (with lower attenuation as a marker of fat quality [11]) and liver attenuation (a surrogate of hepatic steatosis [12]), with previously described methods (13).
Written informed consent was obtained from all study participants, with Institutional Review Board approval at Boston University, Massachusetts General Hospital, and the University of Massachusetts.
Study Population and Clinical Assessment in POOL
POOL is an ongoing, prospective research registry of overweight and obese youth and young adults, 2–25 years old (Boston Children’s Hospital, Boston, MA). Subjects were local residents who were overweight or obese at study entry (BMI more than or equal to age/sex-specific 85th percentile on Centers for Disease Control and Prevention growth charts for those <18 years of age or ≥25 kg/m2 for those ≥18 years of age). Informed consent to participate was obtained from a parent or legal guardian for minors (<18 years of age; with participant assent) or from adult participants. Clinical and demographic data were collected, including height and weight (measured twice to the nearest 0.1 cm and kg, respectively). Total body fat was measured using a plethysmographic method that uses whole-body densitometry to determine fat and fat-free mass. Insulin was measured using an electrochemiluminescence immunoassay (Roche Elecsys/Cobas immunoassay analyzer; CV 1.1–4.9%). HOMA-IR was calculated as specified above. Blood for plasma specimen storage was collected in the fasting state in lithium heparin-containing tubes, centrifuged immediately at 4°C, then transferred into cryovials, and frozen at −80°C for long-term storage.
Quantification of Plasma Extracellular Circulating ex-RNAs
Detailed methods for quantification of ex-RNAs in FHS have been published by our group (14). In the initial study in FHS, plasma small RNA sequencing was performed in 40 FHS participants to determine which ex-RNAs were abundantly and reliably expressed in human plasma (14). A plasma microRNA (miRNA) was chosen to be included for validation in the full FHS Offspring Exam Cycle 8 cohort if it was expressed at >10 reads per kilobase transcript per million reads mapped by sequencing. Because of the novelty and limited understanding of the other (non-microRNA) ex-RNA targets, we included all expressed small nucleolar RNAs (snoRNAs) and Piwi-interacting RNAs (piRNAs). We subsequently included only those plasma ex-RNAs detectable in at least 100 FHS participants for this analysis (as determined from the final analytic cohort specified below; N = 2,317). Of the 2,822 plasma samples from the FHS Offspring Exam Cycle 8 in FHS, 59 (2%) subjects were excluded because of laboratory error (e.g., inaccurate volume of plasma pipetted, N = 31; poor protein precipitation performance, N = 23; or potential contamination, N = 5), resulting in 2,763 subjects. We subsequently excluded individuals who were not fasting at the time of the blood draw for at least 8 h (N = 54), individuals without insulin or fasting blood glucose measured (N = 2), and individuals with diabetes, as defined above (N = 390), yielding a final analytic cohort of 2,317 study participants.
Finally, based on our analyses of ex-RNAs in FHS, we selected two miRNAs (miR-122 and miR-192) to analyze in a separate study of 90 obese/overweight young participants (POOL). Of note, miRNA quantification in POOL was performed by our group in a separate project with a separate set of 90 ex-RNA targets. We chose to direct our analysis in POOL toward those ex-RNAs associated with metabolic phenotypes in FHS. Therefore, we only analyzed miR-122 and miR-192 in this POOL. (piRNAs and snoRNAs were not assessed in POOL.)
Metabolite Profiling in POOL
We performed polar metabolite profiling as previously described (15) (across 82 metabolites). Metabolites were normalized to internal standards. Metabolites below detection limit were counted as “0” in analyses.
Statistical Analysis
Clinical and demographic data are presented as mean and SD, with appropriate tests for intergroup comparison (Wilcoxon for continuous and χ2 for categorical). In the absence of formal glucose tolerance testing performed alongside ex-RNA measurements, we defined the degree of IR by plasma insulin level (16) (our primary IR measure); HOMA-IR was used as a secondary measure of IR. We recognize that these are crude measures of IR, but they are rapidly available and used in clinical practice. As described above, we included 391 ex-RNAs (297 miRNAs, 36 snoRNAs, and 58 piRNAs) in our analysis (Supplementary Table 1). Of note, because any given ex-RNA was not necessarily detectable in every FHS participant, models for insulin or HOMA-IR had a different number of study subjects for each ex-RNA (denoted in regression models in results). Of note, we specifically chose not to perform imputation (or set below–detection limit ex-RNA expression to “23,” the highest PCR cycle number possible on the Biomark system) to avoid bias.
