Abstract
Nucleotide-binding and oligomerization domain (NOD)-like receptors NOD1 and NOD2 are cytosolic innate immune receptors that recognize microbial peptidoglycans. Whilst studies have addressed the role of NOD proteins in innate immune responses, little attention has been given to their impact on the developing adaptive immune system. We have assessed the roles of NOD1 and NOD2 deficiency on T cell development in mice. Our results demonstrate that NOD1 and NOD2 promote the positive selection/maturation of CD8 SP thymocytes in a thymocyte intrinsic manner. TCR-mediated ERK phosphorylation is significantly reduced in the absence of NOD proteins, but RIP2 is not involved in CD8 SP thymocyte selection or ERK signaling. Commensal bacteria free animals have thymocyte maturation defects and exogenous NOD ligands can enhance thymocyte maturation in culture. These results raise the intriguing possibility that abnormal lymphocyte responses observed in NOD-dependent inflammatory diseases are not driven solely by microbial signals in the gut, but may also involve intrinsic lymphocyte defects resulting from impaired CD8 T cell thymic development.
Introduction
Nucleotide-binding and oligomerization domain (NOD)1 and NOD2 are intracellular innate immune sensors for the bacterial peptidoglycan derived molecules γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP) and muramyl dipeptide (MDP), respectively (1, 2). Their expression is normally associated with myeloid and non-lymphoid cell types. Upon ‘sensing’ these ligands via leucine rich repeat domains (LRR)(3, 4), dimerization occurs via the oligomerization domain, leading to caspase-recruitment domain (CARD) dependent interaction with the serine threonine kinase RIP2 (Receptor interacting protein 2) (5–7). The peptidoglycan ligand activated NOD: RIP2 complex promotes the potent activation of NFκB, leading to the production of inflammatory mediators and chemokines (8).
NOD1 and NOD2 are important for epithelial and myeloid cell signaling in response to microbial products from a variety of bacteria, including Mycobacterium sp, Salmonella enterica serovar Typhimurium, Helicobacter sp, and Listeria monocytogenes (9). In addition to signaling the presence of pathogens, the sensing of commensal microbial products via NOD molecules appears to be important for an increasingly diverse range of biological functions. These include intestinal lymphoid tissue genesis (10), the generation or maintenance of functional Paneth cells in the intestine (11, 12), and systemic neutrophil priming at sites distal to the gut (13).
Polymorphisms in NOD2 are associated with two important human inflammatory diseases, Crohn’s disease and Blau syndrome (14–16). Whilst the amino acid changes in the LRR domains of NOD2 that are associated with Crohn’s disease reduce MDP ligand driven signaling, Blau syndrome associated sequence changes appear to constitutively activate NFκB, at least when expressed in epithelial cell lines (5, 17). NOD1 and NOD2 can associate with the autophagy related (Atg) molecule ATG16L1 to promote autophagic double membrane restriction of invasive bacteria (18). As hypofunctional polymorphisms of other autophagy genes are also associated with Crohn’s Disease (19, 20), it appears that inappropriate control of bacteria by autophagy and inappropriate inflammatory responses driven by NFκB and MAPK are linked to this chronic inflammatory disease (21).
The resulting ileal inflammation in Crohn’s disease consists mainly of T lymphocyte infiltrates that are associated with high levels of IFNγ and IL-17 expression (22). During their development in the thymus, all α/β TCR T cells, including α/β TCR intraepithelial lymphocytes (IELs) are subject to positive selection at the CD4+CD8+ (DP) stage of differentiation (23). The signaling generated by newly rearranged αβ T cell receptors (TCR) interacting with self peptide MHC (pMHC) complexes expressed on cortical thymic epithelial cells (cTEC) promotes positive selection that initiates the differentiation of DPs into MHCII restricted CD4 single positive (CD4 SP) or MHCI restricted CD8 SP thymocytes (see Reviews (24, 25)).
Mature human T lymphocytes express both NOD1 and NOD2 (26, 27) and a CD4 T cell intrinsic function for mouse NOD2 has been reported (28). Although both NOD1 and NOD2 transcripts have been detected in mouse thymic tissue, it is unknown if NOD1 and NOD2 play roles in developing lymphocyte signaling (29, 30). Here we provide evidence that both NOD1 and NOD2 function intrinsically in thymocytes to promote CD8 SP thymocyte positive selection. Furthermore, we identify a RIP2-independent pathway where NOD proteins are required for efficient TCR-mediated ERK phosphorylation. Finally we show that NOD ligands can promote thymocyte maturation in culture, and that thymocytes from germ free mice show maturation defects.
Materials and Methods
Mice
Wild-type C57BL/6J (B6) and congenic B6.SJL-PtprcaPepcb mice were obtained from the Jackson Laboratories. Ripk2−/−(7), Nod1−/−(1), Nod2−/−(12), and Nod1−/−Nod2−/− animals were obtained from R. Ulevitch. Ripk2−/− and all Nod deficient animals were backcrossed over ten times to a Scripps C57BL/6 colony originally established from C57BL/6J mice before experimental use. H-Y (31), and OT-I Tap1−/− (32) TCR Tg animals were kind gifts from N. Gascoigne. Animals were bred and maintained under specific pathogen-free conditions unless stated. Five weeks old germfree and conventionally housed C57BL/6J animals were from Kew Bioservices, WEHI. Animal procedures were performed as approved by TSRI Institutional Animal Care and Use Committee, the AMREP Animal Ethics Committee, and the WEHI Animal Ethics Committee.
Flow cytometry
Single cell suspensions from spleen, thymus and blood were used for flow cytometry. For surface staining, fluorophore-labeled mAbs specific for CD4, CD8, CD69, CD24 (HSA), CD45.1, CD45.2, H-Y transgenic TCR (T3.70), Vα2, and TCRβ were obtained from BD, Biolegend or eBioscience. For intracellular staining, cells were fixed with 1.5% paraformaldehyde, permeabilized with methanol, and stained for intracellular phospho-Stat5 (clone 47, BD). Samples were acquired on a LSRII flow cytometer and analyzed using FlowJo software (Tree Star).
Quantitative PCR
Total RNA was isolated from sorted thymocyte populations using RNAeasy miniprep kits (Qiagen) and contaminating genomic DNA was removed by DNAse-I digestion. CDNA was generated using Superscript III (Life Technologies) and the PCR reactions were set up using QuantiTect SYBR Green PCR Kits (Qiagen) with the annealing temperature per cycle set to 56°C on a Stratagene MX3005P PCR machine. Quantification of Nod1 and Nod2 mRNA was normalized with concomitant amplification of b-Actin mRNA in each quadruplicate sample. The following primers were used: NOD1s: 5’-ccaagcctgacaaggtccgaaag, NOD1as: 5’-gaatgagctgggaaggggagaag, NOD2s: 5’-gggtttctgagccagtacgagtg, NOD2as: 5’-ctgacgtgctgtagaaggaaggc, Actins: 5’-cattgctgacaggatgcagaagg, Actinas: 5’-actctggcttgctgatccacat.
Generation of Mixed Bone Marrow Chimeras
T cell depleted bone marrow from non-transgenic or TCR transgenic B6.SJL-CD45.1 and Nod1−/−Nod2−/− mice were mixed at a 1:1 ratio and injected intravenously into lethally irradiated (1000 rad) non-transgenic B6.SJL-CD45.1 recipients. Six to 10 weeks after injection, chimeras were analyzed.
Ca2+ flux
Ca2+ flux measurement was performed as described(33). Single cell thymocyte suspensions from WT mice were loaded for 10 minutes at 37°C with CFSE, washed, then mixed at a 1:1 ratio with unlabeled Nod1−/−Nod2−/− cells. After loading with Indo-1-AM (2mM, Molecular Probes), cells were washed, then stained for 20 minutes on ice with biotinylated antibodies specific for CD3 (145-2C11) and CD4 (RM4.4), and PerCP-Cy5.5-conjugated anti-CD4 (GK1.5), and PE-Cy7-conjugated anti-CD8 (53-6.7, all from Biolegend). Cells were prewarmed to 37°C and streptavidin was added to crosslink biotinylated antibodies. Maximum Ca2+-flux was achieved by the addition of ionomycin (500ng/ml; Calbiochem). The mean fluorescence ratio of Indo-1 violet to Indo-1 blue was calculated using FlowJo software (Tree Star).
OT-I TCR Tg Tap1−/− stimulation assay
To avoid cell signaling triggers by purification, total unpurified 5×106 thymocytes from WT or Nod1−/−Nod2−/− OT-I TCR Tg Tap1−/− mice were cultured in RPMI 1640 with 10% Fetal Calf Serum, 100U/ml Penicillin, 0.1mg/ml Streptomycin, 55µM 2-mercaptoethanol (GIBCO) 1mM Na pyruvate, 1x MEM Non-Essential Amino Acids (GIBCO). For 15 hours at a set concentration of ionomycin (0.2µg/ml (Sigma), varying concentrations of PMA (ranging from 0.3ng/ml to 0.05ng/ml) (Sigma) and 10µg/ml of MDP or TriDAP (Invivogen) were added to the culture in 24 well plates. After culture with stimuli (+/− NOD ligands) for 15 hours, cells were washed and cultured a further 29 hours at 37°C in growth media alone (total culture time 44 hours). Cells were stained with antibodies specific for CD4, CD8, CD69 and HSA, and live cells (7AAD-) were analyzed using an LSRII flow cytometer (BD).
Thymocyte CD69 upregulation assay
Thymocytes from H-Y TCR transgenic WT (CD45.1+), Ripk2−/− (Cy5-labeled) and Nod1−/−Nod2−/− mice (CD45.2+) were mixed at a 1:1:1 ratio and cultured for 5h with non-transgenic C57BL/6 splenocytes and increasing concentrations of Smcy peptide (KCSRNRQYL, GenScript). Cells were then stained with antibodies specific for CD45.2, CD4, CD8, CD69 and H-Y Tg TCR (T3.70), and were acquired on an LSRII flow cytometer. Gates defining CD69+ versus CD69− populations were determined with thymocytes incubated without Smcy peptide.
