Abstract
Lymphangioleiomyomatosis (LAM) is a multisystem disorder of women, characterized by cystic degeneration of the lungs, renal angiomyolipomas (AML), and lymphatic abnormalities. LAM lesions result from the proliferation of benign-appearing, smooth muscle-like LAM cells, which are characterized by loss of heterozygosity (LOH) of one of the tuberous sclerosis complex (TSC) genes. LAM cells are believed to migrate among the involved organs. Because of the apparently metastatic behavior of LAM, we tried to isolate LAM cells from body fluids. A cell fraction separated by density gradient centrifugation from blood had TSC2 LOH in 33 of 60 (55%) LAM patients. Cells with TSC2 LOH were also found in urine from 11 of 14 (79%) patients with AML and in chylous fluid from 1 of 3 (33%) patients. Identification of LAM cells with TSC2 LOH in body fluids was not correlated with severity of lung disease or extrapulmonary involvement and was found in one patient after double lung transplantation. These studies are compatible with a multisite origin for LAM cells. They establish the existence of disseminated, potentially metastatic LAM cells through a relatively simple, noninvasive procedure that should be valuable for molecular and genetic studies of somatic mutations in LAM and perhaps other metastatic diseases.
Keywords: loss of heterozygosity, metastasis, CD235a, angiomyolipoma, smooth muscle cells
Lymphangioleiomyomatosis (LAM) is a multisystem disease of women, characterized by cystic degeneration of the lungs, renal angiomyolipomas (AML), and lymphatic lesions, e.g., thoracic and abdominal lymphangioleiomyomas (1–3). It occurs as a sporadic disease or with tuberous sclerosis complex (TSC), which can be inherited as an autosomal dominant disorder involving multiorgan hamartomas, in which patients frequently develop lung and kidney lesions pathologically and genetically similar to those seen in LAM (4–6). LAM lesions are marked by proliferation of abnormal-appearing smooth muscle-like cells (LAM cells) that have loss of heterozygosity (LOH) (7) and inactivating mutations (8, 9) in one of the two TSC genes. Most mutations have been described in TSC2 (16p13), with mutations in the TSC1 gene (9q34) being less frequent. In LAM lung lesions, LOH or other somatic mutations in either TSC gene have been reported (7–9). Renal AML contain abnormal blood vessels and adipose cells in addition to the smooth muscle-like LAM cells. All three types of AML cells appear to be neoplastic, based on LOH as the second genetic hit (10). LOH at the TSC2 locus was also detected in lymph nodes from a LAM transplant patient (7).
Because somatic mutations in lung and kidney lesions of the same patient appeared to be identical, it was hypothesized that LAM lesions at different sites have a common origin (8). Recurrent LAM cells within donor lungs after transplantation were of recipient origin (11, 12), suggesting metastatic spread. Given the metastatic potential of the benign-appearing LAM cells, we hypothesized that they might be present in body fluids from LAM patients. LAM cells in tissue are heterogeneous in appearance, ranging from small and round or spindle-shaped to large and epithelioid (1). Marker proteins described to date, such as α-smooth muscle actin, vimentin, and desmin, are not unique to LAM cells, and specific cell-surface markers have not been reported. LAM cells do differ from normal smooth muscle cells by the presence of gp100, a 100-kDa glycoprotein found in melanoma that reacts with a monoclonal antibody, HMB45 (13), but percentages of HMB45-positive LAM cells varied from 17% to 67% in biopsies (2), with an inverse relationship between percentages of cells positive for proliferating cell nuclear antigen and HMB45 positivity (14). We therefore decided on genotype as a criterion for identification of LAM cells. We looked for LAM cells in blood and other body fluids, in particular, those derived from involved organs (urine/kidney) or vessels (chyle/lymphatics). Isolation of cells from blood was performed by using the OncoQuick density gradient system that is designed to prepare samples enriched in circulating cancer cells (15). We report here the isolation of neoplastic LAM cells, identified by TSC2 LOH, from the blood and other body fluids of LAM patients.
Materials and Methods
Patients and Sample Collection. Blood, urine, expectorated chyle, or pleural or abdominal chylous fluids were collected from 61 LAM patients admitted to the National Institutes of Health Clinical Center between 2002 and 2003. The diagnosis of LAM was based on high-resolution computed tomography (CT) scans or tissue biopsy (protocol 95-H-0186). Blood or urine samples were also obtained from 25 healthy women with no known history of LAM (protocol 96-H-0100). Both protocols were approved by the National Heart, Lung, and Blood Institute Institutional Review Board. Informed consent was obtained from all participants. Blood samples (25 ml) were fractionated by density-gradient centrifugation at 1,600 × g on OncoQuick columns (Greiner Bio-One, Longwood, FL) (15). The procedure yields three fractions: sedimented blood cells; a low-density fraction in which colorectal cancer (15) and LAM (Table 1) cells were identified; and a fraction above that contains platelets. Cells in urine and chylous fluids were sedimented at 200 × g for 10 min, then washed twice in PBS.