Our first step was to identify ex-RNAs associated with IR. We constructed age-, sex-, and BMI-adjusted linear models to measure association of log-transformed insulin (primary) or HOMA-IR (secondary) with each ex-RNA (mean-centered and standardized). Given the multiple models constructed (one model for each ex-RNA), we used a false discovery rate (FDR) correction using the Benjamini-Hochberg method with a threshold of 0.05 (using PROC MULTTEST in SAS) pooling raw P values for all ex-RNAs together.
We next quantified the association of candidate ex-RNAs (associated with IR) with CT-defined regional adiposity and adipokines using linear models. Our primary analysis was based on imaging data available from the population used in the miRNA-insulin/HOMA relationships above (N = 2,317). In addition, we performed a sensitivity analysis to maximize population size (and power to detect association) across individuals with available imaging, anthropometric, or biochemical measures in the overall cohort (N = 2,763, including diabetes). In addition to BMI and waist circumference, we included several imaging-based measures of regional adiposity: visceral and subcutaneous fat volume (log-transformed), visceral-to-subcutaneous fat volume ratio (a measure of propensity to store fat viscerally, log-transformed), hepatic attenuation (a measure of hepatic steatosis), and visceral and subcutaneous fat attenuation (a measure of fat “quality” and adipose tissue metabolic function). Moreover, we examined the relationship of our candidate ex-RNAs with circulating adiponectin, IL-6, TNFRII, and triglyceride-to-HDL ratio (all biomarkers log-transformed). Of note, for IL-6 and TNFRII, 116 subjects (from the overall set of assayed samples) had samples run twice; we retained the higher value for this analysis arbitrarily. We adjusted all models for age and sex. As in previous models, we used an FDR correction to guard against multiple hypothesis testing.
To examine the role of selected ex-RNAs associated with IR in FHS on cardiometabolic dysfunction in a younger population, we studied the association of two candidate ex-RNAs consistently associated with IR-based phenotypes in FHS (miR-122 and miR-192) with IR in 90 obese/overweight individuals from POOL. We next analyzed association between miR-122 and insulin, independent of age, sex, BMI, and circulating metabolite profiles (as defined by principal components [PCs] of 81 polar metabolites, using varimax rotation).
All statistics were performed with SAS 9.3 software (SAS Institute, Cary, NC) or R (R Project, www.rproject.org) with a two-tailed P value <0.05 (with appropriate FDR thresholds, as noted) considered statistically significant.
Pathway Analysis and Network Visualization
Sixteen miRTarBase identifiers served as input to the Pathway Finder bioinformatics tool, which compares miRNA lists against a table of pathways, their miRNA elements, and the miRNAs that target their protein elements. The strategy of preannotating pathways with targeting miRNA is described by Godard and van Eyll (17). The pathways were sourced from WikiPathways (18). The code to generate the lookup table, the tables themselves, and the Pathway Finder tool are all freely available as open source code at https://github.com/nrnb/mirna-pathway-finder. The output of the tool is a ranked list of pathways with miRNA–protein target event counts. Gene identifier mapping was performed using BridgeDb databases (19) derived directly from Ensembl release 83. The mappings of miRNA-Entrez Gene targets were extracted from miRTarBase version 6.1 (20).
Sixteen miRTarBase identifiers were entered into the Target Interaction Finder tool, which compares miRNA lists against an XGMML representation of the miRTarBase database produced by CyTargetLinker (21). The database contains experimentally validated miRNA–gene target interactions. The tool outputs a new XGMML file that focused on target interactions involving the input list of miRNA. The XGMML file was then imported into Cytoscape (22) for further filtering and visualization. A gene list from the “insulin signaling” pathway was used to perform a selection within the complete miRNA gene target network, and first neighbors were also selected. This subnetwork was extracted as a representation of the mixed targeting events by the 16 miRNAs and 69 insulin signaling pathway genes.
Results
Baseline Characteristics of the FHS Offspring and POOL Cohorts
Selected clinical, demographic, and regional adiposity characterization of our analytic sample in the FHS Offspring Exam Cycle 8 (N = 2,317) and POOL study (N = 90) are shown in Table 1. Our cohort was elderly (mean age 66 years old), 56% female, and overweight (median BMI 27.7 kg/m2). The POOL cohort had a mean age of 15.5 years (range 4.6–25.5 years; 60% female), without diabetes, and with an average BMI percentile of 97% (mean BMI 33.8 kg/m2).
Table 1.