Immunoblot analysis
Protein lysates from thymocytes were made with Nonident P-40 lysis buffer (50mM Tris, 150mM NaCl, 5mM EDTA, 1% NP-40) supplemented with protease inhibitors (Roche). Soluble fractions were boiled in SDS-containing loading buffer before electrophoresis. Gels were transferred to nitrocellulose membrane and blocked with 0.5% BSA in Tris-buffered saline with Tween 20. Blots were probed with Rabbit anti pErk (Cell Signaling Technologies) and anti rabbit antibodies and signal was detected with the enhanced chemiluminescence system (Pierce) Blots were then stripped and reprobed with total anti Erk antibody.
Statistics
Statistical differences were calculated with the Student’s two-tailed t-test with the assumption of different variances and a confidence level of 95%.
Results
Nod1−/−Nod2−/− mice show decreased CD8SP thymocyte cell numbers
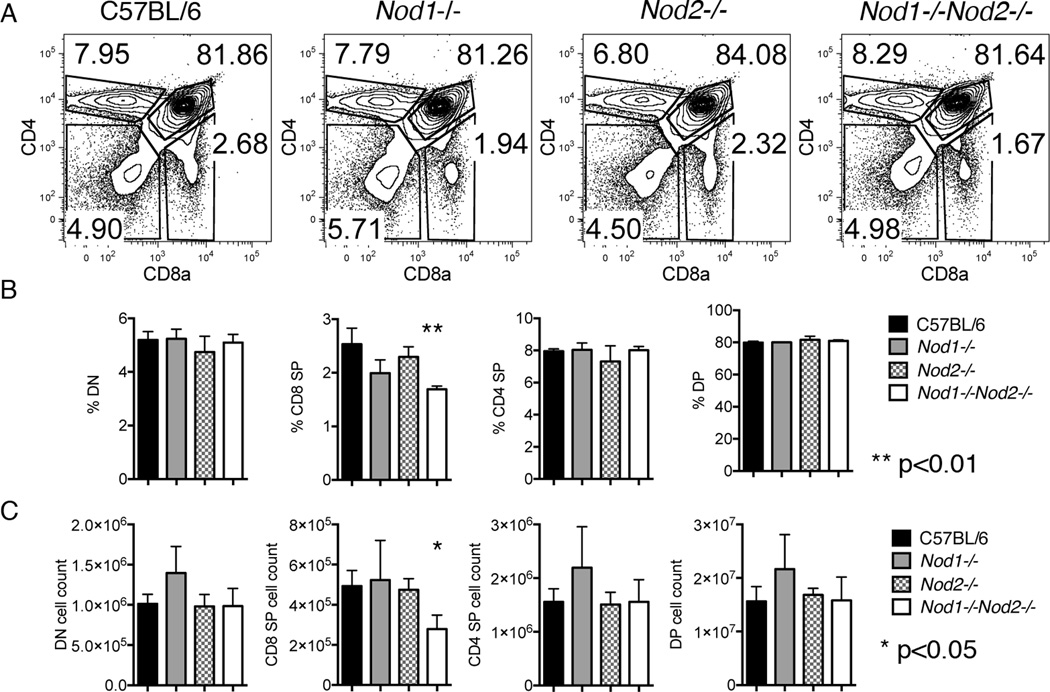
Flow cytometry was used to assess any effects the lack of NOD1 and or NOD2 expression might have on the composition of thymocyte subsets. An initial screen of three animals of either Nod1−/−, Nod2−/− or Nod1−/−Nod2−/− animals revealed a statistically significant reduction of CD8 single positive (CD8SP) cells in the thymii of Nod1−/−Nod2−/− mice (Figure 1). Thymocyte subset characterization from thymii where only NOD1 or NOD2 was deficient did not reveal a statistically significant role for either single gene in CD8SP proportion (Figure 1B) or thymocyte numbers (Figure 1C). The findings indicate that both NOD1 and NOD2 expression is required for the generation or maintenance of normal CD8SP thymocyte numbers.
Figure 1. Thymic subset characterization of thymii from Nod1−/− Nod2−/− or Nod1−/−Nod2−/− mice.
(A) Representative flow cytometry plots from the thymus of C57BL/6 , Nod1−/−, Nod2−/−, and Nod1−/−Nod2−/−mice (labeled above) stained for CD4 and CD8a. Numbers in plots represent percentages of depicted populations. Total proportions (panel B) and numbers (panel C) of thymocyte subpopulations of C57BL/6 (black), Nod1−/−(grey), Nod2−/−(grey hatched), and Nod1−/−Nod2−/− mice (white) are shown. Bars represent mean +/− SD. Statistical analysis with paired t tests was performed. Data represent three animals per group.
The defect in CD8 SP numbers in Nod1−/−Nod2−/− thymocytes is T cell intrinsic
NOD1 and NOD2 are expressed in epithelial and hematopoietic lineages (29, 34, 35), both of which play crucial roles in T cell selection in the thymus. To determine if the reduction in CD8 SP thymocytes observed in Nod1−/−Nod2−/− thymii was due to a defect intrinsic to thymocytes or altered dendritic or thymic epithelial cell function, mixed bone marrow chimeras were generated (Supplementary Figure 1 A , B). An equal number of T cell depleted bone marrow cells from wild type (WT) (B6.SJL-PtprcaPepcb expressing CD45.1) and Nod1−/−Nod2−/− mice (expressing the CD45.2 allele) were transplanted into lethally irradiated WT or Nod1−/−Nod2−/− mice and thymocyte subsets analyzed nine weeks later. Irrespective of the genotype of the recipient mouse, a statistically significant reduction in the proportion of Nod1−/−Nod2−/− derived CD8 SP thymocytes was observed in these chimeras. Thus WT thymic epithelial and hematopoietically derived cells (including thymocytes and dendritic cells) fail to rescue the defect afflicting Nod1−/−Nod2−/− CD8 SP cell numbers, indicating that the CD8 SP thymocyte defect is intrinsic to Nod1−/−Nod2−/− thymocytes. Consistent with an intrinsic thymocyte defect, both Nod1 and Nod2 mRNA were detected by quantitative PCR in CD4SP and CD8SP, but not in DP thymocytes (Supplementary Figure 1C,D).
Nod1−/−Nod2−/− mice have reduced positive selection and CD8 SP thymocyte maturation
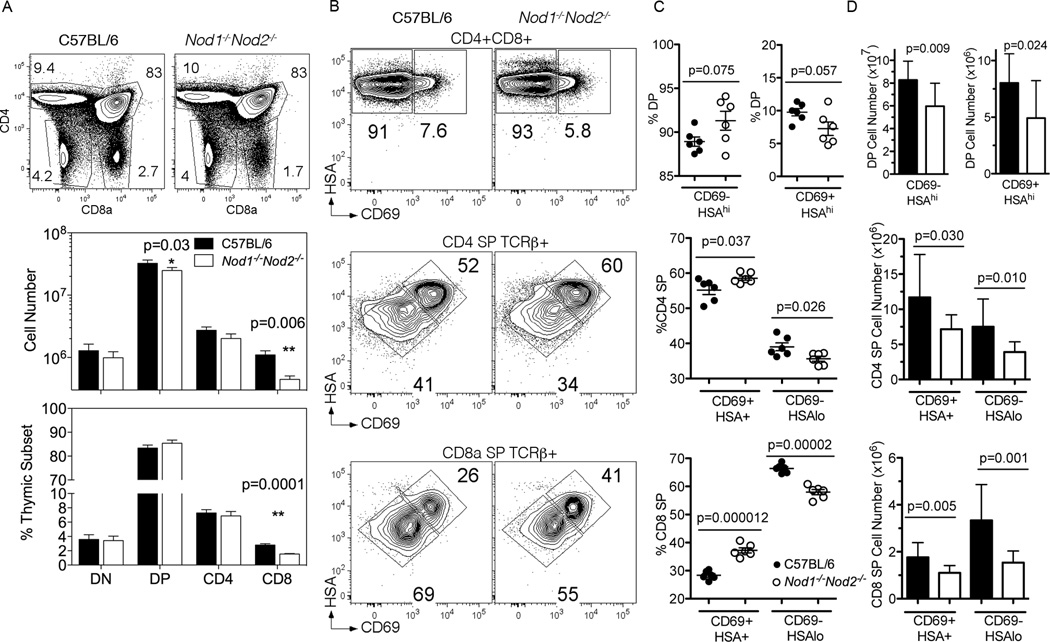
A more thorough analysis of thymic development from a larger cohort of Nod1−/−Nod2−/− mice revealed a two fold decrease in number (Fig 2A middle) and proportion (Fig 2A lower) of CD8 single positive (CD8SP) cells in the thymii of Nod1−/−Nod2−/− mice. A small but significant decrease (25%) in CD4+ CD8+ double positive (DP) cell numbers was also noted. In contrast, the proportions and numbers of double negative (DN) and CD4 single positive (CD4SP) subsets were not significantly different from those in C57BL/6 mice.
Figure 2. Thymic maturation of CD8 SP thymocytes is less efficient in the absence of Nod1 and Nod2.
(A) Representative flow cytometry plots from the thymus of C57BL/6 (left column) and Nod1−/−Nod2−/−mice (right column) stained for CD4 and CD8a. Numbers in plots represent percentages of depicted populations. Total numbers (middle panel) and proportions (lower panel) of thymocyte subpopulations of C57BL/6 (black) and Nod1−/−Nod2−/− mice (white) are shown. Bars represent mean +/− SD. Statistical analysis with paired t tests was performed. Data represent pooled results from three independent experiments with a total of 13 mice per group analyzed. (B) Representative flow cytometry plots from gated thymocyte subsets of CD4+CD8+ (DP) (top row), CD4 SP TCRβ+ (middle row) and CD8 SP TCRβ+ thymocytes (bottom row) from C57BL/6 (left column) and Nod1−/−Nod2−/−mice (right column). Total thymocytes were stained for CD4 and CD8a, and gated thymocyte populations were stained for HSA and CD69. Numbers in plot represent percentages of depicted population. Proportions (C) and numbers (D) of thymocyte subpopulations of C57BL/6 (black) and Nod1−/−Nod2−/− mice (white). Bars represent the mean +/− SD. Statistical analysis with two-tailed unpaired t tests was performed. (C) Data representative of one of two independent experiments with 6 mice per group. (D) Data represent pooled results from two independent experiments with a total of 11 mice per group.
