Summary
As exosomes are emerging as a new mode of intercellular communication, we hypothesized that the payload contained within exosomes is shaped by somatic evolution. To test this, we assayed the impact on primary CD8+ T cell function, a key mechanism for anti-tumor immunity, of exosomes derived from three melanoma-related cell lines. While morphologically similar, exosomes from each cell line were functionally different, as B16F0 exosomes dose-dependently suppressed T cell proliferation. In contrast, Cloudman S91 exosomes promoted T cell proliferation and Melan-A exosomes had a negligible effect on primary CD8+ T cells. Mechanistically, transcript profiling suggested that exosomal mRNA is enriched for full-length mRNAs that target immune-related pathways. Interestingly, B16F0 exosomes were unique in that they contained both protein and mRNA for Ptpn11, which inhibited T cell proliferation. Collectively, the results suggest that upregulation of PTPN11 by B16F0 exosomes to tumor infiltrating lymphocytes would bypass the extracellular control of the immune checkpoints.
Keywords: exosomes, intercellular communication, tumor immunology, immune checkpoints, PTPN11
INTRODUCTION
Similar to defending against pathogens, organizing host immunity against a malignancy involves engaging a variety of immune cell types through a complex network of intercellular interactions. These immune cells can recognize malignant cells as mutations acquired during oncogenesis provide a source of tumor-associated antigens (Matsushita et al., 2012). However, an emerging idea derived from evolutionary biology is that tumors alter this network of interactions to construct a tissue niche that favors malignant cell survival (Laland et al., 2014; Klinke, 2015). Elements of this altered network include inflammatory cytokines that suppress anti-tumor immunity and an increased prevalence of regulatory T cells and myeloid-derived suppressor cells (Gabrilovich and Hurwitz, 2014). Yet, pre-clinical mouse models suggest that there is a dynamic element to establishing an immunosuppressive environment within a tumor. For instance, the ability of an anti-CTLA-4 therapy to control the growth of a variant of the B16 model of metastatic melanoma depends on time such that delivering an anti-CTLA-4 therapy up to 4 days after tumor implantation eliminates tumors while waiting 12 days abrogated the therapeutic effect (van Elsas et al., 1999). Given that the quality of the immune response external to the tumor microenvironment is similar at the different time points, the ability of lymphocytes to enter the tumor and kill malignant cells diminishes with time. Collectively, these results suggest three points. First, the process of mutation and selection associated with oncogenesis should shape the composition of proteins secreted by malignant cells, similar to selecting and editing of tumor antigens. Second, the observed dynamics suggest that the proteins secreted by malignant cells should accumulate within the tumor microenvironment. Third, these secreted proteins help create an immunosuppressive microenvironment in the tumor. To test this hypothesis, we used mass spectrometry-based proteomics to identify proteins secreted into media conditioned by B16F0 cells, that is the B16F0 secretome, and found that the majority of secreted proteins were associated with exosomes.(Kulkarni et al., 2012)
As an emerging mode of intercellular communication, exosomes are membrane vesicles that are derived from the luminal membranes of multivesicular bodies and are released constitutively through exocytosis by both normal and malignant cells (Yanez-Mo et al., 2015). While exosomes can potentially transfer a variety of functional molecules including proteins (Peinado et al., 2012), lipids, and coding and non-coding RNAs (Valadi et al., 2007; Taylor and Gercel-Taylor, 2008; Skog et al., 2008; Pegtel et al., 2010; Ekstrom et al., 2012; Batagov and Kurochkin, 2013; Wu et al., 2015) between cells to influence cell behavior, their biological roles remain controversial. As exosomes are one of a number of extracellular vesicles, inconsistent nomenclature and isolation methods that result in an unknown mixture of extracellular vesicles fuels the controversial nature of the exosome field (Raposo and Stoorvogel, 2013; Mathivanan et al., 2010). Differences in size distributions is one characteristic that can be used to distinguish between the different types of extracellular vesicles. Exosomes are uniformly distributed in size with reported average diameters ranging between 30 nm to 200 nm in size (Yanez-Mo et al., 2015; Taylor and Gercel-Taylor, 2011; Sokolova et al., 2011). Alternatively, samples can also contain extracellular vesicles released during cell death, which range from 10 to 1000 nm and are heterogenously distributed in size (Wu et al., 2015). Yet, storing exosomes under certain conditions prior to imaging can shrink and fragment exosomes, which makes it impossible to establish the purity of the exosome samples (Sokolova et al., 2011; Wu et al., 2015). Without establishing the purity of a sample containing exosomes, inferring biological function of exosomes from experimental results derived using uncharacterized vesicle samples, which provide a single bulk measure averaged over a population, is questionable. Nonetheless, examples from the literature illustrate the potential role that extracellular vesicles could play to mediate intercellular communication (e.g., (Wieckowski et al., 2009; Pegtel et al., 2010; Ekstrom et al., 2012; Melo et al., 2015; Peinado et al., 2012; Schuler et al., 2014)).
Modes for intercellular communication are shaped by transport mechanisms, such as diffusion and convection. In the context of extracellular vesicles, the relative importance of these different transport mechanisms is influenced by vesicle size. Particles with a diameter of less than 50 nm are easily transported out of a tissue into the draining lymphatics while the extracellular matrix sterically hinders the movement of particles 200 nm in diameter and larger within the tissue (Tang et al., 2013; Irvine et al., 2013). Given the importance of size in transport processes, we hypothesize that exosomes provide a general mechanism for intercellular communication within tissues and that the payload contained within exosomes becomes edited during oncogenesis to alter host immunity. To test this hypothesis, we characterized exosomes freshly isolated from three different cell models related to melanoma with a particular emphasis on their potential impact on T cell function. Specifically, we focused on the B16F0, a non-immunogenic cell model of malignant melanoma; Cloudman S91, an immunogenic model of melanoma; and Melan-A, an immortalized melanocyte cell line (Overwijk and Restifo, 2001; Peter et al., 2001). The mouse species from which these cell lines were derived have different propensities for oncogenesis. Specifically, DBA/2 mice are sensitive and C57Bl/6 mice are resistant to skin oncogenesis using carcinogens (DMBA plus TPA) (Angel et al., 2003), which were used to generate the Cloudman S91 and B16 cell lines, respectively. Similar to editing of tumor antigens (Matsushita et al., 2012), inbred strains that are resistant to oncogenesis would have strong selective pressure to maintain normal tissue homeostasis. Our hypothesis implies that malignant cells that arise in resistant mouse strains, such as the B16 model, develop multiple mechanisms to escape from this selective pressure that are not present in sensitive mouse strains, like the Cloudman S91 model, or normal melanocytes, such as the Melan-A model. Moreover, escape from this selective pressure would manifest as an ability to suppress T cell function.
RESULTS
Exosomes are secreted by both normal and malignant melanocytes
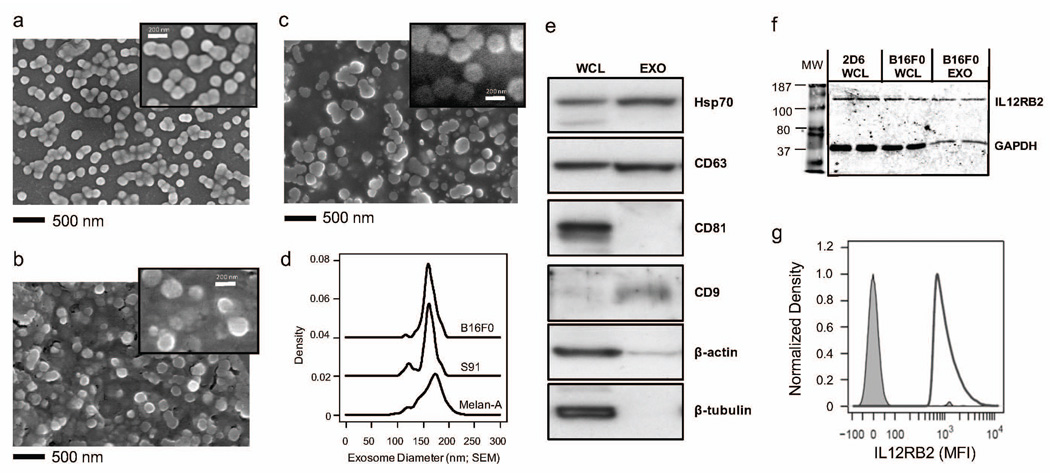
Previously, we found that the majority of proteins secreted by B16F0 cells were associated with exosomes (Kulkarni et al., 2012). To validate this observation, we isolated extracellular vesicles from in vitro cell cultures of mouse melanoma cells, B16F0 and Cloudman S91, and from immortalized melanocytes, Melan-A, using a differential centrifugation protocol. As cells secrete exosomes in addition to a number of other extracellular vesicles, we used scanning electron microscopy (SEM) to image directly the morphology and size distribution of these isolated extracellular particles. The extracellular particles appeared round with frequent membrane connections between the nanoscaled vesicles (see Figure 1 and Supplemental Figure S1). The sizes of the extracellular vesicles were uniformly distributed with similar diameters among the cell lines (B16F0: 163 ± 13 nm (n = 66), Cloudman S91: 160 ± 18 nm (n = 62), Melan-A: 166 ± 22 nm (n = 123)). Western blot analysis of samples derived from B16F0 vesicles indicated the presence of common markers of exosomes, such as Hsp70, CD63, and CD9 (Figure 1e), while CD81, β-actin and β-tubulin were absent. Similar results for Cloudman S91 and Melan-A exosomes are shown in Figure 6b. Given that microvesicles, apoptotic vesicles and necrotic vesicles exhibit heterogeneous distributions that include smaller sized vesicles under EM (Wu et al., 2015) and that these vesicles contain β-actin, we concluded that the isolated extracellular vesicles secreted by all three cell lines were exosomes and that they have similar sizes. Moreover, the size of these exosomes implies that steric interactions limit movement through the extracellular space thereby creating local concentration gradients.
Figure 1. Mouse melanoma and immortalized melanocytes release exosomes.
Extracellular vesicles were isolated from media conditioned by B16F0 cells (a), Cloudman S91 (b), and Melan-A cells (c) and imaged using SEM (bar indicates 500 nm used for a, b and c). SEM images were representative of more than 3 replicates. A higher magnification is shown as inset in panels (see Figure S1). (d) The diameters of the extracellular vesicles from B16F0 (black solid line, N = 66), Cloudman S91 (black dotted line, N = 62), and Melan-A cells (gray shadowed, N = 123) were estimated from the SEM images using ImageJ and summarized using a density distribution, where the density distributions were offset vertically for clarity. (e) Immunoblotting analysis of common exosome markers, where 20 µg of total protein was loaded in each lane (WCL, whole cell lysate; Exo, exosome lysate). (f) Western blot analysis identified IL12RB2 as being present in B16F0 exosomes, B16F0 cells and 2D6 T cells. GAPDH was a loading control and whole cell lysate from 2D6 T cells was used as a positive control for IL12RB2 expression. (g) The presence of IL12RB2 on the surface of exosomes was detected by flow cytometry using IL-12RB2 mAbs-PE. Unstained exosomes were used as a negative control (gray shaded). Results were representative of three replicates.