Clinical and biochemical characteristics in our study
| Variable | FHS Offspring Cohort |
POOL Youth Cohort |
||
|---|---|---|---|---|
| N | Value | N | Value | |
| Age (years) | 2,317 | 65.8 ± 8.9 | 90 | 15.5 ± 4.8 |
| Female sex, n (%) | 2,317 | 1,307 (56) | 90 | 54 (60) |
| Current smoking, n (%) | 2,314 | 191 (8) | — | — |
| BMI (kg/m2) | 2,313 | 27.7 ± 5.1 | 90 | 33.8 ± 10.0 (percentile: 97 ± 3) |
| Waist circumference (cm) | 2,302 | 97.1 ± 14.2 | — | — |
| Systolic blood pressure (mmHg) | 2,315 | 128 ± 17 | 90 | 110 ± 11 |
| Diastolic blood pressure (mmHg) | 2,313 | 74 ± 10 | 90 | 66 ± 8 |
| Glucose (mg/dL) | 2,317 | 100 ± 9 | 90 | 80.2 ± 6.9 |
| Biochemical indices* | ||||
| Triglycerides (mg/dL) | 2,317 | 113 ± 63 | 90 | 92.3 ± 55.6 |
| HDL cholesterol (mg/dL) | 2,316 | 59 ± 18 | 90 | 47.0 ± 11.5 |
| Insulin (pmol/L) | 2,317 | 69 ± 46 | 90 | 17.8 ± 13.3 |
| Hemoglobin A1c, % (mmol/mol) | 2,316 | 5.6 ± 0.3 (38) | — | — |
| IL-6 (pg/mL) | 2,246 | 2.58 ± 3.04 | — | — |
| TNFRII (pg/mL) | 2,314 | 2,592 ± 1,033 | — | — |
| Adiponectin (ng/mL) | 1,812 | 10.6 ± 6.3 | — | — |
| HOMA-IR | 2,317 | 2.51 ± 1.82 | 90 | 3.59 ± 2.95 |
| C-reactive protein | 2,315 | 3.2 ± 7.3 | 90 | 4.5 ± 9.6 |
| Regional adiposity | ||||
| Percent body fat (%) | — | — | 90 | 38.4 ± 10.3 |
| Liver attenuation (HU) | 1,089 | 65.9 ± 9.0 | — | — |
| Subcutaneous fat volume, cm3 | 1,061 | 2,961 ± 1,299 | — | — |
| Visceral fat volume, cm3 | 1,061 | 1,970 ± 1,022 | — | — |
| Subcutaneous fat attenuation (HU) | 1,061 | −101 ± 4.7 | — | — |
| Visceral fat attenuation (HU) | 1,061 | −93.7 ± 4.5 | — | — |
Data are mean ± SD unless otherwise noted. HU, Hounsfield units.
*All biochemical indices were measured at the 8th examination, except for adiponectin, which was measured at the 7th examination. Timing of adiposity measures discussed in text.
Identification of ex-RNAs Associated With IR and IR-Related Adiposity Phenotypes in FHS
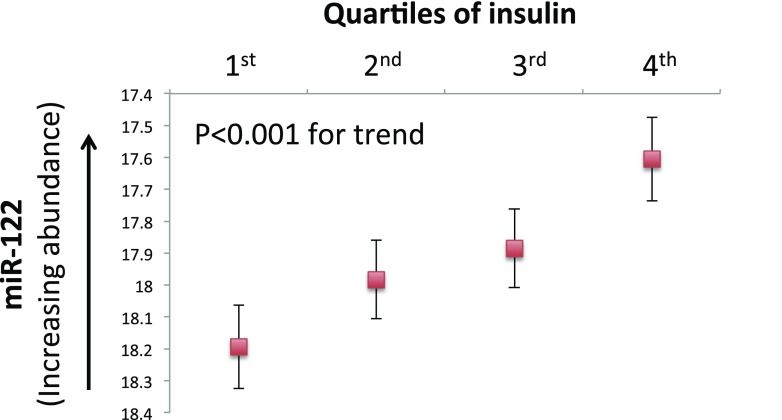
We constructed age-, sex-, and BMI-adjusted linear models to identify ex-RNAs associated with IR. From the overall panel of 391 ex-RNAs included in our analysis, we identified 16 miRNAs, 1 piRNA, and 1 snoRNA associated with insulin (Table 2), our primary outcome. An additional two ex-RNAs were associated with HOMA-IR, our secondary outcome. Of note, we observed a stepwise increase in plasma abundance of miR-122 across quartiles of insulin after adjustment for age, sex, and BMI (Fig. 1).
Table 2.
