Abstract
After foodborne transmission of the facultative intracellular bacterial pathogen Listeria monocytogenes, most of the bacterial burden in the gut is extracellular. However, we previously demonstrated that intracellular replication in an as yet unidentified cell type was essential for dissemination and systemic spread of L. monocytogenes. Here, we show that the vast majority of cell-associated L. monocytogenes in the gut were adhered to Ly6Chi monocytes, a cell type that inefficiently internalized L. monocytogenes. With bone marrow-derived in vitro cultures, high multiplicity of infection (MOI) or the use of opsonized bacteria enhanced uptake of L. monocytogenes in CD64-negative monocytes, but very few bacteria reached the cell cytosol. Surprisingly, monocytes that had up-regulated CD64 expression in transition towards becoming macrophages fully supported intracellular growth of L. monocytogenes. In contrast, “inflammatory” monocytes that increased CD64 expression in the bone marrow of BALB/c/By/J mice prior to L. monocytogenes exposure in the gut did not support L. monocytogenes growth. Thus, contrary to the perception that L. monocytogenes can infect virtually all cell types, neither naïve nor inflammatory Ly6Chi monocytes served as a productive intracellular growth niche for L. monocytogenes. These results have broad implications for innate immune recognition of L. monocytogenes in the gut and highlight the need for additional studies on the interaction of extracellular, adherent L. monocytogenes with the unique subsets of myeloid-derived inflammatory cells that infiltrate sites of infection.
Introduction
L. monocytogenes is a facultative intracellular bacterial pathogen that causes foodborne disease in humans. The primary virulence strategy of L. monocytogenes is thought to be the ability to invade mammalian cells. L. monocytogenes survive and replicate inside a wide variety of cell types including epithelial cells (1), endothelial cells (2), hepatocytes (3), lymphocytes (4), cardiomyocytes (5), and neurons (6). L. monocytogenes induce uptake into non-phagocytic epithelial and endothelial cells using internalin A (InlA) and internalin B to interact with the mammalian receptors, E-cadherin and c-Met, respectively (7). The pore-forming toxin listeriolysin O can promote uptake of L. monocytogenes during membrane repair of certain epithelial cells (8), and other surface proteins and adhesins have also been implicated in the invasion of mammalian cells (9–11).
For myeloid-derived phagocytic cells, both ontogeny and activation status dictate whether a cell type can support intracellular replication of L. monocytogenes. For example, L. monocytogenes can grow in the cytosol of macrophages, but pre-treatment with inflammatory cytokines such as IFN-γ or TNF-α renders the cells bactericidal by efficiently retaining L. monocytogenes in the phagocytic vacuole (12,13). In contrast, neutrophils readily kill L. monocytogenes regardless of activation status (14,15). L. monocytogenes are less efficient at escaping from the vacuoles of bone marrow-derived, GM-CSF cultured dendritic cells (16,17). However, those cells do not closely resemble the conventional dendritic cell subsets observed in vivo (18) so it is not yet clear whether L. monocytogenes replicate in true dendritic cells.
Despite the species name “monocytogenes”, which refers to a robust monocytosis first observed in rabbits (19), there is little published data describing the direct interaction of L. monocytogenes with monocytes. An early study suggested that mononuclear cells isolated from human peripheral blood could slowly take up adherent L. monocytogenes and kill the bacteria, but the cells were only divided into two subsets: neutrophils and non-neutrophils (20). More recently, Drevets et al. showed that most of the L. monocytogenes-associated cells in the blood after i.v. infection of mice were Ly6Chi monocytes (21), and that only cells with an altered phenotype that appeared late (72 h) after lethal injection could efficiently internalize the bacteria (22). Monocytes are produced in the bone marrow, and rapid egress into the bloodstream during inflammation is dependent on expression of the chemokine receptor CCR2 (23). Subsequent extravasation of Ly6Chi monocytes into peripheral tissues is mediated by adhesion molecules such as CD11b, CD62L, and ICAM-1 (24). It was long thought that all bloodborne monocytes differentiated into tissue macrophages, however, recent studies indicate that subsets of monocytes can migrate through tissues and transport antigen to draining lymph nodes without differentiating into macrophages (25,26).
In the process of identifying infected cell types in the gut during foodborne listeriosis in susceptible BALB/c/By/J mice, we unexpectedly found that monocytes were by far the major cell type associated with L. monocytogenes during the early stages of infection. This prompted us to better characterize the phenotype of monocytes that infiltrated gut tissues and to determine the exact nature of their interaction with L. monocytogenes. We show here that neither naïve monocytes cultured in vitro, nor inflammatory monocytes isolated from L. monocytogenes-infected MLN serve as a productive replicative niche for L. monocytogenes despite the prevailing dogma that L. monocytogenes can invade and replicate in nearly all cell types.
Materials and Methods
Bacteria
L. monocytogenes EGDe and an isogenic ΔinlA mutant were provided by Cormac Gahan (Univ. College Cork). The mouse-adapted (InlAm) derivatives L. monocytogenes SD2000, SD2710 (constitutive GFP), and SD2001 (vector control) were described previously (27). L. monocytogenes EGDe was transformed with pGJ-cGFP (27) to create L. monocytogenes SD2610 and pIMC3kan (28) to create L. monocytogenes SD2901. L. monocytogenes were grown in Brain Heart Infusion (BHI) broth shaking at 30°C to early stationary phase, aliquoted, and stored at 80°C.
Mice
Female BALBc/By/J (BALB) mice were purchased from The Jackson Laboratory (Bar Harbor, ME) at 4 weeks of age. Mice were housed in a specific-pathogen free facility with a 9 AM to 7 PM dark cycle and were 6–9 weeks old when used for infections. All procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee.
Foodborne infection
Frozen aliquots of L. monocytogenes were thawed, incubated statically in BHI for 1.5 h at 30°C, washed with PBS, and then suspended in a mixture of PBS and salted sweet cream butter (2:3 ratio). A 2–3 cm piece of white bread (Kroger) saturated with L. monocytogenes was fed to mice near the onset of their dark cycle as described previously (29,30). Unless indicated otherwise, each mouse was fed 108 CFU of L. monocytogenes.
Ex vivo isolation of MLN and intestinal LP cells
All MLN were collected from each mouse, cut into 4 pieces each, and placed in 4 ml of RPMI 1640 (Invitrogen 21870) with 20 mM HEPES and 5% FBS. Collagenase type IV (300 U/ml; Worthington) and DNase I (120 U/ml; Worthington) were added and the nodes were digested for 30 min at 37°C shaking (250 rpm) in a 50-ml conical tube with a sterile 2 cm stir bar. Large intestines (cecum and colon) were flushed with 8 ml cold CMF buffer (Ca2+/Mg2+-free HBSS/10 mM HEPES/25 mM sodium bicarbonate/2% FBS) and then everted using a sterile weaving needle with button thread (31). Mucus was removed by shaking in a 50 ml conical tube with 25 ml CMF for 1 min. Epithelial cells were removed and the LP cells were isolated from the interface of a 44%/70% Percoll gradient as described previously (29).
Flow cytometry
Antibodies specific for CD16/CD32 (93), CD45 (30-F11), F4/80 (BM8), CD11c (N418), CD11b (M1/70), Ly6G (1A8-Ly6g), B220 (RA3-6B2), cKit (2B8), MHC-II (M5/114.15.2), IgG2a (eBr2a), CD3 (145-2C11), CD49b (DX5) from eBioscience; Ly6C (HK1.4), Ly6G (1A8), CD64 (X54-5/7.1) from BioLegend; and E-cadherin (36/E-Cadherin) from BD Biosciences were used. Data were acquired using an iCyt Synergy and analyzed with FlowJo (Tree Star); negative gating controls shown are FMOs (grey histograms); MFI refers to mean fluorescence intensity. Sorted cells had an average purity of 96% and were recovered for at least 30 min at 37°C in media with 20% FBS. For intracellular cytokine staining, cells were incubated in Brefeldin A (3 μg/ml) for 4 h at 37°C in 7% CO2, fixed and permeabilized (BD Cytofix/Cytoperm), and stained with either NOS2 (CXNFT; eBioscience) or IFNγ (XMG1.2; BioLegend) antibodies.
