Abstract
Purpose
To report the clinical and genetic findings of one family with autosomal recessive cone dystrophy (CD) and to identify the causative mutation.
Methods
An institutional study of three family members from two generations. The clinical examination included best-corrected Snellen visual acuity measurement, fundoscopy, the Farnsworth D-15 color vision test, a full-field electroretinogram (ERG) that incorporated the International Society for Clinical Electrophysiology of Vision standards and methodology, fundus autofluorescence (FAF) and infrared (IR), and spectral-domain optical coherence tomography (SD-OCT). Genetic findings were achieved with DNA analysis using whole exome sequencing (WES) and Sanger sequencing.
Results
The proband, a 9-year-old boy, presented with a condition that appeared to be congenital and stationary. The clinical presentation initially reflected incomplete congenital stationary night blindness (icCSNB) because of myopia, a decrease in visual acuity, abnormal oscillatory potentials, and reduced amplitudes on the 30 Hz flicker ERG but was atypical because there were no clear electronegative responses. However, no disease-causing mutations in the genes underlying icCSNB were identified. Following WES analysis of family members, a homozygous splice-site mutation in intron 3 of TTLL5 (c.182–3_182–1delinsAA) was found cosegregating within the phenotype in the family.
Conclusions
The distinction between icCSNB and CD phenotypes is not always straightforward in young patients. The patient was quite young, which most likely explains why the progression of the CD was not obvious. WES analysis provided prompt diagnosis for this family; thus, the use of this technique to refine the diagnosis is highlighted in this study.
Introduction
Tubulin tyrosine ligase-like 5 (TTLL5; Gene ID: 19963; OMIM: 612268) is a 32-exon gene that encodes the largest protein of 13 members of the tubulin tyrosine ligase-like (TTLL) super-family (Figure 1) [1-4]. TTLL5 was found to be expressed in the heart and skeletal muscle [5] but shows expression in other tissues, such as the brain [6] and the eye (Hs.709609). The TTLL5 protein includes a core tubulin tyrosine ligase (TTL) domain in its N-terminus and a coactivator interaction domain (CID) followed by three receptor interaction domains (RID) at its C-terminus [2,4]. Defects in the TTLL5 gene have been reported to affect the correct function of sperm flagella [4], as well as trigger cone-dominated retinopathy, as cone-rod or cone dystrophy (CRD or CD) [7,8]. The latter represents a heterogeneous group of inherited retinal diseases characterized by primary cone and secondary rod photoreceptor degeneration or no rod photoreceptor degeneration involvement leading to progressive loss of central vision associated with photophobia and color vision abnormalities in childhood or early adult life, due to cone degeneration. Subsequently, patients experience dim light vision disturbances and constriction of the peripheral visual field, due to rod degeneration leading to a severe loss of vision and complete blindness in some cases [9]. In contrast, incomplete congenital stationary night blindness (icCSNB) is represented largely by nonprogressive functional defect, in which the signal transmission from rod and cone photoreceptors to the adjacent ON and OFF bipolar cells is affected, while the structure of the photoreceptors is globally maintained [10]. However, due to several overlapping clinical features of CRD and icCSNB, these disorders can be confounded.
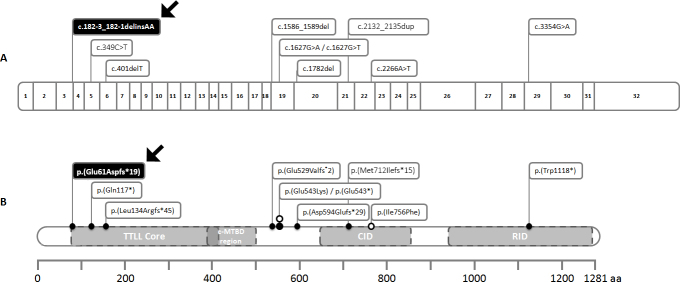
Figure 1.
TTLL5 gene and reported mutations. A: Schematic drawing of exon organization and the corresponding location of mutations in the TTLL5 gene. B: The TTLL5 protein structure shows the predicted consequence of the reported mutations in relation to the domain organization, composed of a core tubulin tyrosine ligase-like (TTLL) domain, a multivalent microtubule recognition region (c-MTBD), a cofactor interaction domain (CID), and a receptor interaction domain (RID). Black arrows point out the mutations reported in this paper, white dots represent missense mutations, and black dots represent nonsense mutations. Amino acids (aa).
