Abstract
Pathogenic bacteria have developed complex and diverse virulence mechanisms that weaken or disable the host immune defense system. IdeS (IgG-degrading enzyme of Streptococcus pyogenes) is a secreted cysteine endopeptidase from the human pathogen S. pyogenes with an extraordinarily high degree of substrate specificity, catalyzing a single proteolytic cleavage at the lower hinge of human IgG. This proteolytic degradation promotes inhibition of opsonophagocytosis and interferes with the killing of group A Streptococcus. We have determined the crystal structure of the catalytically inactive mutant IdeS-C94S by x-ray crystallography at 1.9-Å resolution. Despite negligible sequence homology to known proteinases, the core of the structure resembles the canonical papain fold although with major insertions and a distinct substrate-binding site. Therefore IdeS belongs to a unique family within the CA clan of cysteine proteinases. Based on analogy with inhibitor complexes of papain-like proteinases, we propose a model for substrate binding by IdeS.
Keywords: Streptococcus pyogenes, Mac-1
The bacterium Streptococcus pyogenes is an important human pathogen and the causative agent for a variety of diseases, including pharyngotonsillitis, impetigo, scarlet fever, septicemia, necrotizing fasciitis, and streptococcal toxic shock syndrome (1). The survival of S. pyogenes in its human host depends on its ability to avoid the actions of the innate and adaptive immune response. IgG plays a key role in human immune defense by specifically recognizing invading microorganisms and mediating their killing by professional phagocytes and the complement system. Proteolytic degradation of Igs is a common mechanism used by many pathogenic bacteria, and it is clear that proteinases are important bacterial virulence factors, enabling the bacteria to colonize and circumvent host defense systems (2, 3). S. pyogenes has evolved a specific enzyme to deal with opsonizing IgG antibodies. This enzyme, designated IdeS (IgG-degrading enzyme of S. pyogenes), is a newly discovered, secreted proteinase that specifically cleaves the heavy chain of IgG (4). Biochemical evidence confirms that the single Cys residue in IdeS constitutes the active-site residue and that IdeS must be classified as a cysteine proteinase, as suggested (5, 6). Extracellular bacterial cysteine proteinases have also been characterized from other pathogenic bacteria such as Clostridium histolyticum, Porphyromonas gingivalis, and Staphylococcus aureus (2). These cysteine proteinases exhibit broad substrate specificity and are consequently expressed as proenzymes, requiring proteolytic processing to convert into their mature forms (7–14).
IdeS (also named Mac-1, ref. 15) is unique among prokaryotic cysteine proteinases, as the protein is secreted in its mature form and does not require processing or activation to exert its enzymatic activity (4). IdeS is extremely specific for IgG, which is hydrolyzed in the hinge region after glycine residue 236 in both heavy chains. Until now, to our knowledge, no other substrate for IdeS had been identified (4, 6). In addition, IdeS is not affected by the typical cysteine proteinase inhibitor E-64 (4, 6), which is in sharp contrast to the described prokaryotic cysteine proteinases (16–19), and suggests a unique catalytic property of IdeS. Like the classical streptococcal cysteine proteinase, SpeB (streptococcal pyrogenic exotoxin B), IdeS contains an RGD motif (4, 14, 15), which is involved in the interaction of IdeS with vitronectin (αVβ3) and platelet receptors (αIIβ3) (21), suggesting additional, yet unknown properties of the enzyme. Thus, IdeS represents an atypical cysteine proteinase with unique biochemical features, suggesting that the enzyme might represent an additional family of cysteine proteinases. A clear understanding of its high degree of specificity and its catalytic properties requires the availability of an experimental structure as provided by x-ray crystallography.
In the present study, we determined the 3D structure of the IdeS-C94S mutant by multiwavelength anomalous diffraction (MAD) at a resolution of 1.9 Å. The overall structure of IdeS, along with a detailed analysis of its catalytic site, is described. Comparison of the IdeS-C94S mutant structure with other cysteine proteinase family members provides insight into the interactions that most likely occur between IdeS and its sole substrate IgG.
Materials and Methods
Expression and Purification of IdeS. The coding sequence corresponding to amino acid residues 38–339 of S. pyogenes IdeS-C94S (numbered from the start of the signal sequence) was subcloned into the expression vector pET28b (Novagen). For overexpression in Escherichia coli, the recombinant plasmid was transformed into the bacterial strain Rosetta [DE3] (Novagen). Protein expression was induced at 37°C with 1 mM isopropyl-β-d-thiogalactoside. After further growth for 4 h the bacteria were pelleted, resuspended in 50 mM phosphate (pH 7.5), 300 mM sodium chloride, and 5 mM imidazole, and lysed by sonication. The resulting soluble fraction was subsequently loaded onto a nickel-NTA agarose column (Qiagen, Hilden, Germany). The protein was eluted by using an imidazole step gradient and further purified by gel filtration.
For selenomethionine (SeMet) incorporation, the methionine auxotroph E. coli strain B834[DE3] (Stratagene) harboring the recombinant plasmid was grown in minimal medium containing 0.3 mM dl-SeMet. SeMet-labeled IdeS was expressed and purified under identical conditions as the native enzyme. MS confirmed the quantitative replacement of five Met positions by SeMet.