In vitro cell culture
Bone marrow-derived monocytes (BMMO) were generated as described previously (32). Macrophages used in Fig. 2 were derived from BMMO cultures by transferring lightly-adherent cells on day 5 of culture into 96-well-flat-bottom dishes; cells were allowed to adhere for 3 h before infection. Caco-2 cells (provided by Terrence Barrett, UK) were cultured in DMEM with 10% FBS.
FIGURE 2.
Ly6ChiCD11b+ monocytes are the primary L. monocytogenes-infected cell type in the gut 48 hpi. (A) Surface marker expression on P1 cells harvested from the MLN of infected mice; gray histograms are FMO controls. (B) Mean values (±SD) for GFP expression by CD11b+ vs CD11bneg P1 cells (n=4). (C) Representative dot plots show the percentage of CD45+ cells in the large intestine LP that were Ly6ChiCD11b+; graph shows total (±SEM) number of Ly6Chi monocytes in the LP of uninfected (0 hpi; n=4) or infected (48 hpi; n=8) mice. (D) Approximately 1% of the Ly6ChiCD11b+ monocytes in the large intestine LP were GFP+ (n=8 mice). (E–G) Mice were fed 109 CFU of wild type L. monocytogenes EGDe and MLN populations were subset 48 hpi as shown in Fig. 1A. The total number of each population in the MLN (E), the proportion of total GFP+ cells in each population (F), and the CD11b phenotyping of the P1 cells (G) are shown. Data shown in panel F were analyzed by one-way ANOVA with Tukey’s multiple comparisons test. Data in panels B & G were analyzed by unpaired two-tailed student’s t-test.
In vitro infection
Aliquots of L. monocytogenes were incubated statically in BHI for 1.5 h at 37°C and then suspended in sterile PBS. Sorted cells (105) were seeded in 96-well round-bottom ultra-low attachment plates (Corning), infected for 1 h in suspension, and then washed 3 times with pre-warmed HBSS. For assays using adherent cells (BMMΦ or Caco-2), plates were centrifuged at 300 x g for 5 minutes after the addition of L. monocytogenes to synchronize infection. Total cell-associated CFU was determined by lysing cells in sterile water and plating serial dilutions on BHI agar. For intracellular L. monocytogenes, cells were incubated in RP-10 with 10 μg/ml gentamicin for 20 minutes at 37°C in 7% CO2, then washed once, lysed and plated. Adherent L. monocytogenes were calculated by subtracting the number of intracellular L. monocytogenes from the total cell-associated CFU. In some experiments, L. monocytogenes were opsonized prior to infection by incubating in Ca2+/Mg2+-free HBSS with 10% normal mouse serum for 30 min at 37°C. Serum was obtained by collecting whole blood from the hearts of naïve uninfected BALB mice into serum separator tubes (BD Microtainer®).
Phagocytosis assay
Cells were incubated with FluoSpheres® biotinylated 1 μm latex beads (ThermoFisher) at a 2:1 ratio in RP-10 for 1 h at 37°C in 7% CO2. Cells were washed three times with cold buffer (Ca2+/Mg2+-free HBSS/1% FBS/1 mM EDTA) and then the surface-stained with specific antibodies and streptavidin-PE (eBioscience). Some cells were pretreated with either 20 μg/ml cytochalasin D (Sigma) or vehicle (DMSO) for 30 min prior to incubation with beads.
Microscopy
For Diff-Quik® (Dade-Behring) staining, cells were spun onto Superfrost slides (VWR) for 6 min. at 600 rpm using a Cytospin and fixed in methanol 5 sec, followed by staining in solution I for 10 sec, and solution II 5 sec. Cells were dried and mounted with Permount® under glass coverslips. Cells were visualized using a Zeiss Axio Imager.Z1 with a 100x/1.4NA PlanApo oil immersion objective and analyzed with AxioVision software.
For differential “in/out” staining of L. monocytogenes, cells were washed 3 times with cold buffer (Ca2+/Mg2+-free HBSS/1% FBS/1 mM EDTA) and then incubated with Difco Listeria O Antiserum Poly (BD Biosciences) (1:10) in PBS with 3% BSA for 20 min on ice. The cells were washed and then incubated with goat anti-rabbit IgG-Texas Red® (ThermoFisher) for 20 min on ice. Cells were spun onto poly-L-lysine-coated Superfrost slides (VWR) for 6 min at 600 rpm using a Cytospin. Dried slides were formalin fixed at 4°C for 10 min, washed with PBS, and mounted under coverslips with ProLong® Diamond antifade (Molecular Probes). For F-actin staining, cells were spun onto slides, air-dried, formalin-fixed for 10 min, washed 3 times with PBS, and then permeabilized in TBS-T (TBS/0.1% Triton X-100/1% BSA, pH=8.8) for 15 min at room temp. Texas Red®-X Phalloidin (ThermoFisher) was added for 20 min at room temp followed by 8 washes in TBS-T, and 8 washes with TBS alone. Cells were visualized using a Zeiss Axio Imager.Z1 with a 100x/1.4NA PlanApo oil immersion objective and analyzed with AxioVision software. Each slide was analyzed independently by two different investigators.
ELISA
Femurs and tibias were flushed with a total of 0.5 ml cold RPMI, and the bone marrow collected was centrifuged at 300 × g for 6 min. Serum was isolated from blood using serum separator tubes (BD Microtainer®). Bone marrow supernatants and serum were stored at −80°C. IFN-γ and IL-12 (p70) concentrations were determined using Ready-SET-Go!® ELISA kits (eBioscience). IL-18 concentrations were determined using capture antibody (clone 74) at 4 μg/ml, a biotin-labeled detection antibody (clone 93-10C) (1:2000), and rIL-18 standards ranging from 15–2000 pg/ml (MBL).
Tissue CFU
MLN (5–7 per mouse) were processed as described previously (29). Femurs and tibias were flushed with 10 ml cold RPMI and 10% of the volume was added to sterile water and plated on BHI agar.
Statistics
Unless indicated otherwise, mean values ± SD are shown in all panels and pooled data from at least two separate experiments are shown. Statistical analysis was performed using Prism for Macintosh (version 6; Graph Pad). P values of <0.05 were considered significant and are indicated as follows: *, P<0.05; **, P<0.01; ***, P<0.001; ****, P<0.0001.
