Abstract
Molecular chaperones are thought to inhibit off-pathway interactions such as aggregation from occurring without influencing the on-pathway formation of native structure. Here, we present a mechanism whereby the family of PapD-like chaperones, which are involved in the formation of adhesive pili in pathogenic bacteria, function by suppressing aggregation while simultaneously catalyzing the folding of subunits that make up the pilus. We also show that the Arg-8 residue, invariant in the cleft of all known PapD-like chaperones, makes up part of the active site of the chaperone. The data argue for a temporal mechanism of catalyzed folding. The terminal carboxylate group of a pilus subunit anchors to the active site of the chaperone by hydrogen bonding. This bonding spatially fixes the COOH terminus of the subunit in the correct context for β-sheet formation, using the edge of the NH2-terminal domain of the chaperone as a nucleation site.
Keywords: PapD, PapE, PapD–PapE complex, aggregation
Many Gram-negative pathogenic bacteria assemble adhesive fibers called pili on their surface by a mechanism termed “donor strand complementation/exchange” through the chaperone/usher pathway (1–4). All pili assembled by chaperone/usher pathways are composed of a variety of subunit types having specialized functions, all of which possess incomplete Ig-like folds demarcated by the absence of a COOH-terminal seventh β-strand (1, 3, 5). In the pilus fiber, the Ig fold of every subunit is completed by an NH2-terminal extension donated by a neighboring subunit in a process known as donor strand exchange (3, 6). Recent studies have begun to reveal that pili have multiple functions that participate in complex host–pathogen interactions, which ultimately can determine the outcome of the infectious process (7–9).
The P pilus system is one of the best-characterized fibers assembled by the chaperone/usher pathway and is produced by pyelonephritic strains of Escherichia coli (10, 11). The P pilus contains a specialized two-domain adhesin at its tip that is called “PapG” (12). The NH2-terminal domain of PapG recognizes a globoside host receptor with stereochemical specificity, which is a critical event in pathogenesis (12). The COOH-terminal domain is a pilin domain that functions to link the PapG adhesin to the tip of the pilus fiber by the donor strand-exchange mechanism. The two-domain structure of the PapG adhesin likely represents a paradigm for adhesive pili assembled by chaperone/usher pathways (12). PapF serves as an adaptor protein, donating an N-terminal extension to complete the Ig fold of the pilin domain of PapG, thus connecting it to a tip fibrillum structure, which is composed of repeating subunits of PapE arranged in an open helical configuration (13, 14). PapK connects the tip fibrillum to the pilus rod, which is composed of repeating subunits of PapA arranged in a right-handed helical configuration (13–15).
In the P pilus system, PapD is the periplasmic chaperone and PapC is the outer membrane usher. Periplasmic chaperones, like PapD, are small (25 kDa) two-domain molecules with the domains oriented together in the shape of a boomerang (16, 17). The chaperones bind to and form stable complexes with each of the pilus subunits to facilitate pilus biogenesis (18, 19). The structural basis of how periplasmic chaperones interact with pilus subunits has been defined by x-ray crystallography (1, 3, 20). The absence of a seventh G β-strand results in a deep groove on the surface of the pilin that exposes its hydrophobic core because of the incomplete Ig-like fold. The terminal carboxylate group of the subunit hydrogen-bonds to Arg-8 and Lys-112 in the cleft of the chaperone (21). Arg-8 and Lys-112 are invariant in the cleft of all known PapD-like chaperones, of which there are hundreds of members (17, 22), and mutations in these residues completely abolish chaperone function (14, 21, 23) (Fig. 3B Inset). The edge G1 β-strand, which lines the cleft side of the NH2-terminal domain of the chaperone, fills the subunit groove and partially completes the Ig fold of the subunit in the process of donor strand complementation (24). The chaperone remains bound to the subunit to prevent subunit aggregation at the wrong time and place in the cell (18). Chaperone–subunit complexes are then specifically targeted to the usher, facilitating the donor strand-exchange process, whereby the G β-strand of the chaperone is exchanged with the free NH2-terminal strand of another subunit (13, 25). The orientation of the G β-strand changes with respect to the subunit from a parallel orientation to the subunit F strand when the G strand is donated by the chaperone to an antiparallel orientation when the G strand is donated by a neighboring subunit (20). This change in orientation is accompanied by a collapse of the subunit from a more expanded conformation when bound to the chaperone to a more collapsed state when bound to a subunit's NH2-terminal strand (Fig. 4). It is therefore only after this exchange process that the subunit actually completes its folding pathway (20).