After self-antigen/MHC induced positive selection by cTEC, DP thymocytes upregulate CD69 and begin to migrate to the medulla where these cells are then subject to negative selection (reviewed (36)). A subsequent reduction in the expression of Heat Stable Antigen (HSA) and CD69 on SP thymocytes signals the onset of maturation prior to exit from the thymus into the periphery. Analysis of DP CD69 expressing cells from NOD deficient animals revealed a small decrease in the proportion and number of the CD69+ population (Figure 2B). When the proportion (Figure 2C) and number (Figure 2D) of CD4 SP and CD8 SP thymocytes in C57BL/6 and Nod1−/−Nod2−/− mice that had completed maturation (CD69−HSAlo) were examined, both populations were reduced in Nod1−/−Nod2−/− animals, although the impact was greater on CD8 SP cells. These findings indicate that the selection/maturation of SP thymocytes in Nod1−/−Nod2−/− animals is impaired, and that CD8 SP thymocytes are more dependent upon NOD1 and/or NOD2 for efficient completion of maturation after positive selection.
NOD deficiency alters the positive selection of H-Y TCR Tg thymocytes
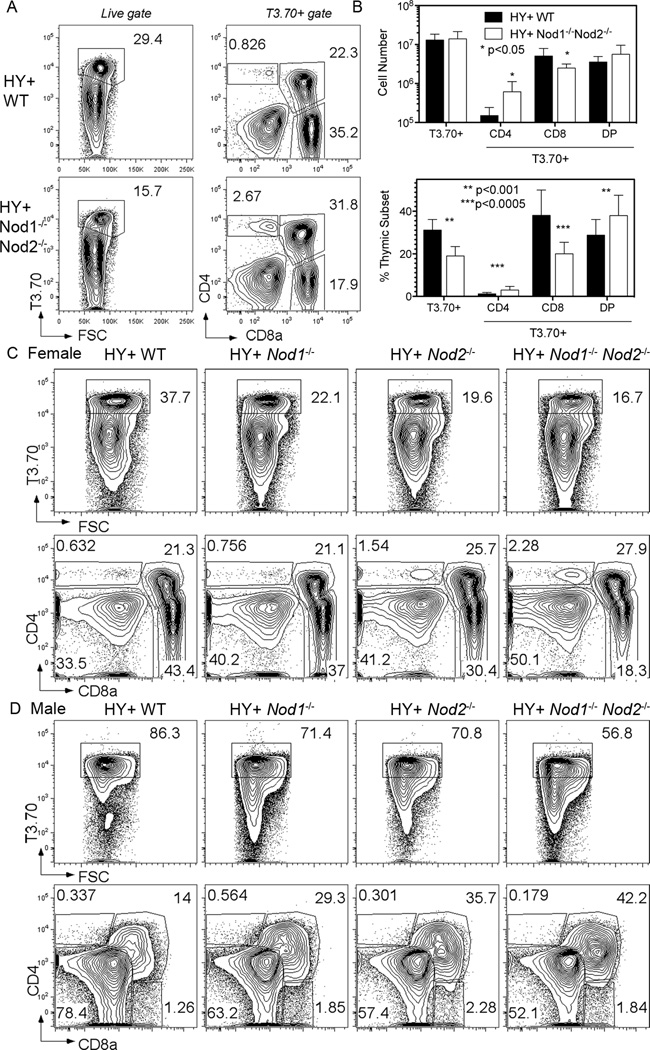
To confirm that NOD1 and/or NOD2 play roles in the positive selection of MHCI restricted T cells, thymocyte subset development in Nod1−/−Nod2−/− animals expressing the H-Y T cell receptor transgene (TCR Tg) was examined. The H-Y TCR recognizes a peptide from the protein encoded by the male chromosome–encoded Smcy gene presented by H-2Db (31). Thymii from female H-Y TCR Tg Nod1−/−Nod2−/− animals exhibit reduced positive selection of H-Y TCR+ (T3.70+) CD8 SP thymocytes when compared with H-Y TCR Tg NOD sufficient thymii (Figure 3A). As had been observed in non-transgenic thymic populations, a significant two-fold reduction in the proportion and number of CD8 SP T3.70+ cells was seen in Nod1−/−Nod2−/− mice (Figure 3B). A significant increase in proportion (2.5 fold) and number (4 fold) of T3.70+ CD4SP cells was also observed in the thymii of H-Y TCR Nod1−/−Nod2−/− animals (Figure 3B). This may be due to increased endogenous Vα rearrangement.
Figure 3. Both NOD genes contribute to positive and negative selection of H-Y TCR Tg thymocytes.
(A) Representative flow cytometry plots from the thymus of H-Y TCR Tg WT (top row) and H-Y TCR Tg Nod1−/−Nod2−/− (bottom row) female mice. The left column shows the percentage of thymocytes expressing the H-Y TCR (T3.70+ cells), whilst the right shows the percentages of T3.70+ thymocyte subpopulations after staining for CD4 and CD8a.
(B) Total numbers (upper graph) and proportions (lower graph) of T3.70+ cells and T3.70+ subpopulations in the thymus of H-Y TCR Tg WT (black bars) and Nod1−/−Nod2−/− (white bars) female mice. Bars represent the mean +/− SD. Statistical analysis with unpaired t tests was performed. Data represent pooled results from three independent experiments with a total of 9 mice per group analyzed.
(C and D) Representative flow cytometry plots from the thymus of H-Y TCR Tg WT (first column), Nod1−/− (second column), Nod2−/− (third column) and Nod1−/−Nod2−/− (fourth column) female (C) and male (D) mice. Upper row shows the percentage of T3.70+ thymocytes, whilst the lower row shows the percentages of T3.70+ thymocyte subpopulations after staining for CD4 and CD8a. Data are representative results of three mice per group analyzed.
Both Nod1 and Nod2 contribute to positive and negative selection of H-Y TCR Tg thymocytes
To determine whether Nod1 or Nod2 were responsible for the positive selection defects observed in Nod1−/−Nod2−/− animals, we examined H-Y TCR Tg mice that were deficient for either Nod1 or Nod2 (Figure 3C). For the H-Y TCR Tg expressing thymocytes, both single deficient Nod1−/− and Nod2−/− female mice showed small decreases in the proportion of CD8 SP cells. As animals deficient for both Nod genes (Nod1−/−Nod2−/−) displayed greater defects in positive selection (Figure 3C), we concluded that both Nod1 and Nod2 promote positive selection signals. Compared to WT H-Y TCR Tg thymocytes, a lower proportion of Nod1−/− and Nod2−/− thymocytes express the H-Y TCR. As there were also increases in CD4 SP thymocytes expressing the H-Y TCR in these mice (Fig 3B, C), it is likely that NOD deficient thymocytes may not efficiently exclude rearrangements of their endogenous TCR loci. Bone marrow chimeras generated using equal proportions of WT and NOD1 or NOD2 deficient H-Y TCR Tg bone marrow revealed that the defects in positive selection contributed by the loss of NOD1 or NOD2 were thymocyte intrinsic (Supplementary Figure 2A). Collectively, these data indicate that both Nod1 and Nod2 play intrinsic roles in promoting positive selection of CD8 T cells expressing the H-Y TCR Tg.
To determine if the absence of NOD proteins also influenced H-Y TCR mediated negative selection, thymocytes were analyzed from male WT, Nod1−/−, Nod2−/−, and Nod1−/−Nod2−/− H-Y TCR Tg animals. As expected, thymocytes from WT H-Y TCR Tg male mice showed a depletion of DP cells and reduced thymic cellularity (Figure 3D). While Nod1−/− and Nod2−/− deficient mice both showed an increase in proportion and number of T3.70+ DP cells, a feature of impaired negative selection in this model, the greatest defect in negative selection was observed in mice deficient for both Nod genes. A proportion of male NOD deficient thymocytes also demonstrated lower levels of H-Y TCR expression, perhaps indicative of ongoing TCR rearrangement, or internalization of the TCR. Importantly, the bone marrow chimera experiments confirmed that the negative selection defect was also intrinsic to NOD deficient thymocytes (Supplementary Figure 2B). We conclude that both Nod1 and Nod2 play intrinsic roles that promote thymocyte negative selection in the H-Y TCR transgenic model.
Lineage diversion of H-Y TCR CD4−CD8− cells in the periphery is impaired by the loss of NOD function
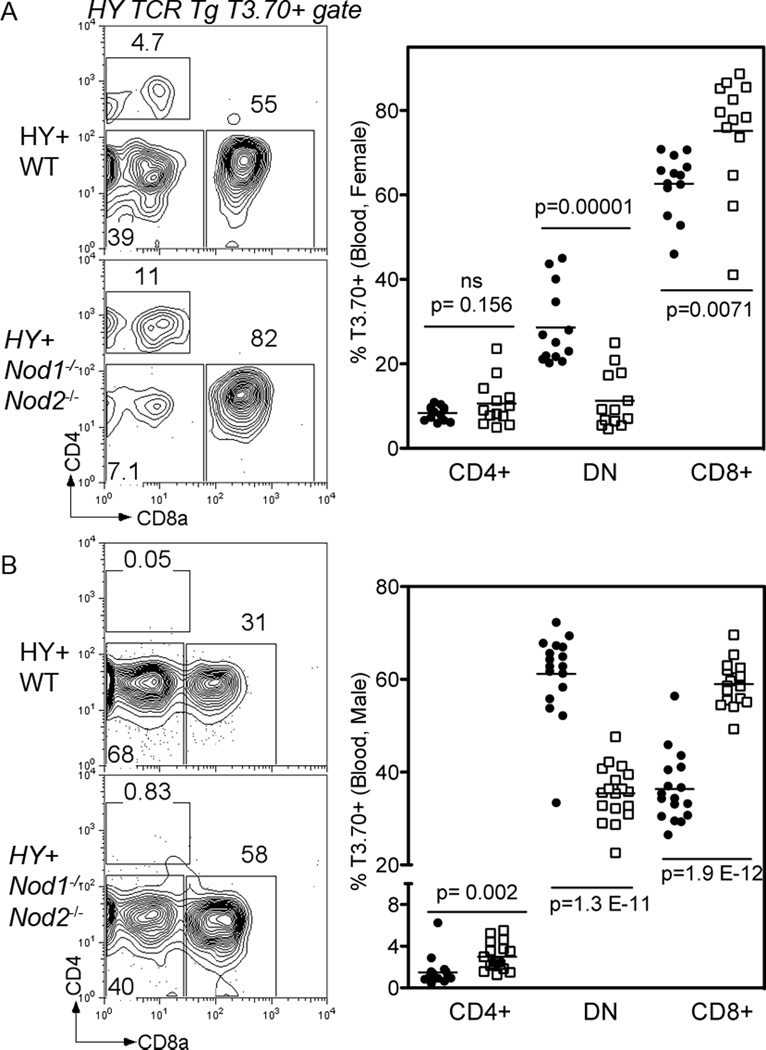
The premature expression of transgenic TCRαβ on CD4−CD8− double negative (DN) thymocytes can alter T cell development, such that it is thymic dependent but MHC independent, a scenario that resembles the γδ T cell developmental pathway. In the periphery, these unusual T cells are identified by expression of the transgenic α/β TCR without CD4 or CD8 co-receptor expression (37, 38). In the case of WT H-Y TCR Tg mice, elegant lineage fate mapping experiments have demonstrated that the majority of T3.70+ CD4−CD8− DN T cells in the periphery of either female or male H-Y TCR Tg animals have bypassed the typical TCRαβ RORγt+ CD4+CD8+ (DP) stage in thymic development (39). An examination of peripheral blood from H-Y TCR Tg Nod1−/−Nod2−/− mice revealed a stark decrease in the proportion of T3.70+ CD4−CD8− DN cells when compared to WT H-Y TCR Tg controls. This decrease coincides with a corresponding increase in the proportion of T3.70+ CD8+ cells in the blood of H-Y TCR Tg Nod1−/−Nod2−/− female (Figure 4 A) or male mice (Figure 4 B). This finding indicates that NOD1 and/or NOD2 are expressed at DN stages of thymocyte development, and can promote lineage diversion in response to the premature expression of a transgenic TCR.