Results
Ly6Chi monocytes are the primary L. monocytogenes-infected cell type in the MLN
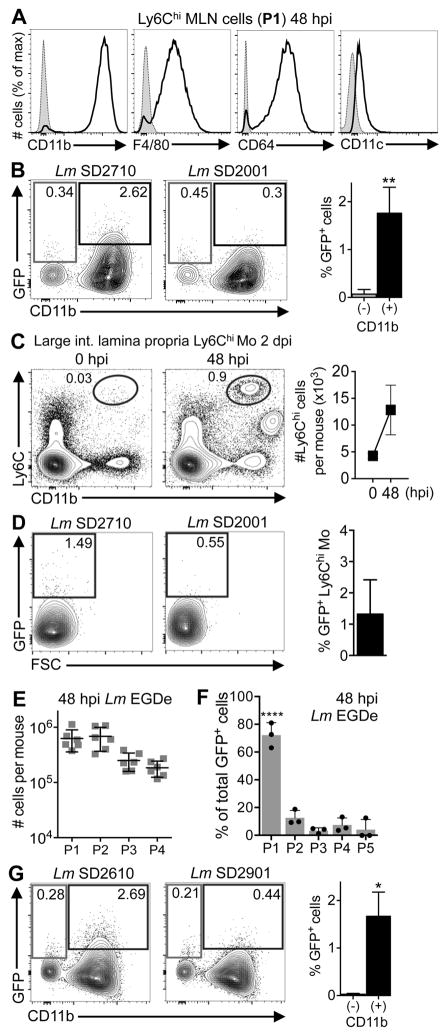
To identify infected phagocytes in the gut, mice were fed mouse-adapted L. monocytogenes that constitutively expressed GFP and MLN cells were analyzed 48 h post-infection (hpi). Previous work showed that 48 hpi was the earliest time point at which L. monocytogenes was consistently detected in the lymph nodes of all infected mice (33). Myeloid cells were broadly subset into the following populations: Ly6Chi (P1), Ly6Ghi (P2), Ly6CloCD11chi (P3), and Ly6CloCD11b+ (P4) (Fig. 1A). The remainder of the cells, which were mainly lymphocytes, were analyzed as P5. As shown in Fig. 1B, the number of Ly6Chi P1 cells and Ly6Ghi P2 cells in the MLN increased 100-fold within 48 h of infection, indicating that these cells were part of the early inflammatory infiltrate. In contrast, the total number of P3 and P4 cells (mostly macrophages and dendritic cells) did not change considerably during the course of the infection (Fig. 1B).
FIGURE 1.
Identification of L. monocytogenes-infected cells in the MLN using a flow cytometric approach. (A) Gating scheme used to subset MLN populations (P1, P2, P3, P4, P5) in mice fed 108 CFU of mouse-adapted L. monocytogenes. (B) Total number (±SEM) of cells in the MLN of uninfected (0 hpi) or infected mice (n=5). (C) Representative dot plots show how thresholds for GFP were set by comparison to cells from mice infected with L. monocytogenes that lacked GFP (vector control strain). (D) Average proportion (±SD) of total GFP+ cells in each population. Pooled data from 5 mice infected in two separate experiments was analyzed by one-way ANOVA with Tukey’s multiple comparisons test.
GFP fluorescence was analyzed by comparison with cells isolated from mice fed an isogenic L. monocytogenes strain that lacked GFP (Fig. 1C). The vast majority (~80%) of all GFP+ cells identified in the MLN 48 hpi were the Ly6Chi cells in the P1 gate (Fig. 1D). As the infection progressed, association with other cell types increased, but P1 remained the largest population of GFP+ cells. Most of the P1 cells were Ly6Chi monocytes based on high expression of CD11b, intermediate F4/80 and CD64, and low CD11c (Fig. 2A). A minor proportion (~5–8%) of the P1 cells lacked CD11b, F4/80, and CD64 and expressed intermediate levels of CD11c and B220 (data not shown), a phenotype consistent with plasmacytoid dendritic cells. However, as shown in Fig. 2B, only the Ly6ChiCD11b+ monocytes, and not the plasmacytoid dendritic cells, were GFP+.
Ly6Chi cells also infiltrated the lamina propria (LP) of the large intestine 2 dpi (Fig. 2C) and approximately 1% of these cells were associated with GFP+ L. monocytogenes (Fig. 2D). To confirm that the composition of the inflammatory infiltrate was not altered due to the use of murinized L. monocytogenes (34), we performed similar analyses using mice fed wild type L. monocytogenes EGDe. As shown in Fig. 2E, similar numbers of P1, P2, P3, and P4 cells were found in the MLN. Furthermore, the predominate fraction of L. monocytogenes-infected (GFP+) cells was Ly6Chi monocytes (Fig. 2F), and not the plasmacytoid DC (Fig. 2G). Therefore, at 48 hpi, the earliest time point L. monocytogenes can be detected in the MLN, the vast majority of the L. monocytogenes-associated cells were infiltrating Ly6Chi monocytes.
Listeria do not efficiently invade the cytosol of cultured monocytes
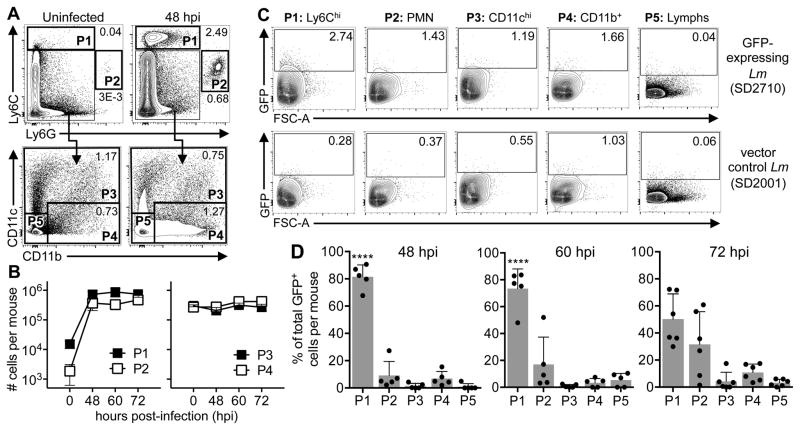
The flow cytometric approach shown in Fig. 1 demonstrated that L. monocytogenes associated with monocytes, but did not prove that the bacteria could survive and replicate in the cells. To test this, we cultured bone marrow cells with M-CSF (Fig. 3A), generating a mixture of cells that displayed the characteristic “waterfall of differentiation” (35) from Ly6C+CD64neg monocytes (blue), Ly6C+CD64+ transitioning cells (orange), and Ly6CnegCD64hi macrophages (black). Diff-Quik staining of sorted cells (Fig. 3B) revealed a classic kidney-shaped nucleus in the monocytes, cytoplasmic vesicles in the larger macrophages, and an intermediate morphology for the transitioning cells.
FIGURE 3.
L. monocytogenes inefficiently invade cultured CD64neg monocytes. (A) Bone marrow cultured with M-CSF for 4 days generated three CD117− populations: monocytes (blue), transitioning cells (orange), and macrophages (black). Representative dot plots indicate the average purity of each population after sorting. (B) Diff-Quik staining of sorted cells. (C) Sorted monocytes, transitioning cells, and macrophages were infected with L. monocytogenes SD2710 and the average number (±SD) of adherent CFU after washing was determined 1 hpi. (D) Total number (±SD) of intracellular (gentamicin10-resistant) L. monocytogenes SD2710 1 h after infection of sorted cells at a MOI of 0.5 (data from one of two separate experiments is shown). (E) Total number of intracellular (gent-resistant) L. monocytogenes associated with triplicate wells of either macrophages (BMMΦ) or Caco-2 cells 1 hpi. (F) E-cadherin expression on BM-derived cells 4 days after in vitro culture. (G) Control plots for phagocytosis assay. Green boxes indicate cells that internalized all associated beads. (H) Percent complete phagocytosis (FITC+PE−) for each cell type 1 h after incubation with beads. (I–J) Sorted monocytes were infected with L. monocytogenes SD2710 at a MOI of 14 for 90 min., washed 3 times, and then stained with Lm-specific antibodies. (I) Representative images of an infected monocyte show both green intracellular bacteria and yellow extracellular bacteria after merging green and red channels. (J) Number of intracellular L. monocytogenes per monocyte with or without opsonization. (K) Representative images of sorted cells infected with L. monocytogenes SD2710, fixed 5 hpi, and stained with phalloidin (red). Arrowheads indicate actin “tails”. (L) Sorted cells were infected for 5 h at low MOI (L) or high MOI with or without opsonization (M) Data from one of two separate experiments is shown; panels D & E were analyzed by two-tailed unpaired student’s t-test. Scale bars, 10 μm (B) or 2 μm (I & K).