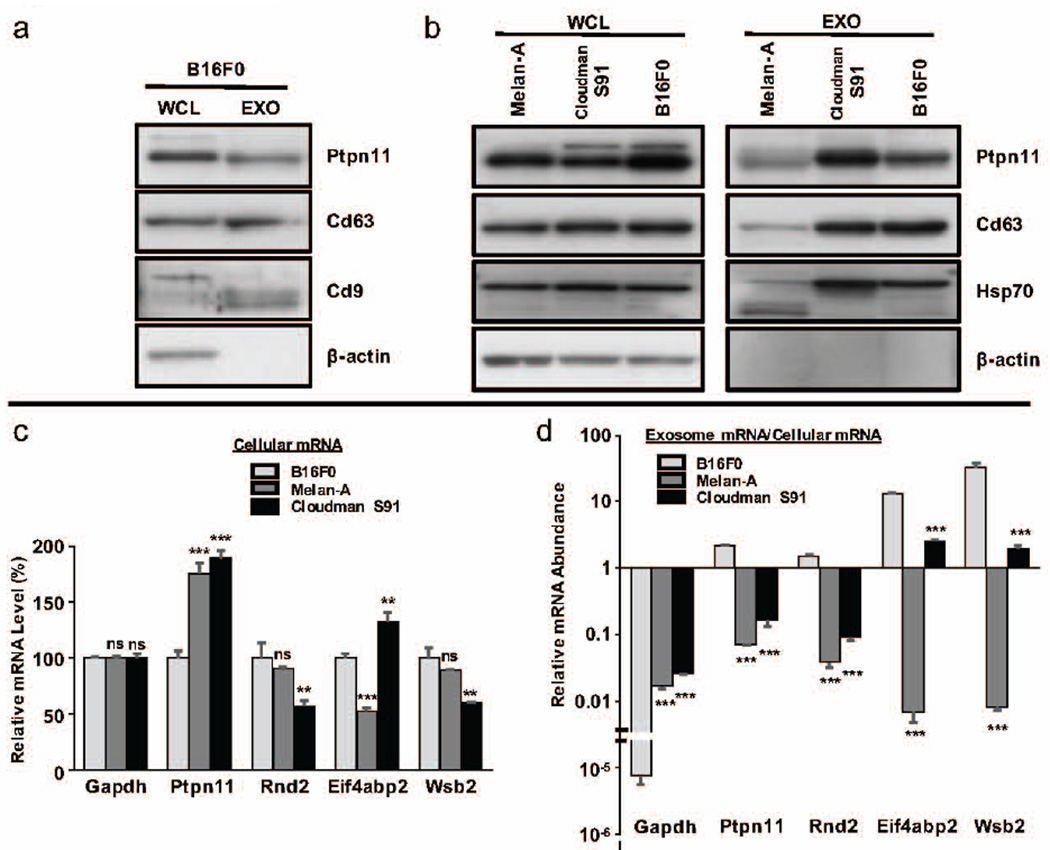
Figure 6. Exosomes from B16F0 but not Cloudman S91 or Melan-A cell lines contained PTPN11 protein and were enriched for PTPN11 mRNA.
(a) Presence of PTPN11 and the exosome markers CD63 and CD9 were assayed in whole cell lysates (WCL) and exosome (EXO) samples from B16F0 cells. (b) The presence of PTPN11 and the exosome markers CD63 and HSP70 were assayed in whole cell lysates and exosome samples from Melan-A, Cloudman S91, and B16F0 cell lines. In Panels (a) and (b), β-actin was included as a negative control for the exosome samples and 20 µg of total protein was loaded in each lane. (c and d) RT-PCR was used to quantify within equally loaded cDNA samples the abundance of transcripts associated with Gapdh, Ptpn11, Rnd2, Eif4abp2, and Wsb2 in cells (c) and in exosomes (d). In (c), relative gene expression is reported using ΔΔCt, where the numerator is the ΔCt for the indicated gene relative to Gapdh in the sample and the numerator is the ΔCt for the average Cts of the indicated gene relative to Gapdh in the B16F0 sample. In (d), relative gene expression is reported using ΔCt for the indicated gene in the exosome sample relative to the cell sample. Statistical differences were assessed using Students t-test, where ** and *** correspond to p < 0.05 and p < 0.001, respectively, for a sample compared to the corresponding B16F0 sample.
Exosomes can transport transmembrane receptors between cells (Al-Nedawi et al., 2008). Previously, we observed that the B16F0 cell line overexpressed one part of the Interleukin-12 (IL12) receptor, IL12RB2 (Kulkarni et al., 2012). Here, we hypothesized that exosomes released by the B16F0 cells also contain IL12RB2, which then could be delivered to recipient cells. In comparing B16F0 exosome lysates with whole cell lysates, Western blot analysis revealed a single band with an apparent molecular weight of 130 kDa for samples from B16F0 cells as well as B16F0 exosomes (see Figure 1f). As a positive control, we also included whole cell lysates from 2D6 T cells, an IL12 responsive cell line (Klinke et al., 2012). Although, the results should be considered as qualitative as the distribution of common loading controls, like GAPDH, between cells and exosomes are unknown. As IL12RB2 may be encapsulated within the exosomes rather than expressed on the surface, flow cytometry revealed that B16F0 exosomes expressed IL12RB2 on the surface (Figure 1g). Collectively, the results suggest that B16F0 cells release exosomes with an average diameter of 163 nm that contain IL12RB2 on the surface. While one might infer that B16F0 exosomes could then deliver IL12RB2 to recipient cells thereby increasing receptor abundance, subsequent experiments with primary CD8+ T cells revealed that such an inference was not so straightforward.
B16F0 exosomes inhibited multiple aspects of an antigenic response in primary CD8+ T cells
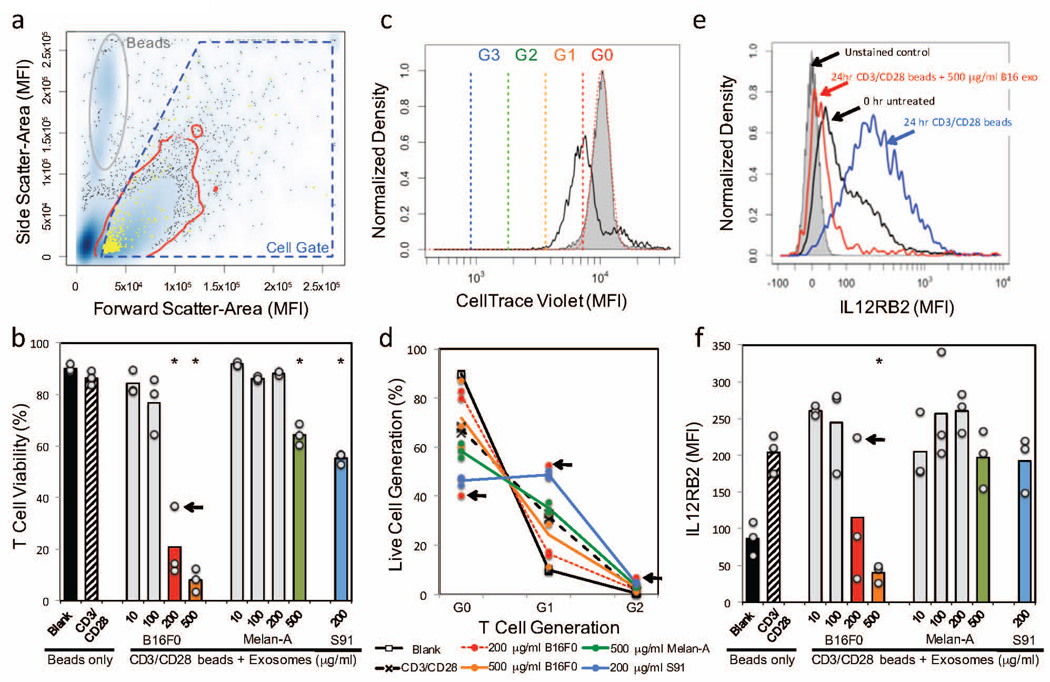
As all three cell lines secrete similarly sized exosomes, exosomes secreted by these three cell lines would similarly accumulate within the local extracellular space. It follows then that if exosomes contribute to local immunosuppression, any differential effect is related to the information that is packaged within these extracellular particles. Given the different propensities for these cell lines to form tumors in immunocompetent mice, our central hypothesis implies that B16F0 exosomes should inhibit anti-tumor immunity while exosomes derived from Melan-A or Cloudman S91 cells should not. Given the importance of CD8+ T cells within the tumor microenvironment in promoting tumor regression (Tumeh et al., 2014; Herbst et al., 2014; van Elsas et al., 1999), we directly assessed whether exosomes influence T cell function by characterizing the impact of B16F0, Cloudman S91, and Melan-A exosomes on primary CD8+ T cells in vitro, as 160 nm particles are likely to have poor tissue penetrance. First, we isolated transgenic CD8+ T cells from mouse splenocytes, of which 94.6% were positive for CD3+ CD8+ and 71.8% were considered to be naïve CD8+ T cells (CD3+ CD8+ CD62L+ CD44−). After isolation, the primary CD8+ T cells were all stained by CellTrace Violet, cultured in media containing IL12 and anti-IL4 mAb to promote a Tc1 phenotype, and activated with anti-CD3/CD28 beads, where blank beads where used as a negative control. In addition, CD8+ T cells activated with anti-CD3/CD28 beads were also stimulated with different concentrations of exosomes freshly isolated from B16F0, Cloudman S91, and Melan-A cells. After 24 hours, cell viability, cell proliferation, and IL12RB2 expression were assayed under the different experimental conditions by flow cytometry (Figure 2). Viability was > 85% in T cells activated with either blank or anti-CD3/CD28 beads (Figure 2b).
Figure 2. B16F0 exosomes inhibit multiple aspects of primary CD8+ T cell response to antigen stimulation.
Primary CD8+ T cells were isolated and stained by CellTrace Violet at 0hr, stimulated for 24 hrs with anti-CD3/CD28 beads and exosomes derived from B16F0, Cloudman S91, and Melan-A cells. (a) Flow cytometric events were gated into cells (blue polygon) and dead/dying cells were excluded based on Live/Dead staining (yellow). (b) The percentage of viable cell events were compared between CD8+ T cells stimulated for 24 hrs with anti-CD3/CD28-conjugated beads (CD3/CD28 beads) and different concentrations of exosomes derived from B16F0, Cloudman S91, and Melan-A cells. (c) Cell proliferation was assessed using a shift in CellTracer Violet, as assayed by flow cytometry. Cells at 0 hours were used as a reference condition (gray shaded) and cells stimulated for 24 hours with CD3/CD28 beads were a positive control (black curve). Colored lines indicate intensity thresholds associated with progressive cell proliferation. (d) The relative T cell proliferation for the indicated experimental conditions are summarized, where individual data points are shown and lines indicate the average response. (e) IL12RB2 was assayed in CD8+ T cells by flow cytometry, where representative histograms for the indicated experimental conditions are shown. (f) The median IL12RB2 staining results were compared between CD8+ T cells stimulated with CD3/CD28 beads and different concentrations of exosomes derived from B16F0, Cloudman S91, and Melan-A cells. Results were obtained in biological triplicate. A significant difference between exosome-treated and the positive control was assessed using a two-sided Students t-test assuming unequal variance (* p < 0.01). One replicate at the 200 µg/ml B16F0 exosome condition was considered an outlier, as indicated by the arrow in panels b, d, and f.
