Abstract
New developments in macromolecular neutron crystallography have led to an increasing number of structures published over the last decade. Hydrogen atoms, normally invisible in most X-ray crystal structures, become visible in neutron structures. Using X-rays allows one to see structure, while neutrons allow one to reveal the chemistry inherent in these macromolecular structures. A number of surprising and sometimes controversial results have emerged from recent neutron structures; because it is difficult to see or predict hydrogen atoms in X-ray structures, when they are seen by neutrons they can be in unexpected locations with important chemical and biological consequences. Here we describe examples of chemistry seen with neutrons for the first time in biological macromolecules over the past few years.
Introduction
As the most common element in biological systems, hydrogen (H) plays a central role in chemical interactions and reactions that underlie life processes. Being able to visualize H atoms in a three-dimensional context is important for understanding this chemistry. Over the years neutron crystallography has been developed at experimental neutron user facilities as a technique for directly visualizing H atoms at an atomic level in and around biological macromolecules and polymers, and therefore for determining how they participate in chemical bonds and electrostatic interactions, how they are transferred during the chemical reactions catalysed by enzymes, and how they are moved during charge transport. Although X-ray crystallography has been used to elucidate over 80000 macromolecular structures, very few of them are well enough resolved to see individual H atoms. Therefore neutron crystallography represents a powerful tool for probing deeper into biological systems and revealing mechanistic information; it can be said that using X-rays allows one to see structure, and using neutrons allows one to more clearly see the chemistry in that structure.
Although still few in number, with around 0.1 % of PDB entries, macromolecular neutron structures have had a long-lasting impact in their respective fields. The now-classic set of neutron diffraction studies published over 30 years ago on the serine protease trypsin, which showed a doubly protonated histidine in the catalytic triad, has served as a benchmark for the power of neutron crystallography to elucidate enzyme mechanisms.1, 2 A more recent specific example of high impact is a neutron crystallographic study of the enzyme diisopropyl fluorophosphatase (DFPase), which can rapidly detoxify chemical warfare agents such as sarin, cyclosarin, and tabun. 3 The results supported a new mechanistic understanding that helped guide the engineering of variants of the enzyme that show enhanced detoxification properties against a range of nerve agents. 4,5 Another example is the series of neutron studies of the crystalline fibrous plant material cellulose which provided detailed understanding of its structure, H-bonding, and response to chemical treatment.6-9 These results guided the development of optimized methods of chemically pretreating nonedible and renewable biomass for the production of biofuels and bioproducts. 10,11
Current state-of-the-art macromolecular neutron crystallography beam lines allow researchers to study biological structures larger than 40 kDa and unit cell volumes greater than 106 Å3, 12,13 to collect data from single crystals and polycrystalline fibres to atomic resolution, 14-18 to use crystals smaller than 0.1 mm3, 19 and to complete data sets within a few hours. 20 This represents a significant advance in capability over the past 15 years, and is the result of several technical advances. These include the development of neutron image plate detectors 21 and quasi-Laue techniques 22 at reactor neutron sources, large 3He position sensitive detectors and time-of-flight Laue techniques 23 at spallation neutron sources, deuteration methods for proteins, 24 and advanced computational methods for structure refinement. 25-27 Consequently, the field is going through a period of rapid growth. In order to meet the demand for more access to macromolecular neutron crystallography beam lines, several new ones are being built at existing sources, and are being planned for next generation accelerator-based spallation sources. Some of these beam lines will have more powerful capabilities that will allow increasingly complex biological systems to be studied and smaller samples to be feasible.
Here we describe examples of chemistry seen with neutrons for the first time in biological macromolecules over the past few years. More general reviews of results from neutron macromolecular crystallography have been published elsewhere. 28-31 We describe the first observation of a hydronium ion, differentiating between types of H-bonds, and intrinsically disordered H-bonding networks. Furthermore, unexpected pKa values of amino acid side chains have been observed in neutron structures, revealing unforeseen catalytic mechanisms, an emerging understanding of the role of water in stabilizing the different functional forms of DNA, and the elucidation of proton transfer pathways and the identification of activated water hydroxyl ions. This trend of unexpected results will undoubtedly continue as the field grows.
Neutrons and X-rays
The brightness of both modern laboratory and synchrotron sources makes X-ray crystallography the method of choice for experimentally determining the structures of biological macromolecules and polymers. However, the dependence of X-ray scattering on atomic electron number means that it is difficult to locate light atoms such as H in those structures. With just one electron per atom, H is all but invisible, and with no electrons protons (H+) are completely invisible. X-ray crystallography has the potential to reveal the positions of H atoms at ultra-high resolution (1.0 Å or better). In practice, however, even in the highest resolution macromolecular X-ray structures, only a limited number of H atom positions can be experimentally determined, typically in well-ordered regions at the core of proteins. 32 Mobile H atoms of mechanistic interest, such as those often found in enzyme active sites, are usually invisible in even the best-resolved structures.