First, we tested the ability of L. monocytogenes to adhere to each of these three cell types. Sorted cells were infected at various MOI for one hour, washed extensively and then plated for CFU. As shown in Fig. 3C, all three cell types displayed a dose-dependent association with L. monocytogenes. To assess invasion, the sorted cells were infected at a MOI of 0.5 for 1 h, washed, and then treated with gentamicin for 20 min. prior to plating. As expected, a large proportion of the inoculum invaded the Ly6CnegCD64hi macrophages (Fig. 3D). However, few gentamicin-resistant CFU were recovered from the monocytes, suggesting that either L. monocytogenes inefficiently invaded or were unable to survive inside the cells. Interestingly, there was a significantly higher number of gentamicin-resistant L. monocytogenes in the transitioning cells (Fig. 3D), indicating that monocytes can become a replicative niche for L. monocytogenes prior to becoming bona fide macrophages.
The L. monocytogenes surface protein InlA promotes invasion of non-phagocytic cells after interacting with its mammalian receptor, E-cadherin (36,37) and it was previously suggested that InlA could also enhance the invasion of macrophage cell lines (38). We examined the ability of InlA-deletion mutant (ΔinlA) L. monocytogenes to invade CD64+ macrophages, but found that InlA was not required for invasion (Fig. 3E). However, macrophages express multiple receptors that can promote uptake of bacteria, so it is possible that the loss of one ligand would not greatly alter invasion rates. Cultured CD64+ macrophages expressed high levels of E-cadherin, and the transitioning cells expressed intermediate levels of E-cadherin (Fig. 3F). In contrast, cultured monocytes did not express any detectable E-cadherin on the cell surface. Thus, the level of E-cadherin on the cultured cells correlated directly with invasion efficiency.
To evaluate phagocytic capacity, we incubated the cultured cells with biotin-conjugated, green fluorescent beads for one hour and then counterstained with PE-conjugated streptavidin. To promote complete phagocytosis, a low bead-to-cell ratio of 0.5 was used, resulting in ~1% FITC+ monocytes (Fig. 3G), which were then analyzed for PE expression. As expected, pre-treatment with cytochalasin D, an inhibitor of actin dynamics, reduced the proportion of cells with only internalized beads (green gate; FITC+PE−) and increased the percentage of cells that had adherent beads (FITC+PE+). As shown in Fig. 3H, there was no significant difference in the percentage of beads internalized by cultured monocytes, transitioning cells, or macrophages. This suggested that monocytes should be capable of internalizing L. monocytogenes, even if they lack receptors to enhance phagocytosis. To find out if L. monocytogenes could invade monocytes, cultured Ly6C+CD64neg cells were exposed to GFP-expressing bacteria at a higher MOI for a longer period of time (90 min.). The cells were then washed extensively and stained with TexasRed® conjugated Listeria-specific antibodies. Because the cells were not permeabilized, only extracellular bacteria bound the antibody, allowing us to use microscopy to differentiate between intracellular L. monocytogenes (green) and adherent, extracellular organisms (yellow) (Fig. 3I). Approximately half of the 258 monocytes we examined contained at least one intracellular bacterium at this time point (data not shown). Pre-treatment of the bacteria with normal mouse serum did not change the percentage of cells that contained intracellular L. monocytogenes (not shown); however, opsonization did cause an increase in the number of bacteria found inside each monocyte (Fig. 3J).
To track the fate of internalized L. monocytogenes in each cell type, sorted cells were infected in vitro for 1 h, washed and then incubated for an additional 4 h in media containing gentamicin. The cells were then stained with phalloidin and microscopy was used to co-localize GFP+ bacteria with cytosolic actin “tails” (Fig. 3K). Five hours after infection at low MOI, very few monocytes were associated with L. monocytogenes; however, cytosolic L. monocytogenes with actin tails were observed in both the transitioning cells and the macrophages (Fig. 3L). Increasing the MOI to promote enhanced invasion of the monocytes resulted in cytosolic localization of L. monocytogenes in 12% of the monocytes, compared to 79% for macrophages (Fig. 3M). Opsonization of the bacteria did not change the intracellular fate in monocytes, but did result in decreased numbers of actin tails in the cytosol of macrophages. Together, these data suggested that monocytes can take up L. monocytogenes, albeit less efficiently than transitioning cells or macrophages, but that escape to the cytosol was an infrequent occurrence.
Monocytes that infiltrate the MLN have a partially differentiated and partially activated phenotype
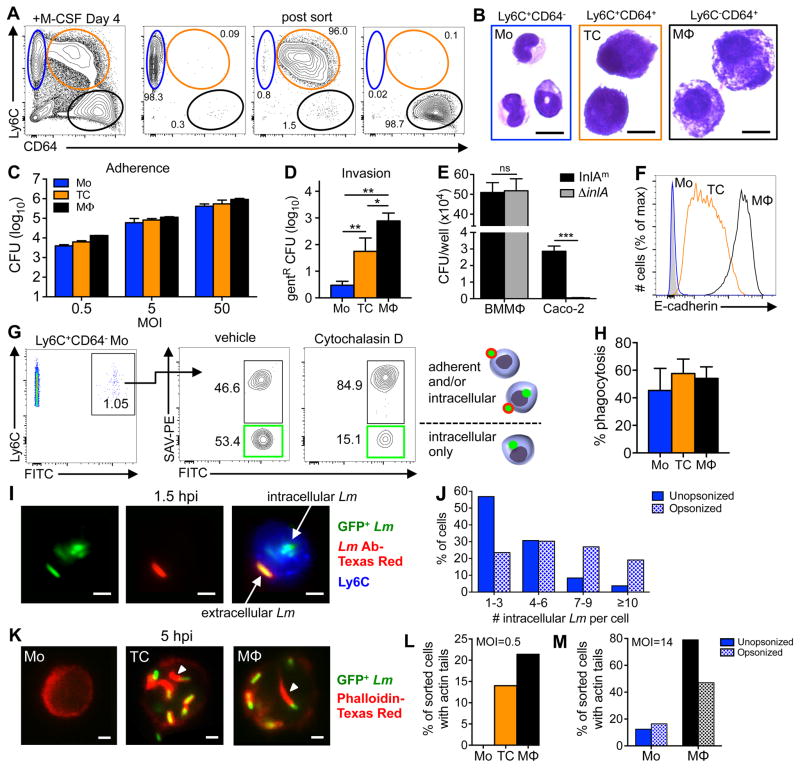
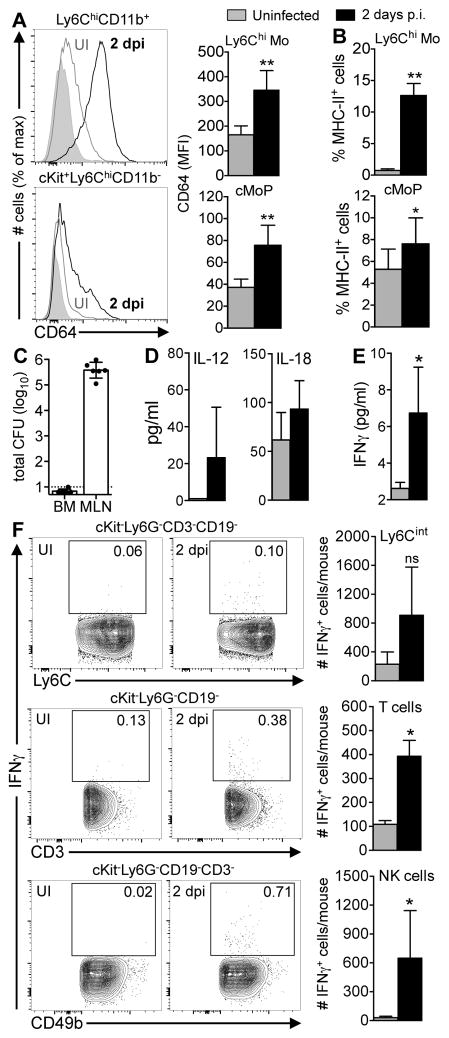
Expression of CD64 (FcγR1) and down-regulation of Ly6C are commonly used surface phenotypes that signify progression of monocytes through the differentiation pathway towards becoming macrophages (39). In a naïve animal, the largest number of monocytes are found in the bone marrow, but the few Ly6ChiCD11b+ cells present in the MLN were negative for CD64 or expressed only low levels when analyzed directly ex vivo (Fig. 4A), similar to the phenotype of cultured monocytes. However, “inflammatory” monocytes recruited to the MLN 2 dpi uniformly expressed increased levels of CD64 (Fig. 4A). The small size and the shape of the nuclei of these cells was suggestive of a monocyte morphology (Fig. 4B), but the increased expression of E-cadherin was suggestive of a transitioning cell (Fig. 4C). Many of the monocytes displayed increased expression of MHC-II on the cell surface (Fig. 4D) and a subset of the cells were producing iNOS (Fig. 4E). However, the phagocytic capacity of inflammatory monocytes sorted from the MLN was only half that of naïve Ly6Chi monocytes that had yet to leave the bone marrow of uninfected mice (Fig. 4F). Thus, Ly6Chi inflammatory monocytes that infiltrated the MLN during infection had a unique, partially differentiated and partially activated phenotype that did not precisely resemble either monocytes or macrophages cultured in vitro or naïve monocytes analyzed directly ex vivo.