Table 1. Detection of LAM cells added to whole blood.
| Nuclei with one signal, %
|
|||
|---|---|---|---|
| NV no. | LAM cells | TSC1 | TSC2 |
| 1 | 0 | 0 | 8 |
| 1 | + | 4 | 46 |
| 2 | + | 0 | 34 |
Blood (25 ml) from two normal volunteers (NV) without or with 1 × 106 cultured LAM cells added was fractionated by using OncoQuick. The low-density fraction was hybridized with fluorescent TSC1 and TSC2 probes. For each sample, 50 nuclei were counted.
PCR Analysis of LOH. Genomic DNA was isolated from fractionated or sedimented cells by using the PicoPure DNA Extraction kit (Arcturus, Mountain View, CA) or from whole blood by using the PureGene kit (Gentra Systems, Minneapolis). DNA was preamplified with random 15-mers as described in ref. 7 and then specifically amplified by using primers flanking microsatellite loci D16S521, Kg8, D16S663, and D16S291 on chromosome 16p13. Antisense primers were labeled with fluorescent dyes (6-FAM or HEX, Invitrogen). Primer sequences were obtained from the Genome Database (www.gdb.org). PCR was performed in 12.5-μl mixtures containing 10 mM Tris·HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 300 μM each dNTP (deoxynucleoside triphosphate), 0.8 μM primers, and 0.5 units of AmpliTaq Gold (Applied Biosystems), using a Stratagene Robocycler with initial denaturation at 95°C for 2 min, followed by 35 cycles of 95°C (45 sec), 55°C (45 sec), and 72°C (1 min). PCR products were analyzed on a 3100 Genetic Analyzer (Applied Biosystems). QLOH was determined by comparing the ratio of fluorescence intensities of each allele in putative LAM (L) cells to that in whole blood (N) from the same patient: (L1/L2)/(N1/N2) (16, 17), where L1 is the diminished allele. QLOH values of 0.6 or less were scored as LOH.
Fluorescent in Situ Hybridization (FISH). Sedimented cells were fixed in methanol:glacial acetic acid (3:1), and the cell suspension was transferred dropwise to microscope slides. The TSC1 probe, RP11-295G24, a 199-kb BAC clone encompassing the TSC1 gene, was obtained from Research Genetics (Huntsville, AL). The TSC2 probe, CDEB9, a 32-kb cosmid clone spanning the 5′ UTR through exon 13 of the TSC2 gene, was generously provided by David Kwiatkowski (Brigham and Women's Hospital, Boston). Probes were labeled directly by using Spectrum Green (TSC1) or Spectrum Orange (TSC2) (Vysis, Downers Grove, IL). Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Images obtained by using a BX40 fluorescence microscope (Olympus, Melville, NY) fitted with an Orca ER digital charge-coupled device camera (Hamamatsu, Bridgewater, NJ) were merged and analyzed by using openlab 3.0 software (Improvision, Lexington, MA). Samples analyzed were chosen from those that had been assayed by PCR, but fluorescent signals were counted by observers who were unaware of the genotyping results. Metaphase preparations of normal human leukocytes were controls to verify that probes hybridized to correct chromosomal locations.
Immunofluorescence Analysis. Cell pellets from either the low-density cell fraction from blood or 20 ml of urine were suspended in 400 μl of 0.1% BSA in PBS, sedimented onto glass slides in a Cytospin 2 centrifuge (Shandon, Pittsburgh), fixed in 4% paraformaldehyde, and permeabilized in 0.1% Triton X-100 in Ca2+- and Mg2+-free PBS. Cells were stained with antibodies against α-smooth muscle actin (1:100) or pan-cytokeratin (1:100) conjugated, respectively, to Cy3 or FITC (Sigma) DAPI (VECTASHIELD, Vector Laboratories). Urine cells were inspected with a Zeiss LSM510 laser scanning confocal microscope, and blood cells were inspected with a Zeiss Axiophot microscope.
Fluorescence-Activated Cell Sorting (FACS). Fractionated cells were stained with monoclonal antibodies directed against glycophorin A (CD235a) and leukocyte common antigen (CD45) (BD Biosciences) conjugated to FITC and APC, respectively. Fluorescent signals were collected by using logarithmic amplifiers. Cell sorting was performed on a flow cytometer (MoFlo, DakoCytomation). Data acquisition, analysis, and compensation were performed by using summit software (DakoCytomation).
Immunohistochemistry. Immunohistochemical studies on 29 LAM formalin-fixed, paraffin-embedded lung specimens were performed to confirm the expression and the localization of glycophorin A in LAM cells. A mouse antibody against CD235a protein (1:50; BD Biosciences) was used, and immunohistochemical staining was performed as described in ref. 16.
Statistical Analysis. We used Fisher's exact test to evaluate the relationship between extent of tissue involvement in LAM and LOH in disseminated cells. P ≤ 0.05 was considered significant.