15N-labeled samples of IdeS-C94S for use in NMR experiments were purified accordingly from Rosetta [DE3] cells grown in M9 minimal medium containing 0.5 g/liter 15NH4Cl.
Crystallization and Data Collection. Crystals of both IdeS-C94S and its SeMet-substituted form were obtained at 20°C by a sitting drop vapor diffusion technique with a mixture of 1.5 μl of protein (7 mg/ml) in buffer containing 5 mM Mops, 100 mM NaCl (pH 8.0), 0.02% azide, and the same volume of reservoir solution [23% polyethylene glycol 2000 monomethyl ether/100 mM sodium acetate, pH 4.5/250 mM ammonium sulfate]. Crystals grown under these conditions have the typical dimensions of 1.0 × 0.3 × 0.02 mm and belong to the orthorhombic space group P21212 with unit cell dimensions a = 63.37 Å, b = 86.83 Å, and c = 57.66 Å. With one molecule of IdeS (36.2 kDa) per asymmetric unit the solvent content was 45.5%.
Before flash-cooling, the crystals were transferred into 2 μl of reservoir solution supplemented with 5% d(-)-2,3-butanediol and immediately frozen in the cryostream.
All data sets were collected at 100 K on the wiggler beamline BW6 at the Deutsches Elektronen Synchrotron, Hamburg, Germany by using a MAR Research (Norderstedt, Germany) 165-mm charge-coupled device detector.
MAD data were collected on a single crystal of the SeMet-incorporated IdeS-C94S at three wavelengths corresponding to the peak of the selenium K absorption edge (λ1), the high energy remote point (λ2), and the inflection point (λ3). Wavelengths were chosen from the x-ray fluorescence spectrum of the same crystal. In addition, a native data set was collected from a single unsubstituted crystal at λ = 1.05 Å.
Each data set was independently integrated and scaled by using denzo and scalepack (22). Data processing and refinement statistics are given in Tables 1 and 2.
Table 1. Data collection.
| IdeS-C94S, Se-Met-labeled | IdeS-C94S | |||
|---|---|---|---|---|
| Wavelength, Å | λ1 = 0.97910 | λ2 = 0.95 | λ3 = 0.97935 | 1.05 |
| Resolution, Å | 20-1.9 | 20-1.9 | 20-1.9 | 20-1.9 |
| Total reflections | 210,883 | 146,498 | 139,042 | 186,153 |
| Unique reflections | 46,131 | 44,826 | 44,731 | 24,440 |
| Completeness, % (final shell, %) | 99.7 (98.6) | 96.6 (84.7) | 96.7 (82.9) | 99.3 (99.4) |
| Rsymm (final shell)* | 6.0 (28.8) | 5.6 (31.6) | 5.8 (30.7) | 6.3 (26.6) |
| I/σ(I) (final shell) | 24.5 (4.1) | 19.2 (2.7) | 17.7 (2.6) | 28.4 (6.3) |
| Wilson B, Å2 | 21.67 | 22.08 | 21.59 | 18.58 |
| MAD-phasing statistics | ||||
| Phasing power isomorphous (anomalous)† | 0.861 (2.185) | -(0.851) | 1.019 (1.190) | |
| Figure of merit (postsolvent flattening) | 0.599 (0.915) | |||
| Rcullis isomorphous (anomalous) | 0.738 (0.587) | -(0.851) | 0.645 (0.854) | |
Rsymm = ΣhΣi |Ii(h) - 〈I(h)〉|/ΣhΣi Ii(h).
Phasing power FHc/E for the isomorphous case and 2FHc″/E for the anomalous case, where FHc is the calculated heavy atom structure factor and E is the rms lack of closure.
Table 2. Refinement statistics.
| Resolution range, Å | 19.86-1.93 |
| Rcryst/Rfree,* % | 19.56/23.47 |
| rms deviation from ideal geometry | |
| Bonds, Å | 0.0086 |
| Angels, ° | 1.37 |
| Protein atoms | 2,280 |
| Solvent | 206 water, 1 sulfate ion |
| Average B factor, Å2 | 24.3 |
Rcryst = Σ|Foh - Fch|/ΣFoh, where Foh and Fch are the observed and calculated structure factor amplitudes for reflection h, respectively. Rfree computed by using randomly selected 8% of the data that were excluded throughout refinement.
Structure Determination and Refinement. The structure of IdeS-C94S mutant was determined by the MAD phasing method. Atomic positions for four of the five Se atoms in the asymmetric unit were obtained by using shelxd (23) and refined by using sharp (24) to a final figure of merit of 0.915 after solvent flattening. The one undetected Se site was later revealed to be located in the highly flexible N-terminal region.