Functionally, exosomes derived from the different cell lines had different effects on primary CD8+ T cells. In terms of cell viability, B16F0 exosomes dose-dependently decreased cell viability with an EC50 equal to 136 µg/ml while the Cloudman S91 exosomes had an EC50 equal to 200 µg/ml. Melan-A exosomes had a minimal effect on cell viability, where viability was decreased to 65% only at the highest concentration (500 µg/ml). In terms of cell proliferation, CellTrace Violet was used to quantify the extent of T cell proliferation under the different experimental conditions (Figure 2c). In primary CD8+ T cells activated with blank beads, 89.7±0.2% of the population was comprised of generation 0 (G0) cells after 24 hours (Figure 2d). Cells proliferated upon activation using anti-CD3/CD28 beads, where 67.3±1.3% of the population was comprised of G0 cells and 31.6±1.2% of the population had proliferated once (G1). Interestingly, B16F0 exosomes inhibited cell proliferation (81.2±2.2% G0/16.4±1.1% G1 for 200 µg/ml, p-value < 0.01 versus anti-CD3/CD28 beads alone) and Cloudman S91 promoted cell proliferation (46.5±1.7% G0/48.7±1.6% G1/4.4±0.3% G2, p-value < 0.0001). Again, the response to Melan-A exosomes plus anti-CD3/CD28 beads was not significantly different than from anti-CD3/CD28 beads alone, with a subtle increase in cell proliferation seen only at the 500 µg/ml dose (58.5±2.8% G0/ 35.1±2.2% G1/3.3±0.7% G2, p-value < 0.01 versus anti-CD3/CD28 beads alone). In terms of promoting a Tc1 phenotype, adding IL12 and anti-IL4 mAb upregulated IL12RB2 over the 24 hour culture conditions (compare black versus blue curves in Figure 2e). Despite the fact that B16F0 exosomes contain IL12RB2, B16F0 exosomes dose-dependently decreased IL12RB2 with an EC50 equal to 171 µg/ml. In contrast, the Cloudman S91 and Melan-A exosomes had no effect on IL12RB2 expression in primary CD8+ T cells (Figure 2f). The presence of IL12RB2 in B16F0 exosomes and the dose-dependent suppression of IL12RB2 by B16F0 exosomes are both mechanisms by which B16F0 cells can deprive tumor infiltrating lymphocytes (TILs) of endogenous IL12, which is an important driver of type 1 polarization. The presence of IL12RB2 on the surface of exosomes released into the tumor microenvironment can sequester IL12 while delivering an exosomal payload to TILs that downregulates IL12RB2 expression would inhibit TILs response to IL12. Overall, the primary CD8+ T cell studies suggest that B16F0 exosomes impact T cell function by inhibiting cell viability, decreasing cell proliferation, and inhibiting the polarization of CD8+ T cells towards a type 1 phenotype. In contrast, exosomes derived from the other two melanocyte cell lines had very different effects. Cloudman S91 exosomes inhibit cell viability but promote cell proliferation and had no effect on Tc1 polarization, while exosomes from Melan-A cells had a negligible functional impact on primary CD8+ T cells.
Exosomes contain intact mRNAs that exhibit differential enrichment relative to the parent cells
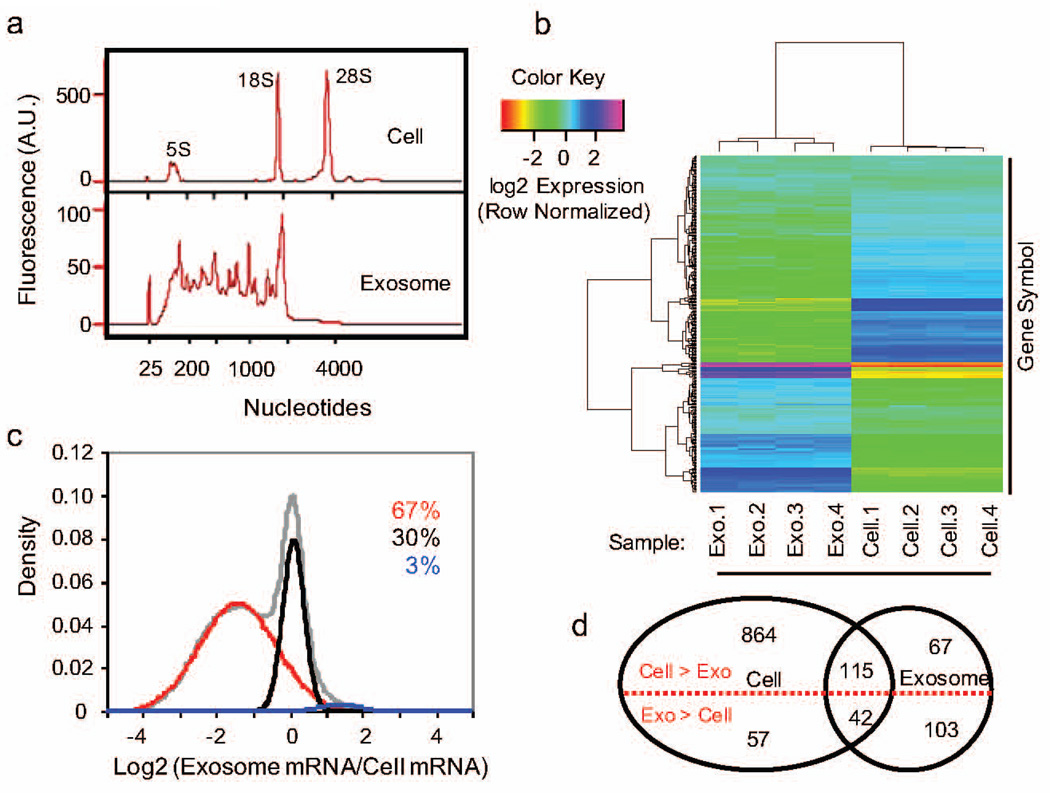
To gain mechanistic insight into these functional differences, we first re-analyzed the mass spectrometry results from the B16F0 secretome.(Kulkarni et al., 2012) Using the 39 identified secretome proteins, Enrichr pathway enrichment results were largely inconclusive with Antigen Processing and Presentation (KEGG HSA04612, p-value < 0.005) providing the most significant pathway. As exosomes also contain RNA, we shifted focused towards coding RNA as a potential mechanism for the observed behavior. Specifically, we used an exon-level cDNA Affymetrix microarray to quantify mRNA transcripts in B16F0 exosomes and the parental B16F0 cells. First, we used an Agilent Bioanalyzer for on-chip-electrophoresis to assess the quality and the distribution in length of RNA in samples isolated from B16F0 cells and freshly isolated B16F0 exosomes (Figure 3a). In contrast to the predominant peaks associated with ribosomal RNAs (5S, 18S, and 28S) in the samples from B16F0 cells, the exosome samples had RNA that was broadly distributed between the 120 and 1800 nucleotides in length and were negative for 28S ribosomal RNA. Affymetrix microarrays were then used to quantify differential expression of mRNA between B16F0 cells and exosomes. Hierarchical clustering illustrates differences among the different samples versus among the probed genes (Figure 3b). Overall the samples were similar between replicates of the same RNA source and different between the cell and exosome samples. In comparing the ratio of average expression between exosome and cell samples, we found that 67% mRNAs probed were more prevalent in the cell, 30% of the mRNAs were equally distributed between the cell and exosome, and 3% of the mRNAs had a higher abundance in the exosomes (Figure 3c, P < 0.01 versus the negative control probesets). Using a Bonferroni correction for multiple hypothesis testing, the distribution in transcripts that were significantly above background is summarized by a Venn diagram. For pathway enrichment analysis, we focused on 145 mRNAs for genes that were more abundant in B16F0 exosomes (Figure 3d, P <1e-9 versus the negative control probesets).
Figure 3. B16F0 exosomes contain mRNAs that are differentially expressed relative to parental cells.
(a) The distribution in RNA contained within the parental B16F0 cells (top panel) and B16F0 exosomes (bottom panel) was quantified using microfluidic electrophoresis (Agilent 2100 Bioanalyzer). (b) Hierarchical clustering of mRNA detected above background (p < 0.01) from four B16F0 exosomes and four B16F0 cell samples using Affymetrix Exon-level cDNA microarrays. (c) The overall distribution in exosome versus cellular abundance of mRNAs (gray curve) was deconvoluted into three normally distributed populations: mRNAs enriched in cells (red curve 67% of total), mRNAs equally distributed between cell and exosomes (black curve 30% of total), and mRNAs enriched in exosomes (blue curve 3% of total). (d) Using a more stringent gene call (p < 1e-9), a Venn diagram summarizes the number of mRNA genes differentially expressed between cell and exosome samples.
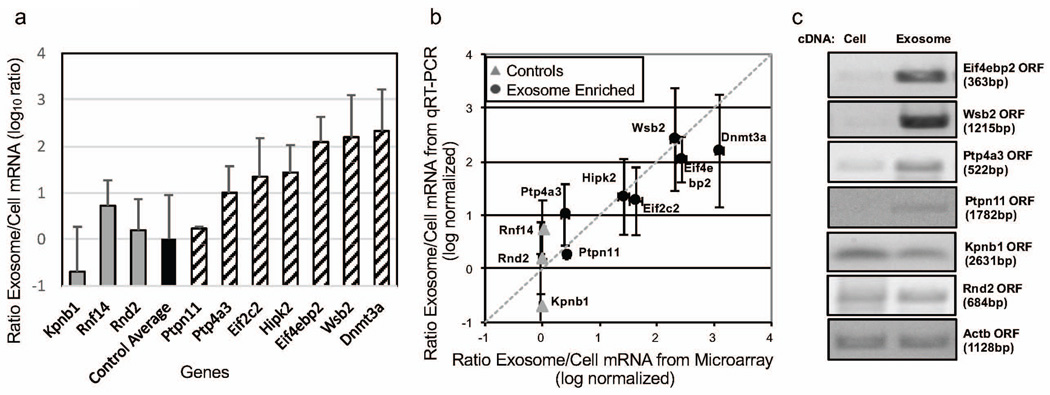
To validate the cDNA microarray results, we amplified a subset of the mRNA using quantitative RT-PCR (Figure 4a and b). As endogenous loading controls for exosomes are unclear, we selected three genes to serve as loading controls, Rnf14, Rnd2 and Kpnb1, that had equal abundance between the cell and exosome samples based on the microarray results. In addition, we selected an additional subset of genes that appeared to be more abundance in exosomes: Ptp4a3, Ptpn11, Hipk2, Eif2c2, Wsb2, Eif4ebp2 and Dnmt3a. Overall, the qRT-PCR results were consistent with the cDNA microarray results in assaying the relative abundance between cells and exosomes for the different genes as the correlation coefficient was 0.922 (Figure 4b). We also used semi-quantitative PCR to confirm that the exosomal mRNAs were intact transcripts rather than fragmented (Figure 4c). From the transcripts observed in both B16F0 cells and exosomes shown in panels a and b of Figure 4, we amplified seven open reading frames (ORFs): Ptpn11, Eif4ebp2, Wsb2, Ptp4a3, Kpnb1, Rnd2 and Actb. Similar to the qRT-PCR results, ORFs of the three loading control genes, Kpnb1, Rnd2 and Actb, were equally abundant in the two samples and mRNA for the genes, Ptpn11, Eif4ebp2, Wsb2 and Ptp4a3 were enriched in the B16F0 exosome samples. Collectively, the gene expression results suggest that mRNAs are selectively packaged into exosomes and that the mRNAs are intact ORFs.