Figure 4. NOD deficiency alters lineage diversion in the H-Y TCR Tg model.
(A and B) Representative flow cytometry plots (left) from peripheral blood of H-Y TCR Tg WT (upper row and filled circles) and Nod1−/−Nod2−/− (bottom row and white squares) female (A) and male (B) mice. Right panels show the proportions of T3.70+ CD4 SP, CD4-CD8- (DN) and CD8 SP T cells in the blood. Data is pooled from multiple experiments. A total of 13 female and 17 male mice of each group were analyzed by the unpaired t test.
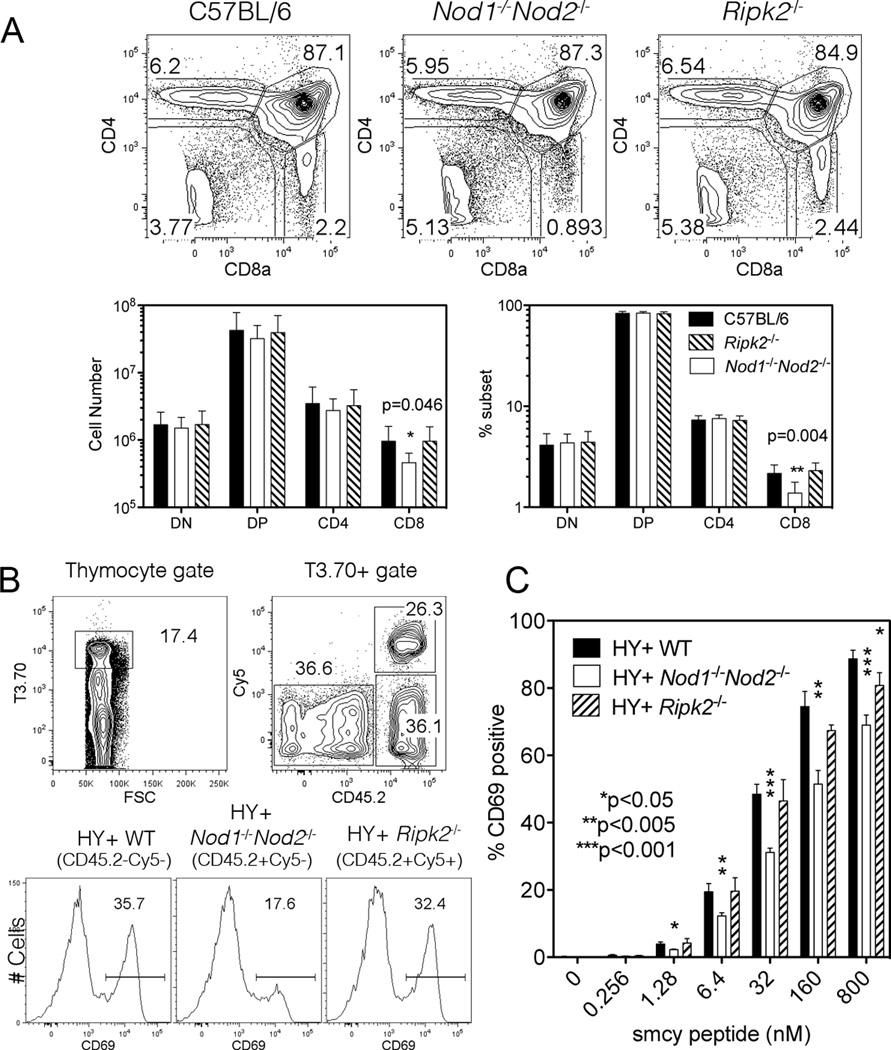
The CD8 SP thymocyte population is normal in Ripk2−/− mice
RIP2, also known as RICK, is the CARD domain containing kinase that interacts with and activates the NFκB pathway in myeloid cells after NOD1 and NOD2 mediated sensing of bacterially derived ligands (7). However Ripk2−/− mice have normal T cell subsets and show no unique role for RIP2 in peripheral T cell survival or effector function (40–42). A limited analysis of thymocyte development in Ripk2−/− mice prompted a comparison of thymocyte populations from C57BL/6, Nod1−/−Nod2−/− and Ripk2−/− mice to determine if the altered positive selection of CD8 SP thymocytes we identified in Nod1−/−Nod2−/− mice is mediated by RIP2 (Figure 5A). No significant differences were noted between C57BL/6 and Ripk2−/− thymocyte populations indicating that the NOD1 and NOD2 signaling pathway that promotes CD8 SP thymocyte positive selection and maturation is independent of the effector kinase RIP2.
Figure 5. Ripk2−/− thymocytes show normal CD8 frequencies and activation.
(A) Representative flow cytometry plots from the thymus of C57BL/6 (left plot), Nod1−/−Nod2−/− (middle plot) and Ripk2−/− (right plot) mice stained for CD4 and CD8a. Numbers in plots represent percentages of depicted populations. Total numbers (lower left) and percentages (lower right) of thymocyte subpopulations of C57BL/6 (black), Nod1−/−Nod2−/− (white) and Ripk2−/− mice (striped) are shown. Bars represent the mean +/− SD. Statistical analysis with unpaired t tests was performed. A representative of two independent experiments is shown (n=3 mice per group/ experiment analyzed).
(B and C) H-Y TCR Tg thymocytes from WT (CD45.2−Cy5−), Ripk2−/− (CD45.2+Cy5+) and Nod1−/−Nod2−/− (CD45.2+Cy5−) female mice were cultured with female C57BL/6 stimulator spleen cells pulsed with different concentrations of the H-Y peptide Smcy. (B) Representative flow cytometry plots where cells were gated on T3.70+ cells (left plot), followed by identification of the three different thymocyte populations according to their CD45.2 vs. Cy5 staining (right plot). Individual thymocyte populations were stained for CD69 upregulation. Histogram plots show the percentage of CD69+ H-Y TCR Tg WT (left), Nod1−/−Nod2−/− (centre), and Ripk2−/− (right) thymocytes after 5h-culture with 32nM of the H-Y peptide Smcy. (C) CD69 upregulation in H-Y TCR Tg WT (black), Nod1−/−Nod2−/− (white) and Ripk2−/− (striped) T3.70+ CD8+ thymocytes stimulated with increasing concentrations of Smcy peptide. Data are representative of two independent experiments with 3 mice per group analyzed. Bars represent the mean +/− SD. *Refers to statistical significance between H-Y TCR Tg WT and mutant thymocytes using the unpaired t test.
Nod1−/−Nod2−/− thymocytes show normal IL-7/STAT5 and Ca2+ signaling
To identify which RIP2 independent signaling pathway might be defective in Nod1−/−Nod2−/− thymocytes, we assessed signaling pathways known to be important for thymocyte positive selection. Although controversial (43), a role for IL-7 cytokine signaling in the selection of CD8 thymocytes has been reported (44). Indeed, conditional deletion of STAT5 in the CD8 compartment leads to a two-fold reduction in CD8 SP thymocytes, a phenotype similar to that observed in Nod1−/−Nod2−/− mice (44). We cultured C57BL/6 and Nod1−/−Nod2−/− thymocytes with or without IL-7 for 15 minutes and assessed STAT5 phosphorylation by intracellular flow cytometry staining (Supplementary Figure 3A). No differences were detected between C57BL/6 and Nod1−/−Nod2−/− thymocytes in IL-7 mediated STAT5 phosphorylation, indicating that the proximal IL-7 signaling pathway appears to be intact.
Given calcium signaling plays an important role in regulating positive selection events in the thymus (45, 46), we tested if this pathway was defective in Nod1−/−Nod2−/− thymocytes (Supplementary Figure 3B). No obvious defects were observed in the magnitude or timing of the Ca2+ flux response of Nod1−/−Nod2−/− thymocytes after crosslinking of CD3 and CD4. Collectively, we found that IL-7 and Ca2+ signaling appear to be intact in Nod1−/−Nod2−/− thymocytes.