FIGURE 4.
Inflammatory monocytes analyzed directly ex vivo have a partially differentiated and partially activated phenotype. Mice were fed 108 CFU of Lm SD2710 (n=6) or left uninfected (UI; n=3) and MLN were analyzed 48 h later. (A) Representative histogram and MFI (±SD) of CD64 expression on Ly6G−Ly6ChiCD11b+ monocytes. (B) Diff-Quik staining (100x) of inflammatory monocytes sorted from MLN 2 dpi; scale bar, 10 μm. (C) Representative E-cadherin expression on monocytes. (D) Representative histogram and MFI (±SD) of MHC-II expression. (E) Percentage of iNOS-producing Ly6Chi monocytes in the MLN. (F) Uptake of fluorescent beads by Ly6Chi monocytes from the bone marrow of uninfected mice or MLN of infected mice (n=6).
Inflammatory monocytes are activated prior to egress from the bone marrow
Askenase et al. recently showed that systemic circulation of IL-12, produced in response to intestinal infection with the intracellular parasite Toxoplasma gondii, resulted in changes to Ly6Chi monocytes while the cells were still in the bone marrow, before they infiltrated the intestinal lamina propria (40). Likewise, we noticed during the course of these studies that Ly6Chi monocytes in the bone marrow had an altered phenotype during L. monocytogenes infection. Two days after foodborne transmission, nearly all of the Ly6Chi monocytes in the bone marrow expressed moderate levels of CD64 and 50–60% of the cKit+Ly6ChiCD11bneg monocyte progenitors in the bone marrow had also up-regulated CD64 (Fig. 5A). Likewise, about 10% of the mature monocytes still present in the bone marrow of infected mice had increased levels of MHC-II compared to uninfected mice (Fig. 5B). These data suggested that a stimulus present in the bone marrow was altering the differentiation of monocytes during infection compared to the steady state.
FIGURE 5.
Inflammatory monocytes are activated prior to egress from the bone marrow. Bone marrow (BM) from uninfected mice (UI; grey bars) or mice fed 108 CFU of Lm SD2710 (black bars) was analyzed 2 dpi (n=6). (A) CD64 expression on Ly6Chi monocytes (Mo) and common monocyte progenitors (cMoP) (56). (B) Mean percentage (±SD) of MHC-II+ Ly6Chi Mo and cMoP. (C) BM harvested from both femurs and tibias was plated and L. monocytogenes CFU were determined 2 dpi (n=6). (D) Mean concentration (±SD) of IL-12 and IL-18 in mouse sera (n=4). (E) Mean concentration of IFNγ (±SD) in the bone marrow without in vitro stimulation (n=4). (F) Representative contour plots of bone marrow cells showing the percentage of IFNγ+ cells. Bar graphs to the right indicate mean number (±SD) of IFNγ+ Ly6Cint, T cells, or NK cells (n=4). Two-tailed Mann-Whitney tests were used for statistical analysis.
High dose i.v. infection leads to the presence of L. monocytogenes in the bone marrow (41,42) so it was feasible that the bacteria could directly activate monocytes. To test this, mice were fed L. monocytogenes and 2 days later, the marrow from both femurs and tibias was collected and plated for CFU. As shown in Fig. 5C, only one out of six mice had detectable L. monocytogenes, and that mouse had only a single CFU present in the total marrow collected. In contrast, more than 100,000 total CFU were recovered from the MLN of those same mice at that time point. Although we did not detect live L. monocytogenes in the bone marrow, it was possible that L. monocytogenes replicating in the gut could cause systemic circulation of infection-induced cytokines IL-12 and/or IL-18 that could then stimulate IFNγ production in the bone marrow. We found small increases in both serum IL-12 and serum IL-18 during foodborne infection, but neither of these changes were statistically significant (Fig. 5D). Nonetheless, the concentration of IFNγ detected in the bone marrow increased about 3-fold during infection (Fig. 5E). Multiple cell types present in the bone marrow were actively secreting IFN-γ during L. monocytogenes infection (Fig. 5F), suggesting that the IFN-γ was produced locally. Together, these observations suggested that intestinal infection generated systemic mediators that caused both developing and mature Ly6Chi monocytes in the bone marrow to have an inflammatory phenotype, prior to recruitment to L. monocytogenes-infected tissues.
L. monocytogenes adhere to, but do not efficiently invade inflammatory monocytes
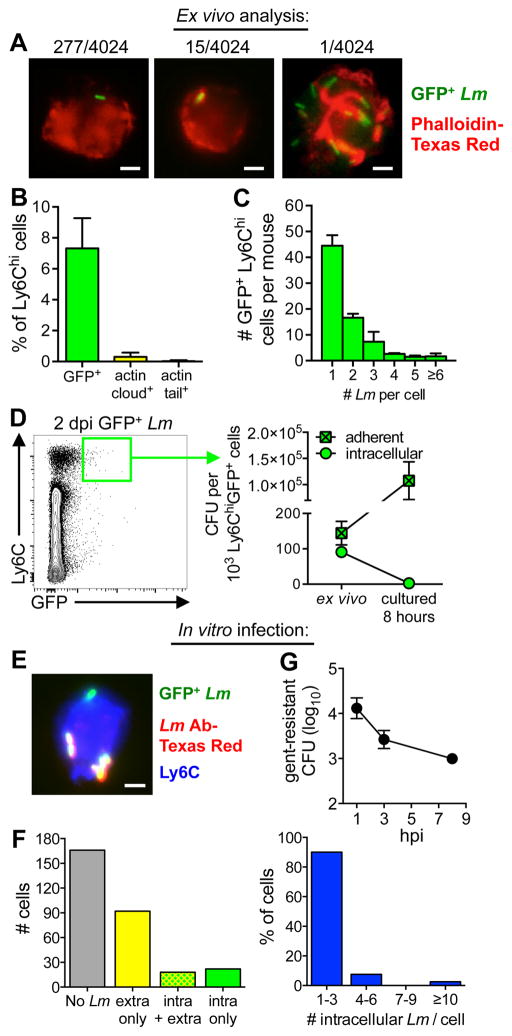
To find out if L. monocytogenes invaded inflammatory monocytes in vivo, Ly6Chi cells were sorted from the MLN two days after mice were fed GFP+ L. monocytogenes and the cells were examined microscopically (Fig. 6A). Approximately 7% of the 4,024 sorted monocytes examined (pooled data from 6 different mice) were associated with GFP+ L. monocytogenes (Fig. 6B). The flow cytometric approach used previously identified only ~2% of the Ly6Chi cells as GFP+ (Fig. 1C). The 3.5-fold difference in these results is likely due to issues with autofluorescence that led us to apply a rigorous threshold to the bulk population in order to definitively label a cell as GFP+ by flow cytometry. The majority of monocytes associated with GFP+ L. monocytogenes had only one or two bacteria per cell (Fig. 6C). Fifteen of the monocytes (0.4%) contained L. monocytogenes surrounded by an actin “cloud” (43) and only one cell (0.02%) had actin tails. However, since the sort purity of the Ly6Chi cells recovered from each mouse was only ~97–99%, it possible that the cell containing L. monocytogenes co-localized with actin was a contaminating macrophage-like cell.