Ex-RNAs associated with IR
| Candidate ex-RNAs | Insulin (log-transformed) |
HOMA-IR (log-transformed) |
|||
|---|---|---|---|---|---|
| N | Estimated β | P value | Estimated β | P value | |
| miR-122-5p | 2,198 | 0.041 | 1.68 × 10−8 | 0.046 | 2.98 × 10−9 |
| miR-16-5p | 2,278 | 0.022 | 1.68 × 10−3 | 0.024 | 1.54 × 10−3 |
| miR-191-5p | 2,225 | 0.033 | 8.40 × 10−9 | 0.037 | 2.56 × 10−9 |
| miR-192-5p | 1,725 | 0.047 | 3.13 × 10−5 | 0.053 | 1.32 × 10−5 |
| miR-194-5p | 2,023 | 0.031 | 3.25 × 10−5 | 0.033 | 3.52 × 10−5 |
| miR-197-3p | 2,013 | — | — | 0.038 | 2.00 × 10−3 |
| miR-19b-3p | 2,230 | 0.034 | 3.08 × 10−5 | 0.037 | 2.58 × 10−5 |
| miR-24-3p | 2,220 | — | — | 0.032 | 1.00 × 10−3 |
| miR-301b-3p | 1,419 | 0.027 | 5.57 × 10−5 | 0.029 | 6.93 × 10−5 |
| miR-30d-5p | 2,221 | 0.030 | 1.26 × 10−4 | 0.033 | 7.39 × 10−5 |
| miR-320a | 2,208 | 0.029 | 1.07 × 10−3 | 0.035 | 2.87 × 10−4 |
| miR-320b | 1,665 | −0.015 | 2.87 × 10−4 | −0.016 | 1.98 × 10−4 |
| miR-342-3p | 2,246 | 0.037 | 1.09 × 10−4 | 0.045 | 1.48 × 10−5 |
| miR-4446-3p | 2,104 | −0.032 | 8.84 × 10−5 | −0.033 | 1.43 × 10−4 |
| miR-486-5p | 2,268 | 0.028 | 2.98 × 10−4 | 0.030 | 3.25 × 10−4 |
| miR-574-3p | 1,917 | 0.030 | 4.28 × 10−4 | 0.035 | 1.35 × 10−4 |
| miR-616-5p | 1,089 | 0.039 | 1.37 × 10−3 | 0.040 | 2.34 × 10−3 |
| miR-664b-3p | 1,678 | 0.014 | 1.95 × 10−5 | 0.015 | 2.23 × 10−5 |
| piRNA-48383 | 1,799 | −0.025 | 1.62 × 10−3 | −0.027 | 2.14 × 10−3 |
| snoRNA-1210 | 1,531 | 0.015 | 1.74 × 10−3 | 0.016 | 1.56 × 10−3 |
All models were adjusted for age, sex, and BMI. We accounted for multiple hypothesis testing with an appropriate prespecified 5% FDR threshold. N denotes number of observations in each model. An estimated β that is listed as “—” represents an ex-RNA that did not pass FDR. Each estimated β is change in log insulin or HOMA-IR per twofold increase (one PCR cycle change) in plasma ex-RNA concentration.
Figure 1.
Age, sex, and BMI-adjusted plasma abundance of miR-122 across quartiles of circulating insulin. Comparisons across all quartiles were statistically significant (after Bonferroni correction for type 1 error), except 1st vs. 2nd quartile and 2nd vs. 3rd quartile.
We next measured association among 18 insulin-associated ex-RNAs with metabolic phenotypes to begin to discern potential mechanisms by which these functional biomolecules may promote IR. As noted in research design and methods, for the analysis of miRNA–phenotype associations, we included individuals from our primary population (N = 2,317, no diabetes) and in a sensitivity analysis (across all N = 2,763 participants) in whom imaging or biochemical indices were available. The results of these models are shown in Supplementary Table 2A. In our primary analysis (from cohort excluding diabetes), we found consistent associations between a greater plasma abundance of miR-122 and greater BMI (P = 1.32 × 10−7), waist circumference (P = 8.41 × 10−6), visceral fat quantity (P = 5.72 × 10−7) and quality (P = 3.92 × 10−6), and lower liver attenuation (P = 2.51 × 10−5), but not subcutaneous fat (P = 0.005; did not survive FDR) or quality (P = 0.26). In addition, greater miR-122 was associated with increased TNFRII and triglyceride-to-HDL ratio. miR-122 was not significantly associated with adiponectin after FDR (β = −0.024 for log-adiponectin; P = 0.004; did not survive FDR).