Results
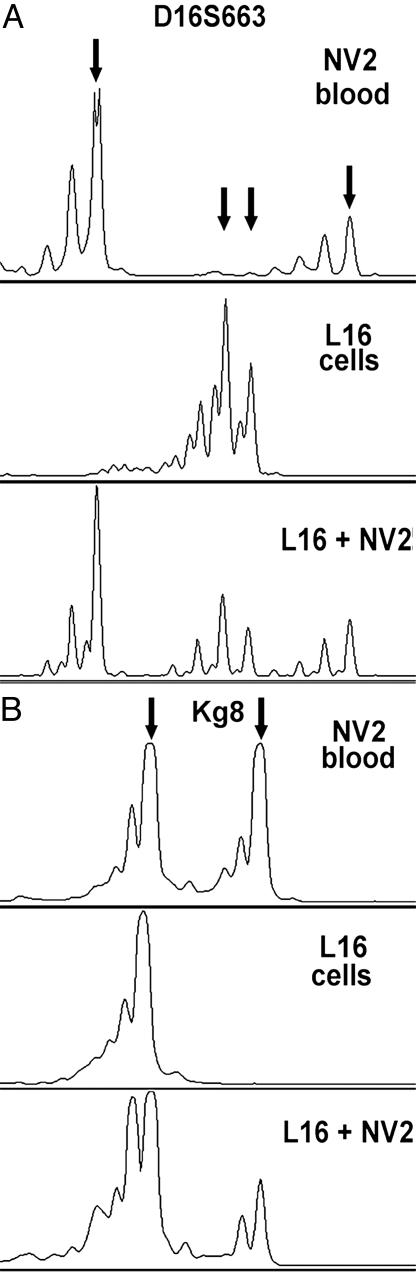
Detection of Exogenous LAM Cells Added to Blood. To determine the feasibility of recovering LAM cells from blood by using the OncoQuick density gradient procedure, we mixed 25 ml of blood from healthy volunteers with 1 × 106 L16 cells cultured from LAM lungs removed at transplantation. PCR microsatellite analysis of the low-density cell fractions revealed genotypes consistent with the presence of LAM cells (Fig. 1A). The L16 cells are homozygous at the Kg8 marker, whereas blood cells from healthy volunteer NV2 are heterozygous (Fig. 1B). An allele ratio of 0.5 was calculated by comparing peak height ratios in the mixture to that in NV2. The observed allele ratio is consistent with one-third of the cells in the low-density cell fraction being homozygous, or LAM cells. Fluorescent in situ hybridization (FISH) analysis of the same sample showed 34% TSC2 allelic loss, likewise consistent with that estimate (Table 1).
Fig. 1.
PCR analysis of a mixture of LAM cells and blood after density-gradient fractionation. (A) Allele peaks for both L16 LAM cells and NV2 blood are detected with marker D16S663. (B) Addition of L16 LAM cells that are homozygous for marker Kg8 to blood that is heterozygous for Kg8 changes the ratio of allele peak heights from 1 to 0.5. Arrows indicate positions of alleles.
Criteria for Identifying a Circulating LAM Cell in Blood. Because the low-density cell fractions contain normal cells whose DNA would be amplified by PCR along with that from LAM cells lacking a TSC2 allele, it was critical to establish threshold values for determining the presence of LOH in mixed cell populations. We defined QLOH as the ratio of fluorescence intensities of the lost allele to the retained allele in cells in the low-density cell fraction compared with that ratio in whole blood. We calculated QLOH values for six chromosome 16p13.3 markers in low-density blood fractions from 25 healthy volunteers and in cells from urine from 17 healthy volunteers compared with matched buffy coat DNA. These values ranged from 0.7 to 1.0 (mean 0.92, SD 0.1, n = 74). Using the mean minus three SD, our threshold value was 0.62. Therefore, patient samples with QLOH values below 0.62 were considered to have lost heterozygosity.
For FISH, the presence of 6.7% or more of cells with only one TSC1 signal or of 20% with only one TSC2 signal was scored as allelic deletion. These threshold values are based on the mean frequency of false positive detection in 19 cases of normal human lymphocytes plus three SD (mean TSC1 = 0.016, SD = 0.017; mean TSC2 = 0.071, SD = 0.044).
Neither smooth muscle actin nor gp100, the melanosomal protein that reacts with monoclonal antibody HMB45, was detected in low-density cell fractions from normal volunteers by immunofluorescent staining (data not shown). However, RT-PCR detected gp100 transcripts in blood from 2 of 15 healthy volunteers, indicating that gp100 is not an appropriate marker for this study because of the potential for false positives.
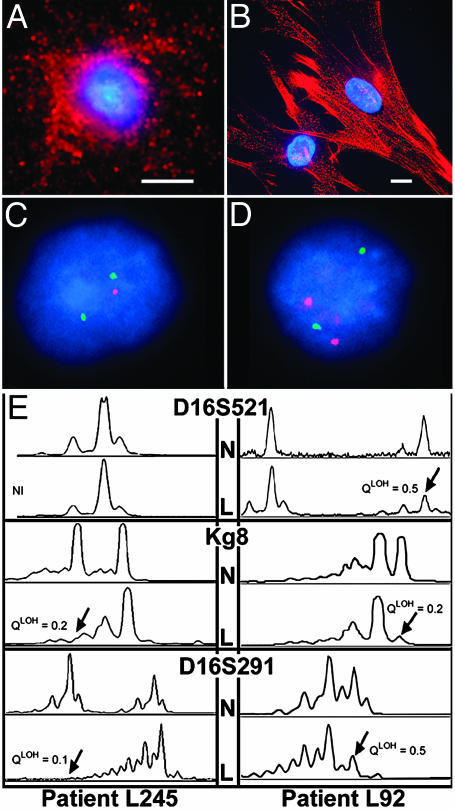
Characterization of Circulating LAM Cells in Blood. We collected the low-density cell fractions from blood of 48 patients with LAM. Of seven stained for smooth muscle actin, each contained cells immunoreactive with a smooth muscle actin antibody (Fig. 2A), but, as might be expected, the staining pattern differed from the stress fibers seen in LAM cells in culture (Fig. 2B).