On the other hand neutrons interact with the nuclei of atoms through the strong interaction. 33 The scattering length is a measure of how efficiently a given atom diffracts neutrons, analogous to the X-ray form factor. H atoms have a relatively strong and negative scattering length (−3.74 fm) which means that they appear as negative troughs in nuclear density maps. The coherent neutron scattering length of deuterium (D, +6.67 fm) is even stronger and is comparable to that of heavier atoms in macromolecules such as C, N, and O, as well as transition metal elements found in biological systems. H atoms that have been substituted by D appear as strong positive peaks in nuclear density maps. H atoms have a large incoherent scattering cross section, which contributes to the scattering background and attenuates the diffracted intensities. Exchanging H with D helps to minimize these effects. Protein crystals and biological fibers are therefore usually prepared or soaked in D2O solutions to replace accessible and chemically labile H atoms (typically H bound to N or O atoms) with D atoms without modifying the molecular structure. Neutrons are most efficiently used in macromolecular crystallography when they are applied to answer outstanding scientific questions that require information about the location of H atoms, after thorough study using X-rays. X-ray data can be combined with neutron data in recently developed methods for structure refinement exploiting the complementarity of these data to provide more accurate and complete macromolecular structures. 25-27
These scattering properties make neutron crystallography a powerful technique for locating H and H+ (D and D+) and for distinguishing between H and D. 28-31 The former can be used to determine the protonation states of certain amino acid side chains such as Arg, Asp, Glu, His, Lys, Tyr, Ser, Thr and Tyr (and therefore whether they are charged or uncharged); the orientation of amide groups in Asn and Gln side chains; the orientation of hydroxyl groups in Ser, Thr, and Tyr residues; the conformation of methyl groups; the detailed solvent structure in and around biological macromolecules including the coordination of functionally important water molecules; the H-bonding pattern in biological polymers and proteins and the nature of those H-bonds. H/D exchange can be used as a tool for studying solvent accessibility and macromolecular dynamics, complementary to NMR techniques; for identifying isotopically labeled features in ab initio phasing approaches; for identifying the hydrophobic protein core that constitutes the minimal folding domain. Neutrons can also be used to unambiguously distinguish between water and its ions, between water molecules and metal atoms, between different species of metals, and between O and N.
Unusual chemical bonds involving hydrogen
H-bonds have functional properties that have long been recognized as essential for biological processes. 34 Importantly, H-bonds are dynamic and can be easily formed and broken under physiological conditions. Because H atoms are difficult to see using X-rays, the presence of H-bonds are normally identified in X-ray crystallographic structures of biological macromolecules and polymers by distance and geometric criteria between two electronegative heavier atoms, one of which is generally covalently bound to H and called the donor. The visibility of H (or D) atoms using neutron crystallography provides direct information about the geometry of this arrangement. Analysis of a database of this information indicates that in biological systems, H-bonds are rarely co-linear, and that they are usually weak compared to covalent and ionic bonds but stronger than van der Waals interactions. 35 Recent neutron studies have provided evidence for H-bonds that show a remarkable variety in their strength and nature (Table 1).
Table 1.
Distances involved in unusual H-bonds discussed in the text. “Type” refers to the type of H-bond (a resonance-assisted H-bond is given by RAHB, a short ionic H-bond by SIHB, and a low barrier H-bond by LBHB). All distances are in Å. D refers to the donor atom, A to the acceptor atom, and H refers to the H or D atom.
| Type | D | A | D-A | D-H | H-A | Reference |
|---|---|---|---|---|---|---|
| RAHB | O5 | Thr175 | 2.63 | 1.21 | 1.42 | 36 |
| Oγ1 | ||||||
| SIHB | His57 | Asp102 | 2.60 | 0.96 | 1.65 | 36 |
| Nδ1 | Oδ2 | |||||
| SIHB | Tyr42 | OOpCA | 2.52 | 0.96 | 1.65 | 42 |
| LBHB | Glu46 | O OpCA | 2.56 | 1.21 | 1.37 | 42 |
One such study was of the serine protease, elastase, with an inhibitor (FR130180) bound. 36 Elastase is of biomedical importance because of its implication in a number of pathological conditions; it can degrade virtually all human connective tissue by proteolysis of peptide bonds. Although there are different types of serine proteases they are characterized by a His57-Asp102-Ser195 (elastase amino acid residue numbering) catalytic triad, and likely operate under a common mechanism.
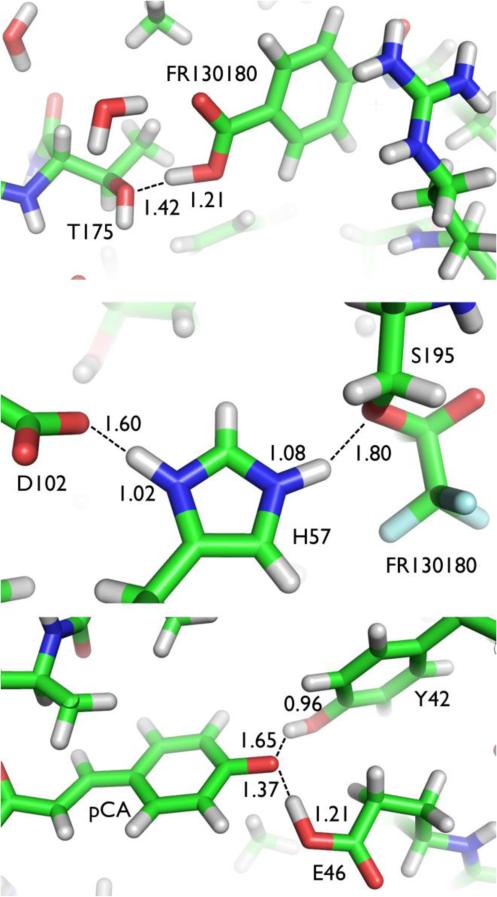
One short H-bond seen in the neutron structure of elastase occurs between the Thr175 hydroxyl and the protonated carboxyl group of the benzoic acid of the bound inhibitor (Figure 1). The D of the carboxyl group is 1.21 Å and 1.42 Å apart from the O5 atom of the inhibitor and the Oγ1 of Thr175, respectively. The O-D distance in the protonated carboxyl group is significantly longer than normal (1.21 Å vs. 0.96 Å) and together with the short H-bond distance (1.42 Å) is consistent with a resonance-assisted H-bond (RAHB). The presence of this RAHB may contribute to the effectiveness of the inhibitor.