FIGURE 6.
Inflammatory monocytes do not productively support intracellular growth of L. monocytogenes. (A–C) Ly6Chi cells were sorted from the MLN 2 d after infection with Lm SD2710 and visualized directly ex vivo. (A) A total of 4,024 cells from 6 different mice (200–800 cells/mouse) were visualized. Representative images and the total number of Lm-associated (GFP+), actin cloud+ cells or actin tail+ cells observed are shown. (B) Mean percentage (±SD) of Lm-associated monocytes per mouse (n=6). (C) Average number (±SD) of L. monocytogenes per GFP+ cell. (D) GFP+ Ly6Chi cells were sorted from the MLN 2 dpi (green gate) and the mean number of adherent or intracellular (gent10-resistant) CFU (±SD) was determined directly ex vivo or after being cultured for 8 h in media with or without gentamicin. (E-F) Ly6Chi cells (1 × 105/well) were sorted from the MLN 2 dpi with L. monocytogenes SD2001 and infected in vitro for 90 minutes with L. monocytogenes SD2710 at a MOI of 2. (E) Representative image for “in/out” differential staining shows a cell with both intracellular and extracellular L. monocytogenes; scale bar, 2 μm. (F) A total of 300 cells were visualized; left graph indicates the number of uninfected cells (gray bar), cells with only extracellular Lm (yellow bar), cells with both intracellular and extracellular Lm (green/yellow bar), or cells with only intracellular bacteria (green bar). Graph on the right indicates the number of intracellular L. monocytogenes observed per cell. (G) Intracellular growth assay performed on sorted GFPneg Ly6Chi cells (5 × 104/well) infected in vitro for 1 h at MOI of 2.
To verify that L. monocytogenes could inefficiently invade, but not survive within inflammatory monocytes, GFP+ Ly6Chi cells were sorted from the MLN 2 dpi (Fig. 6D) and the number of intracellular and extracellular L. monocytogenes associated with those cells was determined by incubating the sorted cells with or without gentamicin for 20 minutes and then lysing and plating. Due to the small number of cells recovered in this experiment, the sort purity was not determined. As shown in Fig. 6D, ~38% of the total cell-associated CFU was intracellular (gentamicin-resistant) directly ex vivo. A portion of the sorted cells were further incubated for 8 hours with or without gentamicin to find out if intracellular (gentamicin-resistant) L. monocytogenes would replicate, or if only the adherent CFU increased over time. As shown in Fig. 6D, the total number of adherent CFU in each well increased ~500-fold. However, no intracellular (gentamicin-resistant) CFU were detected 8 hours after plating the sorted cells.
To further assess the ability of inflammatory monocytes to support intracellular growth, we sorted cells from the MLN of mice infected with L. monocytogenes lacking GFP and then exposed the cells in vitro to GFP-expressing bacteria for 90 min. and performed differential staining to identify intracellular and extracellular L. monocytogenes (Fig. 6E). As shown in Fig. 6F, 40 of the 300 cells visualized (13%) contained at least one intracellular bacterium, with most harboring 1–3 intracellular bacteria per cell. Thus, invasion of the inflammatory monocytes was less efficient than observed for naïve cultured monocytes (Fig. 2), consistent with the reduced phagocytic capacity of these cells (Fig. 4F). The number of gentamicin-resistant intracellular L. monocytogenes in the inflammatory monocytes steadily decreased over time (Fig. 6G). Thus, the majority of the Ly6Chi inflammatory monocytes recruited to the MLN and analyzed directly ex vivo had only adherent L. monocytogenes, and the few bacteria that invaded these cells did not survive.
Discussion
The results presented here highlight two important findings. First, within 48 hours of foodborne infection, the majority of myeloid-derived cells in the MLN are “inflammatory monocytes” that have been pre-activated in the bone marrow prior to L. monocytogenes exposure. These cells did not precisely resemble any cell type that can be cultured directly from bone marrow using only growth factors such as CSF-1 or GM-CSF, or the cells that are present in the steady state in an uninfected animal. Second, although L. monocytogenes is equipped for intracellular growth, the majority of cell-associated L. monocytogenes in the gut following foodborne infection were extracellular (27), presumably adhered to monocytes, a cell type that inefficiently internalized L. monocytogenes. Very few L. monocytogenes were associated with cells expressing markers typical of classical macrophages and DC. This observation has implications for innate immune recognition of L. monocytogenes, because there are a large number of genes that differ in expression level in monocytes compared to macrophages (44).
It was previously reported that InlA/E-cadherin interaction was important for invasion of macrophage-like cell lines (38); however, we found that ΔinlA L. monocytogenes were internalized efficiently in both bone marrow-derived macrophages (Fig. 3E) and THP-1 cells (data not shown). Interaction with E-cadherin could enhance uptake of L. monocytogenes in macrophages, but may not be required because the cells express a variety of other receptors that can trigger phagocytosis, including Gp96 and scavenger receptors class A (9,45). In contrast, monocytes lack all three of these receptors based on data presented here and previous studies (46,47). We propose that L. monocytogenes can readily adhere to monocytes and that this attachment is mediated primarily by non-specific bacterial adhesins or pili. However, because they lack sufficient expression of surface receptors that can trigger cytoskeletal rearrangements to promote particle uptake, few adhered L. monocytogenes are internalized by monocytes unless they are opsonized by specific antibodies or complement.
The microscopy studies presented here suggest that the few L. monocytogenes that do invade monocytes cannot escape into the cytosol. This is in agreement with Raybourne et al. who suggested that human blood monocytes could not support the growth of L. monocytogenes over time (48). It is possible that activity of the pore-forming toxin LLO is impaired in monocytes. The kinetics of phagosome acidification in murine monocytes has not been tested. However, the phagosomes of freshly-isolated human monocytes acidified to a pH of only 5.7 to 5.9 after phagocytosis of live E. coli (49) and LLO has a pH optimum of ~5.5 (50). Westcott et al. further showed that vacuoles in murine monocyte-derived GM-CSF-cultured DC acidified at a slower rate than macrophage phagosomes (17). Thus, it is possible that delayed or reduced acidification of the phagosome could reduce the efficiency of L. monocytogenes escape in monocytes.
It is likely that at least some of the Ly6Chi monocytes we analyzed ex vivo were in the process of differentiating into “Tip-DCs” as approximately 20–25% of the inflammatory monocytes were already producing iNOS. The nomenclature of “Tip-DCs” has been debated (51); it may be inaccurate to define “Tip-DCs” as a subset of dendritic cells, but it is clear that the production of TNF-α and iNOS by these cells is critical for clearance of L. monocytogenes (52). In agreement with our study, Shi et al. showed that CCR2+Ly6Chi cells surrounded foci of infection in the liver following i.v. infection, and very few of those monocyte-derived cells harbored viable L. monocytogenes (53).
Despite the predominate monocyte infiltrate in the gut, the total number of Ly6CloCD64+ macrophages did not change significantly during the first three days following foodborne L. monocytogenes infection. This suggests that monocytes recruited to the MLN were not differentiating into classical macrophages during this timeframe. This finding is in agreement with Bain et al., who used a DSS-induced model of colitis to show that maturation of Ly6Chi monocytes recruited to the intestinal LP was disrupted during inflammation (54). In addition, Rydström and Wick showed that the differentiation of Ly6Chi monocytes was inhibited following exposure to Salmonella, a process that was dependent on MyD88 signaling (55).