Fig. 2.
Characterization of cells in the low-density fraction from blood. Representative cells from blood (A) or grown from LAM lungs (B) that reacted with Cy3-conjugated monoclonal antibodies (red) against smooth muscle actin are shown. Nuclei were stained with DAPI (blue). (Scale bar, 20 μm.) FISH is shown in C and D. (C) A LAM cell with only one TSC2 (red) allele and two copies of TSC1 (green). (D) An interphase nucleus with a normal, disomic pattern of two TSC1 and two TSC2 signals. (E) Representative PCR analyses of chromosome 16p13.3 microsatellite markers D16S521, Kg8, and D16S291 in two LAM patients. The upper traces show the peripheral blood (N) and the lower traces the low-density fraction (L) from the same patient. Patient L245 is not informative for marker D16S521. Arrows indicate the diminished peaks.
We analyzed nuclei by dual-color interphase FISH in 20 of the 48 blood samples, as well as 3 urine and 2 chylous fluid cell preparations. One nucleus with allelic loss of TSC2 in a cell containing two TSC1 (Fig. 2C) and one normal nucleus (Fig. 2D) are shown. By using this approach, allelic loss of TSC2 was detected in 15 (75%) of the samples (Table 2). Allelic loss of TSC1 was not detected, consistent with previous reports that most genetic alterations in LAM were in the TSC2 gene (7, 9, 18).
Table 2. Detection of TSC2 allelic loss by FISH.
| Single signal, %
|
TSC2 status
|
||||
|---|---|---|---|---|---|
| Case no. | Fluid | TSC1 | TSC2 | FISH | PCR |
| L38 | Blood | 2 | 34 | Loss | LOH |
| L61 | Blood | 2 | 8 | No | No |
| L70 | Blood | 4 | 26 | Loss | LOH |
| L71 | Blood | 4 | 24 | Loss | LOH |
| L83 | Blood | 2 | 8 | No | No |
| L118 | Blood | 2 | 32 | Loss | LOH |
| L126 | Blood | 0 | 32 | Loss | NI |
| L132 | Blood | 0 | 40 | Loss | LOH |
| L160 | Blood | 2 | 12 | No | No |
| L161 | Blood | 4 | 42 | Loss | NI |
| L163 | Blood | 2 | 24 | Loss | LOH |
| L169 | Blood | 2 | 14 | No | NI |
| L198 | Blood | 2 | 24 | Loss | LOH |
| L208 | Blood | 6 | 22 | Loss | LOH |
| L214 | Blood | 2 | 32 | Loss | LOH |
| L232 | Blood | 5 | 20 | Loss | NI |
| L232 | Blood | 2 | 44 | Loss | NI |
| L239 | Blood | 4 | 30 | Loss | LOH |
| L262 | Blood | 4 | 12 | No | No |
| L306 | Blood | 4 | 20 | Loss | LOH |
| L25 | Urine | 4 | 16 | No | No |
| L38 | Urine | 6 | 30 | Loss | LOH |
| L169 | Urine | 4 | 34 | Loss | NI |
| L83 | Ascites | 4 | 28 | Loss | No |
| L285 | Pleural | 0 | 16 | No | No |
Samples were selected at random from 60 LAM cases. The number of nuclei scored for single signals (FISH) was 40 for L132 and L232, 100 for L83 ascites, and 50 for all others. The presence of 20% or more of cells with only one TSC2 signal was scored as allelic loss (Loss), based on the mean of false positive detection in normal human lymphocytes plus three SD beyond the mean (n = 19). Blood (n = 25) and urine (n = 17) samples from normal volunteers showed no LOH in the PCR assay. Results of the PCR assay are reported as LOH, no LOH (No), or NI (non-informative).
We tested the low-density cell fractions for LOH by PCR amplification of microsatellite markers on chromosome 16p13.3. Representative examples of LOH for three markers from two patients are shown (Fig. 2E). Of the 48 fractionated LAM patient samples, 12 were noninformative because of either insufficient material or homozygosity of tested markers. We observed LOH in 22 cases, which is 61% of informative samples (46% of all samples).
Table 2 presents the data for 20 samples that were analyzed by both FISH and PCR for detection of somatic TSC2 deletions. FISH identified deletion in 15 (75%) and PCR in 11 (55%). Remarkably, FISH and PCR results were concordant in 18 samples, even in low-density cell fractions that by FISH analysis contained cells with two alleles as the major component. In the remaining five cases, FISH allowed designation of an allelic loss status when PCR was not informative.