CD69 upregulation after H-Y TCR stimulation requires NOD1 and NOD2
To directly compare the signaling capability of the mutant thymocytes to an agonist ligand, WT (CD45.1+), Nod1−/−Nod2−/− and Ripk2−/− H-Y TCR Tg female thymocytes were mixed and co-cultured with C57BL/6 splenocytes that had been pre-incubated with varying concentrations of the Smcy peptide (Figure 5B). The H-Y TCR expressing thymocyte populations were first identified by T3.70 reactivity and the different H-Y TCR Tg thymocytes were distinguished by flow cytometry using the following strategies: The H-Y TCR Tg Ripk2−/− thymocytes were labeled with Cy5 prior to culture; WT H-Y TCR Tg cells were identified by lack of expression of CD45.2, while H-Y TCR Tg Nod1−/−Nod2−/− thymocytes were Cy5− CD45.2+. After stimulation with splenocytes presenting the agonist Smcy peptide, antigen activated thymocytes normally begin to upregulate CD69 in a peptide concentration dependent manner. When compared to H-Y TCR Tg WT and H-Y TCR Tg Ripk2−/− thymocytes, the proportion and degree of CD69 induction after antigenic stimulation was significantly reduced on H-Y TCR Tg Nod1−/−Nod2−/− thymocytes (Figure 5B). The reduction in CD69 upregulation by H-Y TCR Tg Nod1−/−Nod2−/− thymocytes was observed over a range of agonist peptide concentrations (Figure 5C). At the highest doses of agonist peptide, we observed a small defect in the responding Ripk2−/− thymocytes, although this was not as pronounced as the defect in Nod1−/−Nod2−/− thymocyte responses. These results indicate that the upregulation of CD69 expression after H-Y TCR agonist stimulation is dependent on Nod1 and/or Nod2 expression.
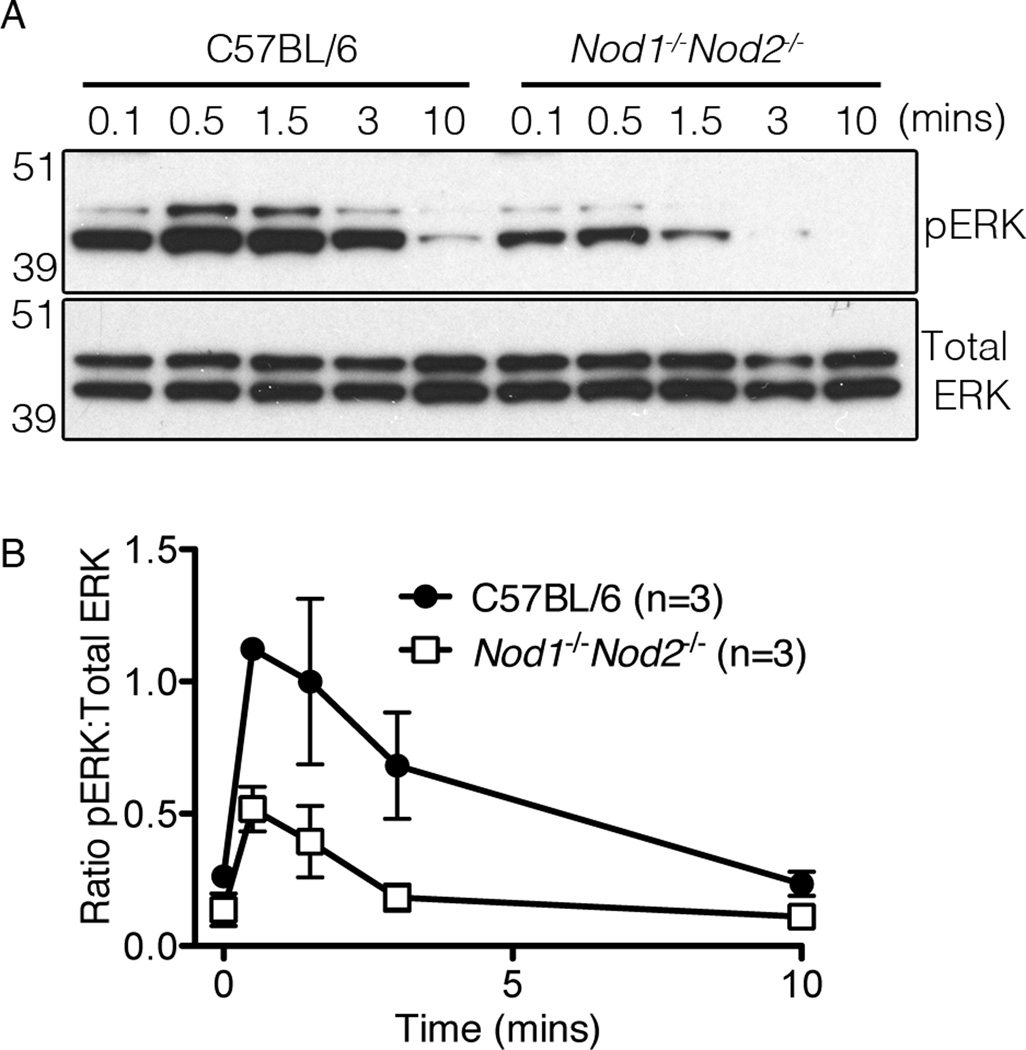
NOD1 and NOD2 promote TCR-mediated ERK phosphorylation
CD69 expression is regulated by the transcription factor AP-1, which is downstream of the Ras/ERK pathway (47). To ascertain if the impaired upregulation of CD69 in TCR stimulated NOD-deficient thymocytes was due to a defect in ERK activation, non-transgenic WT and Nod1−/−Nod2−/− thymocytes were first incubated with biotinylated anti-CD4 and anti-CD3 antibodies on ice, washed, then stimulated with 37°C RPMI media containing streptavidin over a time course of ten minutes. The amounts of phosphorylated ERK1/2 and total ERK1/2 were then determined by western blotting (Figure 6). We consistently found reduced levels of phosphorylated ERK1/2 in TCR activated Nod1−/−Nod2−/− thymocytes, summarized here by the ratio of phospho-ERK1/2 to total ERK1/2 protein (Figure 6B). In contrast, no differences in ERK1/2 phosphorylation were detected between C57BL/6 and Ripk2−/− stimulated thymocytes (Supplementary Figure 4). We conclude that Nod1 and Nod2, but not Ripk2 contribute to the TCR induced activation of the ERK pathway in thymocytes.
Figure 6. NOD deficiency reduces TCR-mediated ERK activation.
(A) Immunoblot analysis of phosphorylated (pERK) and total ERK1/2 in lysates of TCR-stimulated C57BL/6 (left) and Nod1−/−Nod2−/− (right) thymocytes. Thymocytes were bound with biotinylated anti-CD3 and anti-CD4 followed by stimulation with streptavidin over time. After pERK was detected, the membranes were stripped and re-probed with an antibody detecting total ERK1/2. Molecular weight standards are indicated on the left. Data are representative of three independent experiments. (B) Densitometry graph of the ratio of pERK to total ERK levels after stimulation. After measuring the densitometry of pERK to total ERK expression in the same lane/ time point over three experiments, the graph plots mean values +/− SD.
Lack of NOD1 and NOD2 affects signaling of pre-selection thymocytes
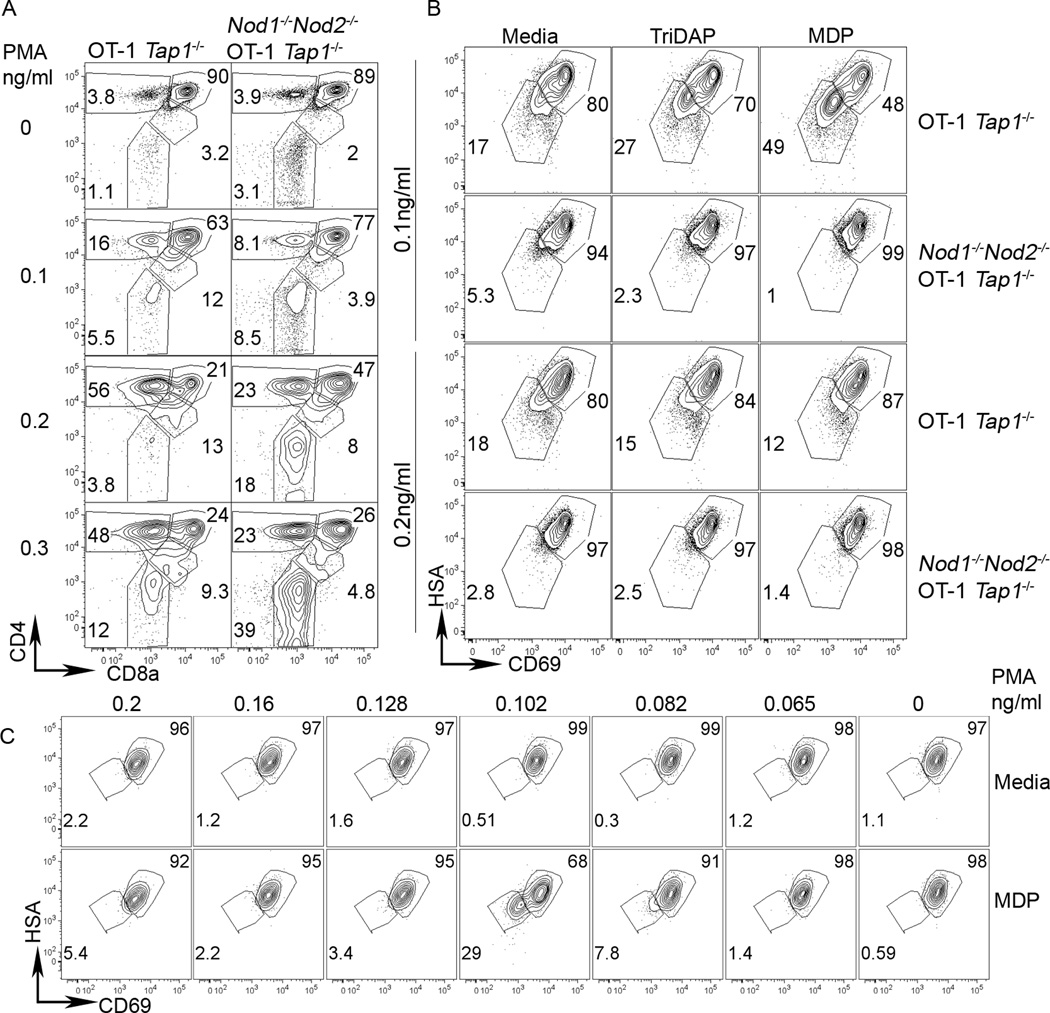
Mouse DP and SP thymocyte subsets have different magnitudes of responses to TCR cross-linking (48, 49). Although unlikely, it is possible that the reduced CD69 up-regulation (as observed on agonist stimulated H-Y TCR Tg Nod1−/−Nod2−/− thymocytes) and ERK phosphorylation within TCR cross-linked Nod1−/−Nod2−/− thymocytes reflects a reduced number of positively selected mature thymocytes rather than a signaling defect per se. To study a homogenous population of thymocytes that had not undergone any selection dependent signaling, we utilized OT-I TCR Tg Tap1−/− mice where the absence of Tap1 prevents DP thymocytes from receiving a MHCI dependent selection signal (48). Both WT and NOD deficient OT-I TCR Tg Tap1−/− thymocytes comprise 90% DP with no CD8SP cells when cultured with media alone (top row Figure 7A). Stimulation of these pre-selection thymocytes with 0.2µg/ml ionomycin and a range of PMA concentrations revealed that WT OT-I TCR Tg Tap1−/− thymocytes (left panels) appeared to be more sensitive to stimulation by PMA than their NOD deficient counterparts (as determined by the proportion of CD4+ cells that had down regulated CD8a on their surface). We conclude that the absence of Nod1 and Nod2 reduces pre-selection thymocyte responses to signals downstream of the TCR as mimicked by PMA and ionomycin.
