Abstract
Arp2/3 complex is thought to be the primary protrusive force generator in cell migration by controlling the assembly and turnover of the branched filament network that pushes the leading edge of moving cells forward. However, mouse fibroblasts without functional Arp2/3 complex migrate at rates similar to wild-type cells, contradicting this paradigm. We show by correlative fluorescence and large-scale cryo-tomography studies combined with automated actin-network analysis that the absence of functional Arp2/3 complex has profound effects on the nano-scale architecture of actin networks. Our quantitative analysis at the single-filament level revealed that cells lacking functional Arp2/3 complex fail to regulate location-dependent fine-tuning of actin filament growth and organization that is distinct from its role in the formation and regulation of dendritic actin networks.
Keywords: Large-scale cellular cryo-tomography, actin networks, quantitative automated analysis, correlative imaging
INTRODUCTION
Cell migration plays a vital role in a variety of essential cellular processes such as immune surveillance and inflammation as well as in pathological processes like cancer and thrombosis. Rearrangement of the actin cytoskeleton is an integral feature of eukaryotic cellular processes including movement, adhesion, endocytosis and establishment of polarity (Krause and Gautreau, 2014). The actin cytoskeleton is characterized by numerous different structures each tuned to perform particular cellular functions. The assembly of the actin cytoskeleton, through nucleation of individual actin filaments from a cytoplasmic pool of actin monomers, is primarily controlled by two classes of actin nucleators (1) the evolutionarily conserved Arp2/3 complex with its seven mandatory constituent proteins and (2) formin family proteins (Campellone and Welch, 2010; Blanchoin and Michelot, 2012). In vitro, the most remarkable difference between these two classes of actin nucleators lies in the morphology of the actin network assembled: whereas formin proteins assemble long, unbranched actin filaments, and typically remain processively associated with the fast-growing end of the actin filament (Pruyne et al., 2002; Zigmond et al., 2003), the Arp2/3 complex produces a dendritic, branched actin network by nucleating new actin filaments (Mullins et al., 1998; Blanchoin et al., 2000) at an angle of about 78° from existing actin filaments (Volkmann et al., 2001; Rouiller et al., 2008; Vinzenz et al. 2012). One prevailing hypothesis in the field is that Arp2/3 complex-nucleated actin networks provide the protrusive force that propels the cell forward. The most compelling evidence supporting this hypothesis came from in vitro reconstitution studies of Listeria motility propelled by actin polymerization (Welch et al., 1997; Loisel et al., 1999). The visualization of dendritic actin networks at the lamellipodia of moving keratocytes and fibroblasts helped to extend this hypothesis to cell migration (Svitkina and Borisy, 1999; Yang and Svitkina, 2011).
Since the initial proposal that Arp2/3 complex is the primary protrusive force generator in cell migration a decade ago, some studies cast doubts on whether Arp2/3 complex is truly important for cell motility (Di Nardo et al., 2005; Strasser et al., 2004). Recent studies indicate that specific actin structures are controlled by the concerted action of multiple nucleators (Block et al., 2012; Breitsprecher et al., 2012; Chesarone and Goode, 2009). In addition, work using genetic or RNAi-mediated depletion confirmed that Arp2/3 complex is essential in lamellipodia formation in fibroblasts (Suraneni et al., 2012; Wu et al., 2012; Nicholson-Dykstra and Higgs, 2008), but cells lacking Arp2/3 complex retain the ability to form filopodia-like protrusions and are able to move (Suraneni et al., 2012; Nicholson-Dykstra and Higgs, 2008). Neither chemical inhibition (Nolen et al., 2009) nor RNAi knockdown experiments (Di Nardo et al., 2005; Steffen et al., 2006) have been fully conclusive in terms of defining the functions of Arp2/3 complex, because a background of as little as 10% functional Arp2/3 complex appears to be sufficient to fully or partially support Arp2/3-dependent functions in the cell (Nolen et al., 2009). These observations raise the question of what exactly is the role of the Arp2/3-based actin nucleation in cell migration.
