Abstract
Alzheimer’s disease causes a progressive dementia that currently affects over 35 million individuals worldwide and is expected to affect 115 million by 2050 (ref. 1). There are no cures or disease-modifying therapies, and this may be due to our inability to detect the disease before it has progressed to produce evident memory loss and functional decline. Biomarkers of preclinical disease will be critical to the development of disease-modifying or even preventative therapies2. Unfortunately, current biomarkers for early disease, including cerebrospinal fluid tau and amyloid-β levels3, structural and functional magnetic resonance imaging4 and the recent use of brain amyloid imaging5 or inflammaging6, are limited because they are either invasive, time-consuming or expensive. Blood-based biomarkers may be a more attractive option, but none can currently detect preclinical Alzheimer’s disease with the required sensitivity and specificity7. Herein, we describe our lipidomic approach to detecting preclinical Alzheimer’s disease in a group of cognitively normal older adults. We discovered and validated a set of ten lipids from peripheral blood that predicted phenoconversion to either amnestic mild cognitive impairment or Alzheimer’s disease within a 2–3 year timeframe with over 90% accuracy. This biomarker panel, reflecting cell membrane integrity, may be sensitive to early neurodegeneration of preclinical Alzheimer’s disease.
We enrolled 525 community-dwelling participants, aged 70 and older and otherwise healthy, into this 5-year observational study. Over the course of the study, 74 participants met criteria for amnestic mild cognitive impairment (aMCI) or mild Alzheimer’s disease (AD) (Online Methods); 46 were incidental cases at entry, and 28 phenoconverted (Converters) from nonimpaired memory status at entry (Converterpre). The average time for phenoconversion to either aMCI or AD was 2.1 years (range 1–5 years). We defined three main participant groups in this paper: aMCI/AD, Converter and Normal Control (NC). The participants with aMCI and mild AD were combined into a single group (aMCI/AD) because this group was defined by a primary memory impairment, and aMCI is generally thought to reflect the earliest clinically detectable stage of AD. The aMCI/AD group included the Converters after phenoconversion. The Converters were included at two time points, prior to phenoconversion (Converterpre), when memory was not impaired, and after phenoconversion (post), when memory was impaired and they met criteria for either aMCI or AD. The NC group was selected to match the whole aMCI/AD group on the basis of age, education and sex. In the third year of the study, we selected 53 participants with either aMCI or AD for metabolomic and lipidomic biomarker discovery. Included in this aMCI/AD group were 18 Converters. We also selected 53 matched cognitively normal control (NC) participants. For the Converters, blood from both time 0 (at entry to the study) and after phenoconversion was used; for the other subjects, blood from the last available visit was used. We used an internal cross-validation procedure to evaluate the accuracy of the discovered lipidomics profile in classifying 41 additional subjects, consisting of the remaining subset of 21 participants with aMCI/AD, including 10 Converters, and 20 matched NC participants (Supplementary Table 1 and Supplementary Fig. 1).
The aMCI/AD, Converter and NC groups were defined primarily using a composite measure of memory performance (the decline in Zmem for the Converters (Cpre versus Cpost) is shown Fig. 1a). In addition, composite measures of other cognitive abilities (Supplementary Fig. 2) and measures of memory complaints and functional capacities were compiled (Supplementary Tables 2 and 3). The discovery and validation groups did not differ on clinical measures (F(4,170) = 1.376, P = 0.244) or on any composite z-score (F(5,169) = 2.118, P = 0.066), demonstrating the general equivalence of the participants used for the discovery and validation phases of the biomarker analysis.
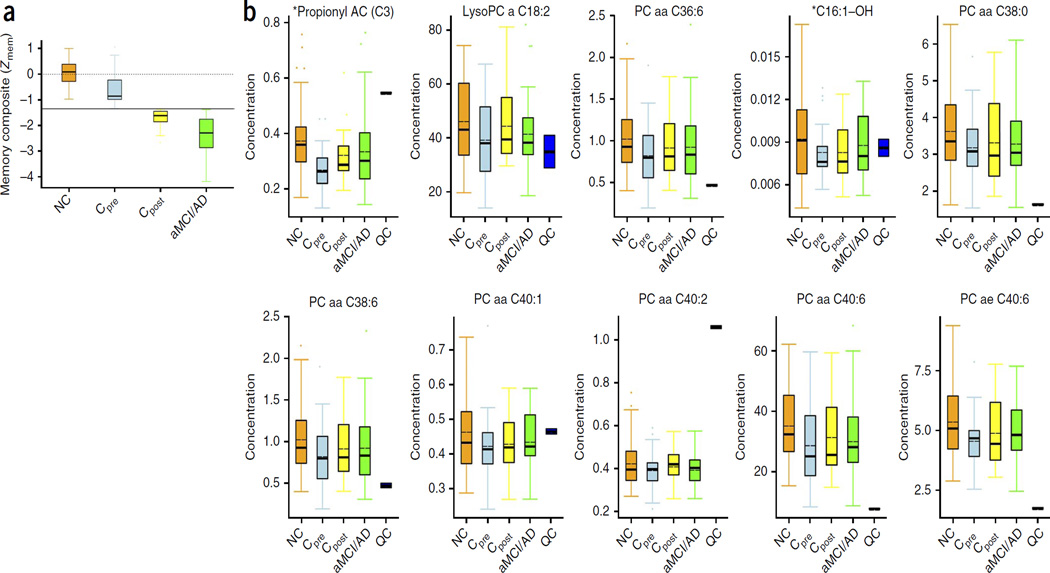
Figure 1.