The resulting phases were used as input for the arp-warp (25) automated model building procedure. Of the 323 amino acid residues, 280 were automatically built into four chains. At this stage, first electron density maps were inspected and correct side chains were manually built by using the program o (26). The refinement procedure against native diffraction data was performed by using cnsv1.1 (27) and included individual B factor refinement, simulated annealing, and positional refinement by using the parameters of Engh and Huber (28). Water molecules were inserted automatically and inspected manually. Iterative rounds of model building and refinement led to a final R factor of 19.56% (Rfree = 23.47%; calculated from 8% of the reflections omitted from refinement) by using all data in the resolution range between 20 and 1.9 Å.
procheck (29) revealed no disallowed ϕ/ψ angles and excellent stereochemistry for the final model that comprises 291 residues. Structural alignments were performed with lsqman (30), and figures were prepared with molscript, (31) bobscript (32), raster3d (33), grasp (34), and pymol (www.pymol.org).
Results
Overall Structure of IdeS. The proteolytically active form of the cysteine proteinase IdeS from S. pyogenes overexpressed in E. coli undergoes proteolytic degradation, thus for crystallization the catalytically essential Cys-94 was mutated to serine (C94S). This active-site mutant retained its ability to bind IgG, which was tested by NMR binding studies (data not shown).
The structure of IdeS-C94S was determined by the MAD phasing method at a resolution of 1.9 Å.
The final model includes residues 49–339, one crystallographically defined sulfate ion, and 206 water molecules. No electron density was visible for the N-terminal 33 residues consisting of the His tag, the thrombin cleavage site, and 12 residues within the IdeS sequence, and these residues were not included in the model. With an R factor of 19.56% (Rfree = 23.47%) and a rms deviation from the ideal values in bond lengths and bond angles of 0.0086 Å and 1.37°, respectively, the model is of good quality.
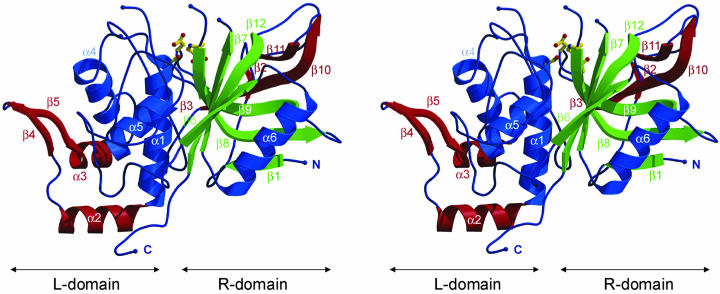
The structure of IdeS has an α/β fold with 12 β-strands and six α-helices. Despite the lack of significant sequence identity, the crystal structure shows that IdeS resembles the typical structural features of the papain superfamily of cysteine proteinases. The polypeptide chain is folded into two distinct domains that interact with one another through an extended polar interface. The marked incision between the two domains represents the active site cleft (on “top” of the molecule, Fig. 1).
Fig. 1.
Stereo ribbon diagram of IdeS-C94S. Compared to papain, additional structural elements are drawn in red, analogue α-helices are shown in blue, and β-strands are shown in green. The catalytic triade residues at the enzyme active site are drawn as in ball-and-stick form.
A structural homology search with dali (35) showed that IdeS is structurally most similar to cathepsin B. A total of 127 amino acid residues of IdeS are topologically equivalent as defined by the program lsqman (30). After optimal superposition of these 127 residues, the α-carbons have a rms deviation value of 1.88 Å. Taking this protein as a representative family member of papain-like cysteine proteinases, all of the major secondary structural elements are also present in IdeS in approximately similar positions and orientations. Outside of this conserved core, IdeS has substantial insertions relative to cathepsin B and other papain family members. Adopting the nomenclature introduced for papain, the left-hand L domain (Fig. 1) is formed mainly by the amino-terminal half of the polypeptide chain up to Leu-220. Exceptions are residues 49–85, which form two β-strands (β2 and β3) and a major loop insertion within the right-hand R domain. Similarly to papain the last 10 carboxyl-terminal residues traverse back to the L domain.
As in other family members the R domain consists of a large twisted antiparallel β-sheet (β6, β7, β8, β9, and β12), which is followed by helix α6. Additionally, there are two β-strands, β10 and β11, within this domain. Further on, there is a large insertion of two helices (α2 and α3) and two β-strands (β4 and β5) in the L domain of the IdeS structure.
Despite the approximately coincident overall fold to cathepsin B, the connecting segments between the secondary structure elements show significant variability. IdeS is considerably larger (339 residues compared to 254 in cathepsin B and 212 in papain, ref. 36), and most of the large structural deviations are caused by residue insertions.
Unlike papain and cathepsin B, IdeS does not contain disulfide bonds and is synthesized without a propeptide. Additionally, IdeS contains an RGD motif at amino acids 214–216, which is located on a surface loop within the L domain as a turn connecting β4 and β5. This motif is important for recognition by integrins and is also present in the streptococcal cysteine proteinase SpeB (streptococcal pyrogenic exotoxin B) (14, 21).
On top of the molecule (Fig. 1) at each side of the active-site cleft are two loops (residues 165 and 166 and 256–259) without appropriate electron density that are considered as conformationally flexible and have been omitted from the model.