Initially, mainly based on in vitro studies, it was hypothesized that TTLL5 plays an important role in the polyglutamylation of primary cilia [11-14]. Immunolocalization studies performed by Sergouniotis et al. in human and murine retinas placed TTLL5 in rods and cones, at the base of the connecting cilium, between the basal body and the ciliary axoneme (or outer segment). This staining was more noteworthy in the cones than in the rods [7]. Furthermore, in contrast to rods, cones are characterized by open membrane disks that lack the complete sheath of the plasma membrane, and in the deficiency of TTLL5, one could foresee more severe damage to cones than to rods [7]. However, mice that lacked functional TTLL5 showed changes in tubulin glutamylation levels only in sperm and no phenotype related to retinal function [4]. Therefore, disruption of tubulin glutamylation does not explain the retinal phenotype. Interestingly, recently, a probable connection between TTLL5 and the functional variant of the retinitis pigmentosa GTPase regulator (RPGR), RPGRORF15, in the photoreceptor cilia was reported [15,16]. It was suggested that TTLL5 binds the basic domain (BD) of RPGRORF15 with its CID, exclusive to TTLL5 among other members of the TTLL super-family. Therefore, TTLL5 is most likely responsible for the glutamylation of RPGRORF15 in its glutamic acid-glycine-rich repetitive region. These findings render TTLL5 a noteworthy disease-causing candidate gene of cone-dominated retinal dystrophies. The most recent reports associate nine presumed loss-of-function mutations in TTLL5 (c.349C>T, p.Gln117*; c.401del, p.Leu134Argfs*45; c.1586_1589del, p.Glu529Valfs*2; c.1627G>T, p.Glu543*; c.1627G>A, p.Glu543Lys; c.1782del, p.Asp594Glufs*29; c.2132_2135dup, p.Met712Ilefs*15; c.2266A>T, p.Ile756Phe; c.3354G>A, Trp1118*) with retinal dystrophies (Figure 1) [7,8]. Although missense and truncating mutations were linked to an isolated retinal phenotype [7], only truncating mutations appear to disrupt the functions of photoreceptors and spermatozoa [8], thus rendering no apparent genotype or phenotype correlation. Moreover, it was found that Rpgr null mice exhibit a similar phenotype compared to Ttll5 mutant mice and are characterized by photoreceptor degeneration and opsin mislocalization [15,17-20]. Bearing in mind the involvement of TTLL5 in CRD and CD and that the respective protein interacts with RPGRORF15, it is important to include this gene in the molecular study of patients affected with retinal dystrophy in general. Here, we identified in a previously misdiagnosed icCSNB case a splice-site mutation in intron 3 of TTLL5 (c.182–3_182–1delinsAA). Reinvestigation of the clinical data corrected the diagnosis to CD.
Methods
Clinical diagnosis and sample collection
Research procedures were conducted in accordance with institutional guidelines and the Declaration of Helsinki; institutional review board approvals were obtained from the participating universities and the national ministry of health of the participating center. Before genetic testing, informed consent was obtained from all family members. Ophthalmic examinations were performed on all subjects. This examination included best-corrected Snellen visual acuity measurement, fundoscopy, the Farnsworth D-15 color vision test, a full-field electroretinogram (ERG) that incorporated the International Society for Clinical Electrophysiology of Vision standards and methodology previously described [21-23], fundus autofluorescence (FAF) and infrared (IR), and spectral-domain optical coherence tomography (SD-OCT).
Sanger sequencing of icCSNB known and candidate genes
The coding exonic and flanking intronic regions of the known genes underlying icCSNB (CACNA1F NM_005183.3, OMIM 300110; CABP4 NM_145200.3, OMIM 608965; CACNA2D4 NM_172364.4, OMIM 608171) [10,24-27] and the candidate genes (ELFN1 NM_001128636.2, OMIM 614964 [28]; GNB3 NM_001297571.1, OMIM 139130 [29,30]) were directly Sanger sequenced on the index patient. PCR conditions are available upon request.
Whole exome sequencing
Exons of genomic DNA samples were captured using Agilent in-solution enrichment methodology (SureSelect SureSelect XT Clinical Research Exome, Agilent, Massy, France) with their biotinylated oligonucleotides probes library (SureSelect XT Clinical Research Exome - 54 Mb, Agilent), followed by paired-end 75 bases massively parallel sequencing on the Illumina HiSeq 4000 system (Illumina, San Diego, CA) [31]. Base calling was achieved using the Illumina Real-Time Analysis software sequence pipeline (2.7.3) with default parameters. Bioinformatics analysis of the resultant sequencing data was done using the IntegraGen constitutional DNA pipeline V2 (IntegraGen, Evry, France).
To rapidly identify the pathogenic variant, WES was applied to all three members of this consanguineous family (Figure 2). Filtering approaches were subsequently applied to identify candidate mutation(s). Referenced variants that occurred homozygously or heterozygously with a minor allele frequency (MAF) >0.005 in dbSNP137, HapMap [32], 1000 Genomes [33], and the NHLBI Exome Sequencing Project Exome Variant Server (EVS) [34] were removed [35-37].