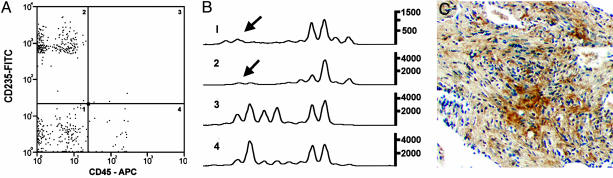
Cell Sorting Improves Detection of LOH in Blood by PCR. Because detection of LOH by microsatellite PCR might be enhanced if numbers of normal blood cells were reduced, we used antibodies against leukocyte common antigen (CD45) and glycophorin A (CD235a) to remove leukocytes and red blood cells, respectively, from 12 low-density cell fractions. Samples were analyzed only by PCR, because the multiple processing steps yielded too few cells for FISH. We observed LOH only in CD45– populations (Fig. 3 A and B), including those that were positive for glycophorin A. In total, we detected LOH in 11 (92%) cell samples separated by FACS after density-gradient fractionation, whereas only 61% were positive for LOH before sorting. No LOH was detected in any sorted cell fractions from healthy volunteers (data not shown).
Fig. 3.
LOH at chromosome 16p13.3 locus in low-density fractions of cells from blood, followed by cell sorting. (A) Representative FACS analysis of cells labeled with monoclonal antibodies conjugated to FITC (CD235a) or APC (CD45). CD45– cells (quadrants 1 and 2) are potentially LAM cells. (B) PCR analyses of microsatellite marker Kg8 in FACS-sorted populations from quadrants shown in A. Arrows indicate positions of missing allele. (C) Photomicrograph showing a positive immunohistochemical reaction for CD235a protein in a LAM nodule (×100).
CD235a reactivity in some LOH-positive cell populations was an unexpected but reproducible result, suggesting the presence of proteins antigenically related to glycophorin A on LAM cells. Lung tissue sections from 29 LAM patients were therefore examined for the presence of reactivity to the CD235a antibody in LAM nodules. Immunoreactivity for CD235a was observed in control red blood cells and in 9 of 29 (31%) LAM patients (Fig. 3C). It was, however, absent in adjacent normal lung and vascular structures.
TSC2 LOH and Disease Characteristics in LAM Patients. Of the 48 LAM patients informative by PCR, 7 also had TSC, and TSC2 LOH was detected in low-density blood cell fractions from 4 of these. Thus, the frequency of detection of somatic deletion was similar in sporadic LAM and TSC-LAM patients.
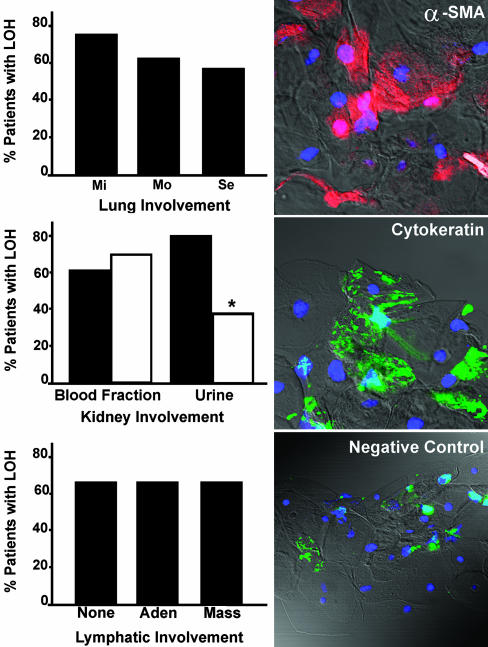
We graded severity of pulmonary disease based on the fraction of lung tissue occupied by cysts and nodules, as estimated on high-resolution CT chest scans (5). The criteria were as follows: normal, no cysts; mild, less than one-third of the lungs involved; moderate, one- to two-thirds of the lungs involved; and severe, more than two-thirds of the lungs involved. There was no statistically significant association between TSC2 LOH in low-density blood cell fractions from 34 informative patients and the extent of lung involvement, although LOH was identified more frequently in patients with mild CT grade (75%) than in moderate (62.5%) and severe cases (57%) (Fig. 4). Identification of LOH in cells from blood or other body fluids may be useful for diagnosis of LAM in patients with mild lung disease and no evident nonpulmonary lesions, which can require additional pathological information to exclude alternative diagnoses.
Fig. 4.
Association between LAM tissue involvement and detection of LOH in blood or urine. (Left) Percentage detection of LOH in low-density cell fractions from bloods of LAM patients with different degrees of lung involvement by high-resolution CT scans. Involvement was rated mild (Mi, n = 12), moderate (Mo, n = 8), and severe (Se, n = 14) (5). Kidney involvement is indicated by the presence (▪) or absence (□) of renal AML and the percentage of 34 LAM patients with LOH in blood or urine. *, P = 0.016 for difference between urine samples from patients with and without AML. Lymphatic involvement evaluated by high-resolution CT scan of chest or abdomen (e.g., Fig. 5D). Masses correspond to lymphangioleiomyomas, which tend to be cystic and filled with chyle when >3 cm in diameter. Data are from patients with no involvement (None, n = 7), adenopathy (Aden, n = 19), and masses (Mass, n = 21). Of the 34 patients represented in all analyses, 65% had LOH in blood samples. (Right) Cytospin preparations of cells from urine were reacted with Cy3-conjugated monoclonal antibodies against smooth muscle actin, FITC-conjugated anti-cytokeratin, or control IgG (negative).