Figure 7. NOD proteins modify signal thresholds in pre-selection thymocytes.
(A) Representative flow cytometry plots from the thymus of WT OT-I TCR Tg Tap1−/− (left plots), Nod1−/−Nod2−/− OT-I TCR Tg Tap1−/− (right plots) mice stained for 7AAD, CD4 and CD8a (top row). For 15 hours, cells were stimulated with 0.2ug/ml ionomycin and a range of PMA concentrations (shown on left). After washing, cells were cultured without stimuli for a further 29 hours before analysis by flow cytometry. Numbers in plots represent percentages of depicted populations after gating on live cells (7AAD- gate). A representative of four independent experiments is shown.
NOD ligands promote maturation of thymocytes receiving selection signals. (B) Thymocytes from WT OT-I TCR Tg Tap1−/− or Nod1−/−Nod2−/− OT-I TCR Tg Tap1−/− mice were cultured as in (A) with 0.1ng/ml PMA (upper two rows) or with 0.2ng/ml PMA (lower two rows) with no ligand (left panels), 10ug/ml Tri-DAP (middle panels) or with 10ug/ml MDP (right panels). Live cells (7AAD-) were stained for HSA and CD69. (C) Titration of PMA dose (shown above) without (upper row) or with MDP (bottom row) reveals CD69 and HSA down regulation occurs when cells are cultured with the NOD2 ligand MDP specifically at 0.1ng/ml PMA.
NOD2 ligands promote thymocyte maturation
Although NOD deficient thymocytes exhibit signaling deficiencies in the absence of exogenous ligands, we tested whether the addition of NOD ligands could further influence thymocyte selection signals. We added 10µg/ml of NOD1 or NOD2 ligands TriDAP or MDP respectively to the in vitro OT-I TCR Tg Tap1−/− cultures for the initial 15-hour culture with PMA and ionomycin. After washing and further culture for 29 hours without added ligands we observed that the majority of thymocytes remained immature (HSAhi CD69+) (Figure 7B). However, thymocytes stimulated with PMA at 0.1ng/ml (the concentration that promotes CD8 differentiation in TCR α−/− models (50, 51)) responded to the presence of ligands by down-regulating HSA and CD69, a read out of maturation. MDP, the ligand for NOD2, promoted thymocyte maturation more robustly than TriDAP, and as expected, thymocytes lacking NOD1 and NOD2 did not respond to the presence of either ligand. Importantly, OT-I TCR Tg Tap1−/− thymocytes cultured at the higher concentration of PMA that promotes CD4 differentiation (0.2ng/ml) appeared to be unaffected by the addition of NLR ligands. The capacity of OT-I TCR Tg Tap1−/− thymocytes to respond to MDP was restricted to an extremely narrow concentration range of PMA co-stimulation (Figure 7C) and appeared to be linked to conditions that promote CD8 rather than CD4 differentiation (50, 51). We conclude that within narrow signaling thresholds, DP thymocytes can respond to NOD ligands, resulting in the promotion of maturation.
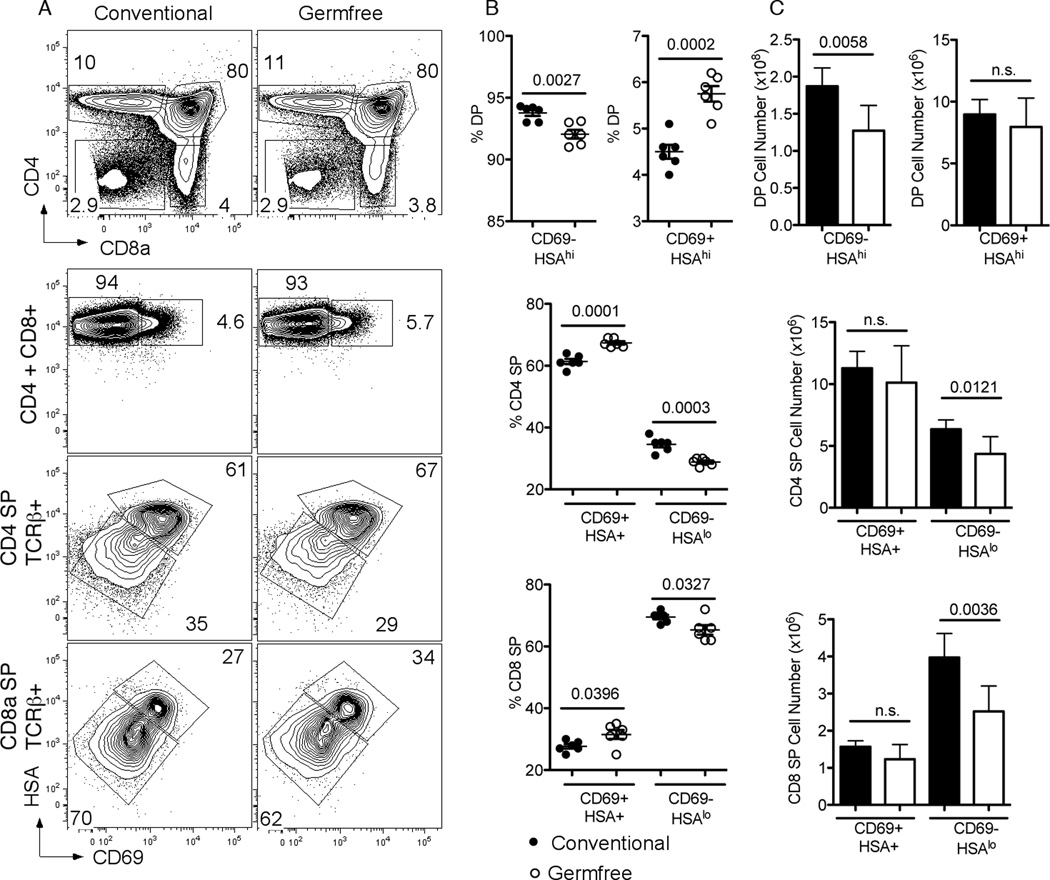
Thymic CD8SP maturation is reduced in germ free mice
The ability of NOD ligands to influence thymocyte development in culture suggested that endogenous commensal organism derived ligands might also influence thymic development. Commensal bacteria and their NLR specific ligands influence neutrophils and the formation of gut lymphoid structures, so thymocyte development from germ free C57BL/6 animals was compared to that from conventionally housed SPF C57BL/6 mice (Figure 8). Consistent with the notion that endogenous bacterial ligands can influence thymocyte development, a decrease in DP cell number and proportion and a 1.5 fold decrease in CD8SP thymocyte numbers was detected when animals were raised in a germ free environment. Analysis of the maturation of the thymocytes from germ free mice revealed a similar pattern to that identified for Nod1−/−Nod2−/− mice. In germ free animals, an increase in proportion of semi-mature SP thymocytes (HSA+CD69+) is observed for both CD4 and CD8 lineages, correlating with a decrease in maturing SP thymocytes (HSAloCD69−) (Fig 8B and C). Importantly, the final maturation step defined by the down-regulation of HSA and CD69 seemed most affected by the lack of commensal organisms, and again seemed to affect the CD8SP thymocyte numbers more so than for CD4SP thymocytes. We conclude that the presence of commensal organisms can promote thymocyte maturation.
Figure 8. Absence of commensal bacteria affects thymocyte maturation.
(A) Representative flow cytometry plots from the thymus of conventionally housed C57BL/6 (left column) and germ-free mice (right column) stained for CD4 and CD8a (top row). Numbers in plots represent percentages of depicted populations. Representative flow cytometry plots of thymocytes stained for HSA and CD69 (bottom three rows). Shown are gated thymocyte subsets of CD4+CD8+ (DP) (top row), CD4SP TCRb+ (middle row), and CD8SP TCRb+ thymocytes (bottom row) from conventionally housed C57BL/6 (left column) and germ-free C57BL/6 mice (right column). Numbers in plot represent percentages of depicted population. Proportions (B) and numbers (C) of thymocyte subpopulations of conventionally housed C57BL/6 (black) and germ-free C57BL/6 mice (white) are shown. Bars represent the mean 6 SD. Statistical analysis with two-tailed unpaired t tests was performed. In (B) and (C), data represent results from six mice per group analyzed.
Discussion
In this study we reveal an unexpected role for the innate sensing molecules NOD1 and NOD2 in CD8 SP thymocyte development. Although there is increasing evidence that pattern recognition receptors play intrinsic roles in augmenting peripheral T cell responses (28, 52, 53), their contribution in the early development of lymphoid cells has not been studied. Our results indicate that both NOD1 and NOD2 promote the positive and negative selection/maturation of CD8 SP thymocytes. Analysis of H-Y TCR transgenic animals where the thymocyte TCR repertoire is artificially restricted revealed non-redundant roles for each NOD molecule in thymocyte selection. In the H-Y TCR Tg experimental model, NOD2 appears to be slightly more important than NOD1 in regulating positive and negative selection. These developmental roles appeared to be additive, in that deficiency of both Nod genes revealed the greatest defects in both positive and negative selection of the HY+ subsets. However roles for either NOD molecule alone in thymic subset development of non-transgenic thymocytes was not observed, perhaps indicating redundancy or other compensatory mechanisms. The TCR signals generated in selecting thymocytes are influenced by interactions with pMHC expressed on antigen presenting cells of hematopoietic and epithelial origin in the cortex and medulla. Despite the expression of NOD1 and NOD2 in epithelial and myeloid cell lineages, data from our bone marrow chimeras point to functional defects in thymic selection when NOD1 and NOD2 are only absent in thymocytes.