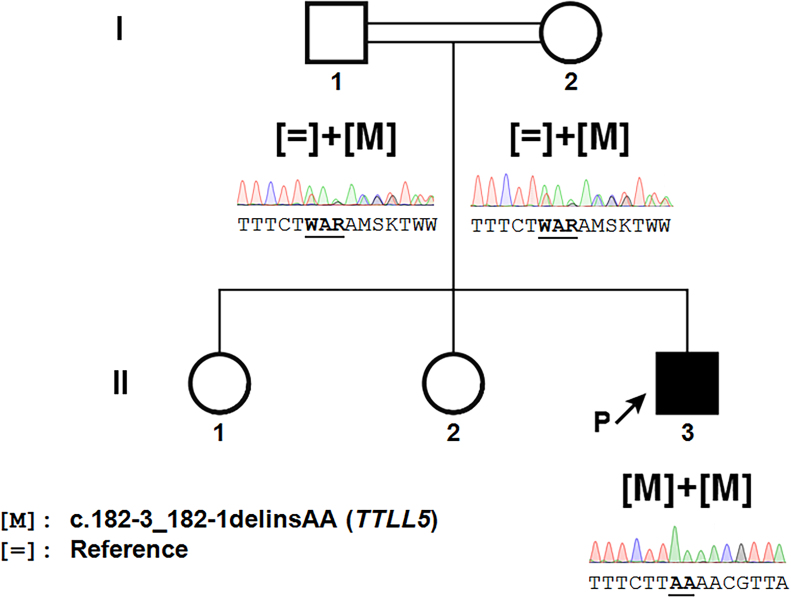
Figure 2.
Novel disease-causing variants in TTLL5 in cone dystrophy: Pedigree of the index patient with disease-causing variants in TTLL5 and cosegregation analysis. The index patient, the proband, is the individual II-3, marked with an arrow. Square symbol=male, round symbol=female, filled symbol=affected, unfilled symbol=unaffected, double line=consanguinity. Underneath the symbols, the sequence electropherograms of the disease-causing variant in TTLL5 is found homozygously in the proband and heterozygously in the parents. The underlined letters correspond to the modifications of the sequence induced by this disease-causing variant.
Molecular validation of the candidate variants
The novel mutation in the TTLL5 gene (c.182–3_182–1delinsAA) was validated using conventional Sanger sequencing according to the manufacturer’s protocols (3730 DNA Analyzer, Applied Biosystems, Weiterstadt, Germany) and tested for cosegregation of the phenotype within the family. However, given that consanguinity among parents was reported for this family, homozygous variants represent the most likely candidate, although this does not completely exclude underlying causative compound-heterozygous mutations [38].
Results
Phenotype and clinical investigations of the affected patient
The index patient was a 9-year-old boy (Figure 2, II-3), corrected for myopia since 3 years of age, who always had difficulty watching television and reading. He never complained of night blindness, peripheral vision loss, and dyschromatopsia, but he had moderate photophobia and wore photochromic glasses. The disease course seemed not progressive. Visual acuity was 20/30 in both eyes with −1.75(−2.25)40° oculus dexter (OD) and −6.25(−1.50)5° oculus sinister (OS). The anterior segments were normal, and the lenses were transparent. The fundus showed a moderate overall depigmentation, absence of the foveal reflex with a slight narrowing of the retinal vessels, and moderate pallor of the optic disc but no atrophy or pigment deposits, which was also confirmed with infrared reflectance images (Figure 3A,B). The retinal autofluorescence images were almost normal except a moderate perifoveal hyperautofluorescence (Figure 3C). Optical coherence tomography (OCT) showed a normal peripheral retina, but we noted bilaterally moderate macular thinning with the ellipsoid zone moderately decreased in the thickness in the temporal part of the fovea (Figure 3D). Color vision testing with Lanthony D-15 HUE color tests showed a bilateral moderate blue-yellow axis of color confusion (not shown). In mixed kinetic and static perimetry, the peripheral isopter IIIc was normal on each side, 90° in temporal and 45° in nasal, but there was a medium decrease in the retinal sensitivity of the central 30°, including in the macular area (Figure 3E). On the ISCEV ERG, the amplitude of the scotopic responses was normal (Figure 3F). However, the photopic responses to a single flash were reduced and showed a moderate electronegative aspect. The 30 Hz flicker responses were reduced to about 30% of the normal value. In the oscillatory potentials (OP), OP1 was absent; in the others, OPs were present but diminished in amplitude (not shown). Due to the reduced photopic responses, the decreased visual acuity, presence of myopia, no obvious disease progression, and absence of night blindness, it was concluded that the patient had an atypical type of icCSNB. Although the name incomplete CSNB is misleading, patients with this disease often reveal the absence of night blindness [10]. Two older sisters had no particular visual difficulties. His parents, both unaffected, were second cousins of Swiss origin. There was no history of this disease in the family. Therefore, the patient was an isolated case, which, given the parental consanguinity, suggested an autosomal recessive mode of inheritance. X-linked heredity could not be excluded, but as the mother had five unaffected brothers, this mode of inheritance was not supported.