Figure 4. Relative mRNA abundance between B16F0 exosomes and cells were consistent between qRT-PCR and microarray analyses.
(a) The abundance of 10 genes (Kpnb1, Rnf14, Rnd2, Ptp4a3, Ptpn11, Eif2c2, Hipk2, Eif4ebp2, Dnmt3a, and Wsb2) in B16F0 exosomes versus B16F0 cells were quantified by quantitative RT-PCR (mean ± s.d., N = 3). The qRT-PCR results were normalized to the average differential abundance of three control genes: Kpnb1, Rnf14, and Rnd2. (b) The relative abundances of mRNAs assayed by qRT-PCR were compared against the relative abundances of mRNAs assayed by cDNA microarray. The dotted line indicates that the two different assays provide the same results for relative abundance. (c) Full-length coding sequences (ORFs) were amplified by semi-quantitative RT-PCR. Equal concentrations of RNA were reverse-transcribed into cDNA and amplified by PCR. After 25 cycles, full-length open-reading frame amplicons were monitored every three cycles and resolved on agarose gel before the amplification was saturated.
B16F0 exosomes deliver a biological payload to T lymphocytes
As a subset of mRNAs were selectively enriched in exosomes, we used the Enrichr pathway enrichment algorithm to identify biological pathways that are associated with mRNAs that are enriched in exosomes. Using 145 enriched mRNAs in B16F0 exosomes, we identified 18 signaling pathways that had positive combined scores (see Supplemental Table S1). Interestingly, several of the pathways are closely associated to the anti-tumor immunity, with the Type I Interferon signaling pathway having the lowest p-value and the IL-2, the T cell receptor, and Type II Interferon signaling pathways all having a positive combined score. One of the challenges with pathway enrichment results is that genes associated with a specific pathway can either promote or inhibit signal transduction. The gene that was common to 12 out of the 18 enriched pathways was Ptpn11. Ptpn11 encodes protein tyrosine phosphatase, non-receptor type 11, also known as SHP2, and negatively regulates a variety of signaling pathways through two tandem Src homology-2 domains.
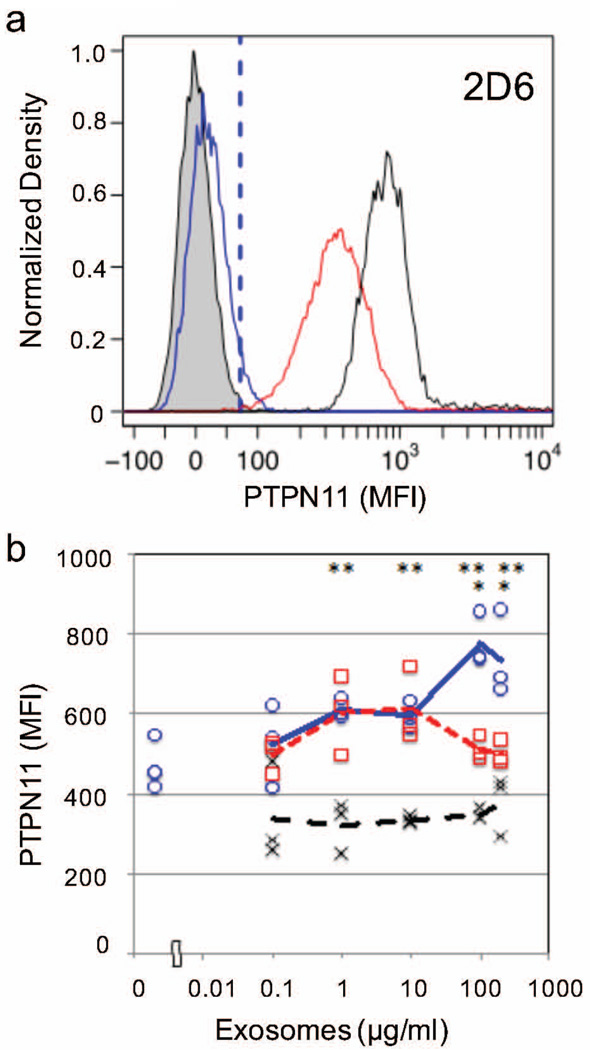
Given the potential role for PTPN11 in negatively regulating Interferon, IL-2, and T cell receptor signaling pathways, we focused next on whether exosomes can deliver a biological payload to upregulate PTPN11 in T lymphocytes. To answer this question, we incubated a type 1 T cell model (2D6) with freshly purified exosomes derived from either B16F0, Cloudman S91, and Melan-A cells and monitored the abundance of PTPN11 in the 2D6 T cells by flow cytometry (Figure 5). Under basal conditions, 2D6 T cells had a median MFI associated with PTPN11 staining equal to 450. Exposing 2D6 T cells to 200 µg/ml of B16F0 exosomes doubled the PTPN11 levels in 2D6 T cells. This increase in PTPN11 levels was dose-dependent and unique to the B16F0 exosomes, as shown in Figure 5b. In contrast, PTPN11 levels in 2D6 T cells stimulated with either Cloudman S91 exosomes or Melan-A exosomes were not significantly different for increasing concentrations of exosomes.
Figure 5. PTPN11 was dose-dependently increased in T lymphocytes when treated with exosomes from B16F0 but not Cloudman S91 or Melan-A cell lines.
(a) Representative flow cytometry results for 2D6 T cells stained using a PE-conjugated PTPN11 mAb following incubation with 200 µg-protein/ml B16F0 exosomes (NanoDrop Assayed) for 30 minutes (black curve). Background level of PTPN11 was assayed in untreated 2D6 cells (red curve). Isotype stained (blue curve) and unstained 2D6 cells (gray shaded) were used as negative controls. (b) The median MFI associated with PTPN11 staining in 2D6 cells following incubation with different sources of exosomes at different concentrations (0.1, 1, 10, 100 and 200 µg-protein/ml denoted on x-axis). The 2D6 cells were incubated with the indicated concentrations of B16F0 exosomes (blue circles), Cloudman S91 exosomes (red squares), and Melan-A exosomes (black crosses). Statistical differences were assessed using Students t-test, * p < 0.05 for B16F0 exosome-incubated cells versus Cloudman S91 exosome-incubated cells, ** p < 0.01 for B16F0 exosome-incubated cells versus Melan-A exosome-incubated cells.
Following from these flow cytometry results, we next assessed whether PTPN11 protein or mRNA were differentially abundant within exosomes from these three cell lines. To assay protein abundance, we probed for PTPN11 in addition to the exosomal markers CD63 and CD9 in lanes equally loaded with whole cell lysates and exosome samples (Figure 6a). All three proteins were present in both the exosome and whole cell lysate samples, while β-actin was absent in the exosome samples. We also noted that CD9 appears to have different isoforms due to the preference for a lower molecular weight isoform in the exosome sample compared to the whole cell lysate. In comparing the three melanoma cell lines, PTPN11 appeared to be most abundant in whole cell lysates from the B16F0 cell line and present in Cloudman S91 and B16F0 exosome samples (Figure 6b). For exosomal protein markers, CD63 and HSP70 were present and β-actin was absent in exosome samples. Generally, immunoblot results for Melan-A exosome samples were similar although the bands were less pronounced. In terms of mRNA, qRT-PCR was used to quantify message for Ptpn11, Rnd2, Eif4abp2, Wsb2, and Gapdh in cells and exosome samples (Panels c and d in Figure 6 and Figure S2). Using equal mRNA loading, the relative expression of the individual genes in each cell line were compared to average abundance of the corresponding gene message in B16F0 cells, where Gapdh expression served as loading controls. While the pattern of relative abundance of message for Ptpn11, Rnd2, Eif4abp2, and Wsb2 were different among the cell lines, Ptpn11 message was 2 times higher in both Melan-A and Cloudman S91 cells compared with B16F0 cells. The narrow distribution in relative Gapdh abundance among the cell lines supports equal loading among the samples. This is important as no clear transcripts in exosome samples exist as loading controls. For instance, Gapdh was at the lower limit of detection in B16F0 samples from exosomes compared to cellular mRNA but was more abundant in exosome samples from Melan-A and Cloudman S91 cell lines (Figure 6d). Interestingly, the abundance of Ptpn11 mRNA among exosome samples exhibited the opposite pattern as the cellular samples, where B16F0 had a factor of 10 greater abundance in exosomes that either the Melan-A or Cloudman S91 samples. In contrast, the abundance of Rnd2, Eif4abp2, and Wsb2 mRNA in exosome samples exhibited similar trends among the cell lines as the cellular samples, where Melan-A cells generally had lower abundance in exosome samples relative to cellular samples. Overall, the immunoblotting and qRT-PCR data was largely consistent with the flow cytometry results presented in Figure 5. Although the 30 minute time point likely reflects protein rather than mRNA delivery, B16F0 exosomes appear to be more efficient in upregulating PTPN11 compared to Cloudman S91 exosomes as the qualitative immunoblotting results suggest that both exosomes contain PTPN11 protein but only B16F0 exosomes are enriched for Ptpn11 mRNA. In short, we found that B16F0 exosomes are different from Melan-A and Cloudman S91 exosomes in that they contain both protein and mRNA for PTPN11 and that they can dose-dependently increase PTPN11 abundance in T cells.