Figure 1.
Special hydrogen bonding in elastase and photoactive yellow protein revealed by neutron crystallography. Top: Resonance-assisted hydrogen bond (RAHB) between Thr175 and inhibitor FR130180 in elastase. Centre: Short ionic hydrogen bond (SIHB) between catalytic residues His57 and Asp102 in elastase. Bottom: Low barrier (LBHB) and short ionic hydrogen bonds (SIHB) in photoactive yellow protein. OpCA...OTyr42 is a SIHB and OpCA...OGlu46 is a LBHB. All distances in Å.
Proteolysis begins with nucleophilic attack on the carbonyl group of the peptide substrate by the Oγ of the catalytic Ser195, resulting in a covalent intermediate. It has been hypothesized that the nucleophilicity of Ser195 is increased, and the transition state is stabilized, by a low barrier H-bond (LBHB) between the side chains of the catalytic His57 and Asp102.37-39 An LBHB has been proposed as a strong nonstandard H-bond with a short donor-acceptor distance and with the H atom more equally shared between donor and acceptors. 40 The H atom may therefore be on average equidistant between donor and acceptor atoms with nearly equal pKa values and a covalent bond-like character. However, in the neutron structure of elastase, the location of D indicates that it is donated by His57 to Asp102 in a short ionic H-bond (SIHB), another strong nonstandard short H-bond but with H localized on the donor atom and with an ionic bond-like character (Figure 1). 41 Both LBHB's and SIHB's are similarly short compared to standard H-bonds, and are therefore difficult to distinguish with X-rays. However, they are formed in very different polar environments. In this application therefore, neutrons have provided unique information on the polar environment around the catalytic triad.
In the neutron structure of photoactive yellow protein (PYP) both a LBHB and a SIHB are observed. 42 PYP is a bacterial photoreceptor with a charged p-coumaric chromophore (pCA) covalently attached to a Cys and sited in a hydrophobic pocket. Absorption of a photon triggers the isomerization of pCA and the subsequent reaction cycle. The H-bonding network near pCA is modulated during the thermal reaction, resulting in proton transfers within the network that are associated with large conformational changes in the protein. X-ray crystallography has shown that the phenolic oxygen atom of pCA (OpCA) is H-bonded to the O atoms of Tyr42 (OTyr42) and Glu46 (OGlu46) side chains of PYP in its ground state. 43 The short distances between OpCA and OGlu46 (2.6 Å) and OpCA and OTyr42 (2.5 Å) suggest they are strong H-bonds.
The locations of D atoms determined in the neutron structures of PYP indicate that OpCA...OTyr42 is a SIHB and that OpCA...OGlu46 is a LBHB (Figure 1). It has been proposed that the isolated negative charge of pCA may be partially stabilized by the delocalization of the negative charge within the LBHB conjugated system. However, despite the unequivocal observation of this LBHB in the neutron structure, it is inconsistent with the results of pKa solution measurements and QM/MM calculations, which suggest that OpCA...OGlu46 should be a SIHB. 43,45 In particular, the pKa of the carboxyl group of Glu46 and pCA in solution have been measured to be 8.8 and 4.25 respectively. The neutron structure suggests that the local environment in PYP decreases the pKa of Glu46 or increases the pKa of pCA in the ground state. In the excited state, an instantaneous change in dipole moment brings about a charge translocation from the phenol ring to the ethylene chain of the pCA and this will result in relaxation of the LBHB into a more standard H bond, liberating pCA for the subsequent molecular events.
In addition to nonstandard strong H-bonds, several weak ones have been observed directly in neutron structures. Over the years, evidence for C-H...O H-bonds has come from infrared spectroscopy and NMR, and their properties have been extensively studied by quantum chemistry. In the neutron structure of the electron transport protein amicyanin, no less than 19 potential C-H...O H-bonds have been identified, many of them contributing to the recognition of the metal centre. 46 Evidence for more (acidic) labile C-H groups has also come from H/D exchange patterns in the atomic resolution neutron structures of cobalamin and crambin.14,15 The case of crambin is particularly interesting because it involves partial exchange of one of the H atoms bound to a Cα (on Gly31), evidence of potential C-H. . .O H-bonding. The ability to clearly orient a number of water molecules surrounding residue Tyr 44 of crambin revealed a putative weak H-bond involving the aromatic π system.
H-bonding has long been thought to play a major role in determining the structure and properties of fibrils of naturally occurring cellulose in plant cell walls, and several putative H-bonding patterns were identified based on distances between hydroxyl groups (secondary alcohols O2, O3, and primary alcohol O6). It was therefore surprising when neutron crystallographic studies revealed a disordered H-bonding system in cellulose. 6-8 This conclusion was based on nuclear density maps showing two locations for each H atom covalently bound to O2 or O6 of the glucose monomers that make up the linear cellulose chains. Together, the two locations for each atom have a total neutron density expected for H in a single location, a disordered condition known as fractional occupancy. Two mutually exclusive H-bonding networks could be drawn based on the alternative locations for the H atoms and it was unclear whether these alternative schemes 1) interconvert dynamically, and perhaps cooperatively, giving a temporally averaged disordered structure, or 2) are static and present in different regions of the microfibril, thus producing a spatially averaged disordered structure.