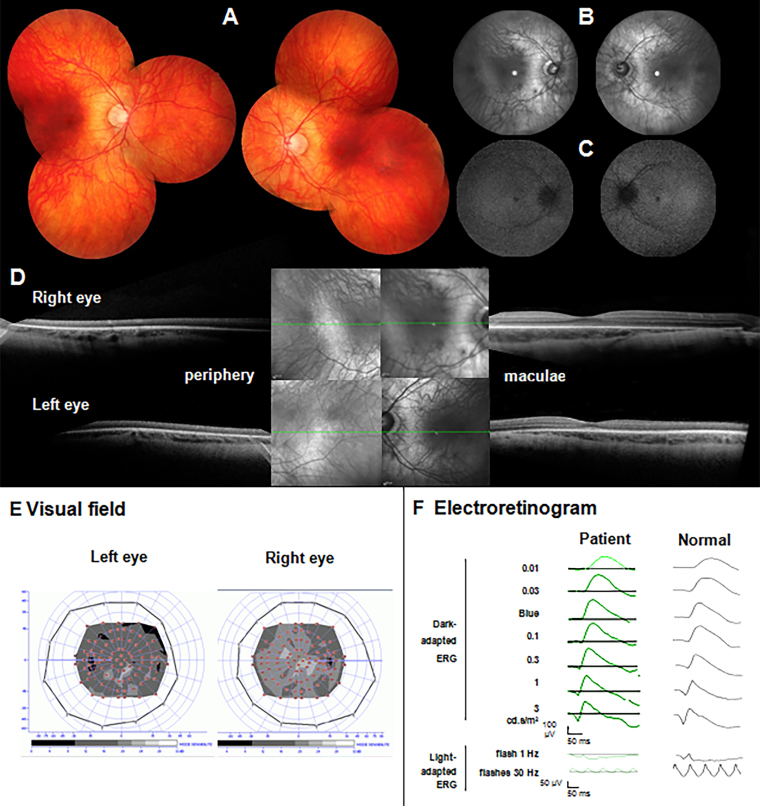
Figure 3.
Clinical investigations of the patient with a homozygous mutation in TTLL5. A: Fundus photographs of the right (left side) and left (right side). There is no macular atrophy or pigment deposits. B, C: The infrared reflectance images (B) and fundus autofluorescence (C) show a moderate increase in perifoveal autofluorescence. D: Optical coherence tomography of the right (up) and left (down) eyes with a selection of peripheral (left) and macular (right) slices for each side show that the periphery is normal while there are subtle changes in the perifoveal area with attenuation of the ellipsoid zone. E: The kinetic (peripheral isopter) and static visual field shows a normal peripheral visual field with a general decrease in retinal sensitivity in the central 30° in accordance with cone dysfunction. F: The International Society for Clinical Electrophysiology of Vision (ISCEV) electroretinogram (ERG) indicates a normal rod function (dark-adapted ERG) while the amplitude values of the cone function (light-adapted ERG) corresponded to 30% of the normal values.
Genotype assessment and mutation cosegregation in the family
Mutation screening showed that none of the known genes underlying icCSNB and the other CSNB candidate genes presented any disease-causing variant. Therefore, WES was performed for the trio, and sequence filtration isolated 15 putative variants (Appendix 1) from 194 insertions/deletions (InDels) and 1,597 single nucleotide variants (SNVs), as the splice-site mutation in intron 3 of the TTLL5 gene, c.182–3_182–1delinsAA (Appendix 2) selected for further cosegregation studies. For all subjects, the overall sequencing coverage of the captured regions was 92% and 88% for 10X and 25X depth of coverage, respectively, resulting in a mean sequencing depth of 109X per base (Appendix 3). Both unaffected parents were found to be heterozygous for this variant, at 40% and 48% in the mother and father, respectively, and the proband was homozygous for this variant, at 99%. The sequencing depth at this position was 132X and 124X in the mother and father, respectively, and 141X for the proband (Appendix 2). This c.182–3_182–1delinsAA mutation in TTLL5 was validated in the index patient and the unaffected parents with direct Sanger sequencing (Figure 2).