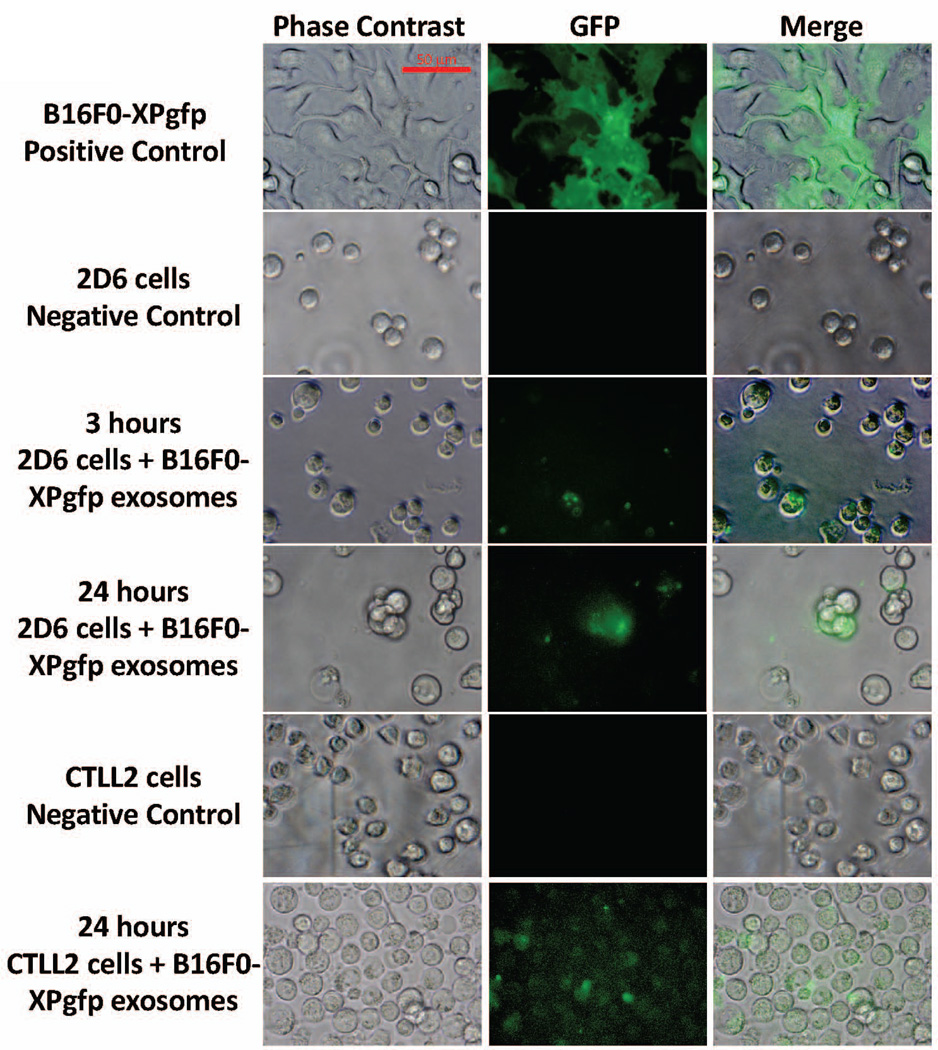
Given that exosomes derived from B16F0 cells had a different pattern of mRNA and protein abundance compared to the other two cell lines, we also asked whether B16F0 exosomes could deliver a biological payload to T cells (Figure 7). Toward this aim, we induced the expression of GFP in B16F0 cells using a lentiviral vector that contains an N-terminal XPack-GFP fusion protein designed to be loaded into exosomes. Exosomes were then isolated from exosome-free media conditioned by these transfected B16F0 cells (B16F0-XPgfp). B16F0-XPgfp exosomes were co-cultured for up to 24 hours with either 2D6 or CTLL-2 cells. In GFP-positive cells, GFP was initially localized but became dispersed within recipient cells within 24 hours. These data suggest that exosomes derived from B16F0 cells can deliver a biological payload to T cells. Taken together, one mechanistic explanation for the selective inhibition of the proliferation of primary CD8+ T cells is that exosomes from B16F0 but not Cloudman S91 or Melan-A cells deliver a biological payload that increases PTPN11 abundance in T cells.
Figure 7. B16F0 exosomes delivered a biological payload to T cells.
B16F0 cells were transfected with a lentivirus-based Xpack-GFP plasmid that targets GFP to exosomes (positive control). Exosomes isolated from B16F0-XPgfp cells were co-cultured with 2D6 (3 and 24 hours) and CTLL2 (24 hours) cells, washed three times with DPBS, and imaged by fluorescence microscopy. Phase contrast and merged images are shown for comparison. 2D6 and CTLL-2 cultured in media alone served as negative controls. Scale bar indicates 50 mum.
Increased PTPN11 inhibited T cell proliferation in response to IL-2
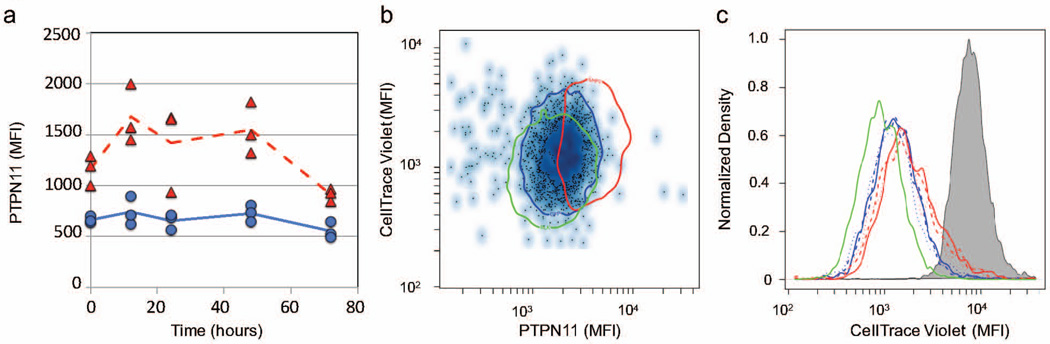
Given the association of local proliferation of CD8+ T cell with tumor regression and the role of PTPN11 in negatively regulating both IL-2 and T cell receptor signaling pathways, we hypothesized that an increase in PTPN11 levels would inhibit T cell proliferation. As a model for CD8+ cytotoxic T lymphocytes that proliferates in response to IL-2 stimulation, we used the CTLL-2 cell line to test whether T cell proliferation was inhibited by an increase in PTPN11 in response to gene transfection using two different Ptpn11 expression plasmids that each encoded a different isoform. The two isoforms differed in that isoform 2 uses an alternative in-frame splice site in the 3 coding region and encodes a slightly shorter protein. As the two expression plasmids containing the two different Ptpn11 isoforms behaved similarly in our hands, the results using the plasmid containing Ptpn11 isoform 1 are shown (Figure 8). Cells exposed to the transfection protocol and transfected with the same plasmid encoding GFP were used as negative controls. Following post-transfection conditioning, PTPN11 transiently increased in CTLL-2 cells transfected with the Ptpn11 plasmid, in comparison with CTLL-2 cells transfected by a plasmid encoding GFP (Figure 8a). Based on these results, we used the 48 hour time point to assay PTPN11 levels and differences in cell proliferation. Using CTLL-2 cells transfected with the plasmids encoding either Ptpn11 or gfp or without a plasmid, we stained the cells using CellTracer Violet (CTV) and stimulated the cells with 2000 U/ml of IL2. After 48 hours, we assayed CTV staining and PTPN11 levels in live CTLL-2 cells associated with the different treatment groups by flow cytometry (Figure 8b and c). In contrast to bulk cell measurements, flow cytometry assays the response of live cell population to PTPN11 expression despite the fact that the efficiency of transfection and plasmid expression may be heterogeneous among cells of the population. The distributions in CTV staining between the different experimental groups were significantly different (Pearsons χ2 test, p < 1e-10). In looking at the distribution in PTPN11 abundance versus CTV staining, an increase in PTPN11 abundance correlated with a decrease in cell proliferation, as measured by an increase in CTV staining (Figure 8b). The decrease in cell proliferation in cells transfected with the plasmid encoding Ptpn11 was consistently observed across the biological replicates (Figure 8c). These results suggested that an increase in PTPN11 alone inhibited CTLL-2 proliferation in response to IL-2 stimulation.
Figure 8. An increase in PTPN11 inhibited the proliferation of CTLL-2 cells in response to IL-2.
(a) At the indicated time points, PTPN11 was assayed in CTLL-2 cells following a 40 hour recovery in complete medium after transient transfection with expression plasmids containing either the gene for GFP (blue circles and solid line) or Ptpn11 (red triangles and dotted line). (b, c) CTLL-2 T cells were transfected with Ptpn11 plasmids (red curves), GFP plasmids (blue curves), or no plasmid control (green curves), recovered for 40 hours in complete medium, stained using CellTrace Violet (CTV), and then stimulated with 2000 U/ml IL-2. Following 48 hours of stimulation with IL-2, the levels of PTPN11 and dilution of CellTrace Violet in live cells were assayed by flow cytometry, where at least 20,000 events were acquired. The results are representative of at least three replicates. (b) Scatter plots summarized the intensity of PTPN11 staining versus CellTrace Violet staining for cells transfected with a GFP plasmid (dots). Similar results for untransfected CTLL-2 cells and cells transfected with a PTPN11 plasmid are shown as a single contour that encloses 95% of the population (green oval: untransfected CTLL-2 cells, blue oval: GFP-transfected CTLL-2 cells, red oval: PTPN11-transfected CTLL-2 cells). (c) Histograms of CellTrace Violet staining in three biological replicates of untransfected (green curves), GFP-transfected (blue curves), and PTPN11-transfected (red curves) observed after 48 hours were compared to CTLL-2 cells fixed at 0 hours (gray shaded curve).
DISCUSSION
Identifying local mechanisms of immunosuppression within the tumor microenvironment is a key barrier for broadening the clinical benefit of immunotherapies for cancer (Ascierto et al., 2013). Similar to the sculpting of tumor antigens during oncogenesis, a related hypothesis is that proteins secreted by malignant cells are shaped by somatic evolution. Malignant cells that then emerge secrete proteins that alter intercellular communication to promote tumor growth. As extracellular vesicles, like exosomes, represent an emerging mode of cell-to-cell communication by delivering proteins and coding and non-coding RNAs to recipient cells, the objective of this study was to test this hypothesis by characterizing the impact of extracellular vesicles derived from three melanoma-related cell lines on T cell function. The three melanoma-related cell lines include the B16F0, the Cloudman S91, and Melan-A. In short, we found that all three cell lines secrete exosomes of a similar size and morphology but exosomes derived from each cell line elicit a different response in primary CD8+ T cells. Exosomes derived from B16F0 cells contain proteins present in the parental cell and intact mRNAs that are differentially packaged into exosomes. Exosomal mRNAs that are enriched in B16F0 exosomes target a number of pathways that are important for anti-tumor immunity, including T cell proliferation (Tumeh et al., 2014), IFN-γ production (Kaplan et al., 1998), and responsiveness to IL12 (Airoldi et al., 2005; Billerbeck et al., 2014). In contrast, Cloudman S91 exosomes increased T cell proliferation over anti-CD3/CD28 antigen presentation beads alone while Melan-A exosomes had a negligible impact on primary CD8+ T cells.
Antibodies to block the PD-1/PD-L1 and CTLA-4 signaling pathways, collectively called immune checkpoint modulators, provide a significant clinical advance in the treatment of melanoma (Hodi et al., 2010; Topalian et al., 2015). Conceptually, immune checkpoints limit the clonal expansion and effector activity of T cells by engaging signaling proteins that inhibit T cell signal transduction. Antibodies against these immune checkpoints enhance immune cell infiltration into and proliferation within the tumor microenvironment (Tumeh et al., 2014). However, pre-clinical studies using the B16 model show that the ability to control tumor growth by inhibiting immune checkpoints decreases as tumors become larger (van Elsas et al., 1999). In comparing exosomes from these three melanoma-related models, exosomes from the two melanoma cell lines contained PTPN11 protein while only B16F0 exosomes were also selectively enriched with Ptpn11 mRNA. Functionally, we found that only B16F0 exosomes were able to increase PTPN11 dose-dependently in recipient cells. While exosomes contain a more complex payload, an increase in PTPN11 alone is able to suppress T cell proliferation. While the exact role that PTPN11 plays in signal transduction remains controversial (Li et al., 2012; Lorenz, 2009), PTPN11 is a phosphatase that can be engaged by both the PD-1/PD-L1 and CTLA-4 signaling pathways to inhibit T cell signaling (Li et al., 2015; Chemnitz et al., 2004; Schneider and Rudd, 2000).