Because renal lesions with the heterogeneous appearance of AML are also diagnostic of LAM, we evaluated concordance between LOH in circulating cells and the presence of AML. LOH was detected in the low-density blood fraction of 63% of LAM patients with AML (Fig. 4) and in 69% without apparent AML. We conclude that LAM cells appear commonly in blood, regardless of the extent of kidney involvement.
We also examined the relationship between extent of lymphatic LAM lesions and detection of LOH in the low-density blood cell fraction. LAM cells were found in the blood of 5 of 7 (71%) patients with no lymphatic involvement (Fig. 4), whereas they were detected in 13 of 19 (68%) patients with adenopathy and in 14 of 21 (67%) patients with lymphangioleiomyomas, the complex chyle-filled masses possibly caused by blockage of lymph vessels at sites of LAM cell (18).
Detection of LAM Cells in Other Body Fluids. To evaluate the possibility that renal tumor cells were shed into the urine, the LOH status of cells from unfractionated 24-h urine samples was investigated. We detected LOH in urine from 11 of 14 (78%) patients with AML but in only 2 of 8 (25%) samples from patients without AML (Fig. 4), a statistically significant difference (P = 0.016). Cells staining for cytokeratin, consistent with shedding of normal epithelial cells, or smooth muscle actin, consistent with the presence of AML cells, were found in urine (Fig. 4). Allelic loss or retention was confirmed in three patients by using FISH (Table 2). Comparison of FISH and PCR data for sedimented cells from three urine cell preparations gave concordant results for two, and, for one sample, PCR was not informative.
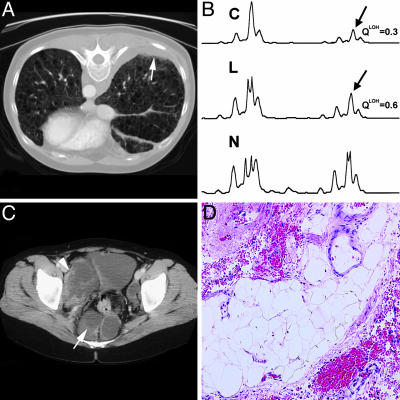
Because chyle is carried in lymphatics, the walls of which may be infiltrated by LAM cells (3) that might be shed, we hypothesized that LAM cells would be readily detected in chyle. We evaluated lymph-derived chylous fluids from three patients. Nuclei from pleural fluid of patient L285 did not show allelic loss of TSC2 (Table 2). Patient L38 presented with a history of expectorating chylous fluid, and chest CT scans showed evidence of pleural effusions (Fig. 5A). Both blood and the expectorated fluid contained cells that had a QLOH ratio consistent with LOH (Fig. 5B).
Fig. 5.
Detection of LOH in body fluids. (A) High-resolution CT scan of the chest with pleural effusion indicated by an arrow. (B) Fluorescent PCR analysis of chromosome 16p13.3 microsatellite marker D16S521 in cells from chyle (C), low-density fraction from blood (L), and whole blood (N). Arrows indicate positions of missing alleles. (C) High-resolution CT scan of the abdomen showing retroperitoneal lymphangioleiomyomas (arrow). (D) Hematoxylin/eosin-stained section of an abdominal tumor biopsy showing fibro-adipose tissue. A and B are from patient L38, and C and D are from patient L83.
Patient L83 presented with extensive abdominal lymphangioleiomyomas (Fig. 5C). We analyzed both chylous ascitic fluid collected from a perirectal mass and a biopsy sample from a pericolonic region that contained tissue surrounding the iliac vessels (Fig. 5D). The biopsy fragments consisted of fibroadipose tissue with no identifiable abnormal smooth muscle cells and did not react with monoclonal antibody HMB45. Allelic loss was observed in 28% of the inspected nuclei (Table 2) from ascitic fluid. However, we failed to detect LOH in the low-density fraction from the blood of L83, either by FISH or by PCR (Table 2). Thus, we were able to detect LAM cells in chylous fluid from lymphatic lesions when LAM cells were not detected in the tissue biopsy or in blood, consistent with the conclusion that ascitic fluid is another potential source of LAM cells.
Discussion
The presence of circulating LAM cells from lung, kidney, or lymphatic sites may identify patients at risk of disease progression or spread. This study proved that specific characteristics, such as chromosomal alterations of the TSC2 locus or staining for smooth muscle actin, which are not present in normal blood cells, permit the identification of disseminated LAM cells. Analyses of the genetic abnormalities in samples of blood or urine or chyle could be a relatively inexpensive and noninvasive means for early diagnosis of disease, as well as perhaps for following its progression and/or response to potential therapy. This study also provides an important link between primary LAM lesions and the process that facilitates dispersion of cells with metastatic potential, which can result in growth of the patient's LAM cells in a transplanted lung (11, 12).