Further neutron studies combined with molecular dynamics and quantum mechanics calculations showed that both static and dynamic disorder is present. 18 Although the chains pack in a well-ordered manner in the core of the cellulose fibers they are more disordered and flexible at the hydrated surfaces. 10 The surface chains interact strongly with water, forming a dense layer of water near the surfaces. This hydration layer will affect the way hydrolyzing enzymes interact with cellulose fibers. A novel statistical mechanical approach was developed to investigate this disorder further, and it was found that its presence can allow for the H-bonding to rearrange and adapt to different environmental conditions, particularly temperature. 47 The adaptive nature of the H-bonding arrangement thus serves to stabilize the plant cell wall fibres and make them more resistant to degradation.
In summary neutron crystallography has provided a wealth of new information on H-bonding in biological systems that could not have been extrapolated from the stereochemistry or distances determined in X-ray studies. Several types of strong, weak and disordered H-bonds have been observed. The presence of H-bonds reflects their electronic environment, and this environment varies dramatically between the surface and the interior of a protein. One of the consequences of the evolution of a particular structure or protein fold is to generate an environment that finely tunes the chemical properties of side chains, such as pKa. Neutrons are one of the only techniques able to probe this effect.
Protonation of amino acid residues
The side chains of polar amino acids can be either charged or uncharged depending on the pH and the pKa of the ionizable functional group. This property allows residues such as Arg, Asp, Cys, Glu, His, Lys, Ser, Thr, and Tyr to play important roles in the chemistry of biological processes. One of the most powerful current applications of macromolecular neutron crystallography is determining protonation states of amino acid residues in the active site of enzymes as a means of addressing reaction mechanisms, with a number of unexpected recent results.
A particularly striking example is the neutron structure of human carbonic anhydrase (HCA II). HCA II is a Zn-metalloenzyme that catalyzes the reversible hydration of CO2 into HCO3− and a proton, and is found prominently in red blood cells and secretory tissues. Proton transport between the active site and bulk solvent is an important part of the reaction and HCA II has become a model enzyme for the study of long-range proton transfer mechanisms in more complex systems, such as ATP synthase, bacteriorhodopsin, and cytochrome c oxidase. Tyr7 is an important residue in the proton transport pathway. Neutron structures determined from crystals grown at pH 7.8 and 10 clearly show that Tyr7 is protonated at the lower pH, and deprotonated and negatively charged at the higher pH. 48, 49 The local chemical environment around Tyr7 has therefore reduced the pKa from its value of 10 in solution to a significantly lower value. Negatively charged Tyr7, visualized directly for the first time in a protein in these studies, acts as a proton sink at high pH and may partly explain the relatively low activity of the enzyme under these conditions.
There have been other examples of amino acids within neutron structures that show pKa values significantly shifted from their values in solution. However, they have tended to involve side chains that might be expected to be charged and uncharged at different stages of the catalytic reaction, e.g. protonated Asp25 in HIV protease, 50 deprotonated Glu166 in ®-lactamase, 51 and deprotonated Lys289 in D-xylose isomerase (XI). 12
Hydroxyl and hydronium ions
Hydronium (H3O+) and hydroxyl (OH−) ions are thought to be crucial in many chemical and biological processes, but they are very reactive and unstable water species that are difficult to distinguish from water molecules using X-rays; hydronium ions, hydroxyl ions, and water molecules all appear as similar peaks at the locations of O atoms in electron density maps. In neutron studies of the catalytic mechanism of XI, protons, hydroxyl ions, and hydronium ions have been directly observed and identified in the active site. 12, 52
XI binds metal cofactors to isomerize several sugars between their aldo and keto isomers and is of significant industrial importance. Magnesium is the physiological metal found at two locations in the active site, M1 and M2. M1 coordinates Glu181, Glu217, Asp245, Asp287 and two water molecules (W2 and W3); M2 coordinates Glu217 (shared with M1), His220, Asp255 (bidentate), Asp257, and a catalytic water species. Neutron structures of XI in the presence and absence of substrate reveal that the catalytic water species is a neutral water molecule. However, in the neutron structure of XI with bound product the catalytic water is clearly shown to be hydroxide, with important mechanistic implications. This represents the first direct observation of a hydroxide ion in a neutron structure.
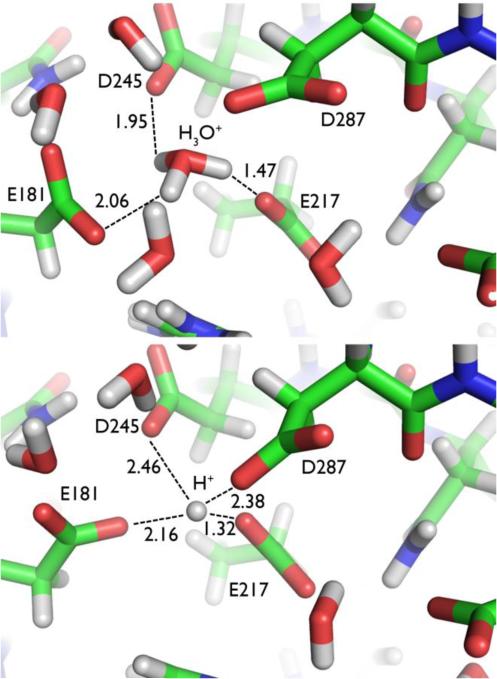
At low pH (<6), the metal cofactors of XI are expelled, and the enzyme becomes inactive. In the neutron structure of XI stripped of its metal cofactors and under normal pH conditions, a hydronium ion (observed as D3O+) takes the place of the metal at M1 (Figure 2). The hydronium ion seems to template the shape and charge of this metal binding site. In the neutron structure at lower pH (5.9), the hydronium ion is dehydrated to a proton (D+) in a trifurcated H-bond, in which the proton is positioned closest (1.32 Å) to, and is thus donated by, Glu217 (Figure 2). The amino acid residues have collapsed around the proton, and the site no longer has the required shape to accept a metal cation. These observations provide an explanation for why the required cofactor metal cations are not bound, and therefore why there is a dramatic decrease in the activity of XI at low pH values. Furthermore, direct visualization of protons, hydronium, and hydroxyl ions in a biological system provides evidence for their ability to interchange under different conditions, a result with broad implications for their possible roles in other biological systems.