Memory composite z-scores and trend plots for the ten-metabolite panel in the discovery phase. (a) Box and whisker plot shows the composite memory z-scores (Zmem) of the combined discovery and validation samples (Supplementary Table 3). The performance of the Converter group (Cpre, Converters at baseline) after phenoconversion (Cpost) is plotted for direct comparison. The plot shows Zmem, as described in Supplementary Table 3. The dotted line centered on 0 represents the median memory composite z-score for the entire cohort of 525 participants, and the black horizontal line represents the cut-off for impairment (−1.35 s.d.). Error bars represent ±s.e.m. As defined, all converters had nonimpaired memory at baseline and impaired memory after phenoconversion. NC, n = 73; Cpre, n = 28; Cpost, n = 28; and aMCI/AD, n = 46. (b) The SID-MRM-MS–based quantitative profiling data was subjected to the nonparametric Kruskal-Wallis test using the STAT pack module (Biocrates). Results are shown for a panel of ten metabolites in the NC group (n = 53), Cpre (n = 18), Cpost (n = 18) and aMCI/AD (n = 35) groups, respectively. The abundance of each metabolite is plotted as normalized concentrations units (nM). The black solid bars within the boxplot represent the median abundance, and the dotted line represents mean abundance for the given group. Error bars represent ± s.d. QC, quality control samples. The P values for analytes between groups were P ≤ 0.05. The two metabolites with P values <0.005 are indicated with an asterisk. Each Kruskal-Wallis test was followed by Mann-Whitney U-tests for post hoc pairwise comparisons (NC versus Cpre and NC versus aMCI/AD). Significance was adjusted for multiple comparisons using Bonferroni’s method (P < 0.025).
We examined 124 plasma samples from the 106 discovery-phase participants for untargeted metabolomic analysis (Online Methods). Metabolomic and lipidomic profiling yielded 2,700 positive-mode features and 1,900 negative-mode features. Metabolites defining the participant groups were selected using the least absolute shrinkage and selection operator (LASSO) penalty8,9. The LASSO analysis revealed features that assisted in unambiguous class separation between the two nonimpaired groups, the Converterpre group and the NC subjects who do not phenoconvert (Table 1). This untargeted analysis revealed considerably lower phosphatidylinositol in the Converterpre group and higher glycoursodeoxycholic acid in the aMCI/AD group compared to the NC group. These metabolites were unambiguously identified using tandem mass spectrometry (Supplementary Fig. 3).
Table 1.
Putative metabolite markers resulting from binary comparison of the study groups
| Metabolite | LASSO coefficient |
Comparison groups | Mode | Mass/charge ratio |
|---|---|---|---|---|
| Phospatidylinositol (18:0/0:0) | ↓ (−0.674) | NC versus Converterpre | NEG | 599.3226 |
| Proline-asparagine dipeptide | ↑ (0.192) | NC versus aMCI/AD | POS | 230.1146 |
| Glycoursodeoxycholic acid | ↑ (0.107) | NC versus aMCI/AD | POS | 450.3196 |
| Malic acid | ↓ (−0.024) | NC versus aMCI/AD | POS | 134.0207 |
The markers were chosen on the basis of significant predictive value as determined by LASSO coefficient analysis. The positive estimated LASSO coefficient suggests elevation in corresponding comparison group (aMCI/AD and Converterpre) compared to NC participants. Arrows indicate upregulation or downregulation in the comparison group as compared to the NC participants. NEG, negative; POS, positive.
The untargeted LASSO analysis revealed amino acids and phospholipids to be potent discriminators of the NC and aMCI/AD groups. Thus, we performed stable isotope dilution–multiple reaction monitoring (MRM) mass spectrometry (SID-MRM-MS) to unambiguously identify and quantify lipids, amino acids and biogenic amines; this would discriminate our groups with emphasis on differences that might predict phenoconversion from NC to aMCI/AD. This targeted analysis revealed significantly lower plasma levels of serotonin, phenylalanine, proline, lysine, phosphatidylcholine (PC), taurine and acylcarnitine (AC) in Converterpre participants who later phenoconverted to aMCI/AD (Table 2).
Table 2.
Difference detection of putative metabolites using SID-MRM-MS
| Metabolite | Fold change | Comparison groups | Mode | P value |
|---|---|---|---|---|
| PC ae C38:4 | ↓ | NC versus Converterpre | POS | 0.00417 |
| Proline | ↓ | NC versus Converterpre | POS | 0.00003 |
| Lysine | ↓ | NC versus Converterpre | POS | 0.0020 |
| Serotonin | ↓ | NC versus Converterpre | POS | 0.0160 |
| Taurine | ↓ | NC versus Converterpre | POS | 0.0030 |
| DOPA | ↑ | NC versus Converterpre | POS | 0.0001 |
| Phenylalanine | ↓ | NC versus Converterpre | POS | 0.00001 |
| Acylcarnitine C7-DC | ↓ | NC versus aMCI/AD | POS | 0.0001 |
The arrows indicate upregulation or downregulation in the comparison group as compared to the NC participants. DOPA, dihydroxyphenylalanine; C7-DC, pimelyl-l-carnitine.