Our goal was to develop markers unique to LAM cells that would distinguish between patients and healthy individuals. To that end, we experimentally defined independent numerical thresholds for detection of LAM cells by FISH and PCR as three SD above mean values for low-density cell fractions from normal volunteers. By using these criteria, FISH and PCR each established the presence or absence of circulating LAM cells in 18 of 19 patients with discordant findings in only one case when both techniques were used and PCR was informative. In our study, 61% of informative (46% of all) LAM samples had detectable levels of microsatellite LOH in low-density blood fractions enriched in LAM cells. Samples that were further purified by removal of blood cells with CD45+ on the surface had detection rates of 92%. Sorting of low-density cell fractions also revealed a subset of cells with LOH that were reactive with CD235a, an antibody to glycophorin A, a sialoglycoprotein present on red blood cells (19). Glycophorin A-like epitopes have been reported on other nonerythroid cells, including normal kidney endothelium, and melanoma and breast cancer cells (20, 21). We found immunoreactivity to CD235a in 31% of the 29 LAM lung samples examined microscopically, suggesting that proteins present in LAM nodules may share epitopes cross-reactive with glycophorin A.
Among our LAM patients, we found a clear correlation between TSC2 LOH identified by PCR and allelic deletion shown by FISH. FISH analysis enabled a determination of allelic deletion in five of six cases that were not informative by PCR. It is intriguing that we were able to obtain QLOH < 0.6 when the percentage of nuclei with one TSC2 signal (FISH) ranged from only 20% to 44%. Similar findings have been reported for highly infiltrative tumors that contain a heterogeneous mixture of stroma, blood vessels, lymphocytes, or other normal cells (22) as well as for QLOH values obtained from experimental mixtures of diploid and aneuploid cells (23). The stringent threshold value for allelic imbalance used in our study underestimates the number of samples that fall into the category of LOH; however, this is preferable to increasing the risk of an erroneous designation of LOH when PCR is used for clinical diagnosis.
It was notable that the frequency of detection of LAM cells in the blood did not parallel the apparent severity of lung pathology. We identified LOH in fractionated blood cells even when the cystic pulmonary lesions were small, consistent with the view that the finding of an allelic imbalance in cells circulating in blood is a valid indicator of LAM. The lower frequency of detection in patients with advanced lung disease may reflect the presence of fewer LAM cells available to be shed as cystic destruction progresses. Renal AML were associated with detection of LOH in 79% in urine samples and only 63% of blood fractions, suggesting that direct shedding into the urine is an important source of the LAM cells found there. Similarly, LAM cells were detected in the blood of 70% of patients with lymphatic disease and were relatively abundant in lymphatic fluid because density-gradient purification was not required for their detection.
RT-PCR of gp100 seemed potentially more powerful than TSC2 LOH as a diagnostic tool for identifying LAM cells in heterogeneous populations. We found, however, an important limitation to its use, i.e., gp100 was detected in blood from two of 15 normal volunteers. In other studies of gp100 expression, although gp100 transcripts were detected in various normal tissues and nonmelanocytic cell lines by RT-PCR, gp100 protein was not detected by Western blot (24) or by immunohistochemistry (25). Our positive findings may reflect the presence of small amounts of gp100 transcripts in normal cells and the high sensitivity of RT-PCR that can amplify such transcripts in tissues where the protein product is not detected.
As we continue to characterize LAM cells isolated from body fluids, we are especially interested to learn how their phenotype may differ from that of LAM cells in tissue lesions, LAM cells associated with other LAM cells of smooth muscle-like appearance, or LAM cells associated with other types of cells in or around LAM nodules and AML. For example, not all LAM nodule cells that are positive for α-smooth muscle actin react detectably with HMB45. The spindle-shaped and epithelioid-appearing cells differ in percentages of HMB45 and PCNA positivity (3). With better understanding of the characteristics of circulating LAM cells, we may find more useful markers for their identification and isolation. Until then, however, demonstration of LOH in an isolated population of these cells from blood can provide a relatively noninvasive and inexpensive means to confirm radiographic diagnosis of LAM, detect the presence of residual disease after lung transplantation or other therapy, and obtain cells for molecular and genetic analysis.
Acknowledgments
We thank the LAM Foundation and Tuberous Sclerosis Alliance for patient referral; Drs. Martha Vaughan, Vincent Manganiello, and Wendy Steagall for critical review and discussion of the manuscript; and Michael Spencer and the National Heart, Lung, and Blood Institute Microscopy Core for technical assistance. T.D. was supported by a grant from the Doris Duke Charitable Foundation.
Author contributions: D.M.C., G.P.-R., and J.M. designed research; D.M.C., G.P.-R., R.D., J.W., and F.K. performed research; T.D. contributed new reagents/analytic tools; D.M.C., G.P.-R., J.P.M., J.W., T.D., and J.M. analyzed data; and D.M.C. and J.M. wrote the paper.
Abbreviations: AML, angiomyolipomas; CT, computed tomography; LAM, lymphangioleiomyomatosis; LOH, loss of heterozygosity; TSC, tuberous sclerosis complex.