Discussion
The patient studied, a 9-year-old boy, presented with a condition that appeared to be congenital and stationary. The clinical presentation initially reflected icCSNB because of myopia, a decrease in visual acuity, abnormal oscillatory potentials, and reduced amplitudes on the 30 Hz flicker ERG but was atypical because there were no clear electronegative responses. However, the subsequent genetic analysis excluded any disease-causing mutations in known genes underlying icCSNB (CACNA1F, CABP4, and CACNA2D4), as well in other candidate CSNB genes (ELFN1 and GNB3). Following WES analysis of the family, a homozygous splice-site mutation in intron 3 of TTLL5 (c.182–3_182–1delinsAA) was found cosegregating within the phenotype in the family. Splice-site prediction software (BDGP: Splice Site Prediction by Neural Network) forecasts the loss of the constitutive acceptor site (c.182–2_182–1) and predicts an alternative constituent acceptor site (nucleotides AG at position c.203_204), with a score of 0.9 by BDGP), which would lead to a truncating mutation p.(Glu61Aspfs*19) or alternatively nonsense mediated mRNA decay (Appendix 4, Figure 1). This will most likely lead to the absence of RPGRORF15 glutamylation and thus lead to the retinal phenotype [15]. Moreover, although in relation to retinal degeneration and pathogenic variants in TTLL5 no strong genotype or phenotype correlation exists (CRD versus CD), patients who present truncating mutations seemed to have a higher probability of reduced fertility [8]. Thus, as the index patient is a boy who possesses a truncating mutation, which is predicted to abolish all functional domains when the mutant protein is formed, it is likely that he will have reduced fertility. This finding may lead to appropriate information provided to the patient at adult age that can eventually be addressed at a fertility clinic.
Recently, defects in the TTLL5 gene were associated with retinal dystrophies that affect primarily cones, leading to either moderate cone dystrophies with onset at adult stages or cone rod dystrophies that start earlier. In total, 14 patients from 11 families have been reported with TTLL5 mutations [7,8]. The following clinical signs were described for some or all of the patients: a variable degree of myopia, dyschromatopsia, reduced visual acuity, high-density concentric perifoveal rings surrounding irregular foveal autofluorescence or hypoautofluorescent patches in the fovea and parafovea combined with irregular autofluorescence outside the foveal region upon fundus autofluorescence imaging, optical coherence tomography abnormalities consistent with photoreceptor loss confined to the foveal region or observed throughout the scan, and mainly altered severely reduced photopic responses due to the cone defect with some patients showing altered scotopic responses due to rod involvement. In contrast, icCSNB is a nonprogressive retinal disorder mainly characterized by the presence or absence of night blindness, decreased visual acuity, photophobia with reduced scotopic b-wave responses and reduced photopic responses, and often high myopia. Due to the reduced photopic responses, the decreased visual acuity, the presence of myopia, no obvious disease progression, and the absence of night blindness, it was concluded that the patient in the study had an atypical type of icCSNB. However, detailed clinical investigation of this patient showed moderate overall depigmentation, a slight narrowing of the retinal vessels, moderate pallor of the optic disc, moderate perifoveal hyperautofluorescence, and bilateral macular thinning with the ellipsoid zone moderately decreased in thickness in the temporal area of the fovea.
Most of the clinical documentation of the previously described patients with TTLL5 mutations was presented for patients older than 30 years old with a progressive disease. In this study, the observation of a young patient (9 years) led to the misdiagnosis of a stationary disease, pointing out the difficulty of making the correct clinical diagnosis of a TTLL5 dystrophy at an early stage without the help of molecular testing. In the present study, WES analysis provided prompt diagnosis for this family; thus, the use of this technique to refine the diagnosis is highlighted. In addition, sequencing of exomes of trios should become an early part of the diagnostic workup of retinal dystrophy. An alternative, rapid, and less bioinformatically demanding method would have been to investigate the patient before WES was performed with targeted next-generation sequencing covering all genes underlying inherited retinal disorders [9].
Acknowledgments
The study was supported by Fondation Voir et Entendre (CZ), Prix Dalloz for “La recherche en ophtalmologie” (CZ), Ville de Paris and Region Ile de France, LABEX LIFESENSES [reference ANR-10-LABX-65] supported by French state funds managed by the Agence Nationale de la Recherche within the Investissements d'Avenir program [ANR-11-IDEX-0004–0], Foundation Fighting Blindness center grant [C-CMM-0907–0428-INSERM04], Agence Nationale de la Recherche (ANR-12-BSVS1–0012–01_GPR179; CZ) and ERC-Synergy HELMHOLTZ (JAS). Authors declare no conflict of interest. The authors are thankful to the patient and family members participating in the study.
Appendix 1.
To access the data, click or select the words “Appendix 1.” Insertions/deletions (InDels) and single nucleotide variants (SNVs) identified by whole-exome sequencing after filtering dbSNP, single-nucleotide polymorphism database; MAF, minor allele frequency; EVS, exome variant server; Eur.Am., European American; Afr.Am., African American; ExAC, exome aggregation consortium; AFR, African; AMR, American; EAS, East Asian; SAS, South Asian; NFE, Non-Finnish European; FIN, Finnish; OTH, Other; SIFT, Sorting Intolerant from Tolerant; UCSC, UCSC Genome Browser.
Appendix 2.