We also observed that fresh extracellular vesicles derived from all three cell lines were uniformly distributed in size with average diameters between 160 and 166 nm. The homogeneous size distributions and the presence of the exosomal markers CD63 and Hsp70 and the absence of β-actin suggest that the isolated extracellular vesicles are exosomes. While typically used as a loading control, β-actin is absent in exosome samples (Gross et al., 2012) but is found in microvesicle samples (Muturi et al., 2013; Muralidharan-Chari et al., 2010) as a result of the outward blebbing of the cell membrane to form microvesicles (Paluch et al., 2005). While our exosomes were larger than the conventional wisdom (e.g., 30 – 100 nm) (Yanez-Mo et al., 2015; Taylor and Gercel-Taylor, 2011), storing exosomes under certain conditions prior to imaging can shrink and fragment exosomes (Wu et al., 2015). Accurate morphology is important as the size can influence whether exosomes can establish concentration gradients within tissues, where the local concentration is increased by the rate of cellular release within a tissue and decreased by transport mechanisms within tissues. While all cells release exosomes, melanoma cells release exosomes at a greater rate compared to normal controls (Logozzi et al., 2009), possibly by a p53-mediated stress response (Yu et al., 2006). The results imply that exosomes recently produced from all three cell types remain within tissues, as their size suggests that steric interactions hinder movement within the extracellular matrix (Tang et al., 2013; Irvine et al., 2013). Similarly, concentration gradients of exosomes provide directional cues for migrating cells within tissues by stabilizing leading-edge protrusions (Sung et al., 2015). Collectively, the results suggest that tumor-derived exosomes can accumulate within the tumor microenvironment to deliver a complex payload that dose-dependently suppresses CD8+ T cell function. Specifically in the case of the B16 model, exosomal upregulation of PTPN11 to tumor infiltrating lymphocytes would bypass the extracellular control of the immune checkpoint pathways that forms the basis of this therapeutic strategy.
Organizing an effective anti-tumor immune response among a variety of cell types involves the relay of information between cells using cytokines like Interleukin-12 (IL12). IL12 locally promotes anti-tumor immunity by enhancing the cytotoxic activity of Natural Killer and CD8+ T cells. Thinking of cancer as an evolutionary process implies that malignant cells develop mechanisms to alter this local network of intercellular communication (Klinke, 2015). In addition to upregulating PTPN11 in lymphocytes, exosomes derived from B16F0 can locally suppress cell response to IL12 in two ways. The first is an indirect mechanism where B16F0 cells secrete exosomes that contain IL12RB2 that could enhance a cytokine sink for endogenous IL12 (Kulkarni et al., 2012). The second is a direct mechanism where B16F0 exosomes dose-dependently inhibited IL12RB2 expression in primary CD8+ T cells. These mechanisms for inhibiting immune cell response to IL12 complement B16F0 release of Wnt-inducible signaling protein 1 (WISP1) that blocks T cell response to IL12 (Kulkarni et al., 2012; Klinke, 2014). Secretion of WISP1, creation of a cytokine sink for IL12, and exosomal delivery of a complex payload, which includes PTPN11, to suppress T cell proliferation, viability, and IL12RB2 expression are some of the additional mechanisms that we identified in the B16 model. While in vivo studies will be necessary to establish the sensitivity of these mechanisms to therapeutic modulation, identifying how the local networks for intercellular communication are changed during oncogenesis is the first step in developing therapeutic strategies to restore the intercellular signaling networks associated with normal tissue homeostasis and to broaden the clinical benefit for existing cancer immunotherapy, such as immune checkpoint modulators.
METHODS
Antibodies and reagents
Cytokines, drugs, kits, and pharmacological inhibitors were obtained from commercial sources and used according to the suppliers recommendations unless otherwise indicated. Western blot Abs: IL12RB2 pAbs (Santa Cruz Biotechnology; clone M-20) and GAPDH mAbs (Cell Signaling Technology; clone 14C10); LI-COR IR secondary Abs (LI-COR Biosciences). Fluorophore-conjugated mAbs were used in flow cytometry: PE-PTPN11 mAb (Santa Cruz Biotechnology; clone B-1), PE-IFN-γ mAb (BD Pharmingen; clone XMG1.2), DyLight 488-DDK mAb (OriGene Technologies; clone 4C5), PE-IL12RB2 mAb (RnD Systems; clone 305719). Isotype control mAbs were rat and mouse mAbs conjugated with PE and APC (BD Biosciences). Blocking reagent was mouse IgG (Jackson Laboratory) in PBSAz for cells (DPBS mixed with 2% FBS and 0.02% sodium azide). Phosflow Lyse/Fix buffer and Perm Buffer III (used in PTPN11 mAbs staining), Cytofix/Cytoperm Fixation/Permeabilization kit (used for IFN-γ mAbs and DDK-protein-tag mAbs staining) were from BD Biosciences. CellTrace Violet (CTV) and Live/Dead Yellow staining were purchased from Invitrogen. PBSAz buffer for exosomes is DPBS mixed with 0.05% BSA and 0.02% sodium azide, which was double-filtered through 0.02 µm filters. CD8a+ T Cell Isolation Kit II, anti-CD3E/anti-CD28 beads for T cell activation (anti-CD3/CD28 beads), APC-CD3E mAb, VioBlue-CD8α mAb, FITC-CD44 mAb, and PE-CD62 mAb were from Miltenyi Biotec Inc.
Mice and primary T cell isolation
Eight-to 12-week-old transgenic B6.Cg-Thy1a/Cy Tg(TcraTcrb)8Rest/J female mice were obtained from Jackson Laboratory. Mice were housed in sterilized microisolator cages in the university vivarium, and facility sentinel animals were regularly screened for specific pathogen agents. All studies were performed in accordance with all federal and institutional guidelines for animal use and were approved by the West Virginia University IACUC. Primary CD8+ T cells were isolated from mouse splenocytes using a magnetic cell sorting CD8a+ T Cell Isolation Kit II and methods described previously (Finley et al., 2011). Enrichment for naïve CD8+ T cells (CD8+, CD3+, CD44−, and CD62L+) was confirmed by flow cytometry.
Cell line culture and stimulation
B16F0, Cloudman S91 (clone M-3), and CTLL-2 cell lines were acquired from American Type Culture Collection (ATCC). An immortalized mouse melanocyte cell line, Melan-A, was provided by V. Hearing (National Cancer Institute, Bethesda, MD, USA) (Le Pape et al., 2008). The TH1 cell model, 2D6, was provided by M. Grusby (Harvard University, Cambridge, MA) and cultured as described previously (Klinke et al., 2012). B16F0 and Cloudman S91 cells were maintained in DMEM (Cellgro/Corning) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone) and penicillin/streptomycin (Gibco). B16F0 cells were also transduced following manufacturer’s instructions with a lentivirus encoding an N-terminal XPack-GFP fusion protein (XPAK530PA-1, System Biosciences, Inc., Mountain View, CA). After lentivirus infection, GFP-positive B16F0 cells (B16F0-XPgfp) were selected for five days in complete medium containing 1.0 µg/ml of puromycin. Melan-A cells were maintained in DMEM supplemented with 10% non-heat-inactivated FBS, 20 mM hydrogen chloride, 10 mM HEPES (Gibco), 100 U/ ml penicillin and 100 µg/ml streptomycin (penicillin/streptomycin), 200 nM 12-o-tetradecanoyl phorbol 13-acetate (TPA, Sigma) and 200 µM phenylthiourea (PTU, Sigma). CTLL-2 cells were maintained in RPMI 1640 medium supplemented with 10% T-STIM without con A containing IL-2 (BD Biosciences), 10% FBS, penicillin/streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 1.5 g/l sodium bicarbonate, 10 mM HEPES, 4ppm β-mercaptoethanol (β-ME; 2 µl in 500 ml medium). Primary CD8+ T cells were cultured in RPMI 1640 supplemented with 10% FBS, penicillin/streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 4 ppm β-ME, 70 U/ml IL-2, 90 pM rmIL-12 (eBiosciences), 5 µg/ml anti IL-4 mAb (eBiosciences), with or without the co-stimulation from anti-CD3/CD28 beads (1:1 cell:bead ratio). All cells were cultured at 37°C in 5% CO2.
Exosome isolation, electron microscopy (EM) imaging and Western blot analysis
Fresh extracellular vesicles were isolated using a staged differential centrifugation protocol from serum-free media conditioned by the indicated cell lines and imaged using either scanning electron microscopy (SEM) or transmission EM (TEM), as described previously (Klinke et al., 2014; Wu et al., 2015). Briefly, extracellular vesicles were isolated from cell-conditioned media using 300×g for 10 minutes to remove cells, 2,600×g for 10 minutes to remove residual cells and debris, 10,000×g for 60 minutes to remove microvesicles, and 100,000×g for 2 hours to collect nano-scaled vesicles in pellets. The resulting pellet was resuspended, washed once in DMEM, and re-pelleted at 100,000×g for 2 hours. Once isolated, nano-scaled vesicles were resuspended in DPBS and kept on ice. Exosome sizes were quantified from the SEM images using ImageJ and summarized as distributions using kernel density estimation in R version 2.15.2. All numbers are reported as mean ± standard deviation, unless otherwise noted. The abundance of exosomal proteins were compared against the respective whole cell lysates by Western blot targeting CD9, CD63, CD81, Hsp70, IL12RB2, PTPN11, and GAPDH, as described previously (Kulkarni et al., 2012). Rabbit anti-CD9, CD63, CD81, and Hsp70 antibodies were obtained from System Biosciences, Inc. (Palo Alto, CA); rabbit anti-mouse GAPDH mAbs (14C10) were purchased from Cell Signaling Technology (Danvers, MA); goat anti-mouse IL12RB2 (M-20), mouse anti-β-actin, and mouse anti-β-tubulin antibodies were from Santa Cruz Biotechnology (Dallas, Texas). For detecting IL12RB2, membranes were probed with secondary Abs conjugated with fluorophores (IRDye800CW donkey anti-goat IgG Abs, IRDye680LT donkey anti-rabbit IgG Abs, LI-COR Biosciences), and imaged using the Odyssey Infrared Imaging System (LI-COR).
Cell stimulation with exosomes
2D6 T cells were first pre-conditioned for 12 h in the absence of IL-12p70 and then stimulated with complete media containing the indicated concentrations of fresh exosomes resuspended in PBS. After 30 min culture, cells were prepared for flow cytometric analysis, as described in (Klinke et al., 2012). Briefly, cells were fixed using Phosflow Lyse/Fix buffer, permeabilized using Perm Buffer III, blocked using mouse IgG, stained using PE-PTPN11 mAb, and suspended in PBSaz for flow cytometry analysis. In addition, GFP was delivered to 2D6 and CTLL-2 cells using fresh exosomes isolated aspectically from B16F0-XPgfp cells. CTLL-2 and 2D6 cells were co-cultured with B16F0-XPgfp exosomes for the indicated times at a concentration of 1.0 mg/ml, washed three times with DPBS, and imaged using a Zeiss Axiovert 40 CFL fluorescent microscope using a standard GFP filter set.