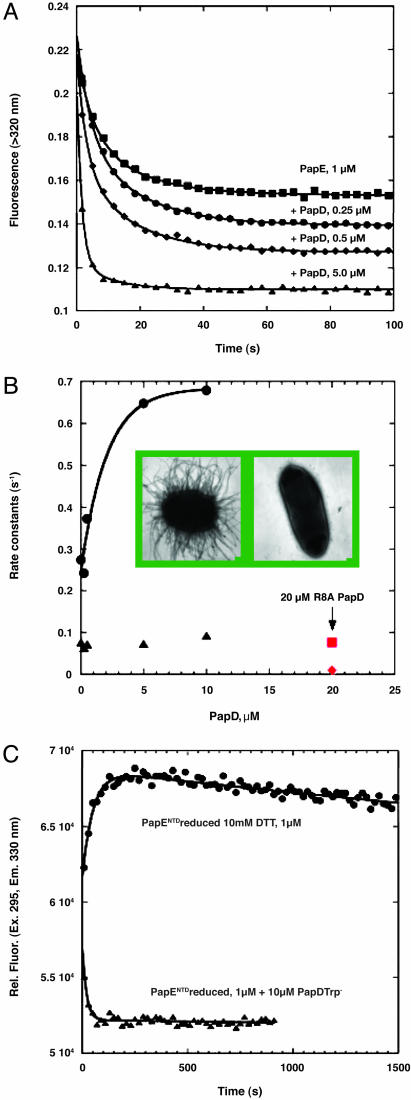
Fig. 3.
The effect of PapD on the refolding properties of PapENTD.(A) Kinetics of refolding of PapENTD after a 1:11 dilution (1 μM final concentration) from 5 to 0.45 M urea in the absence of PapDTrp- (▪) and in the presence of 0.25 (•), 0.5 (♦), and 5 (▴) μM PapDTrp-. The data were fit to a sum of two exponentials, and the fits are the solid lines through the data. (B) The rate constants for the two phases of refolding of PapENTD as a function of concentration of PapDTrp- (• and ▴) and in the presence of 20 μM PapDR8ATrp- (red square and diamond). Inset shows bacteria (MC4100) expressing the pap operon encoded in the plasmid pDH1 (papD-) complemented with wild-type papD in pLS101 (Left) or with papDR8A (Right) (23). (C) Kinetics of refolding of reduced PapENTD (1 μM) in the absence (•) and presence (▴) of PapDTrp- (10 μM). The data for the refolding in the absence of PapDTrp- were fit to a sum of two exponentials, and the data with PapDTrp- were fit to a single exponential and are the solid lines through the data. All experiments were performed in 20 mM Mes/OH (pH 6) at 20°C with 10 mM DTT.
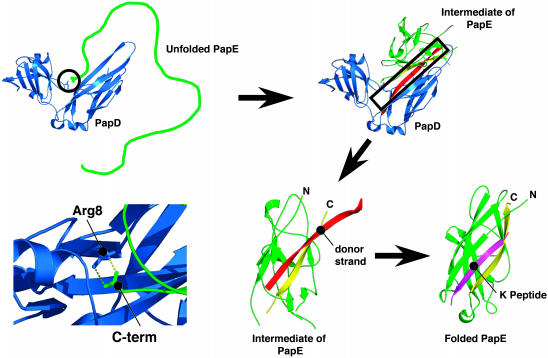
Fig. 4.
Model of refolding of pilus subunits. The figure was generated with pymol. PapD is shown in blue, and the subunit PapE is shown in green. The model shows that the initial interaction between the chaperone and the subunit is the anchoring of the carboxyl group to the guanidinium group of Arg-8. After this, the subunit folds to an intermediate state that is in an expanded conformation. Only after donor strand exchange with the NH2-terminal strand of an incoming subunit [in this case a peptide from the NH2 terminus of PapK (purple)] does the subunit complete its folding pathway (20).