A notable finding of this targeted metabolomic and lipidomic analysis was the identification of a set of ten metabolites, comprising PCs, (PC diacyl (aa) C36:6, PC aa C38:0, PC aa C38:6, PC aa C40:1, PC aa C40:2, PC aa C40:6, PC acyl-alkyl (ae) C40:6), lysophophatidylcholine (lysoPC a C18:2), and acylcarnitines (ACs) (Propionyl AC (C3) and C16:1-OH) that were depleted in the plasma of the Converterpre participants but not in that of the NC group (Fig. 1b). These metabolites remained depleted after phenoconversion to aMCI/AD (Converterspost) and were similar to the levels in the aMCI/AD group.
We then performed targeted quantitative metabolomic and lipidomic analyses using plasma from a separate group of 40 participants as an independent blinded cross-validation, as one sample from the aMCI/AD group was not available for lipidomic analysis. The validation samples were obtained from those clinically defined NC, Converterpre, aMCI/AD subjects. The samples were processed and analyzed using the same SID-MRM-MS technique as in the discovery phase. The targeted quantitative analysis of the validation set revealed similar levels for the ten-metabolite panel (Supplementary Fig. 4) as were observed in the discovery samples (Fig. 1b).
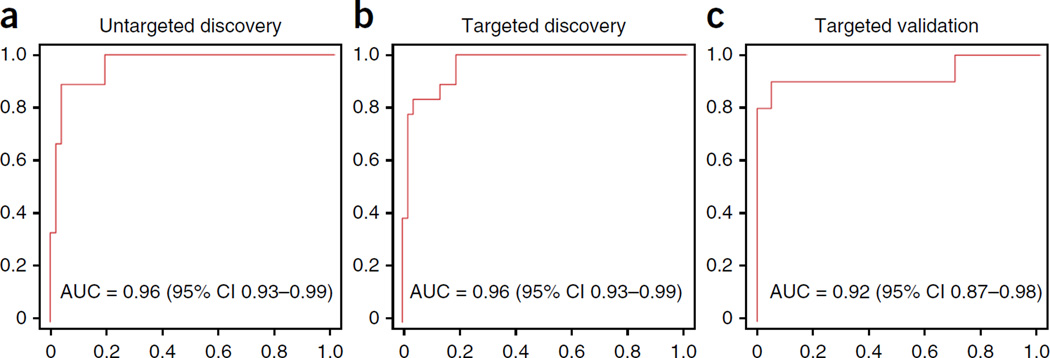
We used the metabolomic data from the untargeted LASSO analysis to build separate linear classifier models that would distinguish the aMCI/AD and Converterpre groups from the NC group. We used receiver operating characteristic (ROC) analysis to assess the performance of the classifier models for group classification. For the Converterpre and NC group classification, the initial LASSO-identified metabolites yielded a robust area under the curve (AUC) of 0.96 (Fig. 2a) and a more modest AUC of 0.83 for aMCI/AD and NC group classification. A separate classifier model using the discovered ten-metabolite panel from the targeted metabolomic analysis classified Converterpre and NC participants with an AUC of 0.96 (Fig. 2b) and an AUC of 0.827 for the aMCI/AD versus NC classification. To validate our biomarker-based group classification, we applied the same simple logistic classifier model developed for the discovery samples to the independent validation samples. The model classified Converterpre and NC participants with an AUC of 0.92 (Fig. 2c) and an AUC of 0.77 for the aMCI/AD versus NC groups. This model yielded a sensitivity of 90% and specificity of 90%, for classifying the Converterpre and NC groups in the validation phase (Fig. 2c).
Figure 2.
ROC results for the lipidomics analyses. (a–c) Plots of ROC results from the models derived from the three phases of the lipidomics analysis. Simple logistic models using only the metabolites identified in each phase of the lipidomics analysis were developed and applied to determine the success of the models for classifying the Cpre and NC groups. The red line in each plot represents the AUC obtained from the discovery-phase LASSO analysis (a), the targeted analysis of the ten metabolites in the discovery phase (b) and the application of the ten-metabolite panel developed from the targeted discovery phase in the independent validation phase (c). The ROC plots represent sensitivity (i.e., true positive rate) versus 1 – specificity (i.e., false positive rate).
We then considered the effects of apolipoprotein E (APOE) genotype on our classification of the Converterpre and NC groups. APOE is involved in lipid metabolism, with the ε4 allele known to be a risk factor for AD. The proportion of ε4 allele carriers was similar in the aMCI/AD (19/69 = 27.5%), NC (17/73 = 23%) and Converter (5/28 = 17%) groups (χ2 = 0.19, P = 0.68, not significant). We repeated the classification analyses using the ten-metabolite model with APO ε4 allele as a covariate. The effect of the ε4 allele was not significant (P = 0.817), and classification accuracy for Converterpre and NC groups changed minimally from an AUC 0.96 to 0.968 (P = 0.992, not significant). Furthermore, a classifier model using only APOE ε4 produced an AUC of 0.54 for classifying the Converterpre and NC groups, implying virtually random classification. These findings indicate that the presumed pathophysiology reflected by the ten-metabolite biomarker panel is orthogonal to APOE-mediated effects.