We found similar results for miR-122 when the entire population was considered (Supplementary Table 2B). In addition, we observed that miR-192 was consistently associated with several cardiometabolic phenotypes, including BMI, waist circumference, and liver attenuation, but not subcutaneous fat. In addition, across the overall population, we observed greater miR-122 and miR-192 associated with lower adiponectin.
miRNAs Associated With IR in the FHS Target-Relevant Signaling Pathways
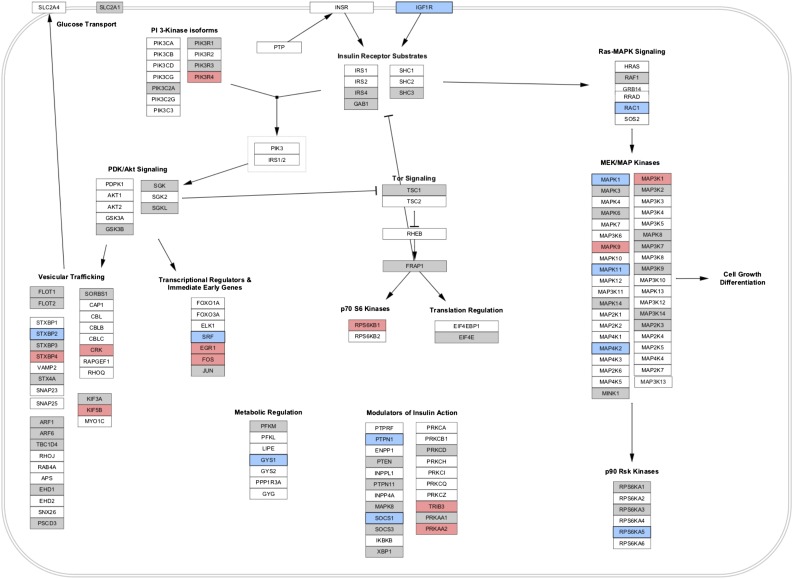
We next performed a pathway analysis to address whether the 16 miRNAs associated with insulin in FHS target pathways were relevant to IR. We identified “insulin signaling” as a pathway targeted by all 16 miRNAs (Supplementary Table 3). Interactions among all 16 target miRNAs and genes in the insulin signaling pathway are shown in Supplementary Fig. 1, suggesting significant cross-targeting of multiple IR-related genes by multiple miRNAs identified by association. In addition, we visualized miRNA gene expression targeting events from selected pathways (as described in research design and methods) to identify functional targets of miR-122 (denoted in blue), miR-192 (denoted in red), and the other 14 miRNAs (denoted in gray) on insulin signaling (Fig. 2 and Supplementary Fig. 2). We selected four specific pathways given their importance in IR: 1) “insulin signaling” (shown in Fig. 2); 2) “factors and pathways affecting insulin-like growth factor signaling”; 3) target of rapamycin signaling; and 4) AMPK signaling (all shown in Supplementary Fig. 2). Of note, several genes targeted by miR-122 had been previously implicated in pathogenesis of IR, including protein tyrosine phosphatase, nonreceptor type 1 (also called PTP1B) (23), mitogen-activated protein (MAP) kinases (24), and AMPK (25).
Figure 2.
Visualization of selected miRNA targeting events on the insulin signaling pathway (from WikiPathways). The genes targeted by miRNA per pathway, as counted in Supplementary Table 3, are visualized in this figure for selected pathways. Pathway was imported into Cytoscape from WikiPathways, and ID mapping was performed to obtain Entrez Gene identifiers for each gene. An intermediate file from the Pathway Finder tool was parsed and imported into Cytoscape to supply the mappings between Entrez Gene and the selected set of miRNAs. A visual style was defined in Cytoscape to highlight any gene targeted by these miRNAs in preferential order: miR-122 (blue), miR-192 (red), and any of the other 14 possible miRNAs (gray). Other selected pathways from WikiPathways are shown in Supplementary Fig. 1.
Given consistent association with IR, adiposity, and pathways involved in IR, we carried forward miR-122 and miR-192 to the POOL cohort.
miR-122 Is Associated With Metabolic Phenotypes in Youth
After adjustment for age, sex, and BMI, a higher miR-122 (but not miR-192) abundance was associated with greater IR, as measured by insulin (β = 0.12 log change per twofold increase in plasma miR-122; P = 0.004) and HOMA-IR (β = 0.12 log change per twofold increase in miR-122; P = 0.006). Neither miR-122 nor miR-192 was associated with hs-CRP or percent body fat in POOL, but they were associated with triglyceride-to-HDL ratio (miR-122: β = 0.14, P = 0.002; and miR-192: β = 0.14, P = 0.02, respectively). We found three PCs that explained 52.5% variance in the polar metabolome assayed. Of note, the first PC was highly loaded on several different metabolites implicated in IR, including leucine, isoleucine, and phenylalanine (branched-chain and aromatic amino acids). To assess whether miR-122 had a metabolism-independent association with IR, we estimated the association of insulin with miR-122, adjusted for age, sex, BMI, and all three metabolite PCs. Greater miR-122 was associated with greater insulin (β = 0.10 log change in insulin per twofold increase in miR-122; P = 0.005), independent of age, sex, BMI, or metabolite profile.