The terms monocyte, macrophage, inflammatory monocyte and inflammatory macrophage have been used variously over the past few decades to describe myeloid-derived cell populations. Although the nomenclature is evolving, more definitive labels for cell subsets will probably require the use of better markers that correlate with cell function, rather than just surface marker expression (44). However, what is clear now is that bone marrow-derived macrophages and DC do not closely resemble the majority of cells that L. monocytogenes encounter in vivo in the gut and that very few L. monocytogenes replicate within phagocytes during the intestinal phase of the infection. Future studies should focus on the interactions of these unique subsets of inflammatory cells with extracellular bacteria, rather than cytosolic bacteria in macrophages, in order to define the earliest innate immune activation events that occur following foodborne L. monocytogenes infection in mice.
Acknowledgments
We thank Jennifer Strange for technical assistance in the UK Flow Cytometry & Cell Sorting Core Facility.
This work was supported by National Institutes of Health grant AI101373.
References
- 1.Gaillard JL, Berche P, Mounier J, Richard S, Sansonetti P. In vitro model of penetration and intracellular growth of Listeria monocytogenes in the human enterocyte-like cell line Caco-2. Infect Immun. 1987;55:2822–9. doi: 10.1128/iai.55.11.2822-2829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drevets DA, Sawyer RT, Potter TA, Campbell PA. Listeria monocytogenes infects human endothelial cells by two distinct mechanisms. Infect Immun. 1995;63:4268–76. doi: 10.1128/iai.63.11.4268-4276.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dramsi S, Biswas I, Maguin E, Braun L, Mastroeni P, Cossart P. Entry of Listeria monocytogenes into hepatocytes requires expression of inIB, a surface protein of the internalin multigene family. Mol Microbiol. 1995;16:251–61. doi: 10.1111/j.1365-2958.1995.tb02297.x. [DOI] [PubMed] [Google Scholar]
- 4.McElroy DS, Ashley TJ, D’Orazio SEF. Lymphocytes serve as a reservoir for L. monocytogenes growth during infection of mice. Microb Pathog. 2009;46:214–21. doi: 10.1016/j.micpath.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alonzo F, 3rd, Bobo LD, Skiest DJ, Freitag NE. Evidence for subpopulations of Listeria monocytogenes with enhanced invasion of cardiac cells. J Med Microbiol. 2011;60:423–34. doi: 10.1099/jmm.0.027185-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dramsi S, Levi S, Triller A, Cossart P. Entry of Listeria monocytogenes into neurons occurs by cell-to-cell spread: an in vitro study. Infect Immun. 1998;66:4461–8. doi: 10.1128/iai.66.9.4461-4468.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cossart P, Pizarro-Cerda J, Lecuit M. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends in Cell Biology. 2003;13:23–31. doi: 10.1016/s0962-8924(02)00006-5. [DOI] [PubMed] [Google Scholar]
- 8.Vadia S, Arnett E, Haghighat AC, Wilson-Kubalek EM, Tweten RK, Seveau S. The pore-forming toxin listeriolysin O mediates a novel entry pathway of L. monocytogenes into human hepatocytes. PLoS Pathog. 2011;7:e1002356. doi: 10.1371/journal.ppat.1002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabanes D, Sousa S, Cebria A, Lecuit M, Garcia-del Portillo F, Cossart P. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J. 2005;24:2827–38. doi: 10.1038/sj.emboj.7600750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burkholder KM, Bhunia AK. Listeria monocytogenes uses Listeria adhesion protein (LAP) to promote bacterial transepithelial translocation and induces expression of LAP receptor Hsp60. Infect Immun. 2010;78:5062–73. doi: 10.1128/IAI.00516-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reis O, Sousa S, Camejo A, Villiers V, Gouin E, Cossart P, Cabanes D. LapB, a novel Listeria monocytogenes LPXTG surface adhesin, required for entry into eukaryotic cells and virulence. J Infect Dis. 2010;202:551–62. doi: 10.1086/654880. [DOI] [PubMed] [Google Scholar]
- 12.Biroum N. Listericidal activity of non-stimulated and stimulated human macrophages in vitro. Clin Exp Immunol. 1977;28:138–45. [PMC free article] [PubMed] [Google Scholar]
- 13.Shaughnessy LM, Swanson JA. The role of the activated macrophage in clearing Listeria monocytogenes infection. Front Biosci. 2007;12:2683–92. doi: 10.2741/2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers HW, Unanue ER. Neutrophils are involved in acute, nonspecific resistance to Listeria monocytogenes in mice. Infect Immun. 1993;61:5090–6. doi: 10.1128/iai.61.12.5090-5096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnett E, Vadia S, Nackerman CC, Oghumu S, Satoskar AR, McLeish KR, Uriarte SM, Seveau S. The pore-forming toxin listeriolysin O is degraded by neutrophil metalloproteinase-8 and fails to mediate Listeria monocytogenes intracellular survival in neutrophils. J Immunol. 2014;192:234–44. doi: 10.4049/jimmunol.1301302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westcott MM, Henry CJ, Cook AS, Grant KW, Hiltbold EM. Differential susceptibility of bone marrow-derived dendritic cells and macrophages to productive infection with Listeria monocytogenes. Cell Microbiol. 2007;9:1397–411. doi: 10.1111/j.1462-5822.2006.00880.x. [DOI] [PubMed] [Google Scholar]
- 17.Westcott MM, Henry CJ, Amis JE, Hiltbold EM. Dendritic cells inhibit the progression of Listeria monocytogenes intracellular infection by retaining bacteria in major histocompatibility complex class II-rich phagosomes and by limiting cytosolic growth. Infect Immun. 2010;78:2956–65. doi: 10.1128/IAI.01027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helft J, Bottcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, Goubau D, Reis e Sousa C. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity. 2015;42:1197–211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Murray E, Webb R, Swann M. A disease of rabbits characterized by large mononuclear leucocytosis, aused by a hitherto undescribed bacillus Bacterium monocytogenes. J Pathol Bacteriol. 1926;29:407–39. [Google Scholar]
- 20.Peterson PK, Verhoef J, Schmeling D, Quie PG. Kinetics of phagocytosis and bacterial killing by human polymorphonuclear leukocytes and monocytes. J Infect Dis. 1977;136:502–9. doi: 10.1093/infdis/136.4.502. [DOI] [PubMed] [Google Scholar]
- 21.Drevets DA, Dillon MJ, Schawang JS, Van Rooijen N, Ehrchen J, Sunderkotter C, Leenen PJ. The Ly-6Chigh monocyte subpopulation transports Listeria monocytogenes into the brain during systemic infection of mice. J Immunol. 2004;172:4418–24. doi: 10.4049/jimmunol.172.7.4418. [DOI] [PubMed] [Google Scholar]