References
- 1.Johnson, S. (1999) Thorax 54, 254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu, S. C., Horiba, K., Usuki, J., Avila, N. A., Chen, C. C., Travis, W. D., Ferrans, V. J. & Moss, J. (1999) Chest 115, 1041–1052. [DOI] [PubMed] [Google Scholar]
- 3.Matsui, K., Tatsuguchi, A., Valencia, J., Yu, Z., Bechtle, J., Beasley, M. B., Avila, N., Travis, W. D., Moss, J. & Ferrans, V. J. (2000) Hum. Pathol. 31, 1242–1248. [DOI] [PubMed] [Google Scholar]
- 4.Costello, L. C., Hartman, T. E. & Ryu, J. H. (2000) Mayo Clin. Proc. 75, 591–594. [DOI] [PubMed] [Google Scholar]
- 5.Moss, J., Avila, N. A., Barnes, P. M., Litzenberger, R. A., Bechtle, J., Brooks, P. G., Hedin, C. J., Hunsberger, S. & Kristof, A. S. (2001) Am. J. Respir. Crit. Care Med. 163, 669–671. [DOI] [PubMed] [Google Scholar]
- 6.Franz, D. N., Brody, A., Meyer, C., Leonard, J., Chuck, G., Dabora, S., Sethuraman, G., Colby, T. V., Kwiatkowski, D. J. & McCormack, F. X. (2001) Am. J. Respir. Crit. Care Med. 164, 661–668. [DOI] [PubMed] [Google Scholar]
- 7.Smolarek, T. A., Wessner, L. L., McCormack, F. X., Mylet, J. C., Menon, A. G. & Henske, E. P. (1998) Am. J. Hum. Genet. 62, 810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carsillo, T., Astrinidis, A. & Henske, E. P. (2000) Proc. Natl. Acad. Sci. USA 97, 6085–6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato, T., Seyama, K., Fujii, H., Maruyama, H., Setoguchi, Y., Iwakami, S., Fukuchi, Y. & Hino, O. (2002) J. Hum. Genet. 47, 20–28. [DOI] [PubMed] [Google Scholar]
- 10.Karbowniczek, M., Yu, J. & Henske, E. P. (2003) Am. J. Pathol. 162, 491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karbowniczek, M., Astrinidis, A., Balsara, B. R., Testa, J. R., Lium, J. H., Colby, T. V., McCormack, F. X. & Henske, E. P. (2002) Am. J. Respir. Crit. Care Med. 167, 976–982. [DOI] [PubMed] [Google Scholar]
- 12.Bittman, I., Rolf, B., Amann, G. & Löhrs, U. (1997) Hum. Pathol. 34, 95–98. [DOI] [PubMed] [Google Scholar]
- 13.Bonetti, F., Chiodera, P. L., Pea, M., Martignoni, G., Bosi, F., Zamboni, G. & Mariuzzi, G. M. (1993) Am. J. Surg. Pathol. 17, 1092–1102. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto, Y., Horiba, K., Usuki, J., Chu, S. C., Ferrans, V. J. & Moss, J. (1999) Am. J. Respir. Cell Mol. Biol. 21, 327–336. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg, R., Gertler, R., Friederichs, J., Fuehrer, K., Dahm, M., Phelps, R., Thorban, S., Nekarda, H. & Siewert, J. R. (2002) Cytometry 49, 150–158. [DOI] [PubMed] [Google Scholar]
- 16.Matsui, K., Takeda, K., Yu, Z.-Y., Travis, W. D., Moss, J. & Ferrans, V. J. (2000) Arch. Pathol. Lab. Med. 124, 267–275. [DOI] [PubMed] [Google Scholar]
- 17.Strizheva, G. D., Carsillo, T., Kruger, W. D., Sullivan, E. J., Ryu, J. H. & Henske, E. P. (2001) Am. J. Respir. Crit. Care Med. 163, 253–258. [DOI] [PubMed] [Google Scholar]
- 18.Avila, N. A., Bechtle, J., Dwyer, A. J., Ferrans, V. J. & Moss, J. (2001) Radiology 221, 415–421. [DOI] [PubMed] [Google Scholar]
- 19.Nakahata, T. & Okumura, N. (1994) Leuk. Lymphoma 13, 401–409. [DOI] [PubMed] [Google Scholar]
- 20.Harvey, J., Parsons, S. F., Anstee, D. J. & Bradley, B. A. (1988) Vox Sang. 55, 104–108. [DOI] [PubMed] [Google Scholar]
- 21.Barsoum, A. L., Czuczman, M. S., Bhavanandan, V. P. & Davidson, E. A. (1984) Int. J. Cancer 34, 789–795. [DOI] [PubMed] [Google Scholar]
- 22.Hampton, G. M., Larson, A. A., Baergen, R. N., Sommer, R. L., Kern, S. & Cavenee, W. K. (1996) Proc. Natl. Acad. Sci. USA 93, 6704–6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paulson, T. G., Galipeau, P. C. & Reid, B. J. (1999) Genome Res. 9, 482–491. [PMC free article] [PubMed] [Google Scholar]
- 24.Brouwenstijn, N., Slager, E. H., Bakker, A. B., Schreurs, M. W., Van der Spek, C. W., Adema, G. J., Schrier, P. I. & Figdor, C. G. (1997) Br. J. Cancer 76, 1562–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jungbluth, A. A., Iversen, K., Coplan, K., Williamson, B., Chen, Y. T., Stockert, E., Old, L. J. & Busam, K. J. (1999) Virchows Arch. 434, 429–435. [DOI] [PubMed] [Google Scholar]