Figure 2.
Hydronium ion and proton in D-xylose isomerase (XI). Top: High pH structure of XI showing coordination of hydronium (D3O+) ion. Bottom: Low pH structure of XI showing coordination of a D+ species.
Identification of a putative catalytic water species as a water molecule rather than a hydroxide was an important result in neutron studies of the catalytic mechanism of the enzyme DFPase. 3, 53 DFPase is a calcium-dependent phosphotriesterase catalyzing the cleavage of a phosphorus - fluorine bond. The nuclear density maps clearly demonstrate that active site residue Asp229 is deprotonated, and that the catalytic calcium ion coordinates an active site water, and not a hydroxide ion. The neutron structure is in agreement with a proposed mechanism with Asp229 acting as a nucleophile, and proceeding through a phosphoenzyme intermediate, and not a metal-activated water molecule that would have been identified as a hydroxide.54 Other features in the structure include an unusual calcium-water coordination in a strained orientation to satisfy H-bonding geometry, and a water-filled central tunnel within which solvent molecules could be accurately oriented based on the nuclear density. The results from the mechanistic and structural studies have allowed for the re-engineering of the enzyme to reverse its stereospecificity, while maintaining nearly full catalytic activity.4,5
Water
Whereas H is the most common element found in biological systems, water is the most common molecule. Biochemical processes occur in aqueous environments with water molecules playing crucial roles. All three atoms of D2O scatter neutrons strongly. D2O molecules have a characteristic boomerang, ellipsoidal or spherical shape in nuclear density maps, depending on their degree of order. On the other hand, D2O molecules are normally observed as spherical electron density centered at the O atom. When X-ray and neutron density maps resulting from joint X-ray and neutron refinement are displayed together, it becomes possible to accurately orient solvent molecules, elucidate ambiguous H-bonding networks, and detect disorder. 25 This approach has yielded a number of surprising results and coordination chemistry.
Water plays a crucial role in stabilizing DNA in different functional conformations (including A-DNA, B-DNA and Z-DNA) and facilitating transitions between these forms. Recent neutron crystallographic studies have produced unexpected results that have led to new insights about the role of water in these structural transitions. Early X-ray crystallographic studies of DNA oligonucleotides resulted in a proposal of hydration in the minor groove of right-handed B-DNA referred to as the zigzag spine. A similar spine of hydration was also seen in the structure of left-handed Z-DNA. 55-59 In the case of a CG dinucleotide repeat in Z-DNA, the hydration spine takes the form of an interstrand water bridge between neighboring O2 atoms of cytosine residues. However, in neutron crystallographic studies of the oligonucleotide d(CGCGCG) this hydration spine is not present; only one of five waters in the spine forms the expected H-bonding pattern. 27
A comparison of the primary hydration spine geometry in the minor groove of B-DNA and Z-DNA, shows that in B-DNA, two T bases are offset from one another, which allows the bridging water molecule to interact with the lone pair electrons on both bases at a distance that is optimal for H-bonding. In Z-DNA, however, the differing geometry leads to more closely stacked C bases, resulting in an overall poor H-bonding geometry with water molecules preferring to form transient H-bonds with the C O2 atoms, producing a disordered minor groove hydration spine.
This unexpected disorder in the minor groove hydration spine in Z-DNA raises an interesting possibility with far reaching implications for our understanding of the basic forces underlying the structural transition between B-DNA and Z-DNA. It is possible that increased disorder in the primary hydration spine represents an increase in entropy that plays a significant role in the thermodynamics of the equilibrium from B-DNA to Z-DNA. This new insight would have been difficult to attain without the combination of neutron crystallographic data and rigorous electrostatics.