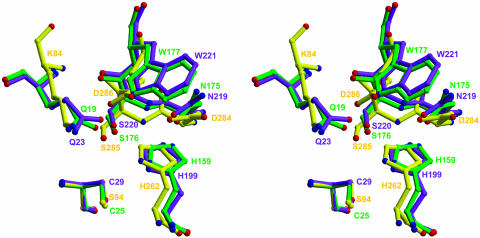
The Mechanism of Catalysis. The active-site region of IdeS resembles that seen in the papain cysteine proteinase superfamily. In the superimposed structures of IdeS, papain, and cathepsin B, the catalytic cysteine (Cys-94–Ser, Cys-25, and Cys-29, respectively) and histidine (His-262, His-159, and His-199) residues align very well. Thus Cys-94–Ser in IdeS is situated at the N-terminal region of helix α1 at the interface between the L and R domains. To demonstrate the high degree of resemblance concerning the overall geometry of the catalytic triad, an additional fitting with respect to the active site was performed (Fig. 2).
Fig. 2.
Comparison of IdeS-C94S (yellow), papain (green) (Protein Data Bank ID code 1POP), and cathepsin B (purple) (Protein Data Bank ID code 1CSB) active sites. The figure was prepared by superposition of papain and cathepsin B on IdeS and fitting the active-site cysteines on residue 94 of IdeS-C94S.
The decrease in the enzymatic activity after mutation of residues Asp-284 and Asp-286 suggests that these amino acids participate in catalysis (37). The structure of IdeS shows that Asp-284 corresponds to residues Asn-175 and Asn-219 of papain and cathepsin B, respectively (Fig. 2) and is linked to His-262 by means of a hydrogen bond. Accordingly, Asp-284 is responsible for the correct orientation of the imidazolium ring of the active-site His-262 and enhances the proteolytic efficiency in this respect. The adjacent Asp-286 is not directly in contact with the active-site residues. However, the substantial decrease in activity caused by the Asp-286–Ala mutation (37) suggests an important role in setting the electrostatic milieu.
In addition, the overlay shows that Lys-84 of IdeS is structurally equivalent to Gln-19 in papain and Gln-23 in cathepsin B. These residues form, together with the amide nitrogen of the active-site cysteine, the oxyanion hole (38–40), which binds to the main-chain carbonyl group of the P1 residue of the substrate. The conformation of Lys-84 is stabilized by a hydrogen bond-salt link formed with Asp-286, which is lost in the Asp-286–Ala mutant. This lacking interaction seems to be the reason for the reduced proteolytic activity (37).
Typically in cysteine proteinases of the CA clan, the aspartic acid of the catalytic triad is shielded by the side chain of a neighboring tryptophan (Trp-177 and Trp-221 in papain and cathepsin B, respectively) (36). In IdeS Asp-286 occupies its position, creating a strongly negatively charged region at the active site (Fig. 2).
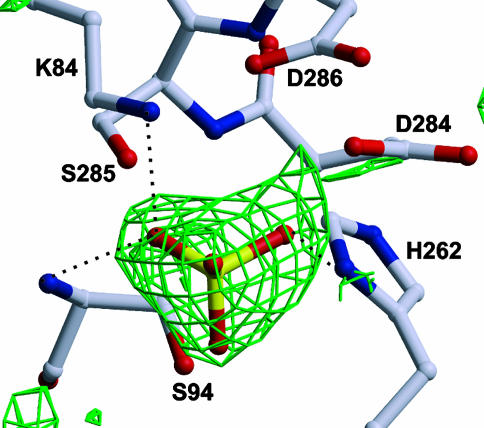
In the crystal structure of IdeS additional electron density is seen at the active site, which we interpret as sulfate ion from the crystallization medium. The sulfate is fixed at this position by three hydrogen bonds formed with Ser-94, His-262, and Lys-84 and very likely masks the oxyanion hole (Fig. 3).
Fig. 3.
Ball-and-stick model of the catalytic triad of IdeS-C94S with the initial Fobs – Fcalc electron density map. This difference in electron density, contoured at 2.5 σ, presumably accounts for a sulfate ion from the crystallization liquid.
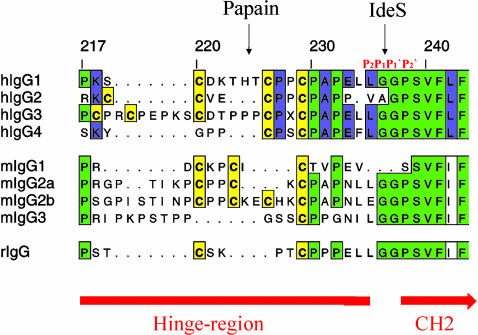
Insights into Substrate Binding Specificity. IdeS is an endopeptidase with a unique high degree of specificity toward IgG, and until today, to our knowledge, no other substrate had been identified (41). The cleavage site within IgG is located in the hinge region between Gly-236 and Gly-237. Thus, an uncommon Leu-Leu-Gly motif occupies the P3, P2, and P1 sites in the human IgG1, IgG3, and IgG4 substrates (Fig. 4) [with P1, P2, P3 and P1′, P2′ designating the substrate residues N/C terminal of the scissile peptide bond, facing the enzyme specificity pockets S1, S2, S3 and S1′, S2′, respectively (42)]. However, a Pro-Val-Ala motif as in human IgG2 is also accepted as substrate (6).