The absence of the seventh C-terminal β-strand in pilus subunits has long been thought to explain the inability of subunits to fold without the help of a periplasmic chaperone (6). Some reports have contested that the chaperone assists in subunit-folding (26, 27). Those reports argued that the subunits fold in a chaperone-independent fashion and that the function of the chaperone was to bind to folded subunits to prevent aggregation. However, those studies (26, 27) did not take into account the effect of the conserved disulfide bond that exists in pilus subunits, and thus the denaturing conditions used were not sufficient to unfold FimH as shown by Barnhart et al. (6). Here, we used conditions that completely denature subunits and reduce the disulfide bond to show that the mechanism by which PapD inhibits aggregation is also part of a mechanism by which PapD acts as a template to catalyze folding. We also demonstrated that the Arg-8 cleft residue serves as part of the active site of the chaperone by maintaining the subunit in the correct register and allowing the chaperone to serve as a rate-enhancing template.
Materials and Methods
Protein Expression and Purification. PapD(WT-6-His)-PapENTD was purified by methods described by Sauer et al. in ref. 20. (PapENTD is an NH2-terminal-deleted form of PapE in which residues 2–12 have been removed.) Buffer for all conditions was 20 mM 2-(N-morpholino)ethanesulfonic acid (Mes) (pH 6.0, Sigma). Urea concentrations were determined with a refractometer. PapDTrp36Phe/Trp128Phe (PapDTrp-) was made by using the splicing by overlap extension technique and was purified as described in ref. 28. Concentrations of PapENTD and PapDTrp- were determined by using extinction coefficients of 13,850 M-1·cm-1 and 13,600 M-1·cm-1, respectively.
Urea Denaturation. The stability of PapD and PapDTrp- was measured as a function of urea in 30 mM Mops/HCl (pH 7.0) at 233 nm by using a J-710 CD spectropolarimeter (Jasco, Easton, MD) at 20°C. Concentrations were 20 and 26 μM for PapD and PapDTrp-, respectively, as determined by amino acid analysis. The stability of PapENTD was measured by using fluorescence at a 295-nm excitation in a spectrofluorometer (Photon Technology International, Lawrenceville, NJ) and by CD in the Jasco J-710. PapENTD, initially in 5 M urea and 20 mM Mes/OH (pH 6.0), was serially diluted into buffer (20 mM Mes/OH, pH 6.0) to 1 μM final concentration or into buffer containing PapDTrp- (also 1 μM final concentration). For CD measurements, PapENTD was added to increasing concentrations of urea at a final protein concentration of 0.5 mg/ml. Buffer baseline and PapDTrp- were recorded along with the samples and subtracted. Urea concentrations at each data point were measured with a refractometer.
Light Scattering. Light scattering of PapENTD was measured by using a spectrof luorometer (Photon Technology International) with excitation and emission wavelengths set at 500 nm. PapENTD (88 μM) and urea (5 M) were diluted into a cuvette (3 ml) with a magnetic stir bar to final concentrations of 8.8 and 0.88 μM, respectively. For experiments with PapDTrp-, the final concentration of PapDTrp- in the refolding buffer was 8.4 μM.
Refolding of PapENTD. Refolding of PapENTD was measured by using manual mixing by fluorescence (excitation wavelength at 295 nm) or by stopped-flow fluorescence spectroscopy. Stopped-flow measurements were recorded with a cutoff filter (320 nm) on a stopped-flow fluorescence apparatus (Applied Photophysics, Surrey, U.K.). Refolding was recorded after a urea jump from 5 to 0.45 M (1:11 dilution) at 20°C to a final concentration of 1 μM.
Refolding in the Presence of PapDTrp-. PapDTrp--catalyzed refolding of PapENTD was measured by using stopped-flow fluorescence spectroscopy, with an excitation wavelength of 295 nm and a cutoff filter of 320 nm. Measurements were recorded in 20 mM Mes/OH (pH 6.0) at 20°C. The concentrations of PapDTrp- and PapDTrp- R8A varied from 3 to 20 μM. Baselines were recorded with experiments of PapD alone and substracted. Refolding of reduced PapENTD (1 μM) was done manually in 20 mM Mes/OH (pH 6) and 10 mM DTT by using a spectrofluorometer (Photon Technology International) with the excitation wavelength set at 295 nm and emission at 330 nm. The final concentration of PapDTrp- was 10 μM. Data were fitted by using kaleidagraph (Synergy Software, Reading, PA).