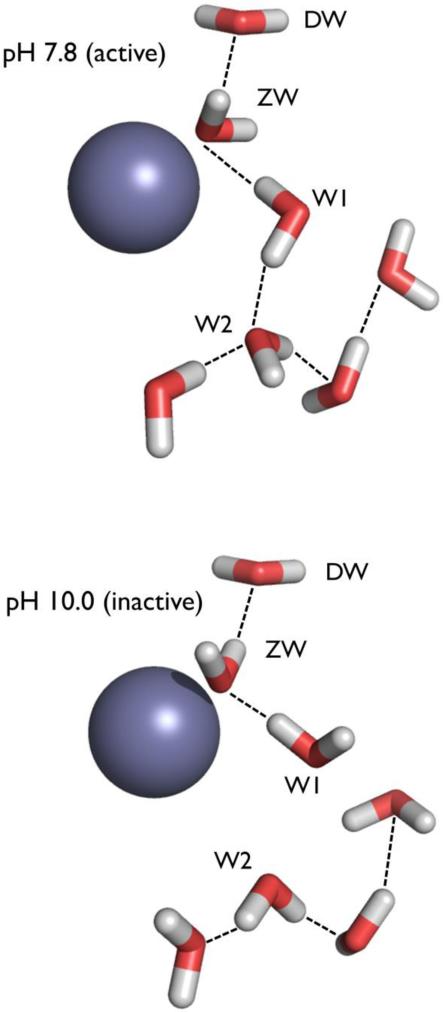
Water also plays a crucial role in the earlier mentioned proton transport associated with catalysis in HCA-II. HCA-II is one of the fastest enzymes known, able to operate at a nearly diffusion-limited kcat / KM due to electrostatic steering, but catalysis has rate limitations and pH sensitivity that are partly due to proton transport. The first step of catalysis is a nucleophilic attack on incoming CO2 by a Zn-bound OH− to produce HCO3−. The second step activates the metal-bound water to OH− through a series of proton transfers. This is thought to occur through a network of well-ordered H-bonded waters (DW, ZW, W1, W2, Figure 3). This network stretches from the metal to the proton shuttling residue, His64, and is H-bonded to several hydrophilic residues lining the active site. Comparison of neutron structures at pH 10.0 and pH 7.8 show water networks that agree in the positions of the O atoms, but differ in the positions of D atoms (i.e. the orientation of water molecules) and therefore H-bonding networks (Figure 3). A change in the orientation of W1 in the pH 10.0 structure interrupts this H-bonding network. The switch between networks suggests possible proton transport pathways through the “electrostatic funnel” leading to bulk solvent, i.e. a change in pH in the crystals has permitted the observation of what might happen during catalysis.
Figure 3.
The water networks in the active site of HCA II determined by neutron crystallography at pH 7.8 (top) and pH 10.0 (bottom). The switch between networks suggests possible proton transport pathways. Observed H-bonds are shown as black lines, active site amino acid residues are omitted for clarity. Zn atoms are in dark grey.
Conclusions
Macromolecular neutron crystallography is undergoing a renaissance. As neutrons are applied to study the chemistry in a growing number of biological systems a trend is emerging of surprising and sometimes controversial results. As the examples discussed here demonstrate, neutrons have recently been used to directly visualize a number of elusive chemical species and features in biological systems for the first time. As we look forward to increasing availability of macromolecular crystallography beam lines, some at more powerful neutrons sources, what should we expect in the future?
We expect to be able to collect data on smaller crystals, as well as crystal with larger unit cell dimensions. This potentially opens up a much larger set of biological molecules and problems that can be addressed. Complexes containing proteins and other biological molecules, such as nucleic acids, can be studied to determine the role of solvent and H-bonding in mediating interactions. Metalloenzymes with unusual clusters that are difficult to define by X-ray methods i.e. those containing electron-rich elements such as Mo and W, also lend themselves to analysis by neutron crystallography, due to the relatively even scattering of elements by neutrons across the periodic table. A number of extremophile Archaea and bacterial species utilize unusual metabolic pathways and rely on novel chemistry to sustain life.
Membrane proteins are a rich, albeit challenging target for neutron crystallography, and can potentially offer insight into the chemical environment within pores and channels, and the molecular basis of ion selectivity. One-half of all drugs targets are membrane proteins, and a detailed understanding of structure and mechanism through neutron crystallography can guide future drug development efforts. Chemical species such as NH3 and NH4+, water species, and ions such as Na+ and K+, can be identified using neutron diffraction.
Experimental H atom positions are useful for computational chemistry, bringing new accuracy into quantum mechanical calculations and molecular dynamics simulations. As neutrons can often clarify otherwise ambiguous H-bonding networks, neutron structures of protein/drug complexes may allow investigators to more accurately predict the strength of protein-ligand interactions and more gauge the entropic and enthalpic contributions to binding energy. One can also envision additional instrumentation at neutron beam lines that can perform spectroscopic measurements during data collection, and the development of robust new stations should take these possibilities into account.
A great deal of chemistry has been seen in the biological structures that have been studied using neutron crystallography over the past few years. The new species and interactions revealed by these few neutron structures offer only a hint of the richness and diversity of chemistry yet to be seen within the vast array of biological systems yet to be studied.
Acknowledgements
This work was partly supported by the Center for Structural Molecular Biology supported by the US Office of Biological and Environmental Research, US Department of Energy, under FWP ERKP752. PL was partly supported by an NIH-NIGMS funded consortium (R01GM071939) between ORNL and LBNL to develop computational tools for neutron protein crystallography. This research used facilities sponsored by the Scientific User Facilities Division, Office of Basic Energy Sciences, US Department of Energy.
Footnotes
‡ Footnotes should appear here. These might include comments relevant to but not central to the matter under discussion, limited experimental and spectral data, and crystallographic data.