To access the data, click or select the words “Appendix 2.” Novel disease-causing variant identified in TTLL5 (NM_015072.4) by whole-exome sequencing after filtering. HOM_mut, homozygous mutated; HTZ, heterozygous mutated.
Appendix 3. Quality control of the whole-exome sequencing.
To access the data, click or select the words “Appendix 3.”
Appendix 4. Predicted Wilt-type coding sequence.
To access the data, click or select the words “Appendix 4.”
References
- 1.Trichet V, Ruault M, Roizes G, De Sario A. Characterization of the human tubulin tyrosine ligase-like 1 gene (TTLL1) mapping to 22q13.1. Gene. 2000;257:109–17. doi: 10.1016/s0378-1119(00)00383-8. [DOI] [PubMed] [Google Scholar]
- 2.Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temurak N, van Dijk J, Boucher D, van Dorsselaer A, Suryavanshi S, Gaertig J, Edde B. Tubulin polyglutamylase enzymes are members of the TTL domain protein family. Science. 2005;308:1758–62. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 3.van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C. A targeted multienzyme mechanism for selective microtubule polyglutamylation. Mol Cell. 2007;26:437–48. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Lee GS, He Y, Dougherty EJ, Jimenez-Movilla M, Avella M, Grullon S, Sharlin DS, Guo C, Blackford JA, Jr, Awasthi S, Zhang Z, Armstrong SP, London EC, Chen W, Dean J, Simons SS., Jr Disruption of Ttll5/stamp gene (tubulin tyrosine ligase-like protein 5/SRC-1 and TIF2-associated modulatory protein gene) in male mice causes sperm malformation and infertility. J Biol Chem. 2013;288:15167–80. doi: 10.1074/jbc.M113.453936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He Y, Simons SS., Jr STAMP, a novel predicted factor assisting TIF2 actions in glucocorticoid receptor-mediated induction and repression. Mol Cell Biol. 2007;27:1467–85. doi: 10.1128/MCB.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch Grau M, Gonzalez Curto G, Rocha C, Magiera MM, Marques Sousa P, Giordano T, Spassky N, Janke C. Tubulin glycylases and glutamylases have distinct functions in stabilization and motility of ependymal cilia. J Cell Biol. 2013;202:441–51. doi: 10.1083/jcb.201305041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sergouniotis PI, Chakarova C, Murphy C, Becker M, Lenassi E, Arno G, Lek M, MacArthur DG. Consortium UC-E, Bhattacharya SS, Moore AT, Holder GE, Robson AG, Wolfrum U, Webster AR, Plagnol V. Biallelic variants in TTLL5, encoding a tubulin glutamylase, cause retinal dystrophy. Am J Hum Genet. 2014;94:760–9. doi: 10.1016/j.ajhg.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedoni N, Haer-Wigman L, Vaclavik V, Tran HV, Farinelli P, Balzano S, Royer-Bertrand B, El-Asrag ME, Bonny O, Ikonomidis C, Litzistorf Y, Nikopoulos K, Yioti G, Stefaniotou M, McKibbin M, Ellingford J, Booth AP, Black G, Toomes C, Inglehearn CF, Hoyng CB, Bax N, Klaver CC, Thiadens AA, Murisier F, Schorderet DF, Ali M, Cremers FP, Andreasson S, Munier FL, Rivolta C. Mutations in the polyglutamylase gene TTLL5, expressed in photoreceptor cells and spermatozoa, are associated with cone-rod degeneration and reduced male fertility. Hum Mol Genet. 2016;25:4546–55. doi: 10.1093/hmg/ddw282. [DOI] [PubMed] [Google Scholar]
- 9.Boulanger-Scemama E, El Shamieh S, Demontant V, Condroyer C, Antonio A, Michiels C, Boyard F, Saraiva JP, Letexier M, Souied E, Mohand-Said S, Sahel JA, Zeitz C, Audo I. Next-generation sequencing applied to a large French cone and cone-rod dystrophy cohort: mutation spectrum and new genotype-phenotype correlation. Orphanet J Rare Dis. 2015;10:85. doi: 10.1186/s13023-015-0300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeitz C, Robson AG, Audo I. Congenital stationary night blindness: An analysis and update of genotype-phenotype correlations and pathogenic mechanisms. Prog Retin Eye Res. 2015;45:58–110. doi: 10.1016/j.preteyeres.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Janke C, Rogowski K, van Dijk J. Polyglutamylation: a fine-regulator of protein function? ‘Protein Modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:636–41. doi: 10.1038/embor.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Dijk J, Miro J, Strub JM, Lacroix B, van Dorsselaer A, Edde B, Janke C. Polyglutamylation is a post-translational modification with a broad range of substrates. J Biol Chem. 2008;283:3915–22. doi: 10.1074/jbc.M705813200. [DOI] [PubMed] [Google Scholar]