Here we present the discovery and validation of plasma metabolite changes that distinguish cognitively normal participants who will progress to have either aMCI or AD within 2–3 years from those destined to remain cognitively normal in the near future. The defined ten-metabolite profile features PCs and ACs, phospholipids that have essential structural and functional roles in the integrity and functionality of cell membranes10,11. Deficits of the plasmalemma in AD have been described previously12. Studies have shown decreased plasma PC levels13 and lysoPC/PC ratios14 and increased cerebrospinal fluid (CSF) PC metabolites in patients with AD15, as well as decreased phosphatidylinositol in the hippocampus16 and other heteromodal cortical regions17. Furthermore, amyloid-β may directly disrupt bilayer integrity by interacting with phospholipids18. ACs are known to have a major role in central carbon and lipid metabolism occurring within the mitochondria11. They have also been associated with regulation, production and maintenance of neurons through enhancement of nerve growth factor production11, which is a known potent survival and trophic factor for brain cholinergic neurons, particularly those consistently affected by AD within the basal forebrain19–21. Decreasing plasma AC levels in the Converterpre participants in our study may indirectly signal an impending dementia cascade that features loss of these cholinergic neuronal populations. We posit that this ten–phospholipid biomarker panel, consisting of PC and AC species, reveals the breakdown of neural cell membranes in those individuals destined to phenoconvert from cognitive intactness to aMCI or AD and may mark the transition between preclinical states where synaptic dysfunction and early neurodegeneration give rise to subtle cognitive changes2.
Most approaches to fluid-based biomarker discovery have focused on amyloid-β1–42 (Aβ42), total tau and phosphorylated tau-181 obtained from CSF. Classification of symptomatic patients versus normal controls or other dementias or conversion from MCI to AD is high22, but the predictive value of these CSF biomarkers in preclinical patients is not as strong, suggesting that these markers may be useful only for confirmation of clinical diagnosis23. Blood-based biomarkers are not routinely used in clinical practice but may be more useful because they are easily obtained with less risk of complication in older adults. Studies focusing on Aβ42 or Aβ42/tau ratios derived from blood have been disappointing24, but recent studies suggest that assessment of the proteome and metabolome in blood may have more promise. One recent study using plasma identified 18 proteins that discriminated subjects with symptomatic AD from normal control subjects with nearly 90% accuracy and predicted conversion from symptomatic MCI to AD with 91% accuracy25. Another cross-sectional study reported 18 plasma biomarkers, many related to inflammation, that correctly classified subjects with symptomatic AD and normal control subjects with a sensitivity and specificity of 85% and an AUC of 93% (ref. 26). The biomarker panel was externally validated in a cohort of normal control subjects and subjects with symptomatic AD with sensitivity and specificity of 80% and an AUC of 85%.
To our knowledge, this is the first published report of a blood-based biomarker panel with very high accuracy for detecting preclinical AD. This metabolic panel robustly identifies (with accuracy above 90%) cognitively normal individuals who, on average, will phenoconvert to aMCI or AD within 2–3 years. The accuracy for detection is equal to or greater than that obtained from most published CSF studies27,28, and blood is easier to obtain and costs less to acquire, making it more useful for screening in large-scale clinical trials and for future clinical use. This biomarker panel requires external validation using similar rigorous clinical classification before further development for clinical use. Such additional validation should be considered in a more diverse demographic group than our initial cohort. We consider our results a major step toward the NIA-AA (National Institute on Aging and Alzheimer’s Association) consensus statement mandate for biomarkers of preclinical AD2.
ONLINE METHODS
Neurocognitive methods
The University of Rochester Research Subjects Review Board and the University of California, Irvine Institutional Review Board each approved a common research protocol for this investigation. Content of informed consent forms was thoroughly discussed with subjects at the time of entry into the study and verbal and written consent was obtained from all subjects, including that for serial neuropsychological testing and blood draws for biomarker evaluation. A total of 525 volunteers participated in this study as part of the Rochester/Orange County Aging Study, an ongoing natural history study of cognition in community-dwelling older adults (Supplementary Note). All participants were community-dwelling older adults from the greater Rochester, NY, and Irvine, CA, communities. Participants were recruited through local media (newspaper and television advertisements), senior organizations and word of mouth. Inclusion criteria included age 70 or older, proficiency with written and spoken English and corrected vision and hearing necessary to complete the cognitive battery. Participants were excluded for the presence of known major psychiatric or neurological illness (including Alzheimer’s disease or MCI, cortical stroke, epilepsy and psychosis) at time of enrollment, current or recent (<1 month) use of anticonvulsants, neuroleptics, HAART, antiemetics and antipsychotics for any reason and serious blood diseases including chronic abnormalities in complete blood count and anemia requiring therapy and/or transfusion. Briefly, we prospectively followed participants with yearly cognitive assessments and collected blood samples following an overnight fast (withholding of all medications) (Supplementary Note). At enrollment, each participant completed detailed personal, medical and family history questionnaires. At baseline and at each yearly visit, participants completed measures assessing activities of daily living, memory complaints, and signs and symptoms of depression and were given a detailed cognitive assessment (Supplementary Table 2).