Conclusions
In a large community-based population of adults, we identify circulating ex-RNAs that are associated with markers of IR and adiposity, independent of age, sex, and BMI. Specifically, miR-122 was consistently related to dysfunctional adiposity phenotypes previously demonstrated to influence downstream cardiometabolic risk, including visceral and hepatic fat and selected adipokines and inflammatory mediators. In a separate cohort of 90 overweight/obese youth without diabetes, we demonstrated that miR-122 was associated with IR independent of metabolite profile (via metabolomics), age, sex, or BMI, suggesting that ex-RNAs may have a role in IR independent of emerging metabolic markers of IR. Based on in silico pathway analyses for the 16 miRNAs found in FHS (of total 18 ex-RNAs), we found that the identified miRNAs targeted several key pathways previously implicated in IR (including mammalian target of rapamycin, insulin signaling, and AMPK), with significant cross-targeting of multiple IR-related genes by multiple miRNAs (Fig. 2). Specifically, miR-122 targeted several genes previously implicated in IR, including genes involved in muscle responses to insulin (e.g., PRKAB1, a subunit of AMPK, a critical regulator of metabolism in IR [26]). Collectively, these findings provide translational support for a role of ex-RNAs (specifically miR-122) in IR across weight class, metabolism, and age, calling for further mechanistic investigation to delineate a role for ex-RNAs in the metabolic architecture of IR.
There is increasing recognition that specific ex-RNAs function in pathways critical to obesity and metabolic disease, including adipocyte differentiation, angiogenesis, hepatic steatosis, oxidative stress, and inflammation. Furthermore, adipocyte-derived miRNAs can mediate cross talk with circulating macrophages (27) or hepatocytes (28), or in muscle tissue (29), altering mRNA expression of key intermediates involved in IR. These findings implicate ex-RNAs as functional biomarkers that may orchestrate high-level transcriptional and metabolic control in humans. Accordingly, there has been a surge in translational investigation in this area, demonstrating involvement of miRNAs in brown/white fat specification and adipose tissue inflammation (30), pancreatic β-cell function (31), and hepatic steatosis (32). Human investigation in both children (6) and adults (3) has demonstrated several miRNAs dysregulated in obesity and progression of cardiometabolic disease, with dynamic changes in miRNAs during weight loss. Despite these important advances, most studies have been limited by sample size, profile preselected miRNA candidates (excluding piRNAs or snoRNAs), and restrict their population of interest to obesity. Given the important differences between animal models of diabetes or obesity and human disease, large-scale human translational data alongside detailed obesity-related phenotypes (e.g., regional adiposity) are critical.
In this report, we identify ex-RNAs associated with IR in the FHS, demonstrating associations with key phenotypes central to IR, including adipokines, inflammation, and regional adiposity, specifically miR-122. Using pathway analysis across all 16 miRNAs with curated pathways (of the total 18 ex-RNAs), we found that these miRNAs targeted genes involved in central pathways of IR, with some genes targeted by multiple miRNAs. These translational findings are in keeping with several prior reports from smaller cohorts on the significance of miR-122 in obesity and IR pathogenesis (33). miR-122 is dynamically regulated during surgical weight loss and is associated with hepatic steatosis (32,34), with a near abolition of circulating miR-122 levels after bariatric surgery (35). Seminal work by Esau et al. (36) demonstrated that direct antisense-mediated silencing of miR-122 caused a reduction in hepatic steatosis, decreased circulating cholesterol levels, and global shifts in lipid metabolism. Taken in concert with our findings of an association between miR-122 and CT-determined hepatic fat in Framingham, these findings suggest a potential role for miR-122 in hepatic steatosis, a major comorbidity involved in IR pathogenesis. In addition, in pathway analyses, miR-122 appeared to target PRKAB1, a member of the AMPK pathway, a regulator of IR in muscle, suggesting that liver-derived miR-122 may target remote metabolically active tissues as an endocrine mediator of disease. Indeed, miR-122 has been found in circulating exosomes (37) that may transfer epigenetic information across tissue types. Finally, the finding of an association between IR and non-miRNA ex-RNAs (e.g., piRNA and snoRNA) is intriguing, as the role of non-miRNA ex-RNAs (e.g., piRNAs) in metabolic diseases is just beginning to be clarified: several non-miRNA species (e.g., piRNAs) have been recently shown to transfer epigenetic information from sperm to egg (38). Ultimately, although these human translational findings are associational in nature, they motivate further mechanistic research into ex-RNA biogenesis and the role of ex-RNAs in cell–cell communication and target organ metabolic signaling.