Here, we examine the effect of the absence of functional Arp2/3 complex on the architecture of actin networks in fibroblast cells at the single-filament scale. We differentiated fibroblasts from isogenic mouse embryonic stem cells with disruption of the ARPC3 gene so that these cells do not express the ARPC3 subunit of Arp2/3 complex (Suraneni et al., 2012). Because Arp2/3 complex can only fulfill its function in the cell when all seven subunits are assembled, this genetic elimination of the ARPC3 subunit ensures that functional Arp2/3 complex is completely absent in these cells. Unexpectedly, these ARPC3−/− fibroblasts were found capable of moving at rates similar to wild-type cells albeit with defects in persistent directional migration (Suraneni et al., 2015). This observation contradicts the existing paradigm that Arp2/3 complex is the primary protrusive force generator in cell migration by controlling the assembly and turnover of the branched filament network that pushes the leading edge of moving cells forward. Light microscopy and fluorescence imaging showed that the relatively smooth lamellipodia at leading edges of spreading wild-type fibroblasts are replaced by large actin-based protrusions in ARPC3−/− fibroblasts that have mDia1 and mDia2 formins at their tips and are enriched with the actin cross-linker fascin (Suraneni et al., 2012; Suraneni et al., 2015), similar to the thin, finger-like filopodia of wild-type cells (Vignjevic et al., 2006; Block et al., 2008). The striking differences between wild-type lamellipodia and these massive filopodia-like protrusions (FLPs) suggest drastic molecular-level reorganization of the actin networks in the absence of functional Arp2/3 complex.
To elucidate the nature of these changes, we investigated the nano-scale architecture of the actin networks in protrusions and along the cell edges of both spreading wild-type and spreading ARPC3−/− fibroblasts. Because the morphologies of the cells in each population can be heterogeneous, depending on whether they were actively moving, we compared the morphology of protrusive cell edges using a defined spreading assay where the cells have a more synchronized cell morphology (Suraneni et al., 2012). We employed correlative light microscopy guided by fascin and actin fluorescence in conjunction with electron cryo-tomography to define the three-dimensional architecture of these actin networks at the single filament level. In the past, ultrastructural studies of actin networks were based on the analysis of small numbers of tomograms or platinum replicas. Here, we analyzed a total of 290 cryo-tomograms using highly efficient automated actin tracing protocols (Xu et al., 2015) to determine differences between the nano-scale organization of these networks in a statistically meaningful way. The use of large-scale automated analysis of actin-network properties in synchronized fibroblast cells thus allowed us to quantitatively compare actin-network types in wild-type and mutant cells. The next largest comparison of actin network properties using cryo-tomography of whole cells used a total of 26 cryo-tomograms for four different conditions and required tweaking of the filament detection algorithm for each individual tomogram (Jasnin et al., 2013).
In addition to a thin lamellipodial zone of Arp2/3-complex mediated dendritic networks, we identified two distinct types of actin bundles as defined by their packing density. These two bundle types were strictly segregated in space, one in filopodia and the other one adjacent to the dendritic networks in the lamella region. In the absence of functional Arp/23 complex not only is the dendritc network absent as expected but also a major re-arrangement of the actin bundles occurs. The two wild-type bundle types are absent and replaced by a single location-independent bundle type with more densely packed, longer actin filaments. The reduced variety of actin bundle types in ARPC3−/− fibroblasts suggests a significant regulatory role of the Arp2/3 complex in location-dependent fine-tuning of actin filament growth and organization that is distinct from its role in the formation and regulation of dendritic actin networks.