Importantly, we did not find a role for RIP2 in the NOD mediated CD8 SP thymocyte developmental pathway, complementing other studies where RIP2 was not required for mature T cell function (40, 42). In a Toxoplasma gondii infection model, a RIP2 independent pathway operating in mature T cells has been reported to involve the NFκB transcription factor c-Rel interacting with NOD2 to promote optimal T cell responses (28). However, as c-Rel plays no role in either positive or negative thymocyte selection in the H-Y TCR Tg model (54) and we found no obvious defects in nuclear NFκB activity in NOD deficient thymocytes (not shown), our study highlights a novel RIP2-independent NOD signal transduction pathway operating intrinsically in selecting CD8 SP thymocytes.
Although it is well established that ERK signaling is important in thymocyte positive selection (55–57), our work demonstrates for the first time that NOD1 and NOD2 promote ERK activation via TCR signaling in thymocytes. In the absence of a link with RIP2, the focus turns to other molecules known to associate with NOD proteins, which include AAMP, Grim-19, ACAP1 and Erbin (58–61).
CD4 thymocytes appear to be more dependent upon ERK signal transduction than CD8 thymocytes (55), yet our data indicates a greater role for NOD1 and NOD2 in CD8 thymocyte development. In TCRα−/− thymocyte pre-selection models, doses of PMA between 0.1- 0.11ng/ml drive differentiation towards a CD8+ lineage, while doses of PMA at 0.2 – 0.22ng/ml drive DP cells towards a CD4SP fate (51). Our results using similar cultures indicate that thymocytes receiving “weaker” CD8 lineage promoting signals are more responsive to the presence of NOD ligands, perhaps explaining the genetic sensitivity of CD8SP thymocytes to Nod1 and Nod2 ablation. However, in addition to the expected lack of response to NLR ligands, NOD deficient thymocytes also exhibit reduced responses to signals generated through the TCR, or mimicked by PMA and Ionomycin. How NOD1 and NOD2 influence the transduction of these signals and if ligand sensing is important in this regard is an exciting question.
While some microbes, including Mycobacteria can infect the thymus, systemic bacterial infections often lead to atrophy of the thymic cortex (62, 63), a process presumably designed to prevent central tolerization of developing thymocytes in the presence of microbial antigens. Whether ligand sensing via LRR domains is regulating NOD1 and NOD2 signaling downstream of the TCR in thymocytes is unknown. Certainly the presence of systemic peptidoglycan ligands derived from commensal bacteria can play a role in neutrophil function and priming via NOD1 (13). Recent work also indicates that in thymic epithelial cells, the expression level of the transcription factor Aire is regulated by commensal bacteria and NOD1 expression (64). Whether peptidoglycan products found in the serum are able to cross the thymus/blood barrier and influence thymocyte maturation is unknown but analysis of germ free mice suggests this to be true. The SLC15A family of peptidyl transporters is important in the transport of NOD peptidylglycan ligands across cellular membranes. In myeloid and dendritic cells, SLC15A3 and SLC15A4 are highly expressed and are important for the transport of endocytosed NOD ligands from endosomes and lysosomes into the cytosol (65, 66). However neither SLC15A3 or SLC15A4 mRNA appear to be expressed at high levels in thymocytes. Intestinal epithelial cells, but not macrophages, use SLC15A1 to transport NOD ligands across the cell membrane (67–69). Interestingly, SP thymocytes express ten fold higher levels of SLC15A1 mRNA than either DP thymocytes or peripheral T cell subsets, suggesting that SP thymocytes could selectively transport NOD1 and NOD2 ligands across the membrane (70).
In summary, our study clearly identifies a novel role for the innate immune sensing molecules NOD1 and NOD2 in promoting positive selection of CD8SP thymocytes. Noncaseating granulomas are one of the diagnostic markers of Crohn’s disease and an increase in proportion of CD8+ T cells is associated with Crohn’s granuloma but not lymphoid aggregates or lymphoid follicles (71). As the peripheral T cell repertoire is influenced by the strength of TCR signals developing cells encounter in the thymus, it is possible that abnormal lymphocyte responses in inflammatory diseases like Crohn’s disease and Blau syndrome may not be solely driven by microbial signals, but may also include contributions of abnormal lymphocyte signaling and repertoire selection.
Supplementary Material
Acknowledgments
We thank A. Barrios, D. Ramsbottom and P. Skog for technical help, K. Sauer, N. Gascoigne, J. Kaye and G. Fu for helpful discussions.
Footnotes
This work was supported by NIH grant A1067403, a National Health and Medical Research Council project grant #1009538 and the Victorian Operational Infrastructure Support Program. A.L.G was supported with an Australian Research Council Future Fellowship and M.M.M. was supported by a Swiss National Science Foundation postdoctoral fellowship. M.O.K. was supported by a NHMRC Career Development Award.
The authors declare no conflict of interest.
References
- 1.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nunez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 2.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 3.Perez LH, Butler M, Creasey T, Dzink-Fox J, Gounarides J, Petit S, Ropenga A, Ryder N, Smith K, Smith P, Parkinson SJ. Direct bacterial killing in vitro by recombinant Nod2 is compromised by Crohn’s disease-associated mutations. PLoS ONE. 2010;5:e10915. doi: 10.1371/journal.pone.0010915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mo JY, Boyle JP, Howard CB, Monie TP, Davis BK, Duncan JA. Pathogen sensing by nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is mediated by direct binding to muramyl dipeptide and ATP. J. Biol. Chem. 2012 doi: 10.1074/jbc.M112.344283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanabe T, Chamaillard M, Ogura Y, Zhu L, Qiu S, Masumoto J, Ghosh P, Moran A, Predergast MM, Tromp G, Williams CJ, Inohara N, Nunez G. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J. 2004;23:1587–1597. doi: 10.1038/sj.emboj.7600175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inohara N, Koseki T, Lin J, del Peso L, Lucas PC, Chen FF, Ogura Y, Nunez G. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J. Biol. Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 7.Kobayashi K, Inohara N, Hernandez LD, Galan JE, Nunez G, Janeway CA, Medzhitov R, Flavell RA. RICK/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416:194–199. doi: 10.1038/416194a. [DOI] [PubMed] [Google Scholar]
- 8.Park JH, Kim YG, McDonald C, Kanneganti TD, Hasegawa M, Body-Malapel M, Inohara N, Nunez G. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 9.Elinav E, Strowig T, Henao-Mejia J, Flavell RA. Regulation of the antimicrobial response by NLR proteins. Immunity. 2011;34:665–679. doi: 10.1016/j.immuni.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, Eberl G. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 11.Petnicki-Ocwieja T, Hrncir T, Liu YJ, Biswas A, Hudcovic T, Tlaskalova-Hogenova H, Kobayashi KS. Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15813–15818. doi: 10.1073/pnas.0907722106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi KS, Chamaillard M, Ogura Y, Henegariu O, Inohara N, Nunez G, Flavell RA. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science. 2005;307:731–734. doi: 10.1126/science.1104911. [DOI] [PubMed] [Google Scholar]
- 13.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat. Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O’Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 15.Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Hafner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nat. Genet. 2001;29:19–20. doi: 10.1038/ng720. [DOI] [PubMed] [Google Scholar]
- 16.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 17.Chamaillard M, Philpott D, Girardin SE, Zouali H, Lesage S, Chareyre F, Bui TH, Giovannini M, Zaehringer U, Penard-Lacronique V, Sansonetti PJ, Hugot JP, Thomas G. Gene-environment interaction modulated by allelic heterogeneity in inflammatory diseases. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3455–3460. doi: 10.1073/pnas.0530276100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. e1631-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 20.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, McArdle W, Strachan D, Bethel G, Bryan C, Lewis CM, Deloukas P, Forbes A, Sanderson J, Jewell DP, Satsangi J, Mansfield JC, Cardon L, Mathew CG. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat. Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fritz T, Niederreiter L, Adolph T, Blumberg RS, Kaser A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut. 2011 doi: 10.1136/gut.2009.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand S. Crohn’s disease: Th1, Th17 or both? The change of a paradigm: new immunological and genetic insights implicate Th17 cells in the pathogenesis of Crohn’s disease. Gut. 2009;58:1152–1167. doi: 10.1136/gut.2008.163667. [DOI] [PubMed] [Google Scholar]
- 23.Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 24.Starr TK, Jameson SC, Hogquist KA. Positive and negative selection of T cells. Annu. Rev. Immunol. 2003;21:139–176. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- 25.Gascoigne NR, Palmer E. Signaling in thymic selection. Curr. Opin. Immunol. 2011;23:207–212. doi: 10.1016/j.coi.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kerns HM, Jutila MA, Hedges JF. The distinct response of gammadelta T cells to the Nod2 agonist muramyl dipeptide. Cell. Immunol. 2009;257:38–43. doi: 10.1016/j.cellimm.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petterson T, Mansson A, Riesbeck K, Cardell LO. Nucleotide-binding and oligomerization domain-like receptors and retinoic acid inducible gene-like receptors in human tonsillar T lymphocytes. Immunology. 2011;133:84–93. doi: 10.1111/j.1365-2567.2011.03414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw MH, Reimer T, Sanchez-Valdepenas C, Warner N, Kim YG, Fresno M, Nunez G. T cell-intrinsic role of Nod2 in promoting type 1 immunity to Toxoplasma gondii. Nat Immunol. 2009;10:1267–1274. doi: 10.1038/ni.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J. Biol. Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 30.Iwanaga Y, Davey MP, Martin TM, Planck SR, DePriest ML, Baugh MM, Suing CM, Rosenbaum JT. Cloning, sequencing and expression analysis of the mouse NOD2/CARD15 gene. Inflamm. Res. 2003;52:272–276. doi: 10.1007/s00011-003-1170-z. [DOI] [PubMed] [Google Scholar]