Fig. 4.
Sequence comparison of the IgG-Fc hinge region and the cleavage site of human, mouse, and rabbit IgG subclasses.
To explore the substrate binding to the active site of IdeS, the two structurally related complex structures of cathepsin B with the peptide inhibitor CA030 (Protein Data Bank ID code 1CSB) and of papain complexed with the inhibitor E64 (Protein Data Bank ID code 1PE6) have been superimposed onto the IdeS structure. The bound inhibitors CA030 and E64 occupy the substrate binding pockets S′ and S, respectively. Based on these structures, the substrate residues P2′-P2 of the cleavage site in IgG1 were modeled into the specificity subsites. The suggested model of substrate binding after geometrical energy minimization is shown in Fig. 5.
Fig. 5.
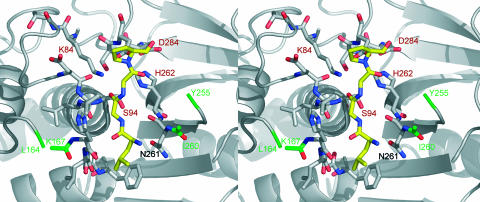
Stereo top view of the active-site cleft of IdeS-C94S, shown together with a tetrapeptide (yellow, carbon; blue, nitrogen; red, oxygen) derived from the natural cleavage sequence in the hinge peptide of IgG. The first and last visible amino acids of the flexible loop regions of IdeS (Leu-164, Lys-167, Ile-260, and Tyr-255) are shown in green. The orientation of the molecule is that of Fig. 6B.
According to this model, the hinge peptide of IgG will be processed in the manner proposed for papain, with its cleavable peptide bond at the active-site Cys-94 thiolate (38). The carbonyl oxygen of residue P1 (Gly-236) is bound in the oxyanion hole, forming hydrogen bonds with the peptide bond amide of Cys-94 and the side-chain nitrogen of Lys-84 (Fig. 5). This orientation directs the amide nitrogen of P1′ residue (Gly-237) close to ND1 of the imidazole of His-262. The putative S1′ subsite in IdeS is not as wide as in papain, and a sterical interference with larger P1′ residues than glycine is conceivable. The preceding amide nitrogen of residue Gly-236 may be fixed by forming a hydrogen bond with the carbonyl oxygen of residue Asn-261 (Fig. 5).
As shown in Fig. 5, a side chain of residue P1 (Gly-236) would point toward the enzyme specificity pocket S1. At this position the side chains of Leu-92 and His-168 block the pocket, molding it relatively narrow and only accessible for amino acids with small side chains like glycine and alanine (Fig. 6C). Nevertheless, the ψ/ϕ angles of Gly-236/Gly-237 in the model are located in favored regions of the Ramachandran plot, and thus alanine would be acceptable at position P1 as in the substrate human IgG2. In contrast, papain renders little selectivity at residue P1, because the side chain points into the bulk solvent (ref. 43 and Fig. 6A).
Fig. 6.
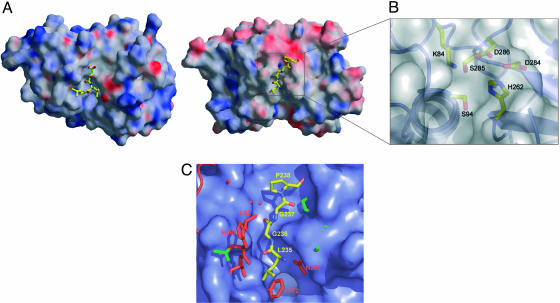
Solid surface comparison of the active site of papain and IdeS. (A and B) The molecular surface colored by electrostatic potential of papain in complex with its inhibitor E64 (A, shown as ball-and-stick form, Protein Data Bank ID code 1PE6) in comparison with IdeS-C94S with the modeled tetrapeptide Pro-Gly-Gly-Leu (B, ball-and-stick form) and a clipping of the active site. To facilitate a comparison both structures are oriented equally and show the top view of the active-site cleft (rotated 90° about the x axis toward the viewer relative to Fig. 1). (C) Overlay of papain (Protein Data Bank ID code 1POP) shown as blue surface and IdeS-C94S shown as ball-and-stick in red with the start and end point of the flexible loops shown in green. The modeled tetrapeptide (ball-and-stick form) is shown in front of the molecular surface of the catalytic cleft (yellow, carbon; blue, nitrogen; red, oxygen). Especially the nonprimed site of the active-site cleft is much narrower in IdeS than in papain.
Most proteinases of the papain superfamily C1 have a preference for hydrophobic side chains at P2. Also IdeS accepts leucine and valine at this position, but bulky aromatic residues as in papain would not fit into the shallow S2 subsite, caused by the presence of Asn-261 within this groove (Fig. 6C). In addition, the S2 pocket is formed mostly by Asn-263, Ser-252, Pro-173, and Phe-172 (Fig. 5).