Results and Discussion
To determine whether the chaperone facilitated subunit-folding, we monitored the changes in the intrinsic fluorescence of Trp-24 in the tip fibrillum subunit PapE during the folding process in the presence and absence of PapD (Fig. 2). A mutant of PapD in which both tryptophans, Trp-36 and Trp-128, were changed to phenylalanine (PapDTrp-) allowed us to monitor solely the fluorescence of Trp-24 in PapE, even in the presence of PapD. The stability of the PapDTrp- mutant toward the denaturant was slightly less than that of the wild-type protein as determined by CD spectroscopy [ΔG° (kcal/mol), m (cooperativity index) = 9.3, 3.3; ΔG°, m (wild type) = 6.2, 1.8 (data not shown) (29, 36)]. However, this mutant was able to complement a papD- mutation in the pap gene cluster to restore pilus assembly [assayed by measuring haemagglutination titers of human red blood cells (haemagglutination titer = 64 in both Trp- and wild-type strains) and by the presence of pili as determined by electron microscopy].
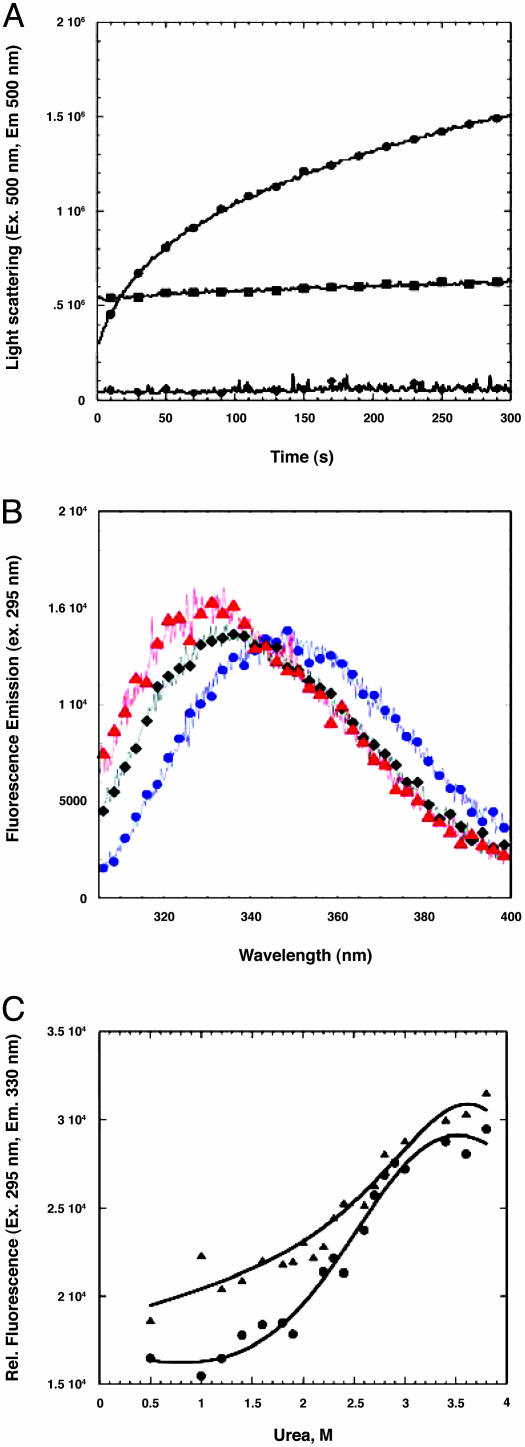
Fig. 2.
Refolding properties of PapENTD. (A) Light scattering after a 1:10 dilution of PapENTD from 5 M urea, in the absence (•) and presence (▪) of 8.4 μM PapDTrp- (final concentration of 8.8 μM subunit) and at 0.88 μM PapENTD (♦) in the absence of PapDTrp-. (B) Emission spectra of PapENTD (1 μM) in 5 (blue circle) and 0.5 (♦) M urea and in the presence of 10 μM PapD (red triangle). Spectra were recorded by using an excitation wavelength of 295 nm. (C) Equilibrium refolding of PapENTD as a function of urea in the absence (▴) and presence (•) of PapDTrp-. Fluorescence was measured by using an excitation wavelength of 295 nm and an emission maxima of 330 nm. All experiments were performed in 20 mM Mes/OH (pH 6) at 20°C.