MATERIALS AND METHODS
Correlative fluorescence and whole cell electron cryo-tomography sample preparation
Fibroblast wild-type and mutant cell cultures
ARPC3+/+ (wild-type) and ARPC3−/− embryonic stem cells (ESCs) were cultured and differentiated into fibroblasts (Suraneni et al., 2012). Briefly, ARPC3+/+ and ARPC3−/− ESCs were separated from irradiated mouse embryonic fibroblasts (iMEFs) by transferring into new dishes every 45 min for 3 hrs. The ESCs were recovered by centrifugation at 1000–1200 rpm for 5 min and used for differentiation. The ESCs were resuspended in ESC medium containing leukemia inhibitory factor (LIF) and plated into flasks or dishes coated with gelatin. The cells were allowed to settle down for 5–6 h, and ESC medium was replaced with fibroblast medium containing 0.33 μM retinoic acid (RA) and cultured for 72 hrs with the medium changed twice in the first 24 hrs. Cells were cultured for 4 more days with daily media changes without RA and undifferentiated ESCs cells were separated from the fibroblasts by consecutive transfers. To synchronize cell morphology (Suraneni et al., 2012), the ARPC3+/+ or ARPC3−/− fibroblasts cells were first detached from the substrate by trypsinization, then allowed to spread for 2 hrs on glass bottom dishes (Mattek) with attached 200-mesh holey carbon-coated finder grids (Quantifoil) pre-coated with 5 μg/ml of bovine fibronectin (SigmaAldrich). Following these 2 hrs, the plates were fixed in light fixative (2% formaldehyde (0.1M Pipes, 1mM EGTA and 1mM MgSO4) for 30 min at 37°C, washed (3x) in PHEM (60mM Pipes, 25mM Hepes, 2mM MgSO4, 10mM EGTA) and 6x washes in complete PBS, all at room temperature.
Fluorescence staining
Cells used for correlative imaging were fixed in light fixative (2% formaldehyde (0.1M Pipes, 1mM EGTA and 1mM MgSO4) for 30 min at 37°C, washed (3x) in PHEM Buffer (60mM Pipes, 25mM Hepes, 2mM MgSO4, 10mM EGTA), followed by a quick wash with 0.05% Tween20 in PHME buffer, 3X washes with PHEM Buffer, and 3X washes with 0.01M Glycine in complete PBS, all at room temperature. The cells were then stained directly with Alexa Fluor 546 Phalloidin (1:500; Molecular Probes, Cat# A22283) for actin filaments for 20 min or incubated with primary antibodies against Fascin (1:50; Millipore, Cat# MAB 3582) overnight at 4°C, washed in 3X washes with 0.01M Glycine in complete PBS and incubated with Alexa488-conjugated goat anti-mouse antibody (1:250) for 1 hr at room temperature, followed for both staining protocols washing (6X) in complete PBS.
Reconstituted fascin/actin-filament assemblies
Fascin/actin-filament assemblies were grown for 8–24 h at 4°C on positively charged lipid layers consisting of a 3:7 wt/wt solution of dilaurylphosphatidylcholine (Avanti Polar Lipids, Inc.) and didodecyldimethylammonium bromide (DDDMA; Acros Organic) dissolved in chloroform (Volkmann et al., 2001). The lipid-surfactant mixture (0.5 μl drop containing 0.5 μg lipids) was layered over the polymerization buffer containing fascin (1 μM) before the injection of actin monomers (0.5 μM). The polymerization buffer contained 20 mM *-phosphate, pH 6.5, 40 mM KCl, 1 mM MgCl2, 1 mM ATP, and 0.2 mM EGTA. Monolayers were transferred to glow discharged (EMS Inc.), 200-mesh copper electron microscopy grids coated with lacey carbon films (EMS Inc). Excess liquid was blotted and the grids were then frozen in liquid N2-cooled ethane, either manually or with Vitrobot IV (FEI Company). Tomographic tilt series of samples suspended over holes were acquired under low-dose conditions using a Tecnai F20 G2 Twin transmission electron microscope (FEI company), equipped with a field emission gun (FEG) operated at 200 kV, a nominal magnification of 29,000, and at 3.5 μm defocus. Tilt angles were ranging from −60° to +60° and increments of 2°. We used the SerialEM tomography package (Mastronarde, 2005), a 626-DH 70 degree cryo-holder (Gatan Inc.), and a Gatan 890 US4000SP 2K x 2K CCD camera (Gatan Inc.). The total exposure per tilt series was between 50 and 70 e−/Å2.