The S3 site of IdeS is able to accommodate nonpolar amino acids such as leucine, proline, or phenylalanine as in the natural substrates human IgG1, IgG2, and IgG4, respectively. However, a description of this pocket in all details is difficult because of the flexibility within the loops that create the S3 site, connecting α4 and α5, because it lacks the stabilizing disulfide bridge as seen in papain and cathepsin B. Nevertheless, it seems possible that the two flexible loop regions within the S2, S3 environment adapt a conformation induced by substrate binding.
Even more difficult is the prediction of the exact position and conformation of the P2′ residue. Most likely it depends on the orientation of the CH2 domain of the Fc fragment, which is located C terminally of the hinge peptide.
To clarify the extensive differences between the specificity subsite of IdeS and other cysteine proteinases of the papain-like family, we prepared an active-site overlay of the papain molecular surface and the IdeS-C94S structure (Fig. 6C). A comparison of the active-site cleft displays the large overall differences among these structures in both surface constitution and charge. Whereas in IdeS the S2′ site is caused by two aspartate residues (D284 and D286) mainly negatively charged, this region is mainly uncharged in papain. Furthermore the S2 and S3 specificity subsites in IdeS are very narrow (Fig. 6 B and C) and do not provide enough space for the P3 residue of E64, which explains the remarkable fact that this typical cysteine proteinase inhibitor is inactive. Probably the above-mentioned features are also responsible for the lack of affinity to natural inhibitors as, for example, cystatin C, kininogen, and related inhibitors (6, 44, 45).
The molecular structure of the substrate binding groove seen in unliganded IdeS-C94S cannot readily explain the high specificity of the enzyme for IgG. Because peptides with a sequence of the IgG hinge region are not significantly hydrolyzed (6), additional exosite binding is likely to contribute to enzyme substrate interaction.
Discussion
The important human pathogen S. pyogenes has developed complex and diverse virulence mechanisms to resist host defense systems. The recently identified protein IdeS is an extracellular cysteine endopeptidase of S. pyogenes that has the ability to inhibit opsonophagocytosis (15) and killing by human polymorphonuclear leukocytes, through proteolytic degradation of IgG (4, 5). The presence of specific antibodies against IdeS in sera of patients with pharyngitis, acute rheumatic fever, and severe invasive disease episodes, and the continuous production of this proteinase during infection (4, 15) indicates that IdeS is an important virulence determinant and a potential therapeutic drug target. The crystal structure of the IdeS-C94S mutant presented here reveals structural similarities with papain-like cysteine proteinases, but also displays several strikingly different features. Nevertheless, the alignment of the 3D structure with that of papain and the structurally most related cathepsin B demonstrates the affiliation of IdeS to clan CA of cysteine proteases, which is characterized by the typical L and R domains (Fig. 1) as seen in papain-like proteinases. Beyond the conserved core of four α-helices and seven β-strands among these enzymes, there are extensive insertions in the IdeS structure particularly within the L domain. The most notable site is the unusual RGD motif, which mediates the binding to vitronectin (αVβ3) and platelet receptors, suggesting an additional function of the enzyme (21). Further uncommon characteristics are the lack of disulfide bridges and the expression as a mature enzyme lacking a prosegment.
IdeS is an endopeptidase, hence the active site at the C-terminal of the substrate scissile bond is not occluded by other parts of the protease as seen in the exopeptidase cathepsin B. Additionally, the fact that the amino acid sequence lacks significant homology to other cysteine proteinases (Fig. 7, which is published as supporting information on the PNAS web site) indicates that IdeS represents a member of a unique family within the CA clan of peptidases. However, the canonical papain fold of IdeS, despite low conservation of the primary sequence, suggests a divergent evolution of these proteinases.
The structural similarity of the active site points to a catalytic mechanism of IdeS resembling that of papain-like enzymes. Thus, it is likely that Cys-94 and His-262 form a thiolate-imidazolium ion pair, where Asp-284 orients the active-site histidine correctly, and Lys-84 contributes to the oxyanion hole. These roles in catalysis are consistent with mutagenesis data for the respective cysteine and histidine residues of IdeS (37). Nevertheless, striking differences exist concerning the type of residues involved in the catalytic mechanism as the oxyanion hole-forming lysine replaces the typical glutamine residue in this position, whereas the histidine-orienting residue, which is in most cases asparagine, is replaced by an aspartate.
Another remarkable feature of IdeS is the high degree of specificity for its substrate IgG. In contrast to structurally homologous cysteine proteinases as streptococcal pyrogenic exotoxin B, cathepsin B, and papain, which have a broad specificity, IdeS specifically cleaves at the lower hinge region of the IgG between two glycines (Gly-236/Gly-237) or less efficiently between glycine and alanine in human IgG2 (Fig. 4). This region of the Fc fragment has also been identified as the binding site for the Fcγ receptor in the Fc–FcγRIII complex (46). Competition between IdeS and FcγRIII for IgG binding can explain the retained ability to inhibit IgG-mediated phagocytosis of the proteolytically inactive IdeS-C94A (21).