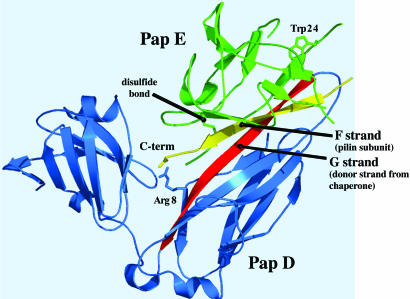
To measure the effect of PapD on PapE-folding, we used PapENTD. Removal of residues 2–12 from PapE has been shown to be sufficient to prevent subunit–subunit associations that occur during donor strand exchange (20). As with the PapD–PapK and FimC–FimH structures, the PapENTD subunit is an Ig-like fold that is incomplete, missing the seventh G β-strand (Fig. 1). The chaperone donates its own G1 β-strand from the NH2-terminal domain to complete the Ig fold of the subunit (20).
Fig. 1.
X-ray structure of PapD(6-His)-PapENTD (20). PapD is in blue, and the subunit PapENTD is in green, with the disulfide and tryptophans highlighted. The G strand of the chaperone that occupies the groove of the subunit is shown in red, and the C-terminal strand of the subunit that binds to Arg-8 in the chaperone cleft is shown in yellow. The figure was generated with pymol (Delano Scientific, San Carlos, CA).
The PapD–PapENTD complex was purified as described in ref. 20, and PapENTD was separated from wild-type PapD in 5 M urea on a nickel chelate column. In 5 M urea, PapENTD is unfolded as determined by both CD and fluorescence, because no further spectral changes occur at urea concentrations up to 8 M (data not shown). Previous attempts to refold P pilus subunits from urea were unsuccessful because of the formation of a visible precipitate (6). Upon dilution of the urea-denatured PapENTD into buffer (1:11, 5–0.45 M urea), we observed a time-dependent increase in light scattering at concentrations above ∼8 μM, indicating aggregation of the subunit (Fig. 2A). However, no increase in light scattering was observed upon dilution of the subunit into buffer containing an equivalent amount of PapDTrp- or when the concentration of PapENTD was lowered to ≤1 μM. The loss of light scattering in the presence of PapD suggests that PapD prevents the aggregation process, consistent with its role as a molecular chaperone.
The light-scattering data at low concentrations of PapENTD suggested that we could monitor PapENTD-refolding at low concentrations (≤1 μM). Fig. 2B shows the spectral changes by fluorescence for PapENTD at 1 μM and indicates that there is a significant change in fluorescence upon dilution to low urea concentrations (excitation wavelength = 295 nm, emission maxima = 350 nm at 5 M urea, emission maxima = 335 nm at 0.5 M urea). These spectral changes are consistent with the formation of native-like structure. There is only a slight further change in the emission maxima in the presence of an equivalent amount of PapD (emission maxima = 330 nm), indicating that the subunit may achieve a more native-like structure in the presence of PapD but that this change is likely to be small. In equilibrium refolding experiments, we recorded the fluorescence after dilution of PapENTD (in 5 M urea) to lower urea concentrations. In these experiments, we observed a weakly cooperative transition, indicating that some structure can form at 1.5 M urea or at lower concentrations (Fig. 2C). In the presence of PapD, the cooperativity of the transition increased, but it was unclear whether this was due to an increase in stability or an effect on the aggregation state of the subunit at lower urea concentrations.
We used stopped-flow fluorescence to monitor the kinetics of folding in the presence and absence of PapD. The kinetics of refolding in the absence of the chaperone (Fig. 3A) indicated that the folding process consisted of two observable phases (t1/2 = 3.7 and 13.5 s, respectively). The kinetics observed in the presence of PapDTrp- (Fig. 3A) revealed that the rate of the faster phase increased initially with PapD concentration (t1/2 = 1.0 s at 5 μM PapD) and then remained virtually unchanged at higher concentrations (up to 10 μM; Fig. 3B). The rate of change of the slower phase remained constant, irrespective of the presence of PapD (Fig. 3B).