Correlative fluorescence and electron cryo-tomography imaging
Light microscopy
Unique markings on the finder grids were used to match the region of interest in both light and electron microscope images. Leading edge and regions of interest were localized using phase imaging and/or epifluorescence using an Eclipse TE2000-U inverted microscope (Nikon Instruments Inc.) while samples were maintained fully hydrated. Images were acquired on a 12-bit cooled CCD camera (Orca; Hamamatsu) controlled by MetaMorph software (Universal Imaging Corp.) using a 40X 0.5 NA Plan Apo phase-contrast objective lens (Nikon).
Epifluorescence images of fixed cells were acquired with a triple band pass dichroic mirror using a 60X/1.4 NA Plan Apo DIC objective lens (Nikon).
Electron cryo-tomography
Grids were manually plunge frozen in liquid ethane at liquid nitrogen temperature and then transferred to the electron microscope sample holder. Using full mapping of the grid, followed by mid range magnification tiling, we systematically bridged the gaps in scales and located the cells of interest in the electron microscope using the markings of the finder grids. Correlative imaging of fluorescently labeled cells was used to verify the content of the actin features of interest. The majority (more than 95%) of the quantitative analysis of network properties was performed on cells that where not subjected to fluorescence imaging. Electron cryo-tomography data were collected from selected regions with a Titan Krios (FEI Company) equipped with Falcon II direct detector imaging device (FEI Company) and a 300KeV field emission gun either at NUS or at SBMRI or on a Polara equipped with K2 Summit detector (Gatan) and energy filter (Gatan) at Caltech. Tomographic tilt series (±65°, every 1.5°) were acquired using Batch Tomo (FEI Company) or SerialEM (Mastronarde, 2005), at an average dose of about 100–120 e−/Å2 and defocus of about 8–10 μm. Magnification was chosen to result in pixel sizes of 4–6 Å in the reconstructions. The fidelity and quality of the data collection was monitored at SBMRI with real-time automatic reconstruction protocols implemented in the CoAn package (Volkmann and Hanein, 1999).
Image reconstruction and post-processing
Tilt series were aligned using the IMOD package (Kremer et al., 1996) with a combination of fiducial-based and patch-based approaches. Three-dimensional densities were generated using the Simultaneous Iterative Reconstruction Technique (SIRT) as implemented in Tomo3D (Agulleiro and Fernandez, 2011). This reconstruction technique tends to show a better definition of actin filaments as well as cell membranes than alternative Fourier or weighted back-projection methods. To decrease the amount of noise and to amend the effects of the missing wedge, we post-processed the reconstructed density in the following way. First, we applied a variant of the non-local means filter (Buades et al., 2005) to each slice of the tomograms individually. This procedure not only suppresses noise in general, it also leads to suppression of the streaking along the Z-axis (Page et al., 2015) that is often observed in tomographic reconstructions due to the missing wedge. As a second step, a light sharpening operation using an unsharpen mask was applied to improve the contrast of the filaments. Finally, reconstructions were binned by a factor of two for further analysis. Surface representations were generated using Chimera (Goddard et al., 2007), and micrograph and tomography slice images were generated using IMOD (Kremer et al., 1996).