- 13.Backer CB, Gutzman JH, Pearson CG, Cheeseman IM. CSAP localizes to polyglutamylated microtubules and promotes proper cilia function and zebrafish development. Mol Biol Cell. 2012;23:2122–30. doi: 10.1091/mbc.E11-11-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–86. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 15.Sun X, Park JH, Gumerson J, Wu Z, Swaroop A, Qian H, Roll-Mecak A, Li T. Loss of RPGR glutamylation underlies the pathogenic mechanism of retinal dystrophy caused by TTLL5 mutations. Proc Natl Acad Sci USA. 2016;113:E2925–34. doi: 10.1073/pnas.1523201113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu X, Fry AM, Tulloch B, Manson FD, Crabb JW, Khanna H, Faragher AJ, Lennon A, He S, Trojan P, Giessl A, Wolfrum U, Vervoort R, Swaroop A, Wright AF. RPGR ORF15 isoform co-localizes with RPGRIP1 at centrioles and basal bodies and interacts with nucleophosmin. Hum Mol Genet. 2005;14:1183–97. doi: 10.1093/hmg/ddi129. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Hong DH, Pawlyk B, Yue G, Adamian M, Grynberg M, Godzik A, Li T. The retinitis pigmentosa GTPase regulator (RPGR)- interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc Natl Acad Sci USA. 2003;100:3965–70. doi: 10.1073/pnas.0637349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson DA, Khan NW, Othman MI, Chang B, Jia L, Grahek G, Wu Z, Hiriyanna S, Nellissery J, Li T, Khanna H, Colosi P, Swaroop A, Heckenlively JR. Rd9 is a naturally occurring mouse model of a common form of retinitis pigmentosa caused by mutations in RPGR-ORF15. PLoS One. 2012;7:e35865. doi: 10.1371/journal.pone.0035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong DH, Pawlyk BS, Shang J, Sandberg MA, Berson EL, Li T. A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3). Proc Natl Acad Sci USA. 2000;97:3649–54. doi: 10.1073/pnas.060037497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong DH, Yue G, Adamian M, Li T. Retinitis pigmentosa GTPase regulator (RPGRr)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J Biol Chem. 2001;276:12091–9. doi: 10.1074/jbc.M009351200. [DOI] [PubMed] [Google Scholar]
- 21.Marmor MF, Fulton AB, Holder GE, Miyake Y, Brigell M, Bach M. ISCEV Standard for full-field clinical electroretinography (2008 update). Doc Ophthalmol. 2009;118:69–77. doi: 10.1007/s10633-008-9155-4. [DOI] [PubMed] [Google Scholar]
- 22.Aleman TS, Lam BL, Cideciyan AV, Sumaroka A, Windsor EA, Roman AJ, Schwartz SB, Stone EM, Jacobson SG. Genetic heterogeneity in autosomal dominant retinitis pigmentosa with low-frequency damped electroretinographic wavelets. Eye (Lond) 2009;23:230–3. doi: 10.1038/eye.2008.264. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson SG, Yagasaki K, Feuer WJ, Roman AJ. Interocular asymmetry of visual function in heterozygotes of X-linked retinitis pigmentosa. Exp Eye Res. 1989;48:679–91. doi: 10.1016/0014-4835(89)90009-2. [DOI] [PubMed] [Google Scholar]
- 24.Strom TM, Nyakatura G, Apfelstedt-Sylla E, Hellebrand H, Lorenz B, Weber BH, Wutz K, Gutwillinger N, Ruther K, Drescher B, Sauer C, Zrenner E, Meitinger T, Rosenthal A, Meindl A. An L-type calcium-channel gene mutated in incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:260–3. doi: 10.1038/940. [DOI] [PubMed] [Google Scholar]
- 25.Bech-Hansen NT, Naylor MJ, Maybaum TA, Pearce WG, Koop B, Fishman GA, Mets M, Musarella MA, Boycott KM. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:264–7. doi: 10.1038/947. [DOI] [PubMed] [Google Scholar]