Although the rate constants for refolding increased, we also observed an increase in the total amplitude of the fluorescence change (Fig. 3A). This increase in amplitude is consistent with a possible loss of aggregation of PapENTD, even at these low protein concentrations. In the presence of increasing PapD, the aggregation goes down, and, at the same time, the rate of folding is increased. Another possibility is that in the absence of PapD, the subunit collapses to an intermediate form, and that in the presence of PapD, a more folded conformation is achieved. The change in the fluorescence spectrum of PapENTD in the presence of PapD (Fig. 2B) is consistent with this hypothesis.
To confirm that the observed changes for refolding were due to a specific effect from PapD, we repeated the above refolding experiments in the presence of an Arg-8 mutant of PapDTrp- in which Arg-8 was changed to alanine (Trp-/Arg8Ala). Arg-8 is an invariant residue within the superfamily of chaperones, and mutation to alanine completely abolishes pilus assembly because of a loss of subunit binding (Fig. 3B Inset) (6, 23). At a final concentration of 20 μM, little to no enhancement in the refolding rate is observed in the presence of the Arg8Ala mutant (Fig. 3B).
PapENTD has a single disulfide bond formed between Cys-15 and Cys-48 in the mature form, and Cys-15 forms part of the A strand, which makes contact with the G1 strand of the chaperone (Fig. 1). This disulfide may provide an artificial nucleus for folding. To determine the importance of the disulfide on the folding of PapENTD, we conducted the same experiments under reducing conditions (5 M urea and 10 mM DTT). The refolding in the reduced state alone is a very slow process, with two phases corresponding to a t1/2 of ∼41 s and ∼7,000 s (Fig. 3C). In the presence of PapDTrp- (10 μM), only a single slow phase is observed, with a t1/2 of 13 s, ∼500 times faster than the slowest phase of the reduced subunit (Fig. 3C).
The results presented here provide strong evidence that the unfolded PapE subunit binds rapidly to the bacterial chaperone PapD and that this interaction both prevents aggregation and catalyzes the formation of the Ig fold. Catalysis occurs irrespective of the presence of the disulfide between Cys-15 and Cys-48, although the rate of refolding was slower when the disulfide bond was reduced. Therefore, it is difficult to know whether disulfide-bond formation in the subunit precedes chaperone binding or whether the disulfide bond forms as a result of folding on the PapD template, perhaps with the assistance of DsbA (30). In either case, PapD increases the rate of refolding of the subunit, and this is likely to be critical for subunits that use the chaperone/usher pathway for their assembly into pili in pathogenic bacteria. Different folding rates or differences in the rates of disulfide-bond formation of different subunits may be important in determining the order of subunit assembly in composite organelles.
The template that the chaperone provides is critically dependent on the Arg-8 residue in the interdomain cleft of PapD, which has been shown to interact with the terminal carboxyl group of the subunits (21) and is essential for chaperone function (Fig. 3B Inset). Previous reports have incorrectly argued that PapD-like chaperones function solely to prevent aggregation (26), although, based on experiments similar to those described here, those same investigators now agree that PapD-like chaperones catalyze folding (31). The consistency in the two reports [ours and that of Vetsch et al. (31)] reflects the likelihood that catalysis of folding by chaperones is a general mechanism in pilus biogenesis in a wide variety of bacterial pathogens (24). However, the work presented here describes the molecular basis of the temporal order of folding, taking into account the formation of the conserved disulfide bond and the function of the invariant Arg-8 residue as an active site in the process that probably works in conjunction with the Lys-112 residue to bind unfolded subunits (21, 23).