The comparison of the specificity subsites with its counterpart in papain (Fig. 6C) reveals the nonprimed site in IdeS to be much narrower (Fig. 6C). This finding explains the minor variety of amino acids accepted in these subsites, and the 10-fold lower affinity of IdeS for IgG2, as well as the reported inactivity of Ides toward mouse, pig, and sheep IgG1 (47). Further on, oligopeptides containing the cleavage sequence Pro-Gly-Gly-Leu are not hydrolyzed by IdeS (6). Therefore, the characteristics of the active-site cleft are not a sufficient explanation for the strict specificity of IdeS, suggesting that more distant regions of the IgG molecule must contribute to form the enzyme substrate complex to allow proteolytic processing. As IdeS binds to the Fc fragment after papain digestion with a similar affinity as to the whole IgG (6), a potential exosite must be located within the CH2 fragment of the Fc portion. The exact area is yet undefined and requires the structure determination of the complex. Still, because IdeS-digested Fc does not bind to the proteinase, both the nonprimed residues P2 and P1 and the hypothetical exosite are likely to contribute to the formation of the enzyme substrate complex.
Previous studies showed that some typical cysteine proteinase inhibitors, as, for example, cystatins and E64 (44, 45), cannot block the proteolytic activity of IdeS (6). Comparison of the crystal structures of IdeS and other proteinases reveals that overall differences between the active-site cleft of IdeS and other papain–like cysteine proteinases are responsible for the inability to affect IdeS activity.
On the other hand, it was shown that pepstatin, an typical aspartic and HIV proteinase inhibitor, is able to inactivate IdeS (47). Originally it was assumed that this feature depends on aspartate residues in the proximity of the active-site cleft. In aspartic proteinases the pepstatin binding is supported mainly by the catalytic dyad aspartate residues, which have to be coplanar to assemble the characteristic hydrogen-bonding network (20). However, the structural analysis of IdeS shows that Asp-284 and Asp-286, involved in the formation of the active-site cleft, cannot adopt this conformation. Consequently, the inhibitor must act on IdeS in another mode as described for various aspartic proteinase/pepstatin complexes (20).
The analysis of the crystal structure of the streptococcal endopeptidase IdeS presented here supports the biochemical evidence that IdeS is a member of a unique family of cysteine proteinases (6). The molecular structure of the substrate-binding groove explains the unusual inhibition profile of the enzyme, and the data support the view that additional exosite binding is likely to contribute to the high specificity of IdeS toward IgG.
Supplementary Material
Acknowledgments
We thank Prof. Wolfram Bode for fruitful discussion and Loyola D'Silva for measuring and analyzing the NMR data. This study was supported by the Swedish Research Council (Projects 7480, 14379, and 14767) and Hansa Medical AB (Lund, Sweden).
Abbreviations: IdeS, IgG-degrading enzyme of Streptococcus pyogenes; MAD, multiwavelength anomalous diffraction; SeMet, selenomethionine.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1Y08).
References
- 1.Cunningham, M. W. (2000) Clin. Microbiol. Rev. 13, 470–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Travis, J. & Potempa, J. (2000) Biochim. Biophys. Acta 1477, 35–50. [DOI] [PubMed] [Google Scholar]
- 3.Travis, J., Potempa, J. & Maeda, H. (1995) Trends Microbiol. 3, 405–407. [DOI] [PubMed] [Google Scholar]
- 4.von Pawel-Rammingen, U., Johansson, B. P. & Björck, L. (2002) EMBO J. 21, 1607–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Pawel-Rammingen, U., Johansson, B. P., Tapper, H. & Björck, L. (2002) Nat. Med. 8, 1044–1045. [DOI] [PubMed] [Google Scholar]
- 6.Vincents, B., von Pawel-Rammingen, U., Björck, L. & Abrahamson, M., Biochemistry, in press. [DOI] [PubMed]
- 7.Elliott, S. D. (1945) J. Exp. Med. 81, 573–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubin, G. (2003) Acta Biochim. Pol. 50, 715–724. [PubMed] [Google Scholar]
- 9.Potempa, J., Sroka, A., Imamura, T. & Travis, J. (2003) Curr. Protein Pept. Sci. 4, 397–407. [DOI] [PubMed] [Google Scholar]
- 10.Witte, V., Wolf, N. & Dargatz, H. (1996) Curr. Microbiol. 33, 281–286. [DOI] [PubMed] [Google Scholar]
- 11.Berge, A. & Björck, L. (1995) J. Biol. Chem. 270, 9862–9867. [DOI] [PubMed] [Google Scholar]