References
- 1.Kossiakoff AA, Spencer SA. Nature. 1980;288:414. doi: 10.1038/288414a0. [DOI] [PubMed] [Google Scholar]
- 2.Kossiakoff AA, Spencer SA. Biochemistry. 1981;20:6462. doi: 10.1021/bi00525a027. [DOI] [PubMed] [Google Scholar]
- 3.Blum M-M, Mustyakimov M, Rüterjans H, Kehe K, Schoenborn BP, Langan P, Chen JC-H. Proc. Natl. Acad. Sci. USA. 2009;106:713. doi: 10.1073/pnas.0807842106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melzer M, Chen JC-H, Heidenreich A, Gäb J, Koller M, Kehe K, Blum M-M. J. Am. Chem. Soc. 2009;131:17226. doi: 10.1021/ja905444g. [DOI] [PubMed] [Google Scholar]
- 5.Research Highlights, Nature Chem. 2010;2:4. [Google Scholar]
- 6.Nishiyama Y, Sugiyama J, Chanzy H, Langan P. J. Am. Chem. Soc. 2003;125:14300. doi: 10.1021/ja037055w. [DOI] [PubMed] [Google Scholar]
- 7.Nishiyama Y, Langan P, Chanzy H. J. Am. Chem. Soc. 2002;124:9074. doi: 10.1021/ja0257319. [DOI] [PubMed] [Google Scholar]
- 8.Jarvis M. Nature. 2003;426:611. doi: 10.1038/426611a. [DOI] [PubMed] [Google Scholar]
- 9.Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperley DC, Kennedy CJ, Jarvis MC. Proc. Natl. Acad. Sci. USA. 2011;108:1195. doi: 10.1073/pnas.1108942108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chundawat SPS, Bellesia G, Uppugundla N, da Costa Sousa L, Gao D, Cheh AM, Agarwal UP, Bianchetti CM, Phillips GN, Jr., Langan P, Balan V, Gnanakaran S, Dale BE. J. Am. Chem. Soc. 2011;133:11163. doi: 10.1021/ja2011115. [DOI] [PubMed] [Google Scholar]
- 11.Langan P, Gnanakaran S, Rector KD, Pawley N, Fox D, Cho DW, Hammel KE. Energy & Environmental Science. 2011;4:3820. [Google Scholar]
- 12.Kovalevsky AY, Hanson L, Fisher SZ, Mustyakimov M, Mason SA, Forsyth VT, Blakeley MP, Keen DA, Wagner T, Carrell HL, Katz AK, Glusker JP, Langan P. Structure. 2010;18:688. doi: 10.1016/j.str.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes RC, Coates L, Blakeley MP, Tomanicek SJ, Langan P, Kovalevsky AY, García-Ruiz JM, Ng JD. Acta Cryst . 2012;F68:1482. doi: 10.1107/S1744309112032447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JC-H, Hanson BL, Fisher SZ, Langan P, Kovalevsky AY. Proc. Natl. Acad. Sci. USA. 2012;109:15301. doi: 10.1073/pnas.1208341109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jogl G, Wang X, Mason SA, Kovalevsky A, Mustyakimov M, Fisher Z, Hoffman C, Kratky C, Langan P. Acta Cryst. 2011;D67:584. doi: 10.1107/S090744491101496X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sawada D, Nishiyama Y, Langan P, Forsyth VT, Kimura S, Wada M. PLoS One. 2012;7:e39376. doi: 10.1371/journal.pone.0039376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sawada D, Nishiyama Y, Langan P, Forsyth VT, Kimura S, Wada M. Biomacromolecules. 2012;13:288. doi: 10.1021/bm201512t. [DOI] [PubMed] [Google Scholar]
- 18.Nishiyama Y, Johnson GP, French AD, Forsyth VT, Langan P. Biomacromolecules. 2008;9:3133. doi: 10.1021/bm800726v. [DOI] [PubMed] [Google Scholar]
- 19.Leal RMF, Teixeira SCM, Blakeley MP, Mitchell EP, Forsyth VT. Acta Cryst. 2009;F65:232. doi: 10.1107/S1744309109002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Munshi P, Chung SL, Blakeley MP, Weiss KL, Myles DAA, Meilleur F. Acta Cryst. 2012;D68:35. doi: 10.1107/S0907444911048402. [DOI] [PubMed] [Google Scholar]
- 21.Niimura N, Karasawa Y, Tanaka I, Miyahara J, Takahashi K, Saito H, Koizumi S, Hidaka M. Nucl. Inst. Meth. Phys. Res. Section A. 1994;349:521. [Google Scholar]
- 22.Niimura N, Minezaki Y, Nonaka T, Castagna JC, Cipriani F, Hoghoj P, Lehmann MS, Wilkinson C. Nat. Struct. Biol. 1997;4:909. doi: 10.1038/nsb1197-909. [DOI] [PubMed] [Google Scholar]
- 23.Langan P, Greene G, Schoenborn BP. J. Appl. Cryst. 2004;37:253. [Google Scholar]
- 24.Shu F, Ramakrishnan V, Schoenborn BP. Proc. Natl. Acad. Sci. USA. 2000;97:3872. doi: 10.1073/pnas.060024697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams PD, Mustyakimov M, Afonine PV, Langan P. Acta Cryst. 2009;D65:567. doi: 10.1107/S0907444909011548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afonine PV, Mustyakimov M, Grosse-Kunstleve RW, Moriarty NW, Langan P, Adams PD. Acta Cryst. 2010;D66:1153. doi: 10.1107/S0907444910026582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fenn TD, Schnieders MJ, Mustyakimov M, Wu C, Langan P, Pande VS, Brunger AT. Structure. 2011;19:523. doi: 10.1016/j.str.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blakeley MP, Langan P, Niimura N, Podjarny A. Curr. Opin. Struct. Biol. 2008;18:593. doi: 10.1016/j.sbi.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blakeley MP. Crystallogr. Rev. 2009;15:157. [Google Scholar]