Primary CD8+ T cells were stained with CellTrace Violet (CTV) and then cultured for 24 hours in complete media containing anti-CD3/CD28 beads at a 1:1 ratio with cells. Primary CD8+ T cells were also stimulated with the indicated concentrations of fresh exosomes. Primary CD8+ T cells cultured in complete media that contained only blank beads at a 1:1 ratio was used as a negative control. At the indicated time points, cells were stained using Live/Dead Yellow, fixed using Phosflow Lyse/Fix buffer, blocked using mouse IgG, stained with PE-IL12RB2 mAb, resuspended in PBSaz and analyzed by flow cytometry. All experiments were performed in biological triplicate.
RNA analysis
Total RNA isolated from B16F0 exosomes and cells by RNeasy Plus kit (Qiagen) was quantified using Nanodrop and analyzed by on-chip-electrophoresis using the Agilent Bioanalyzer. RNA sample (100 ng) with an RNA integrity number value greater than 7 was processed by the Ambion WT Expression Kit. Each reaction yielded between 6 to 9 micrograms of cDNA, whereby 5.5 µg of cDNA was fragmented and labeled with biotin by the GeneChip WT Terminal Labeling Kit (Affymetrix). The efficiency of fragmentation reaction was checked via Agilent Bioanalyzer. The entire reaction of fragmented and biotin-labeled cDNA (50 µl) with added hybridization controls was hybridized to the mouse GeneChip 1.0 ST Exon Arrays (Affymetrix) at 45°C for 17 hours in GeneChip Hybridization Oven 640 (Affymetrix). Mouse GeneChip 1.0 ST Exon Arrays were stained using FS 450_0001 protocol in Affymetrix GeneChip Fluidics Station 450. Phycoerythrin labeling was detected within the Affymetrix GeneChip Scanner 3000 7G plus using 532 nm light and detected by a photomultiplier tube. Expression Console software (Affymetrix) was used to check quality controls of hybridized chips. All chips that passed quality controls were RMA normalized using Expression Console software. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE71610 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE71610).
Quantitative reverse-transcription PCR (q-RT-PCR) was used to validate the cDNA microarray results. RNA samples were purified using an RNeasy mini kit (Qiagen) and reversely transcribed with Superscript III First-Strand (Invitrogen). Quantitative PCR was carried out on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) using Perfecta SYBR Green SuperMix, ROX (Quanta Biosciences). Proprietary primer mixtures were purchased from Qiagen for the corresponding genes: Kpnb1 (QT00153419), Rnf14 (QT00157241), Rnd2 (QT00314216), Ptp4a3 (QT00138243), Eif2c2 (QT01757833), Hipk2 (QT00197890), Eif4ebp2 (QT00144606), Dnmt3a (QT00106519), Wsb2 (QT01747739). Data was analyzed using the delta/delta CT method and the Sequence Detector Software version 2.2 (Applied Biosystems). In addition, the semi-quantitative PCR was used to amplify the full-length coding sequences (ORFs) of the indicated genes from the same cDNAs, in which the amplified DNA products were monitored from certain cycles after the desired fragments appeared, and compared before the amplification was saturated. The primer sequences are listed as follows: Beta-actin forward ATGGATGACGATATCGCTGC, reverse CTAGAAGCACTTGCGGTGCAC; Ptpn11 forward ATGACATCGCGGAGATGGTTTC, reverse TCATCTGAAACTCCTCTGCTGCTG; Kpnb1 forward ATGGAGCTCATAACCATCCTCG, reverse TCAAGCCTGGTTCTTCAGTTTCC; Ptp4a3 forward ATGGCCCGCATGAACCGGC, reverse CTACATGACGCAGCATCTGGTC; Eif4ebp2 forward ATGTCCGCGTCGGCCGGTG, reverse TCAGATGTCCATCTCAAACTGAG; Rnd2 forward ATGGAGGGGCAGAGTGGC, reverse TCACATGAGGTTACAGCTCTTG; and Wsb2 forward ATGGAGGCCGGAGAGGAG, reverse CTAGAAAGTCCTGTATGTGAGG. To compare mRNA enrichment among B16F0, Melan-A, Cloudman S91 cells and exosomes, the following primer pairs were used for real-time quantitative RT-PCR: Gapdh forward TGCACCACCAACTGCTTAGC, reverse GGCATGGACTGTGGTCATGAG; Ptpn11 forward AGAGGGAAGAGCAAATGTGTCA, reverse CTGTGTTTCCTTGTCCGACCT; Rnd2 forward CGCTGCAAGATCGTAGTGGT, reverse CAGAGGCCGGACATTGTCAT; Eif4abp2 forward GGGACGCTGTTCTCCACAAC, reverse TATTGGGCAGATGGCAAGGTG; and Wsb2 forward CTTCTCGCCAGACGGTTCC, reverse GGTGACGTGCCCAGAGTTTC.
PTPN11 transfection and assay of CTLL-2 T cell proliferation
CTLL-2 T cells were transiently transfected with two different PTPN11 expression plasmids with CMV promoters (MC219480 for isoform variant 1, MC219394 for isoform variant 2, OriGene Technologies), and a GFP expression plasmid with a CMV promoter (pmaxGFP Vector from Lonza) using a Nucleofector electroporation 2b device, program L-029 and solution L (Lonza). As a negative control, CTLL-2 cells electroporated with plasmid TE buffer. Following transfection, cells were cultured in calcium-free medium containing 10% FBS at 37°C for 10 minutes and then cultured in complete medium containing 50 ng/ml TPA (PMA; Sigma) to induce gene expression for 40 hours. CTLL-2 cells were then labeled with CTV to record cell proliferation and stimulated with 2000 U/ml IL-2 for the indicated time points. IL-2 stimulated cells were stained with Live/Dead Yellow, fixed, permeabilized, blocked, stained using PE-conjugated PTPN11 mAbs and subjected to flow cytometric analysis. At least two biological replicates of transfection were performed. For each batch of transfected cells, experiments were performed in biological triplicates.
Flow cytometry
Immunostaining of surface proteins on exosomes was performed using methods described previously (Wu et al., 2015). In brief, freshly isolated exosomes were resuspended in PBSAz buffer, blocked using mouse IgG, stained with PE-conjugated IL12RB2 mAb, washed in 20 ml PBSAz and spun down at 150,000×g for 1 hour at 4°C. Stained exosomes were then resuspended in 0.5–1 ml PBSAz supplemented with 0.04% paraformaldehyde (Electron Microscopy Sciences), and stored at 4°C before flow cytometry analysis. For all flow cytometric analyses, FACSAria or LSRFortessa flow cytometers were used (BD Biosciences). The fluorescent intensity for each parameter was reported as a pulse area using 18-bit resolution. Single-stain controls were used to establish fluorescent compensation parameters. Unstained cells were used as negative flow cytometry controls. Flow cytometry data was exported as FCS3.0 files and analyzed using R/Bioconductor software (Klinke and Brundage, 2009).
Statistics
The statistical differences in Western blots and cell proliferation were compared using Students t-test. A p value < 0.05 was considered as statistically significant. Gene transcript abundance was estimated based on the average expression of all core probesets for a gene measured by Affymetrix microarray. The probability of a gene being expressed above background by random chance was estimated using the negative control probesets and a Welch’s t-test with the degrees of freedom calculated using the Welch-Satterthwaite equation. Using at least 5,000 cell events, statistical significance of the distributions in CellTrace Violet MFI was assessed with a probability binning approach whereby the univariate distribution was discretized into 100 bins and the number of events in the negative control and sample bins were compared with a Pearsons χ2 test. To assess statistical significance, a p-value of less than 0.01 was used for estimating the distribution of mRNA expression between cells and exosomes. A p-value of less than 1e-9 was used for pathway enrichment analysis using the Enrichr algorithm (Chen et al., 2013) and incorporates a Bonferroni correction for multiple hypothesis testing.
Supplementary Material
Extracellular vesicles were isolated from media conditioned by B16F0 cells (a and b), Cloudman S91 (c), and Melan-A cells (d) and imaged using electron microscopy. SEM was used to image extracellular vesicles in panels a, c, and d, while TEM was in panel b.
(A) Following reverse transcription with equal amounts of mRNA, cDNAs were loaded in each well for single-plex qPCR. The trace of the reporter signal normalized to the reference dye (Rn) is shown as a function of cycle (circles). Following baseline subtraction, a curve (dotted blue line) was fit to the exponential growth phase and used to estimate the number of cycles (CT) required for Rn to exceed a threshold (dotted green line). The results are representative of at least three replicates for each target. (B) A calibration curve was used to assess the performance of the qPCR assay. Template abundance was varied by diluting cDNA obtained from B16F0 whole cell lysate. Gapdh, Wsb2, and Ptpn11 were assayed in the different dilution samples. CT values below 36 cycles were proportional to the logarithm of the template concentration. The efficiency of the PCR amplification for the different target were between 80 and 120% (Gapdh: 104%, Wsb2: 90%, and Ptpn11: 118%). The standard deviation in CT values suggests that a difference by a factor of two in template concentration could be detected with 99% confidence using a sample size of 3. (C) CT values estimate the abundance of mRNA obtained from whole cell lysates (red circles) versus exosomes (black squares) for each cell line (B16F0, S91, and Melan-A) and target tested (Wsb2, Ptpn11, Gapdh, Eif4abp2, Kpnb1, and Rnd2). ** indicates that the difference in CT between whole cell lysates and exosome samples was significant (p-value < 0.0002).
P-values were computed using the Fisher exact test. The Z-score is a statistical ranking metric derived from running the Fisher exact test for many random gene sets in order to compute a mean rank and standard deviation from the expected rank for each term in the gene-set library and finally calculating a z-score to assess the deviation from the expected rank. Combined score is calculated from p-value and z-score.
Significance.
While exosomes are emerging as a new mode of intercellular communication through the transfer of proteins and RNAs between cells, their role in establishing a malignant tissue niche remains controversial. Here, we observed that tumor-derived exosomes can upregulate PTPN11, which is a phosphatase involved in immune checkpoint pathways, to inhibit T cell proliferation and are sized to accumulate within the tumor microenvironment. More interestingly, the results suggest a mechanism of resistance to immunotherapy whereby upregulation of PTPN11 by B16F0 exosomes to lymphocytes that infiltrate the tumor would bypass the extracellular control of the immune checkpoints.
Acknowledgments
DJK conceived the study; YW, ECM, and WD performed experiments; DJK, WD, and YW analyzed the data; YW and DJK drafted the manuscript; and all authors revised and approved the final version. The authors thank Jason Ware for assistance with RT-PCR. This work was supported by grants from the National Science Foundation (NSF) CAREER 1053490 and the National Cancer Institute (NCI) R15CA123123 and R01CA193473. The flow cytometry and imaging cores were supported by grants from the National Institutes of Health (GM103488/RR032138, RR020866, and OD016165). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NSF, the NCI, or the National Institutes of Health.