- 31.Kisielow P, Teh HS, Bluthmann H, von Boehmer H. Positive selection of antigen-specific T cells in thymus by restricting MHC molecules. Nature. 1988;335:730–733. doi: 10.1038/335730a0. [DOI] [PubMed] [Google Scholar]
- 32.Pircher H, Burki K, Lang R, Hengartner H, Zinkernagel RM. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 1989;342:559–561. doi: 10.1038/342559a0. [DOI] [PubMed] [Google Scholar]
- 33.Fu G, Gascoigne NR. Multiplexed labeling of samples with cell tracking dyes facilitates rapid and accurate internally controlled calcium flux measurement by flow cytometry. J. Immunol. Methods. 2009;350:194–199. doi: 10.1016/j.jim.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertin J, Nir WJ, Fischer CM, Tayber OV, Errada PR, Grant JR, Keilty JJ, Gosselin ML, Robison KE, Wong GH, Glucksmann MA, DiStefano PS. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J. Biol. Chem. 1999;274:12955–12958. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 35.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J. Biol. Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 36.Weinreich MA, Hogquist KA. Thymic emigration: when and how T cells leave home. J. Immunol. 2008;181:2265–2270. doi: 10.4049/jimmunol.181.4.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terrence K, Pavlovich CP, Matechak EO, Fowlkes BJ. Premature expression of T cell receptor (TCR)alphabeta suppresses TCRgammadelta gene rearrangement but permits development of gammadelta lineage T cells. J. Exp. Med. 2000;192:537–548. doi: 10.1084/jem.192.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baldwin TA, Sandau MM, Jameson SC, Hogquist KA. The timing of TCR alpha expression critically influences T cell development and selection. J. Exp. Med. 2005;202:111–121. doi: 10.1084/jem.20050359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Egawa T, Kreslavsky T, Littman DR, von Boehmer H. Lineage diversion of T cell receptor transgenic thymocytes revealed by lineage fate mapping. PLoS ONE. 2008;3:e1512. doi: 10.1371/journal.pone.0001512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall HT, Wilhelm MT, Saibil SD, Mak TW, Flavell RA, Ohashi PS. RIP2 contributes to Nod signaling but is not essential for T cell proliferation, T helper differentiation or TLR responses. Eur. J. Immunol. 2008;38:64–72. doi: 10.1002/eji.200737393. [DOI] [PubMed] [Google Scholar]
- 41.Fairhead T, Lian D, McCully ML, Garcia B, Zhong R, Madrenas J. RIP2 is required for NOD signaling but not for Th1 cell differentiation and cellular allograft rejection. Am J Transplant. 2008;8:1143–1150. doi: 10.1111/j.1600-6143.2008.02236.x. [DOI] [PubMed] [Google Scholar]
- 42.Nembrini C, Reissmann R, Kopf M, Marsland BJ. Effective T-cell immune responses in the absence of the serine/threonine kinase RIP2. Microbes Infect. 2008;10:522–530. doi: 10.1016/j.micinf.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 43.Weinreich MA, Jameson SC, Hogquist KA. Postselection thymocyte maturation and emigration are independent of IL-7 and ERK5. J. Immunol. 2011;186:1343–1347. doi: 10.4049/jimmunol.1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park JH, Adoro S, Guinter T, Erman B, Alag AS, Catalfamo M, Kimura MY, Cui Y, Lucas PJ, Gress RE, Kubo M, Hennighausen L, Feigenbaum L, Singer A. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat Immunol. 2010;11:257–264. doi: 10.1038/ni.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finkel TH, McDuffie M, Kappler JW, Marrack P, Cambier JC. Both immature and mature T cells mobilize Ca2+ in response to antigen receptor crosslinking. Nature. 1987;330:179–181. doi: 10.1038/330179a0. [DOI] [PubMed] [Google Scholar]
- 46.Bhakta NR, Oh DY, Lewis RS. Calcium oscillations regulate thymocyte motility during positive selection in the three-dimensional thymic environment. Nat Immunol. 2005;6:143–151. doi: 10.1038/ni1161. [DOI] [PubMed] [Google Scholar]
- 47.Castellanos MC, Munoz C, Montoya MC, Lara-Pezzi E, Lopez-Cabrera M, de Landazuri MO. Expression of the leukocyte early activation antigen CD69 is regulated by the transcription factor AP-1. J. Immunol. 1997;159:5463–5473. [PubMed] [Google Scholar]
- 48.Davey GM, Schober SL, Endrizzi BT, Dutcher AK, Jameson SC, Hogquist KA. Preselection thymocytes are more sensitive to T cell receptor stimulation than mature T cells. J. Exp. Med. 1998;188:1867–1874. doi: 10.1084/jem.188.10.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Havran WL, Poenie M, Kimura J, Tsien R, Weiss A, Allison JP. Expression and function of the CD3-antigen receptor on murine CD4+8+ thymocytes. Nature. 1987;330:170–173. doi: 10.1038/330170a0. [DOI] [PubMed] [Google Scholar]
- 50.Ohoka Y, Kuwata T, Tozawa Y, Zhao Y, Mukai M, Motegi Y, Suzuki R, Yokoyama M, Iwata M. In vitro differentiation and commitment of CD4+ CD8+ thymocytes to the CD4 lineage, without TCR engagement. Int. Immunol. 1996;8:297–306. doi: 10.1093/intimm/8.3.297. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson B, Kaye J. Requirement for sustained MAPK signaling in both CD4 and CD8 lineage commitment: a threshold model. Cell. Immunol. 2001;211:86–95. doi: 10.1006/cimm.2001.1827. [DOI] [PubMed] [Google Scholar]
- 52.Mercier BC, Cottalorda A, Coupet CA, Marvel J, Bonnefoy-Berard N. TLR2 engagement on CD8 T cells enables generation of functional memory cells in response to a suboptimal TCR signal. J. Immunol. 2009;182:1860–1867. doi: 10.4049/jimmunol.0801167. [DOI] [PubMed] [Google Scholar]
- 53.Gaddis DE, Michalek SM, Katz J. TLR4 signaling via MyD88 and TRIF differentially shape the CD4+ T cell response to Porphyromonas gingivalis hemagglutinin B. J. Immunol. 2011;186:5772–5783. doi: 10.4049/jimmunol.1003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strasser A, Grumont RJ, Stanley ML, Gerondakis S. The transcriptional regulator Rel is essential for antigen receptor-mediated stimulation of mature T cells but dispensable for positive and negative selection of thymocytes and T cell apoptosis. Eur. J. Immunol. 1999;29:928–935. doi: 10.1002/(SICI)1521-4141(199903)29:03<928::AID-IMMU928>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 55.Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–443. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 56.McNeil LK, Starr TK, Hogquist KA. A requirement for sustained ERK signaling during thymocyte positive selection in vivo. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13574–13579. doi: 10.1073/pnas.0505110102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Hollander GA, Gascoigne NR, Palmer E. Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature. 2006;444:724–729. doi: 10.1038/nature05269. [DOI] [PubMed] [Google Scholar]
- 58.McDonald C, Chen FF, Ollendorff V, Ogura Y, Marchetto S, Lecine P, Borg JP, Nunez G. A role for Erbin in the regulation of Nod2-dependent NF-kappaB signaling. J. Biol. Chem. 2005;280:40301–40309. doi: 10.1074/jbc.M508538200. [DOI] [PubMed] [Google Scholar]
- 59.Bielig H, Zurek B, Kutsch A, Menning M, Philpott DJ, Sansonetti PJ, Kufer TA. A function for AAMP in Nod2-mediated NF-kappaB activation. Mol. Immunol. 2009;46:2647–2654. doi: 10.1016/j.molimm.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto-Furusho JK, Barnich N, Xavier R, Hisamatsu T, Podolsky DK. Centaurin beta1 down-regulates nucleotide-binding oligomerization domains 1- and 2-dependent NF-kappaB activation. J. Biol. Chem. 2006;281:36060–36070. doi: 10.1074/jbc.M602383200. [DOI] [PubMed] [Google Scholar]
- 61.Barnich N, Hisamatsu T, Aguirre JE, Xavier R, Reinecker HC, Podolsky DK. GRIM-19 interacts with nucleotide oligomerization domain 2 and serves as downstream effector of anti-bacterial function in intestinal epithelial cells. J. Biol. Chem. 2005;280:19021–19026. doi: 10.1074/jbc.M413776200. [DOI] [PubMed] [Google Scholar]
- 62.Nobrega C, Cardona PJ, Roque S, do Pinto OP, Appelberg R, Correia-Neves M. The thymus as a target for mycobacterial infections. Microbes Infect. 2007;9:1521–1529. doi: 10.1016/j.micinf.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 63.Savino W. The thymus is a common target organ in infectious diseases. PLoS Pathog. 2006;2:e62. doi: 10.1371/journal.ppat.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakajima A, Negishi N, Tsurui H, Kadowaki-Ohtsuji N, Maeda K, Nanno M, Yamaguchi Y, Shimizu N, Yagita H, Okumura K, Habu S. Commensal bacteria regulate thymic Aire expression. PLoS One. 2014;9:e105904. doi: 10.1371/journal.pone.0105904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee J, Tattoli I, Wojtal KA, Vavricka SR, Philpott DJ, Girardin SE. pH-dependent internalization of muramyl peptides from early endosomes enables Nod1 and Nod2 signaling. J. Biol. Chem. 2009;284:23818–23829. doi: 10.1074/jbc.M109.033670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nakamura N, Lill JR, Phung Q, Jiang Z, Bakalarski C, Maziere Ade, Klumperman J, Schlatter M, Delamarre L, Mellman I. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature. 2014;509:240–244. doi: 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- 67.Ingersoll SA, Ayyadurai S, Charania MA, Laroui H, Yan Y, Merlin D. The role and pathophysiological relevance of membrane transporter PepT1 in intestinal inflammation and inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol. 2012;302:G484–G492. doi: 10.1152/ajpgi.00477.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ismair MG, Vavricka SR, Kullak-Ublick GA, Fried M, Mengin-Lecreulx D, Girardin SE. hPepT1 selectively transports muramyl dipeptide but not Nod1-activating muramyl peptides. Can. J. Physiol. Pharmacol. 2006;84:1313–1319. doi: 10.1139/y06-076. [DOI] [PubMed] [Google Scholar]
- 69.Marina-Garcia N, Franchi L, Kim YG, Hu Y, Smith DE, Boons GJ, Nunez G. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J. Immunol. 2009;182:4321–4327. doi: 10.4049/jimmunol.0802197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 71.Oki M, Ohtani H, Kinouchi Y, Sato E, Nakamura S, Matsumoto T, Nagura H, Yoshie O, Shimosegawa T. Accumulation of CCR5+ T cells around RANTES+ granulomas in Crohn’s disease: a pivotal site of Th1-shifted immune response? Lab. Invest. 2005;85:137–145. doi: 10.1038/labinvest.3700189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.