The limitations of our study should be viewed in light of its design. We focused on ex-RNAs commonly abundant in plasma of FHS participants, limiting discovery of low-abundance ex-RNAs. In addition, we included those participants with expressed levels of each miRNA in regressions for insulin (not “imputing” values for miRNAs that were below detection limit). Although this limits conclusions to participants in whom each miRNA was expressed, complex traits like IR are likely influenced by variation in common epigenetic factors. Although CT measures or biomarkers were not present in all FHS participants with ex-RNA quantification (a potential source of bias), the observed associations are strong in the largest cohort studied so far in metabolic disease. Finally, use of gold-standard methods to determine insulin physiology in individuals with diabetes (e.g., clamp) will be important in future studies to extend our results to diabetes.
In conclusion, we identified a group of plasma-circulating ex-RNAs related to IR and related adiposity, inflammatory, and metabolic phenotypes. These associations are independent of sex and BMI and conserved across age, suggesting an age-independent role for ex-RNAs in integrating metabolic inputs in IR. Selected miRNAs associated with IR target genes implicated in IR in muscle and may have a functional, trans-organ role in mediating IR. These results provide large-scale human translational epidemiologic data to support a role for ex-RNAs in IR and its metabolic consequences. Future investigation into specific mechanisms and modulation of ex-RNA biology to reduce the burden of diabetes is warranted.
Supplementary Material
Article Information
Acknowledgments. The authors thank the participants of the POOL and FHS, as well as the countless volunteers and administrative staff without whom this work would not be possible. The authors also thank Dr. Aifeng Zhang and Dr. Fei Wang (Massachusetts General Hospital, Boston, MA) for assistance in experimentation. Data from the ex-RNAs measured in the FHS have been deposited in the National Institutes of Health Database of Genotypes and Phenotypes.
Funding. This work was supported by the American Heart Association (14FTF19940000 to R.S.), the Harvard Catalyst (UL1-TR-001102), the Thrasher Research Fund (to R.S.), the National Institutes of Health (K23-HL-127099), the National Institutes of Health Common Fund Extracellular RNA Communication Consortium (UH3-TR-900921 to J.E.F. and UH3-TR-000901 to S.D.), the Milton Foundation, New Balance Foundation Obesity Prevention Center at Boston Children’s Hospital, and Boston Children’s Hospital (for the POOL study to S.O.). The FHS is funded by National Institutes of Health contract N01-HC-25195.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. R.S., V.M., M.P., M.G.L., and E.M. performed statistical analyses. R.S., V.M., M.P., K.T., S.D., and J.E.F. wrote the manuscript. M.P. and E.F. performed metabolomics analyses. K.T. and J.E.F. performed RNA quantification. K.H. and A.P. performed bioinformatics analyses. D.L. performed data collection within FHS. U.H. performed CT analysis in FHS. S.O. and S.d.F. gathered data and samples in the POOL study. S.D. and J.E.F. jointly supervised the work. All authors provided critical revision of the manuscript. R.S., V.M., S.D., and J.E.F. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc16-1354/-/DC1.
The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University, and its affiliated academic health care centers or the National Institutes of Health.
References
- 1.Neeland IJ, Turer AT, Ayers CR, et al. Dysfunctional adiposity and the risk of prediabetes and type 2 diabetes in obese adults. JAMA 2012;308:1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol 2015;11:276–288 [DOI] [PubMed] [Google Scholar]
- 3.Ortega FJ, Mercader JM, Catalán V, et al. Targeting the circulating microRNA signature of obesity. Clin Chem 2013;59:781–792 [DOI] [PubMed] [Google Scholar]
- 4.Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Surgery-induced weight loss is associated with the downregulation of genes targeted by microRNAs in adipose tissue. J Clin Endocrinol Metab 2015;100:E1467–E1476 [DOI] [PubMed] [Google Scholar]
- 5.Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care 2014;37:1375–1383 [DOI] [PubMed] [Google Scholar]
- 6.Prats-Puig A, Ortega FJ, Mercader JM, et al. Changes in circulating microRNAs are associated with childhood obesity. J Clin Endocrinol Metab 2013;98:E1655–E1660 [DOI] [PubMed] [Google Scholar]
- 7.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med 1975;4:518–525 [DOI] [PubMed] [Google Scholar]