Footnotes
Conflict of Interest
The authors declare that they have no financial/commercial conflicts of interest.
References
- Airoldi I, Di CE, Cocco C, Sorrentino C, Fais F, Cilli M, D’Antuono T, Colombo MP, Pistoia V. Lack of IL12RB2 signaling predisposes to spontaneous autoimmunity and malignancy. Blood. 2005;106(12):3846–3853. doi: 10.1182/blood-2005-05-2034. [DOI] [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat. Cell Biol. 2008;10(5):619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- Angel JM, Caballero M, DiGiovanni J. Identification of novel genetic loci contributing to 12-O-tetradecanoylphorbol-13-acetate skin tumor promotion susceptibility in DBA/2 and C57BL/6 mice. Cancer Res. 2003;63(11):2747–2751. [PubMed] [Google Scholar]
- Ascierto PA, Capone M, Urba WJ, Bifulco CB, Botti G, Lugli A, Marincola FM, Ciliberto G, Galon J, Fox BA. The additional facet of immunoscore: immunoprofiling as a possible predictive tool for cancer treatment. J Transl Med. 2013;11:54. doi: 10.1186/1479-5876-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol. Direct. 2013;8:12. doi: 10.1186/1745-6150-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billerbeck E, Labitt RN, Vega K, Frias-Staheli N, Dorner M, Xiao JW, Rice CM, Ploss A. Insufficient interleukin-12 signalling favours differentiation of human CD4(+) and CD8(+) T cells into GATA-3(+) and GATA-3(+) T-bet(+) subsets in humanized mice. Immunology. 2014;143(2):202–218. doi: 10.1111/imm.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chemnitz JM, Parry RV, Nichols KE, June CH, Riley JL. SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. J. Immunol. 2004;173(2):945–954. doi: 10.4049/jimmunol.173.2.945. [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekstrom K, Valadi H, Sjostrand M, Malmhall C, Bossios A, Eldh M, Lotvall J. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J Extracell Vesicles. 2012;1:18389. doi: 10.3402/jev.v1i0.18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley SD, Gupta D, Cheng N, Klinke DJ. Inferring Relevant Control Mechanisms for Interleukin-12 Signaling within Naive CD4+ T cells. Immunol Cell Biol. 2011;89:100–110. doi: 10.1038/icb.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich DI, Hurwitz AA. Tumor-Induced Immune Suppression: Mechanisms and Therapeutic Reversal. 2. New York, NY: Springer-Verlag; 2014. [Google Scholar]
- Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nature Cell Biol. 2012;14(10):1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- Herbst RS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010 Aug;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine DJ, Swartz MA, Szeto GL. Engineering synthetic vaccines using cues from natural immunity. Nat Mater. 2013;12(11):978–990. doi: 10.1038/nmat3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc. Natl. Acad. Sci. U.S.A. 1998;95(13):7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ. Induction of Wnt-inducible signaling protein-1 correlates with invasive breast cancer oncogenesis and reduced type 1 cell-mediated cytotoxic immunity: a retrospective study. PLoS Comp Biol. 2014;10(1):e1003409. doi: 10.1371/journal.pcbi.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ. Eavesdropping on altered cell-to-cell signaling in cancer by secretome profiling. Mol Cell Oncology. 2016;3(1):e1029061. doi: 10.1080/23723556.2015.1029061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ, Brundage KM. Scalable analysis of flow cytometry data using R/Bioconductor. Cytometry A. 2009;75(8):699–706. doi: 10.1002/cyto.a.20746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ, Cheng N, Chambers E. Quantifying crosstalk among Interferon-γ, Interleukin-12, and Tumor Necrosis Factor signaling pathways within a TH1 cell model. Sci Signal. 2012;5(220):ra32. doi: 10.1126/scisignal.2002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke DJ, Kulkarni YM, Wu Y, Byrne-Hoffman C. Inferring alterations in cell-to-cell communication in HER2+ breast cancer using secretome profiling of three cell models. Biotechnol. Bioeng. 2014 Apr;111:1853–1863. doi: 10.1002/bit.25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni YM, Chambers E, McGray AJR, Ware JS, Bramson JL, Klinke DJ. A quantitative systems approach to identify paracrine mechanisms that locally suppress immune response to Interleukin-12 in the B16 melanoma model. Integrative Biol. 2012;4:925–936. doi: 10.1039/c2ib20053h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laland K, et al. Does evolutionary theory need a rethink? Nature. 2014;514(7521):161–164. doi: 10.1038/514161a. [DOI] [PubMed] [Google Scholar]
- Le Pape E, Wakamatsu K, Ito S, Wolber R, Hearing VJ. Regulation of eumelanin/pheomelanin synthesis and visible pigmentation in melanocytes by ligands of the melanocortin 1 receptor. Pigment Cell Melanoma Res. 2008;21(4):477–486. doi: 10.1111/j.1755-148X.2008.00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jie HB, Lei Y, Gildener-Leapman N, Trivedi S, Green T, Kane LP, Ferris RL. PD-1/SHP-2 inhibits Tc1/Th1 phenotypic responses and the activation of T cells in the tumor microenvironment. Cancer Res. 2015;75(3):508–518. doi: 10.1158/0008-5472.CAN-14-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hsu DD, Wang H, Feng GS. Dual faces of SH2-containing protein-tyrosine phosphatase Shp2/PTPN11 in tumorigenesis. Front Med. 2012;6(3):275–279. doi: 10.1007/s11684-012-0216-4. [DOI] [PubMed] [Google Scholar]
- Logozzi M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE. 2009;4(4):e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz U. SHP-1 and SHP-2 in T cells: two phosphatases functioning at many levels.Immunol. Rev. 2009;228(1):342–359. doi: 10.1111/j.1600-065X.2008.00760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482(7385):400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo SA, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123(10):1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muturi HT, Dreesen JD, Nilewski E, Jastrow H, Giebel B, Ergun S, Singer BB. Tumor and endothelial cell-derived microvesicles carry distinct CEACAMs and influence T-cell behavior. PLoS ONE. 2013;8(9):e74654. doi: 10.1371/journal.pone.0074654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overwijk WW, Restifo NP. B16 as a mouse model for human melanoma. Curr Protoc Immunol, Chapter. 2001 May;20(Unit 20.1) doi: 10.1002/0471142735.im2001s39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch E, Piel M, Prost J, Bornens M, Sykes C. Cortical actomyosin breakage triggers shape oscillations in cells and cell fragments. Biophys J. 2005;89(1):724–733. doi: 10.1529/biophysj.105.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U.S.A. 2010;107(14):6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter I, Mezzacasa A, LeDonne P, Dummer R, Hemmi S. Comparative analysis of immunocritical melanoma markers in the mouse melanoma cell lines B16, K1735 and S91-M3. Melanoma Res. 2001;11(1):21–30. doi: 10.1097/00008390-200102000-00003. [DOI] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H, Rudd CE. Tyrosine phosphatase SHP-2 binding to CTLA-4: absence of direct YVKM/YFIP motif recognition. Biochem. Biophys. Res. Commun. 2000;269(1):279–283. doi: 10.1006/bbrc.2000.2234. [DOI] [PubMed] [Google Scholar]
- Schuler PJ, et al. Human CD4+ CD39+ regulatory T cells produce adenosine upon co-expression of surface CD73 or contact with CD73+ exosomes or CD73+ cells. Clin. Exp. Immunol. 2014;177(2):531–543. doi: 10.1111/cei.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova V, Ludwig AK, Hornung S, Rotan O, Horn PA, Epple M, Giebel B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf B Biointerfaces. 2011;87(1):146–150. doi: 10.1016/j.colsurfb.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Sung BH, Ketova T, Hoshino D, Zijlstra A, Weaver AM. Directional cell movement through tissues is controlled by exosome secretion. Nat Commun. 2015;6:7164. doi: 10.1038/ncomms8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Gabrielson NP, Uckun FM, Fan TM, Cheng J. Size-dependent tumor penetration and in vivo efficacy of monodisperse drug-silica nanoconjugates. Mol. Pharm. 2013;10(3):883–892. doi: 10.1021/mp300684a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008;110(1):13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin Immunopathol. 2011;33(5):441–454. doi: 10.1007/s00281-010-0234-8. [DOI] [PubMed] [Google Scholar]
- Topalian SL, Wolchok JD, Chan TA, Mellman I, Palucka K, Banchereau J, Rosenberg SA, Wittrup KD. Immunotherapy: The Path to Win the War on Cancer - Not Just Another Hallmark. Cell. 2015;161(3):185–186. doi: 10.1016/j.cell.2015.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- van Elsas A, Hurwitz AA, Allison JP. Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granu-locyte/macrophage colony-stimulating factor (GM-CSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J. Exp. Med. 1999;190:355–366. doi: 10.1084/jem.190.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieckowski EU, Visus C, Szajnik M, Szczepanski MJ, Storkus WJ, Whiteside TL. Tumor-derived microvesicles promote regulatory T cell expansion and induce apoptosis in tumor-reactive activated CD8+ T lymphocytes. J. Immunol. 2009;183(6):3720–3730. doi: 10.4049/jimmunol.0900970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Deng W, Klinke DJ. Exosomes: improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst. 2015;140(19):6631–6642. doi: 10.1039/c5an00688k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanez-Mo M, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Harris SL, Levine AJ. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66(9):4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Extracellular vesicles were isolated from media conditioned by B16F0 cells (a and b), Cloudman S91 (c), and Melan-A cells (d) and imaged using electron microscopy. SEM was used to image extracellular vesicles in panels a, c, and d, while TEM was in panel b.
(A) Following reverse transcription with equal amounts of mRNA, cDNAs were loaded in each well for single-plex qPCR. The trace of the reporter signal normalized to the reference dye (Rn) is shown as a function of cycle (circles). Following baseline subtraction, a curve (dotted blue line) was fit to the exponential growth phase and used to estimate the number of cycles (CT) required for Rn to exceed a threshold (dotted green line). The results are representative of at least three replicates for each target. (B) A calibration curve was used to assess the performance of the qPCR assay. Template abundance was varied by diluting cDNA obtained from B16F0 whole cell lysate. Gapdh, Wsb2, and Ptpn11 were assayed in the different dilution samples. CT values below 36 cycles were proportional to the logarithm of the template concentration. The efficiency of the PCR amplification for the different target were between 80 and 120% (Gapdh: 104%, Wsb2: 90%, and Ptpn11: 118%). The standard deviation in CT values suggests that a difference by a factor of two in template concentration could be detected with 99% confidence using a sample size of 3. (C) CT values estimate the abundance of mRNA obtained from whole cell lysates (red circles) versus exosomes (black squares) for each cell line (B16F0, S91, and Melan-A) and target tested (Wsb2, Ptpn11, Gapdh, Eif4abp2, Kpnb1, and Rnd2). ** indicates that the difference in CT between whole cell lysates and exosome samples was significant (p-value < 0.0002).
P-values were computed using the Fisher exact test. The Z-score is a statistical ranking metric derived from running the Fisher exact test for many random gene sets in order to compute a mean rank and standard deviation from the expected rank for each term in the gene-set library and finally calculating a z-score to assess the deviation from the expected rank. Combined score is calculated from p-value and z-score.