For this study, data from the cognitive tests were used to classify our participants into groups for biomarker discovery. We derived standardized scores (z-scores) for each participant on each cognitive test and computed composite z-scores for five cognitive domains (attention, executive, language, memory and visuoperceptual) (Supplementary Table 3). Normative data for z-score calculations were derived from the performance of our participants on each of the cognitive tests adjusted for age, education, sex and visit. To reduce the effect of cognitively impaired participants on the mean and s.d., age-, education-, sex- and visit-adjusted residuals from each domain z-score model were robustly standardized to have median 0 and robust s.d. of 1, where the robust s.d. = IQR/1.35, as 1.35 is the IQR (interquartile range) of a standard normal distribution.
We categorized the participants into groups of subjects with incident aMCI or early AD (combined into one category, aMCI/AD), cognitively NC subjects and those who converted to aMCI or AD over the course of the study (Converters) based on these composite scores. Impairment was defined as a z-score 1.35 below the cohort median. All participants classified as aMCI met recently revised criteria29 for the amnestic subtype of MCI30. We excluded other behavioral phenotypes of MCI in order to concentrate on the amnestic, which most likely represents nascent AD pathology31. All participants with early AD met recently revised criteria for probable AD32 with impairment in memory and at least one other cognitive domain. For the aMCI/AD group, scores on the measures of memory complaints (MMQ) and activities of daily living (PGC-IADL) were used to corroborate research definitions of these states. All Converters had nonimpaired memory at entry to the study (Zmem≥−1.35), developed memory impairment over the course of the study (Zmem≤−1.35) and met criteria for the above definitions of aMCI or AD. To enhance the specificity of our biomarker analyses, NC participants in this study were conservatively defined with Zmem ± 1 s.d. of the cohort median rather than simply ≥−1.35, and all other z-scores ≥−1.35 s.d. (Supplementary Note).
At the end of year 3 of the study, 202 participants had completed a baseline and two yearly visits. At the third visit, 53 participants met criteria for aMCI/AD and 96 met criteria for NC. Of the 53 aMCI/AD participants, 18 were Converters and 35 had incident aMCI or AD. The remaining 53 participants did not meet our criteria for either group and were not considered for biomarker profiling. Some of these individuals met criteria for nonamnestic MCI, and many had borderline or even above average memory scores that precluded their inclusion as either aMCI/AD or NC (Supplementary Fig. 1). We matched 53 NC participants to the 53 aMCI/AD participants based on sex, age and education level. We used blood samples obtained on the last available study visit for the 53 MCI/AD and 53 NC for biomarker discovery. We included two blood samples from each of the 18 Converters, one from the baseline visit (Converterpre) when Zmem was nonimpaired and one from the third visit (Converterpost) when Zmem was impaired and they met criteria for either aMCI or AD. Thus, a total of 124 samples from 106 participants were submitted for biomarker discovery.
We employed internal cross-validation to validate findings from the discovery phase. Blood samples for validation were identified at the end of the fifth year of the study, and all 106 participants included in the discovery phase were excluded from consideration for the validation phase (Supplementary Fig. 1). Cognitive composite z-scores were recalculated based on the entire sample available, and the same procedure and criteria were used to identify samples for the validation phase. A total of 145 participants met criteria for a group: 21aMCI/AD and 124 NC. Of the 21 aMCI/AD, 10 were Converters. We matched 20 NC participants to the aMCI/AD participants on the basis of age, sex and education level as in the discovery phase. In total, 40 participants contributed plasma samples to the validation phase, as 1 aMCI/AD subject’s plasma sample was not able to be used. As before, the 10 Converters also contributed a baseline sample (Converterpre) for a total of 50 samples.
Neurocognitive statistical analyses
The neurocognitive analyses were designed to demonstrate the general equivalence of the discovery and validation samples on clinical and cognitive measures. We used separate multivariate ANOVA (MANOVA) to examine discovery and validation group performance on the composite z-scores and on self-reported measures of memory complaints, memory related functional impairment and depressive symptoms, as well as a global measure of cognitive function. In the first MANOVA, biomarker sample (discovery, validation) was the independent variable and MMQ, IADL, geriatric depression scale and mini-mental state examination were the dependent variables. In the second MANOVA, biomarker sample (discovery, validation) was the independent variable, and the five cognitive domain z-scores (Zatt, Zexe, Zlan, Zmem and Zvis) were the dependent variables. Significance for the two-sided tests was set at α = 0.05, and we used Tukey’s honestly significant difference (HSD procedure for post hoc comparisons. All statistical analyses were performed using SPSS (version 21).
Lipidomics methods
Reagents
Liquid chromatography–mass spectrometry (LC-MS)-grade acetonitrile, isopropanol, water and methanol were purchased from Fisher Scientific (New Jersey, USA). High purity formic acid (99%) was purchased from Thermo-Scientific (Rockford, IL). Debrisoquine, 4-nitrobenzoic acid (4-NBA), Pro-Asn, glycoursodeoxycholic acid andmalic acid were purchased from Sigma (St. Louis, MO, USA). All lipid standards including 14:0 LPA, 17:0 Ceramide, 12:0 LPC, 18:0 Lyso PI and PC(22:6/0:0) were procured from Avanti Polar Lipids (USA).