- 26.Zeitz C, Kloeckener-Gruissem B, Forster U, Kohl S, Magyar I, Wissinger B, Matyas G, Borruat FX, Schorderet DF, Zrenner E, Munier FL, Berger W. Mutations in CABP4, the gene encoding the Ca2+-binding protein 4, cause autosomal recessive night blindness. Am J Hum Genet. 2006;79:657–67. doi: 10.1086/508067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wycisk KA, Zeitz C, Feil S, Wittmer M, Forster U, Neidhardt J, Wissinger B, Zrenner E, Wilke R, Kohl S, Berger W. Mutation in the auxiliary calcium-channel subunit CACNA2D4 causes autosomal recessive cone dystrophy. Am J Hum Genet. 2006;79:973–7. doi: 10.1086/508944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cao Y, Sarria I, Fehlhaber KE, Kamasawa N, Orlandi C, James KN, Hazen JL, Gardner MR, Farzan M, Lee A, Baker S, Baldwin K, Sampath AP, Martemyanov KA. Mechanism for Selective Synaptic Wiring of Rod Photoreceptors into the Retinal Circuitry and Its Role in Vision. Neuron. 2015;87:1248–60. doi: 10.1016/j.neuron.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vincent A, Audo I, Tavares E, Maynes JT, Tumber A, Wright T, Li S, Michiels C, Consortium GNB, Condroyer C, MacDonald H, Verdet R, Sahel JA, Hamel CP, Zeitz C, Heon E. Biallelic Mutations in GNB3 Cause a Unique Form of Autosomal-Recessive Congenital Stationary Night Blindness. Am J Hum Genet. 2016;98:1011–9. doi: 10.1016/j.ajhg.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arno G, Holder GE, Chakarova C, Kohl S, Pontikos N, Fiorentino A, Plagnol V, Cheetham ME, Hardcastle AJ, Webster AR, Michaelides M, Consortium UKIRD. Recessive Retinopathy Consequent on Mutant G-Protein beta Subunit 3 (GNB3). JAMA Ophthalmol. 2016;134:924–7. doi: 10.1001/jamaophthalmol.2016.1543. [DOI] [PubMed] [Google Scholar]
- 31.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, Dermitzakis E, Bonnen PE, Altshuler DM, Gibbs RA, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Yu F, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Gibbs RA, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Dermitzakis E, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Bonnen PE, Gibbs RA, Gonzaga-Jauregui C, Keinan A, Price AL, Yu F, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Schaffner SF, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Pollack S, Price AL, Schaffner SF, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, Kang HM, Jordan D, Leal SM, Gabriel S, Rieder MJ, Abecasis G, Altshuler D, Nickerson DA, Boerwinkle E, Sunyaev S, Bustamante CD, Bamshad MJ, Akey JM, Broad GO, Seattle GO, Project NES. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Audo I, Bujakowska K, Orhan E, El Shamieh S, Sennlaub F, Guillonneau X, Antonio A, Michiels C, Lancelot ME, Letexier M, Saraiva JP, Nguyen H, Luu TD, Leveillard T, Poch O, Dollfus H, Paques M, Goureau O, Mohand-Said S, Bhattacharya SS, Sahel JA, Zeitz C. The familial dementia gene revisited: a missense mutation revealed by whole-exome sequencing identifies ITM2B as a candidate gene underlying a novel autosomal dominant retinal dystrophy in a large family. Hum Mol Genet. 2014;23:491–501. doi: 10.1093/hmg/ddt439. [DOI] [PubMed] [Google Scholar]
- 36.Audo I, Bujakowska K, Orhan E, Poloschek CM, Defoort-Dhellemmes S, Drumare I, Kohl S, Luu TD, Lecompte O, Zrenner E, Lancelot ME, Antonio A, Germain A, Michiels C, Audier C, Letexier M, Saraiva JP, Leroy BP, Munier FL, Mohand-Said S, Lorenz B, Friedburg C, Preising M, Kellner U, Renner AB, Moskova-Doumanova V, Berger W, Wissinger B, Hamel CP, Schorderet DF, De Baere E, Sharon D, Banin E, Jacobson SG, Bonneau D, Zanlonghi X, Le Meur G, Casteels I, Koenekoop R, Long VW, Meire F, Prescott K, de Ravel T, Simmons I, Nguyen H, Dollfus H, Poch O, Leveillard T, Nguyen-Ba-Charvet K, Sahel JA, Bhattacharya SS, Zeitz C. Whole-Exome Sequencing Identifies Mutations in GPR179 Leading to Autosomal-Recessive Complete Congenital Stationary Night Blindness. Am J Hum Genet. 2012;90:321–30. doi: 10.1016/j.ajhg.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeitz C, Jacobson SG, Hamel CP, Bujakowska K, Neuille M, Orhan E, Zanlonghi X, Lancelot ME, Michiels C, Schwartz SB, Bocquet B, Congenital Stationary Night Blindness C, Antonio A, Audier C, Letexier M, Saraiva JP, Luu TD, Sennlaub F, Nguyen H, Poch O, Dollfus H, Lecompte O, Kohl S, Sahel JA, Bhattacharya SS, Audo I. Whole-exome sequencing identifies LRIT3 mutations as a cause of autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2013;92:67–75. doi: 10.1016/j.ajhg.2012.10.023. </jrn>. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benayoun L, Spiegel R, Auslender N, Abbasi AH, Rizel L, Hujeirat Y, Salama I, Garzozi HJ, Allon-Shalev S, Ben-Yosef T. Genetic heterogeneity in two consanguineous families segregating early onset retinal degeneration: the pitfalls of homozygosity mapping. Am J Med Genet A. 2009;149A:650–6. doi: 10.1002/ajmg.a.32634. [DOI] [PubMed] [Google Scholar]