We have elucidated specific temporal events that explain the molecular mechanism by which PapD actively folds subunits. Molecular chaperones are thought to inhibit off-pathway interactions such as aggregation without influencing the on-pathway formation of native structure (32). Proteins such as the prolyl isomerases (33) or protein disulfide isomerases (34) are thought to catalyze folding in vivo, but it is still unclear what the substrates of these proteins are and whether the substrates that have been identified require a catalyst for folding (35). Although PapE was used as a model system in these studies, all subunits depend on PapD for their folding in vivo (28). Fig. 4 presents a model of how we believe the folding of pilus subunits is likely to occur in the presence of PapD. The initial step in the PapD-assisted folding mechanism is the anchoring of the carboxyl group of the subunit in the chaperone cleft through Arg-8 (and likely Lys-112). After this initial step, the F and A strands of PapE “zipper-up” along either side of the G β-strand of the chaperone and form the subunit groove. This folding intermediate, which is in an expanded conformation, provides the correct spatial context for the final folding of the subunit structure that follows (20). The molecular details of how the subunit interacts with the chaperone and the information gleaned from this study highlight the importance of investigating therapeutics that specifically target the chaperone cleft (22).
Acknowledgments
We dedicate this manuscript to the memory of Carl-Ivar Branden, who motivated this study, which began 15 years ago. We thank Craig Smith for his help in generating Fig. 1 and the model in Fig. 4, and the members of both laboratories for stimulating discussions. This work was supported by National Institutes of Health Grants DK13332 (to C.F.), AI29549 (to S.J.H.), AI057160 (to S.J.H.), and AI48689 (to S.J.H.).
Author contributions: J.G.B., C.F., and S.J.H. designed research; J.G.B. and J.S.P. performed research; J.G.B., C.F., and S.J.H. analyzed data; and J.G.B. wrote the paper.
Abbreviations: Mes, 2-(N-morpholino)ethanesulfonic acid; PapDTrp-, PapDTrp36Phe/Trp128Phe.
References
- 1.Choudhury, D., Thompson, A., Stojanoff, V., Langermann, S., Pinkner, J., Hultgren, S. J. & Knight, S. D. (1999) Science 285, 1061–1066. [DOI] [PubMed] [Google Scholar]
- 2.Soto, G. E. & Hultgren, S. J. (1999) J. Bacteriol. 181, 1059–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sauer, F. G., Fütterer, K., Pinkner, J. S., Dodson, K. W., Hultgren, S. J. & Waksman, G. (1999) Science 285, 1058–1061. [DOI] [PubMed] [Google Scholar]
- 4.Sauer, F. G., Mulvey, M. A., Schilling, J. D., Martinez, J. J. & Hultgren, S. J. (2000) Curr. Opin. Microbiol. 3, 65–72. [DOI] [PubMed] [Google Scholar]
- 5.Jacob-Dubuisson, F., Heuser, J., Dodson, K., Normark, S. & Hultgren, S. (1993) EMBO J. 12, 837–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnhart, M. M., Pinkner, J. S., Soto, G. E., Sauer, F. G., Langermann, S., Waksman, G., Frieden, C. & Hultgren, S. J. (2000) Proc. Natl. Acad. Sci. USA 97, 7709–7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulvey, M. A., Lopez-Boado, Y. S., Wilson, C. L., Roth, R., Parks, W. C., Heuser, J. & Hultgren, S. J. (1998) Science 282, 1494–1497. [DOI] [PubMed] [Google Scholar]
- 8.Anderson, G. G., Palermo, J. J., Schilling, J. D., Roth, R., Heuser, J. & Hultgren, S. J. (2003) Science 301, 105–107. [DOI] [PubMed] [Google Scholar]
- 9.Justice, S. S., Hung, C., Theriot, J. A., Fletcher, D. A., Anderson, G. G., Footer, M. J. & Hultgren, S. J. (2004) Proc. Natl. Acad. Sci. USA 101, 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hultgren, S. J., Normark, S. & Abraham, S. N. (1991) Annu. Rev. Microbiol. 45, 383–415. [DOI] [PubMed] [Google Scholar]