Metabolite extraction
Briefly, the plasma samples were thawed on ice and vortexed. For metabolite extraction, 25 µL of plasma sample was mixed with 175 µL of extraction buffer (25% acetonitrile in 40% methanol and 35% water) containing internal standards (10 µL of debrisoquine (1 mg/mL), 50 µL of 4, nitrobenzoic acid (1 mg/mL), 27.3 µl of ceramide (1 mg/mL) and 2.5 µL of LPA (lysophosphatidic acid) (4 mg/mL) in 10 mL). The samples were incubated on ice for 10 min and centrifuged at 14,000 r.p.m. at 4 °C for 20 min. The supernatant was transferred to a fresh tube and dried under vacuum. The dried samples were reconstituted in 200 µL of buffer containing 5% methanol, 1% acetonitrile and 94% water. The samples were centrifuged at 13,000 r.p.m. for 20 min at 4 °C to remove fine particulates. The supernatant was transferred to a glass vial for Ultraperformance liquid chromatography–electrospray ionization quadrupole time-of-flight mass spectrometry (UPLC-ESI-QTOF-MS) analysis.
UPLC-ESI-QTOF-MS–based data acquisition for untargeted lipidomic profiling
Each sample (2 µL) was injected onto a reverse-phase CSH C18 1.7 µM 2.1×100 mm column using an Acquity H-class UPLC system (Waters Corporation, USA). The gradient mobile phase comprised of water containing 0.1% formic acid solution (Solvent A), 100% acetonitrile (Solvent B) and 10% acetonitrile in isopropanol containing 0.1% formic acid and 10 mM ammonium formate (Solvent C). Each sample was resolved for 13 min at a flow rate of 0.5 mL/min for 8 min and then 0.4 mL/min from 8 to 13 min. The UPLC gradient consisted of 98% A and 2% B for 0.5 min and then a ramp of curve 6 to 60% B and 40% A from 0.5 min to 4.0 min, followed by a ramp of curve 6 to 98% B and 2% A from 4.0 to 8.0 min, a ramp to 5% B and 95% C from 9.0 min to 10.0 min at a flow rate of 0.4 mL/min and finally a ramp to 98% A and 2% B from 11.0 min to 13 min. The column eluent was introduced directly into the mass spectrometer by electrospray ionization. Mass spectrometry was performed on a quadrupole time-of-flight (Q-TOF) instrument (Xevo G2 QTOF, Waters Corporation, USA) operating in either negative (ESI−) or positive (ESI+) electrospray ionization mode with a capillary voltage of 3,200 V in positive mode and 2,800 V in negative mode and a sampling cone voltage of 30 V in both modes. The desolvation gas flow was set to 750 l h−1, and the temperature was set to 350 °C. The source temperature was set at 120 °C. Accurate mass was maintained by introduction of a lock-spray interface of leucine-enkephalin (556.2771 [M+H]+ or 554.2615 [M−H]−) at a concentration of 2 pg/µL in 50% aqueous acetonitrile and a rate of 2 µL/min. Data were acquired in centroid MS mode from 50 to 1,200 m/z mass range for TOF-MS scanning as single injection per sample, and the batch acquisition was repeated to check experimental reproducibility. For the metabolomics profiling experiments, pooled quality control (QC) samples (generated by taking an equal aliquot of all the samples included in the experiment) were run at the beginning of the sample queue for column conditioning and every ten injections thereafter to assess inconsistencies that are particularly evident in large batch acquisitions in terms of retention time drifts and variation in ion intensity over time. This approach has been recommended and used as a standard practice by leading metabolomics researchers33. A test mix of standard metabolites was run at the beginning and at the end of the run to evaluate instrument performance with respect to sensitivity and mass accuracy. The overlay of the total ion chromatograms of the quality control samples depicted excellent retention time reproducibility. The sample queue was randomized to remove bias.
Stable isotope dilution–multiple reaction monitoring mass spectrometry
LC-MSmass spectrometry (LC-MS/MS) is increasingly used in clinical settings for quantitative assay of small molecules and peptides such as vitamin D, serum bile acid and parathyroid hormone under Clinical Laboratory Improvement Amendments environments with high sensitivities and specificities34. In this study, targeted metabolomic analysis of plasma samples was performed using the Biocrates Absolute-IDQ P180 (BIOCRATES, Life Science AG, Innsbruck, Austria). This validated targeted assay allows for simultaneous detection and quantification of metabolites in plasma samples (10 µL) in a high-throughput manner. The methods have been described in detail35,36. The plasma samples were processed as per the instructions by the manufacturer and analyzed on a triple-quadrupole mass spectrometer (Xevo TQ-S, Waters Corporation, USA) operating in the MRM mode. The measurements were made in a 96-well format for a total of 148 samples, and seven calibration standards and three quality control samples were integrated in the kit. Briefly, the flow injection analysis tandem mass spectrometry (MS/MS) method was used to quantify a panel of 144 lipids simultaneously by multiple reaction monitoring. The other metabolites are resolved on the UPLC and quantified using scheduled MRMs. The kit facilitates absolute quantitation of 21 amino acids, hexose, carnitine, 39 acylcarnitines, 15 sphingomyelins, 90 phosphatidylcholines and 19 biogenic amines. Data analysis was performed using the MetIQ software (Biocrates), and the statistical analyses included the nonparametric Kruskal-Wallis test with follow-up Mann-Whitney U-tests for pairwise comparisons using the STAT pack module v3 (Biocrates). Significance was adjusted for multiple comparisons using Bonferroni’s method (P < 0.025). The abundance is calculated from area under the curve by normalizing to the respective isotope labeled internal standard. The concentration is expressed as nmol/L. Human EDTA plasma samples spiked with standard metabolites were used as quality control samples to assess reproducibility of the assay. The mean of the coefficient of variation (CV) for the 180 metabolites was 0.08, and 95% of the metabolites had a CV of <0.15.