- 12.Collin, M. & Olsén, A. (2001) EMBO J. 20, 3046–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagawa, T. F., Cooney, J. C., Baker, H. M., McSweeney, S., Liu, M., Gubba, S., Musser, J. M. & Baker, E. N. (2000) Proc. Natl. Acad. Sci. USA 97, 2235–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stockbauer, K. E., Magoun, L., Liu, M., Burns, E. H., Jr., Gubba, S., Renish, S., Pan, X., Bodary, S. C., Baker, E., Coburn, J., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lei, B., DeLeo, F. R., Hoe, N. P., Graham, M. R., Mackie, S. M., Cole, R. L., Liu, M., Hill, H. R., Low, D. E., Federle, M. J., et al. (2001) Nat. Med. 7, 1298–1305. [DOI] [PubMed] [Google Scholar]
- 16.Potempa, J., Dubin, A., Korzus, G. & Travis, J. (1988) J. Biol. Chem. 263, 2664–2667. [PubMed] [Google Scholar]
- 17.Rice, K., Peralta, R., Bast, D., de Azavedo, J. & McGavin, M. J. (2001) Infect. Immun. 69, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Björck, L., Åkesson, P., Bohus, M., Trojnar, J., Abrahamson, M., Olafsson, I. & Grubb, A. (1989) Nature 337, 385–386. [DOI] [PubMed] [Google Scholar]
- 19.Barrett, A. J., Kembhavi, A. A., Brown, M. A., Kirschke, H., Knight, C. G., Tamai, M. & Hanada, K. (1982) Biochem. J. 201, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fujimoto, Z., Fujii, Y., Kaneko, S., Kobayashu, H. & Mizuno, H. (2004) J. Mol. Biol. 341, 1227–1235. [DOI] [PubMed] [Google Scholar]
- 21.Lei, B., DeLeo, F. R., Reid, S. D., Voyich, J. M., Magoun, L., Liu, M., Braughton, K. R., Ricklefs, S., Hoe, N. P., Cole, R. L., et al. (2002) Infect. Immun. 70, 6880–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–319. [DOI] [PubMed] [Google Scholar]
- 23.Schneider, T. R. & Sheldrick, G. M. (2002). Acta Crystallogr. D 58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- 24.de la Fortelle, E. & Bricogne, G. (1997) Methods Enzymol. 276, 472–494. [DOI] [PubMed] [Google Scholar]
- 25.Morris, R. J., Perrakis, A. & Lamzin, V. S. (2002) Acta Crystallogr. D 58, 968–975. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 27.Brünger, A. T., Adams, P. D., Clore, G. M., Delano, W. L., Gros, P., Grossekunstleve, R. W., Iang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 28.Engh, R. A. & Huber, R. (1991). Acta Crystallogr. A 47, 392–400. [Google Scholar]
- 29.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 30.Kleywegt, G. J. & Jones, T. A. (1994) in From First Map to Final Model, eds. Bailey, S., Hubbard, R. & Waller, D. (Science and Engineering Research Council Daresbury Laboratory, Warrington, U.K.), pp. 59–66.
- 31.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 32.Esnouf, R. M. (1999) Acta Crystallogr. D 55, 938–940. [DOI] [PubMed] [Google Scholar]
- 33.Merritt, E. A. & Murphy, M. E. P. (1994) Acta Crystallogr. D 50, 869–873. [DOI] [PubMed] [Google Scholar]
- 34.Nicholls, A., Sharp, K. A. & Honig, B. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]
- 35.Holm, L. & Sander, C. (1994) Proteins 19, 165–173. [DOI] [PubMed] [Google Scholar]
- 36.Musil, D., Zucic, D., Turk, D., Engh, R. A., Mayr, I., Huber, R., Popovic, T., Turk, V., Towatari, T., Katunuma, N. & Bode, W. (1991) EMBO J. 10, 2321–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei, B., Liu, M., Meyers, E. G., Manning, H. M., Nagiec, M. J. & Musser, J. M. (2003) Infect. Immun. 71, 2881–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drenth, J., Kalk, K. H. & Swen, H. M. (1976) Biochemistry 15, 3731–3738. [DOI] [PubMed] [Google Scholar]
- 39.Menard, R., Carriere, J., Laflamme, P., Plouffe, C., Khouri, H. E., Vernet, T., Tessier, D. C., Thomas, D. Y. & Storer, A. C. (1991) Biochemistry 37, 8924–8928. [DOI] [PubMed] [Google Scholar]
- 40.Schroder, E., Phillips, C., Garman, E., Harlos, K. & Crawford, C. (1993) FEBS Lett. 315, 38–42. [DOI] [PubMed] [Google Scholar]
- 41.von Pawel-Rammingen, U. & Björck, L. (2003) Curr. Opin. Microbiol. 6, 50–55. [DOI] [PubMed] [Google Scholar]
- 42.Schechter, I. & Berger, A. (1967) Biochem. Biophys. Res. Commun. 27, 157–162. [DOI] [PubMed] [Google Scholar]
- 43.Zhu, M., Shao, F., Innes, R. W., Dixon, J. E. & Xu, Z. (2004) Proc. Natl. Acad. Sci. USA 101, 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez-Fernandez, M., Barrett, A. J., Gerhartz, B., Dando, P. M., Ni, J. & Abrahamson, M. (1999) J. Biol. Chem. 274, 19195–19203. [DOI] [PubMed] [Google Scholar]
- 45.Salvesen, G., Parkes, C., Abrahamson, M., Grubb, A. & Barrett, A. J. (1986) Biochem. J. 234, 429–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sondermann, P., Huber, R., Oosthuizen, V. & Jacob, U. (2000) Nature 406, 267–273. [DOI] [PubMed] [Google Scholar]
- 47.Agniswamy, J., Lei, B., Musser, J. M. & Sun, P. D. (2004) J. Biol. Chem., in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.