Analysis of actin filament distributions
Automatic actin filament tracing
To extract filament traces from tomographic reconstructions, we employed filament extraction with reduced representation templates (Xu et al., 2015). Reduced representations approximate the target by a small number of anchor points. These anchor points are then used to calculate the scoring function within the search volume (Volkmann, 2004). This strategy makes the approach more robust against noise and against local variations than approaches based on density templates, which need to be carefully tweaked for individual tomograms to allow efficient filament detection (Jasnin et al., 2013). To further enhance the response for filaments, we penalized arrangements with high variance along the long axis of the reduced representation template, thus enhancing the signal when brightness is approximately the same along the long axis. Parameters were adjusted manually to optimize response to actin filaments for a small number (twelve) of test tomograms. These parameters gave near-optimal responses by visual inspection for all additional tomograms without further adjustments.
Calculation of actin-filament packing densities
To calculate estimates of the actin-filament packing densities we performed the following steps. First, we converted the score maps derived from the actin-filament tracing step into binary maps. The threshold was adjusted individually for each tomogram to give an average of 90 Å for the diameter of the actin traces. We then generated overlapping boxes, each of which were fully contained in the respective actin-filament bundle. Next, we calculated the volume fraction of voxels inside the actin filament traces versus those outside for each of the boxes and calculated mean values and standard deviations of these volume fractions for actin filament bundles in wild-type filopodia, in mutant FLPs, and in the cell peripheries of both wild-type and mutant cells. We used Student’s t-test to determine significance of the differences between the mean values.
Calculation of actin-filament length statistics
To estimate the length of individual actin filaments, we first converted each individual actin filament in 30 selected fields of view into a set of coordinates along the filament trace. This operation was carried out manually taking the original densities as well as the score maps into account. Prior to this step, the volume was rotated so that the slicing direction in the field of view coincided with the long axis of the bundles and the majority of filaments were contained in a thin slab of density. Gaps along the trace where handled as the start of a new filament, even if the gap was small and the direction of the two filaments coincided, so the reported lengths are minimum lengths. Once coordinates were extracted, we compiled the distributions of the actin-filament lengths in ARPC3−/− and wild-type fibroblasts using those coordinates. Non-parametric Mann-Whitney and Kolmogorov-Smirnov tests were used to determine whether the difference between these distributions is statistically significant.
RESULTS
Three-dimensional cryo-tomography of whole cells allows quantitative characterization at the single filament scale
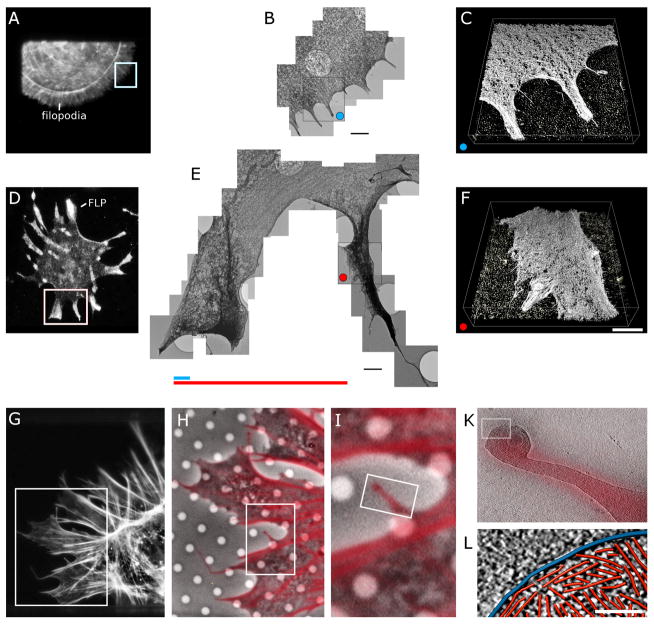
The lengths and widths of FLPs can be orders of magnitude larger than filopodia of wild-type fibroblasts (Fig. 1), consistent with structured illumination microscopy imaging of actin filaments stained with fluorescent phalloidin (Suraneni et al., 2015). While filopodia in wild-type cells are restricted to lengths of 1–2 μm (Fig. 1A–C), FLPs in the mutant cells can be up to 10–15 μm long and more than 5 μm wide (Fig. 1D–F).