Sample size considerations
The signal intensity of the metabolites within similar groups was normally distributed with a standard deviation of 1.5. If the true difference in the Converterpre and NC groups’ mean is twofold, we will have over 90% power to detect differential metabolites at an overall significance level of 5% with Bonferroni’s adjustment using 30 subjects per group.
Lipidomics statistical analyses
The m/z features of metabolites were normalized with log transformation that stabilized the variance, followed by a quantile normalization to make the empirical distribution of intensities the same across samples37. The metabolites were selected among all those known to be identifiable using a ROC regularized learning technique38,39 based on the LASSO penalty8,9 as implemented with the R package ‘glmnet’40, which uses cyclical coordinate descent in a path-wise fashion. We first obtained the regularization path over a grid of values for the tuning parameter λ through tenfold cross-validation. The optimal value of the tuning parameter lambda, which was obtained by the cross-validation procedure, was then used to fit the model. All the features with nonzero coefficients were retained for subsequent analysis. This technique is known to reduce overfitting and achieve similar prediction accuracy as the sparse supporting vector machine. The classification performance of the selected metabolites was assessed using area under the ROC curve (AUC). The ROC can be understood as a plot of the probability of classifying correctly the positive samples against the rate of incorrectly classifying true negative samples. So the AUC measure of an ROC plot is a measure of predictive accuracy. To maintain rigor of independent validation, the simple logistic model with the ten-metabolite panel was used, although a more refined model can yield greater AUC. The validation phase was performed in a blinded fashion such that the sample group was not known by the statistical team.
Supplementary Material
Acknowledgments
We thank E. Johnson and D. Greenia for study coordination; P. Bailie, M. Patel and A. Balasubramanian for assistance with data collection; and A. Almudevar for statistical support. We also thank R. Padilla and I. Conteh for processing the blood samples and R. Singh and P. Kaur for technical assistance in developing the lipidomics data. This work was funded by US National Institutes of Health grants R01AG030753 and DOD W81XWH-09-1-0107 to H.J.F.
Footnotes
Accession codes. Lipodomics data were deposited in the European Bioinformatics Institute MetaboLights database with accession code MTBLS72.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
H.J.F., T.R.M., C.H.K., W.J.H., S.G.F., M.M., M.S.F. and A.K.C. conceived of the study. W.J.H., J.M.H., M.D.N., S.A.R. and C.H.K. recruited participants and provided material support for data collection. M.M., D.J.B. and C.B.P. collected the clinical data. D.R.P., S.G.F. and M.M. derived the cognitive z-score methodology. M.M. completed statistical analysis of the cognitive data. A.K.C., M.S.F., T.R.M. and L.H.M. completed the lipidomics analyses. M.T.T., X.Z. and A.K.C. completed statistical analysis of the lipidomics data. M.M., A.K.C., M.S.F. and H.J.F. wrote the manuscript. All authors edited the manuscript for content.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.World Health Organization. Dementia: a Public Health Priority. Geneva: World Health Organization; 2012. [Google Scholar]
- 2.Sperling RA, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s & Dementia: the Journal of the Alzheimer’s Association. 2011;7:280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hulstaert F, et al. Improved discrimination of AD patients using β-amyloid(1–42) and tau levels in CSF. Neurology. 1999;52:1555–1562. doi: 10.1212/wnl.52.8.1555. [DOI] [PubMed] [Google Scholar]
- 4.Small SA, Perera GM, De La Paz R, Mayeux R, Stern Y. Differential regional dysfunction of the hippocampal formation among elderly with memory decline and Alzheimer’s disease. Ann. Neurol. 1999;45:466–472. doi: 10.1002/1531-8249(199904)45:4<466::aid-ana8>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 5.Klunk WE, et al. Imaging brain amyloid in Alzheimer’s disease with Pittsburgh Compound-B. Ann. Neurol. 2004;55:306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 6.Franceschi C, et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. NY Acad. Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 7.Thambisetty M, Lovestone S. Blood-based biomarkers of Alzheimer’s disease: challenging but feasible. Biomark. Med. 2010;4:65–79. doi: 10.2217/bmm.09.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tibshirani R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B Stat. Methodol. 1996;58:267–288. [Google Scholar]
- 9.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning; Data Mining, Inference, and Prediction. New York: Springer-Verlag; 2008. [Google Scholar]
- 10.van Meer G, de Kroon AI. Lipid map of the mammalian cell. J. Cell Sci. 2011;124:5–8. doi: 10.1242/jcs.071233. [DOI] [PubMed] [Google Scholar]