- 22.Drevets DA, Schawang JE, Mandava VK, Dillon MJ, Leenen PJ. Severe Listeria monocytogenes infection induces development of monocytes with distinct phenotypic and functional features. J Immunol. 2010;185:2432–41. doi: 10.4049/jimmunol.1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol. 2006;7:311–7. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- 24.Lauvau G, Chorro L, Spaulding E, Soudja SM. Inflammatory monocyte effector mechanisms. Cell Immunol. 2014;291:32–40. doi: 10.1016/j.cellimm.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, Ivanov S, Duan Q, Bala S, Condon T, van Rooijen N, Grainger JR, Belkaid Y, Ma’ayan A, Riches DW, Yokoyama WM, Ginhoux F, Henson PM, Randolph GJ. Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity. 2013;39:599–610. doi: 10.1016/j.immuni.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodero MP, Poupel L, Loyher PL, Hamon P, Licata F, Pessel C, Hume DA, Combadiere C, Boissonnas A. Immune surveillance of the lung by migrating tissue monocytes. Elife. 2015;4:e07847. doi: 10.7554/eLife.07847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones GS, Bussell KM, Myers-Morales T, Fieldhouse AM, Bou Ghanem EN, D’Orazio SE. Intracellular Listeria monocytogenes comprises a minimal but vital fraction of the intestinal burden following foodborne infection. Infect Immun. 2015;83:3146–56. doi: 10.1128/IAI.00503-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monk IR, Casey PG, Cronin M, Gahan CG, Hill C. Development of multiple strain competitive index assays for Listeria monocytogenes using pIMC; a new site-specific integrative vector. BMC Microbiol. 2008;8:96. doi: 10.1186/1471-2180-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bou Ghanem EN, Myers-Morales T, D’Orazio SE. A mouse model of foodborne Listeria monocytogenes infection. Curr Protoc Microbiol. 2013;31:9B 3 1–9B 3 16. doi: 10.1002/9780471729259.mc09b03s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bou Ghanem EN, Myers-Morales T, Jones GS, D’Orazio SEF. Oral transmission of Listeria monocytogenes in mice via ingestion of contaminated food. J Vis Exp. 2013;75 doi: 10.3791/50381. 3791/50381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Resendiz-Albor AA, Esquivel R, Lopez-Revilla R, Verdin L, Moreno-Fierros L. Striking phenotypic and functional differences in lamina propria lymphocytes from the large and small intestine of mice. Life Sci. 2005;76:2783–803. doi: 10.1016/j.lfs.2004.08.042. [DOI] [PubMed] [Google Scholar]
- 32.Francke A, Herold J, Weinert S, Strasser RH, Braun-Dullaeus RC. Generation of mature murine monocytes from heterogeneous bone marrow and description of their properties. J Histochem Cytochem. 2011;59:813–25. doi: 10.1369/0022155411416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bou Ghanem EN, Jones GS, Myers-Morlaes T, Patil PN, Hidayatullah AN, D’Orazio SEF. InlA promotes dissemination of Listeria monocytogenes to the mesenteric lymph nodes during food borne infection of mice. PLoS Pathog. 2012;8:e1003015. doi: 10.1371/journal.ppat.1003015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai YH, Disson O, Bierne H, Lecuit M. Murinization of internalin extends its receptor repertoire, altering Listeria monocytogenes cell tropism and host responses. PLoS Pathog. 2013;9:e1003381. doi: 10.1371/journal.ppat.1003381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamoutounour S, Henri S, Lelouard H, de Bovis B, de Haar C, van der Woude CJ, Woltman AM, Reyal Y, Bonnet D, Sichien D, Bain CC, Mowat AM, Reis e Sousa C, Poulin LF, Malissen B, Guilliams M. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur J Immunol. 2012;42:3150–66. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- 36.Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–41. doi: 10.1016/0092-8674(91)90009-n. [DOI] [PubMed] [Google Scholar]
- 37.Lecuit M, Dramsi S, Gottardi C, Fedor-Chaiken M, Gumbiner B, Cossart P. A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 1999;18:3956–63. doi: 10.1093/emboj/18.14.3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawyer RT, Drevets DA, Campbell PA, Potter TA. Internalin A can mediate phagocytosis of Listeria monocytogenes by mouse macrophage cell lines. J Leukoc Biol. 1996;60:603–10. doi: 10.1002/jlb.60.5.603. [DOI] [PubMed] [Google Scholar]
- 39.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, Mazloom AR, Ma’ayan A, Chua WJ, Hansen TH, Turley SJ, Merad M, Randolph GJ C. Immunological Genome. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–28. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Askenase MH, Han SJ, Byrd AL, Morais da Fonseca D, Bouladoux N, Wilhelm C, Konkel JE, Hand TW, Lacerda-Queiroz N, Su XZ, Trinchieri G, Grainger JR, Belkaid Y. Bone-marrow-resident NK cells prime monocytes for regulatory function during infection. Immunity. 2015;42:1130–42. doi: 10.1016/j.immuni.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bruijn MF, van Vianen W, Ploemacher RE, Bakker-Woudenberg IA, Campbell PA, van Ewijk W, Leenen PJ. Bone marrow cellular composition in Listeria monocytogenes infected mice detected using ER-MP12 and ER-MP20 antibodies: a flow cytometric alternative to differential counting. J Immunol Methods. 1998;217:27–39. doi: 10.1016/s0022-1759(98)00080-5. [DOI] [PubMed] [Google Scholar]
- 42.Join-Lambert OF, Ezine S, Le Monnier A, Jaubert F, Okabe M, Berche P, Kayal S. Listeria monocytogenes-infected bone marrow myeloid cells promote bacterial invasion of the central nervous system. Cell Microbiol. 2005;7:167–80. doi: 10.1111/j.1462-5822.2004.00444.x. [DOI] [PubMed] [Google Scholar]
- 43.Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hume DA, Summers KM, Rehli M. Transcriptional Regulation and Macrophage Differentiation. Microbiol Spectr. 2016;4 doi: 10.1128/microbiolspec.MCHD-0024-2015. [DOI] [PubMed] [Google Scholar]
- 45.Ishiguro T, Naito M, Yamamoto T, Hasegawa G, Gejyo F, Mitsuyama M, Suzuki H, Kodama T. Role of macrophage scavenger receptors in response to Listeria monocytogenes infection in mice. Am J Pathol. 2001;158:179–88. doi: 10.1016/S0002-9440(10)63956-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfram L, Fischbeck A, Frey-Wagner I, Wojtal KA, Lang S, Fried M, Vavricka SR, Hausmann M, Rogler G. Regulation of the expression of chaperone gp96 in macrophages and dendritic cells. PLoS One. 2013;8:e76350. doi: 10.1371/journal.pone.0076350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geng Y, Kodama T, Hansson GK. Differential expression of scavenger receptor isoforms during monocyte-macrophage differentiation and foam cell formation. Arterioscler Thromb. 1994;14:798–806. doi: 10.1161/01.atv.14.5.798. [DOI] [PubMed] [Google Scholar]
- 48.Raybourne RB, Roth G, Deuster PA, Sternberg EM, Singh A. Uptake and killing of Listeria monocytogenes by normal human peripheral blood granulocytes and monocytes as measured by flow cytometry and cell sorting. FEMS Immunol Med Microbiol. 2001;31:219–25. doi: 10.1111/j.1574-695X.2001.tb00523.x. [DOI] [PubMed] [Google Scholar]
- 49.Horwitz MA, Maxfield FR. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J Cell Biol. 1984;99:1936–43. doi: 10.1083/jcb.99.6.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geoffroy C, Gaillard JL, Alouf JE, Berche P. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect Immun. 1987;55:1641–6. doi: 10.1128/iai.55.7.1641-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guilliams M, Ginhoux F, Jakubzick C, Naik SH, Onai N, Schraml BU, Segura E, Tussiwand R, Yona S. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat Rev Immunol. 2014;14:571–8. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 53.Shi C, Velazquez P, Hohl TM, Leiner I, Dustin ML, Pamer EG. Monocyte trafficking to hepatic sites of bacterial infection is chemokine independent and directed by focal intercellular adhesion molecule-1 expression. J Immunol. 2010;184:6266–74. doi: 10.4049/jimmunol.0904160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bain CC, Scott CL, Uronen-Hansson H, Gudjonsson S, Jansson O, Grip O, Guilliams M, Malissen B, Agace WW, Mowat AM. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6:498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rydstrom A, Wick MJ. Salmonella inhibits monocyte differentiation into CD11c hi MHC-II hi cells in a MyD88-dependent fashion. J Leukoc Biol. 2010;87:823–32. doi: 10.1189/jlb.0909615. [DOI] [PubMed] [Google Scholar]
- 56.Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, Feuerer M. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013;14:821–30. doi: 10.1038/ni.2638. [DOI] [PubMed] [Google Scholar]