- 8.Mahabadi AA, Massaro JM, Rosito GA, et al. Association of pericardial fat, intrathoracic fat, and visceral abdominal fat with cardiovascular disease burden: the Framingham Heart Study. Eur Heart J 2009;30:850–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutter MK, Wilson PW, Sullivan LM, Fox CS, D’Agostino RB Sr, Meigs JB. Use of alternative thresholds defining insulin resistance to predict incident type 2 diabetes mellitus and cardiovascular disease. Circulation 2008;117:1003–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hivert MF, Manning AK, McAteer JB, et al. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes 2008;57:3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenquist KJ, Pedley A, Massaro JM, et al. Visceral and subcutaneous fat quality and cardiometabolic risk. JACC Cardiovasc Imaging 2013;6:762–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah RV, Allison MA, Lima JA, et al. Liver steatosis and the risk of albuminuria: the multi-ethnic study of atherosclerosis. J Nephrol 2015;28:577–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenquist KJ, Massaro JM, Pedley A, et al. Fat quality and incident cardiovascular disease, all-cause mortality and cancer mortality. J Clin Endocrinol Metab 2015;100:227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freedman JE, Gerstein M, Mick E, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun 2016;7:11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 2015;162:540–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laakso M. How good a marker is insulin level for insulin resistance? Am J Epidemiol 1993;137:959–965 [DOI] [PubMed] [Google Scholar]
- 17.Godard P, van Eyll J. Pathway analysis from lists of microRNAs: common pitfalls and alternative strategy. Nucleic Acids Res 2015;43:3490–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutmon M, Riutta A, Nunes N, et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res 2016;44:D488–D494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Iersel MP, Pico AR, Kelder T, et al. The BridgeDb framework: standardized access to gene, protein and metabolite identifier mapping services. BMC Bioinformatics 2010;11:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 2016;44:D239–D247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutmon M, Kelder T, Mandaviya P, Evelo CT, Coort SL. CyTargetLinker: a cytoscape app to integrate regulatory interactions in network analysis. PLoS One 2013;8:e82160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stull AJ, Wang ZQ, Zhang XH, Yu Y, Johnson WD, Cefalu WT. Skeletal muscle protein tyrosine phosphatase 1B regulates insulin sensitivity in African Americans. Diabetes 2012;61:1415–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CC, Goalstone ML, Draznin B. Molecular mechanisms of insulin resistance that impact cardiovascular biology. Diabetes 2004;53:2735–2740 [DOI] [PubMed] [Google Scholar]
- 25.Ruderman NB, Carling D, Prentki M, Cacicedo JM. AMPK, insulin resistance, and the metabolic syndrome. J Clin Invest 2013;123:2764–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Long YC, Zierath JR. AMP-activated protein kinase signaling in metabolic regulation. J Clin Invest 2006;116:1776–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogawa R, Tanaka C, Sato M, et al. Adipocyte-derived microvesicles contain RNA that is transported into macrophages and might be secreted into blood circulation. Biochem Biophys Res Commun 2010;398:723–729 [DOI] [PubMed] [Google Scholar]
- 28.Sharma H, Estep M, Birerdinc A, et al. Expression of genes for microRNA-processing enzymes is altered in advanced non-alcoholic fatty liver disease. J Gastroenterol Hepatol 2013;28:1410–1415 [DOI] [PubMed] [Google Scholar]
- 29.Wang YC, Li Y, Wang XY, et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia 2013;56:2275–2285 [DOI] [PubMed] [Google Scholar]
- 30.Karbiener M, Scheideler M. MicroRNA functions in brite/brown fat - novel perspectives towards anti-obesity strategies. Comput Struct Biotechnol J 2014;11:101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandez-Valverde SL, Taft RJ, Mattick JS. MicroRNAs in β-cell biology, insulin resistance, diabetes and its complications. Diabetes 2011;60:1825–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Becker PP, Rau M, Schmitt J, et al. Performance of serum microRNAs -122, -192 and -21 as biomarkers in patients with non-alcoholic steatohepatitis. PLoS One 2015;10:e0142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, Hong J, Cao Y, et al. Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur J Endocrinol 2015;172:291–300 [DOI] [PubMed] [Google Scholar]
- 34.Yamada H, Suzuki K, Ichino N, et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta 2013;424:99–103 [DOI] [PubMed] [Google Scholar]
- 35.Wu Q, Li JV, Seyfried F, et al. Metabolic phenotype-microRNA data fusion analysis of the systemic consequences of Roux-en-Y gastric bypass surgery. Int J Obes 2015;39:1126–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Esau C, Davis S, Murray SF, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 2006;3:87–98 [DOI] [PubMed] [Google Scholar]
- 37.Bala S, Petrasek J, Mundkur S, et al. Circulating microRNAs in exosomes indicate hepatocyte injury and inflammation in alcoholic, drug-induced, and inflammatory liver diseases. Hepatology 2012;56:1946–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donkin I, Versteyhe S, Ingerslev LR, et al. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab 2016;23:369–378 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.