- 11.Normark, S., Hultgren, S., Marklund, B. I., Stromberg, N. & Tennent, J. (1988) Antonie Leeuwenhoek 54, 405–409. [DOI] [PubMed] [Google Scholar]
- 12.Dodson, K. W., Pinkner, J. S., Rose, T., Magnusson, G., Hultgren, S. J. & Waksman, G. (2001) Cell 105, 733–743. [DOI] [PubMed] [Google Scholar]
- 13.Dodson, K. W., Jacob-Dubuisson, F., Striker, R. T. & Hultgren, S. J. (1993) Proc. Natl. Acad. Sci. USA 90, 3670–3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuehn, M. J., Heuser, J., Normark, S. & Hultgren, S. J. (1992) Nature 356, 252–255. [DOI] [PubMed] [Google Scholar]
- 15.Bullitt, E. & Makowski, L. (1995) Nature 373, 164–167. [DOI] [PubMed] [Google Scholar]
- 16.Holmgren, A. & Branden, C. I. (1989) Nature 342, 248–251. [DOI] [PubMed] [Google Scholar]
- 17.Hung, D. L., Knight, S. D., Woods, R. M., Pinkner, J. S. & Hultgren, S. J. (1996) EMBO J. 15, 3792–3805. [PMC free article] [PubMed] [Google Scholar]
- 18.Striker, R., Jacob-Dubuisson, F., Frieden, C. & Hultgren, S. J. (1994) J. Biol. Chem. 269, 12233–12239. [PubMed] [Google Scholar]
- 19.Sauer, F. G., Knight, S. D., Waksman, G. & Hultgren, S. J. (2000) Semin. Cell Dev. Biol. 11, 27–34. [DOI] [PubMed] [Google Scholar]
- 20.Sauer, F. G., Pinkner, J. S., Waksman, G. & Hultgren, S. J. (2002) Cell 111, 543–551. [DOI] [PubMed] [Google Scholar]
- 21.Kuehn, M. J., Ogg, D. J., Kihlberg, J., Slonim, L. N., Flemmer, K., Bergfors, T. & Hultgren, S. J. (1993) Science 262, 1234–1241. [DOI] [PubMed] [Google Scholar]
- 22.Lee, Y. M., Almqvist, F. & Hultgren, S. J. (2003) Curr. Opin. Pharmacol. 3, 513–519. [DOI] [PubMed] [Google Scholar]
- 23.Slonim, L. N., Pinkner, J. S., Branden, C. I. & Hultgren, S. J. (1992) EMBO J. 11, 4747–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sauer, F. G., Barnhart, M., Choudhury, D., Knight, S. D., Waksman, G. & Hultgren, S. J. (2000) Curr. Opin. Struct. Biol. 10, 548–556. [DOI] [PubMed] [Google Scholar]
- 25.Thanassi, D. G., Saulino, E. T., Lombardo, M. J., Roth, R., Heuser, J. & Hultgren, S. J. (1998) Proc. Natl. Acad. Sci. USA 95, 3146–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vetsch, M., Sebbel, P. & Glockshuber, R. (2002) J. Mol. Biol. 322, 827–840. [DOI] [PubMed] [Google Scholar]
- 27.Pellecchia, M., Sebbel, P., Hermanns, U., Wüthrich, K. & Glockshuber, R. (1999) Nat. Struct. Biol. 6, 336–339. [DOI] [PubMed] [Google Scholar]
- 28.Lindberg, F., Tennent, J. M., Hultgren, S. J., Lund, B. & Normark, S. (1989) J. Bacteriol. 171, 6052–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bann, J. G., Pinkner, J., Hultgren, S. J. & Frieden, C. (2002) Proc. Natl. Acad. Sci. USA 99, 709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob-Dubuisson, F., Pinkner, J., Xu, Z., Striker, R., Padmanhaban, A. & Hultgren, S. J. (1994) Proc. Natl. Acad. Sci. USA 91, 11552–11556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vetsch, M., Puorger, C., Spirig, T., Grauschopf, U., Weber-Ban, E. U. & Glockshuber, R. (2004) Nature 431, 329–333. [DOI] [PubMed] [Google Scholar]
- 32.Walter, S. & Buchner, J. (2002) Angew. Chem. Int. Ed. 41, 1098–1113. [DOI] [PubMed] [Google Scholar]
- 33.Fischer, G., Tradler, T. & Zarnt, T. (1998) FEBS Lett. 426, 17–20. [DOI] [PubMed] [Google Scholar]
- 34.Freedman, R. B., Klappa, P. & Ruddock, L. W. (2002) EMBO Rep. 3, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matouschek, A., Rospert, S., Schmid, K., Glick, B. S. & Schatz, G. (1995) Proc. Natl. Acad. Sci. USA 92, 6319–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santoro, M. M. & Bolen, D. W. (1988) Biochemistry 27, 8063–8068. [DOI] [PubMed] [Google Scholar]