Figure 1. Correlative fluorescence and electron microscopy of leading edge protrusions of wildtype and ARPC3−/− fibroblasts.
A–C. Wild-type cell stained with AF546 phalloidin.
A. Fluorescence micrograph.
B. Collage of eleven overlapping three-dimensional electron tomograms of the cell protrusions marked by white box in (A). Bar is 1.5 μm.
C. Surface representation of one of the three-dimensional reconstruction of the region marked by a blue dot in (B). Bar is 750 nm.
D–F. ARPC3−/− cell stained with an antibody to fascin.
D. Fluorescence micrograph.
E. Collage of 23 overlapping electron tomograms of the protrusions in the region marked by white box in (D). Bar is 1.5 μm. The colored lines compare the average length of filopodia (blue) in wild-type cells and FLPs (red) in ARPC3−/− cells. The tomograms show the massive difference in the organization of filaments at the leading edge in cells with and without Arp2/3 complex.
F. Surface representation of one of the three-dimensional reconstructions of the FLP region marked by a red dot in (E). Bar is 750 nm.
G–L. Series of fluorescence and electron micrographs of an ARPC3−/− cell stained with AF546 phalloidin ranging from an overview of the cell to individual actin filaments.
G. Fluorescence micrograph of half of the cell.
H. Overlay of high power fluorescence and phase contrast images of the region marked by the rectangular white box in (I).
I. Higher magnification of the region marked by the rectangular white box in (J).
K. Overlay of the fluorescence and an 11.4-nm thick slice through a three-dimensional electron cryo-tomographic reconstruction of the FLP in the bifurcating region marked in (K).
L. Three-dimensional renderings of traced actin filaments (red) and the plasma membrane (blue) in a 3.8-nm slice of the cryo-tomogram in (K). Bar is 100 nm.
To unravel how the lack of functional Arp2/3 complex at the molecular level contributes to this morphology, we used large-scale cryo-tomography of frozen-hydrated whole cells and automated actin-network analysis for defining actin-network properties in three dimensions at the nanometer-scale. We used correlative light and electron cryo tomography (Patla et al., 2010) as an approach to systematically traversing the scale continuum from a few microns to the definition of single actin filament in three dimensions at the nanometer-scale (Fig. 1) in order to verify the correspondence between fluorescence and structural features at the nano-scale. We focused on the organization of actin filament bundles in filopodia of wild-type cells and in FLPs of mutant cells as well as actin bundles at the cell periphery of both cell types.
Fluorescence speckle microscopy showed that the leading edge of motile cells consists of two distinct actin networks, often referred to as lamellipodium and lamellum (Ponti et al., 2004). These cell regions are generally devoid of major organelles and are instead composed of a dense and dynamic network of actin filaments (Pollard and Borisy, 2003) consistent with our observations. The lamellipodium is thought to consist of Arp2/3-complex mediated dendritic actin networks, the lamellum localizes behind the lamellipodium consisting primarily of linear actin bundles (Giannone et al., 2007). The specific characteristics of the lamellipodia and lamella differ substantially between different cell types (Huber et al., 2008) and especially the lamelliopdium can display highly dynamic behavior during different stages of movement (Machacek and Danuser, 2006). Thus, for quantitative analysis, it essential not only to minimize cell-type dependent differences but also to synchronize cell states as much as possible. Here, we use a combination of isogenic cell lines, a specialized spreading protocol (Suraneni et al., 2012) and light fixation to achieve that goal. Because the cells are arrested during spreading, all cell edges are equivalent and synchronized in a defined state of movement with minimal variations. Using this strategy, we collected, reconstructed and analyzed 198 cryo-tomograms from whole, unstained ARPC3−/− mutant cells and 92 cryo-tomograms from whole, unstained wild-type cells.