- 30.Niimura N, Podjarny A. Neutron Protein Crystallography – Hydrogen, protons, and hydration in Biomacromolecules. Oxford University Press Book Series No. 25; p. 2011. [Google Scholar]
- 31.Nishiyama Y, Langan P, Wada M, Forsyth T. Acta Cryst. 2010;D66:1172. doi: 10.1107/S0907444910032397. [DOI] [PubMed] [Google Scholar]
- 32.Howard EI, Sanishvili R, Cachau RE, Mitschler A, Chevrier B, Barth P, Lamour V, Van Zandt M, Sibley E, Bon C, Moras D, Schneider TR, Joachimiak A, Podjarny A. Proteins. 2004;55:792. doi: 10.1002/prot.20015. [DOI] [PubMed] [Google Scholar]
- 33.Sears VF. Neutron News. 1992;3:29. [Google Scholar]
- 34.Jeffrey GA, Saenger W. Hydrogen bonding in biological structures. Springer-Verlag; 1991. [Google Scholar]
- 35.Niimura N, Arai S, Kurihara K, Chatake T, Tanaka I, Bau R. Cell Mol. Life Sci. 2006;63:285. doi: 10.1007/s00018-005-5418-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamada T, Kinoshita T, Kurihara K, Adachi M, Ohhara T, Imai K, Kuroki R, Tada T. J. Am. Chem. Soc. 2009;131:11033. doi: 10.1021/ja9028846. [DOI] [PubMed] [Google Scholar]
- 37.Cleland WW, Kreevoy MM. Science. 1994;264:1887. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 38.Frey PA, Whitt SA, Tobin JB. Science. 1994;264:1927. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]
- 39.Cleland WW, Frey PA, Gerlt JA. J. Biol. Chem. 1998;273:25529. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 40.Hibbert F, Emsley J. Adv. Phys. Org. Chem. 1990;26:255. [Google Scholar]
- 41.Fuhrmann CN, Daugherty MD, Agard DA. J. Am. Chem. Soc. 2006;128:9086. doi: 10.1021/ja057721o. [DOI] [PubMed] [Google Scholar]
- 42.Yamaguchi S, Kamikubo H, Kurihara K, Kuroki R, Niimura N, Shimizu N, Yamazaki Y, Kataoka M. Proc. Natl. Acad. Sci. USA. 2009;106:440. doi: 10.1073/pnas.0811882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anderson S, Crosson S, Moffat K. Acta Cryst. 2004;D60:1008. doi: 10.1107/S090744490400616X. [DOI] [PubMed] [Google Scholar]
- 44.Saito K, Ishikita H. Proc. Natl. Acad. Sci. USA. 2012;109:167. doi: 10.1073/pnas.1113599108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saito K, Ishikita H. Biochemistry. 2012:51. doi: 10.1021/bi201877e. [DOI] [PubMed] [Google Scholar]
- 46.Sukumar N, Mathews FS, Langan P, Davidson VL. Proc. Natl. Acad. Sci. USA. 2010;107:6817. doi: 10.1073/pnas.0912672107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen T, Gnanakaran S. Biophysical Journal. 2009;96:3032. doi: 10.1016/j.bpj.2008.12.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fisher SZ, Kovalevsky AY, Domsic JF, Mustyakimov M, McKenna R, Silverman DN, Langan PA. Biochemistry. 2010;49:415. doi: 10.1021/bi901995n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher SZ, Kovalevsky AY, Mustyakimov M, Silverman DN, McKenna R, Langan P. Biochemistry. 2011;50:9421. doi: 10.1021/bi201487b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adachi M, Ohhara T, Kurihara K, Tamada T, Honjo E, Okazaki N, Arai S, Shoyama Y, Kimura K, Matsumura H, Sugiyama S, Adachi H, Takano K, Mori Y, Hidaka K, Kimura T, Hayashi Y, Kiso Y, Kuroki R. Proc. Natl. Acad. Sci. USA. 2009;106:4641. doi: 10.1073/pnas.0809400106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomanicek SJ, Standaert RF, Weiss KL, Ostermann A, Schrader TE, Ng JD, Coates L. J. Biol. Chem. 2013;288:4715. doi: 10.1074/jbc.M112.436238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kovalevsky AY, Hanson BL, Mason SA, Yoshida T, Fisher SZ, Mustyakimov M, Forsyth VT, Blakeley MP, Keen DA, Langan P. Angew. Chem. Intl. Ed. Eng. 2011;50:7520. doi: 10.1002/anie.201101753. [DOI] [PubMed] [Google Scholar]
- 53.Chen JC-H, Mustyakimov M, Schoenborn BP, Langan P, Blum M-M. Acta Cryst. 2010;D66:1131. doi: 10.1107/S0907444910034013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blum M-M, Löhr F, Richardt A, Rüterjans H, Chen JC-H. J. Am. Chem. Soc. 2006;128:12750. doi: 10.1021/ja061887n. [DOI] [PubMed] [Google Scholar]
- 55.Wang AH-J, Hakoshima T, van der Marel G, van Boom JH, Rich A. Cell. 1984;37:321. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- 56.Egli M, Williams LD, Gao Q, Rich A. Biochemistry. 1991;30:11388. doi: 10.1021/bi00112a005. [DOI] [PubMed] [Google Scholar]
- 57.Bancroft D, Williams LD, Rich A, Egli M. Biochemistry. 1994;33:1073. doi: 10.1021/bi00171a005. [DOI] [PubMed] [Google Scholar]
- 58.Gessner RV, Quickley GJ, Egli M. J. Mol. Biol. 1994;236:1154. doi: 10.1016/0022-2836(94)90018-3. [DOI] [PubMed] [Google Scholar]
- 59.Chatake T, Tanaka I, Umino H, Arai S, Niimura N. Acta Cryst. 2005;D61:1088. doi: 10.1107/S0907444905015581. [DOI] [PubMed] [Google Scholar]