- 11.Jones LL, McDonald DA, Borum PR. Acylcarnitines: role in brain. Prog. Lipid Res. 2010;49:61–75. doi: 10.1016/j.plipres.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Nitsch RM, et al. Evidence for a membrane defect in Alzheimer disease brain. Proc. Natl. Acad. Sci. USA. 1992;89:1671–1675. doi: 10.1073/pnas.89.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer EJ, et al. Plasma phosphatidylcholine docosahexaenoic acid content and risk of dementia and Alzheimer disease: the Framingham Heart Study. Arch. Neurol. 2006;63:1545–1550. doi: 10.1001/archneur.63.11.1545. [DOI] [PubMed] [Google Scholar]
- 14.Mulder C, et al. Decreased lysophosphatidylcholine/phosphatidylcholine ratio in cerebrospinal fluid in Alzheimer’s disease. J. Neural Transm. 2003;110:949–955. doi: 10.1007/s00702-003-0007-9. [DOI] [PubMed] [Google Scholar]
- 15.Walter A, et al. Glycerophosphocholine is elevated in cerebrospinal fluid of Alzheimer patients. Neurobiol. Aging. 2004;25:1299–1303. doi: 10.1016/j.neurobiolaging.2004.02.016. [DOI] [PubMed] [Google Scholar]
- 16.Prasad MR, Lovell MA, Yatin M, Dhillon H, Markesbery WR. Regional membrane phospholipid alterations in Alzheimer’s disease. Neurochem. Res. 1998;23:81–88. doi: 10.1023/a:1022457605436. [DOI] [PubMed] [Google Scholar]
- 17.Pettegrew JW, Panchalingam K, Hamilton RL, McClure RJ. Brain membrane phospholipid alterations in Alzheimer’s disease. Neurochem. Res. 2001;26:771–782. doi: 10.1023/a:1011603916962. [DOI] [PubMed] [Google Scholar]
- 18.Haughey NJ, Bandaru VV, Bae M, Mattson MP. Roles for dysfunctional sphingolipid metabolism in Alzheimer’s disease neuropathogenesis. Biochim. Biophys. Acta. 2010;1801:878–886. doi: 10.1016/j.bbalip.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kordower JH, Fiandaca MS. Response of the monkey cholinergic septohippocampal system to fornix transection: a histochemical and cytochemical analysis. J. Comp. Neurol. 1990;298:443–457. doi: 10.1002/cne.902980406. [DOI] [PubMed] [Google Scholar]
- 20.Kordower JH, et al. The aged monkey basal forebrain: rescue and sprouting of axotomized basal forebrain neurons after grafts of encapsulated cells secreting human nerve growth factor. Proc. Natl. Acad. Sci. USA. 1994;91:10898–10902. doi: 10.1073/pnas.91.23.10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitehouse PJ, Price DL, Clark AW, Coyle JT, DeLong MR. Alzheimer disease: evidence for selective loss of cholinergic neurons in the nucleus basalis. Ann. Neurol. 1981;10:122–126. doi: 10.1002/ana.410100203. [DOI] [PubMed] [Google Scholar]
- 22.Hansson O, et al. Association between CSF biomarkers and incipient Alzheimer’s disease in patients with mild cognitive impairment: a follow-up study. Lancet Neurol. 2006;5:228–234. doi: 10.1016/S1474-4422(06)70355-6. [DOI] [PubMed] [Google Scholar]
- 23.Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat. Rev. Neurol. 2010;6:131–144. doi: 10.1038/nrneurol.2010.4. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry MC. Biomarkers of Alzheimer disease in plasma. NeuroRx. 2004;1:226–234. doi: 10.1602/neurorx.1.2.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray S, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat. Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 26.Doecke JD, et al. Blood-based protein biomarkers for diagnosis of Alzheimer disease. Arch. Neurol. 2012;69:1318–1325. doi: 10.1001/archneurol.2012.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roe CM, et al. Improving CSF biomarker accuracy in predicting prevalent and incident Alzheimer disease. Neurology. 2011;76:501–510. doi: 10.1212/WNL.0b013e31820af900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagan AM, et al. Cerebrospinal fluid tau/β-amyloid42 ratio as a prediction of cognitive decline in nondemented older adults. Arch. Neurol. 2007;64:343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
References
- 29.Albert MS, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petersen RC, et al. Mild cognitive impairment: clinical characterization and outcome. Arch. Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 31.Espinosa A, et al. A longitudinal follow-up of 550 mild cognitive impairment patients: evidence for large conversion to dementia rates and detection of major risk factors involved. J. Alzheimers Dis. 2013;34:769–780. doi: 10.3233/JAD-122002. [DOI] [PubMed] [Google Scholar]
- 32.McKhann GM, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Want EJ, et al. Global metabolic profiling procedures for urine using UPLC-MS. Nat. Protoc. 2010;5:1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- 34.Grebe SK, Singh RJ. LC-MS/MS in the clinical laboratory - where to from here? Clin. Biochem. Rev. 2011;32:5–31. [PMC free article] [PubMed] [Google Scholar]
- 35.Illig T, et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Römisch-Margl WPC, Bogumil R, Röhring C, Suhre K. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics. 2011;7:1–14. [Google Scholar]
- 37.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 38.Ma S, Huang J. Regularized ROC method for disease classification and biomarker selection with microarray data. Bioinformatics. 2005;21:4356–4362. doi: 10.1093/bioinformatics/bti724. [DOI] [PubMed] [Google Scholar]
- 39.Liu Z, Tan M. ROC-based utility function maximization for feature selection and classification with applications to high-dimensional protease data. Biometrics. 2008;64:1155–1161. doi: 10.1111/j.1541-0420.2008.01015.x. [DOI] [PubMed] [Google Scholar]
- 40.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.