Actin networks in ARPC3−/− cells are significantly altered
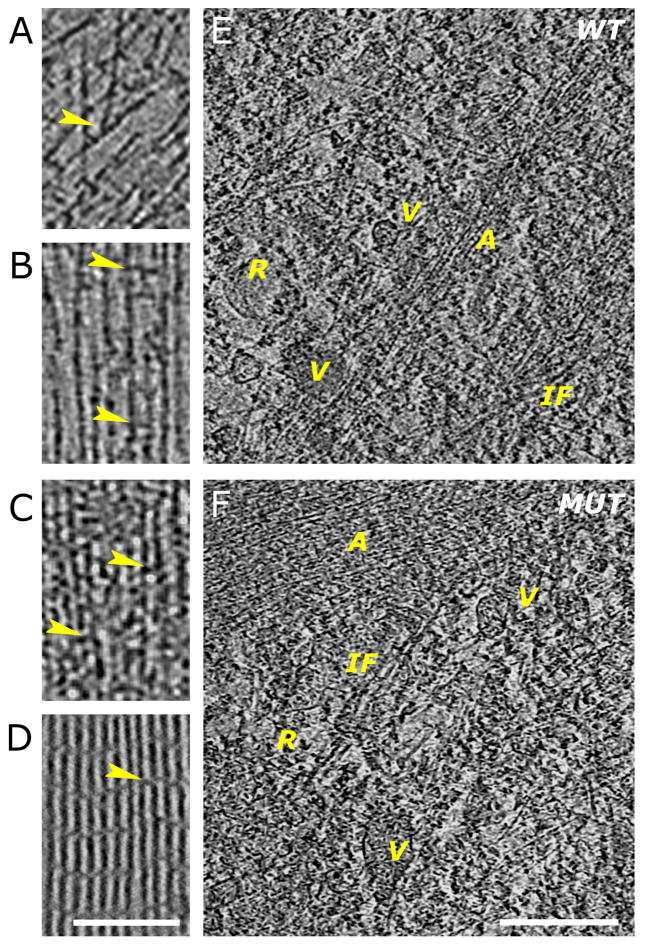
The quality and nanometer-scale resolution of these three-dimensional cryo-tomographic reconstructions allowed us to distinguish individual actin filaments from each other and from other structural elements in the cell and allowed identification of branch junctions and filament cross-linkers at the leading edge (Fig. 2A–D) as well as ribosomes, tentative intermediate filaments and vesicular structures in the interior of the cells (Fig. 2E–F). The level of detail in these tomographic reconstructions enabled high-throughput automated tracing of individual filaments (Figs. 3–5 and supplementary movies) and thus quantitative evaluation of filament packing densities and determination of individual filament lengths.
Figure 2. Comparison of features in wild-type and ARPC3−/− fibroblasts at the nanometer scale.
A–D. Examples showing 3.8-nm thick slices through three-dimensional cyro-tomograms.
A. Region near the leading edge of a wild-type cell showing actin filaments running at different angles and branch junctions (arrowhead).
B. Region in a filopodium of a wild-type cell showing parallel filaments separated by variable distances and with cross-linkers of different sizes (arrowheads).
C. Region of an FLP in an ARPC3−/− mutant fibroblast showing closely packed parallel bundles of filaments with different size cross-linkers (arrowheads).
D. Slice through a three-dimensional cryo-tomogram of a bundle of purified actin filaments cross-linked by fascin. The arrowhead points at a fascin cross-link between the actin filaments. Bar is 200 nm and applies to A–D.
E–F. Features in the Interior region of wild-type (E) and ARPC3−/− (F) fibroblasts. Various vesicular structures (V) with different content and attachements are visible. Tentative intermediate filaments (IF) and ribosomes (R) can also be identified. The actin structures visible (A) are reminiscent of those at the cell edge. They are more loosely organised in the wild-type cells. 7.6-nm slices are shown, the bar is 500 nm.
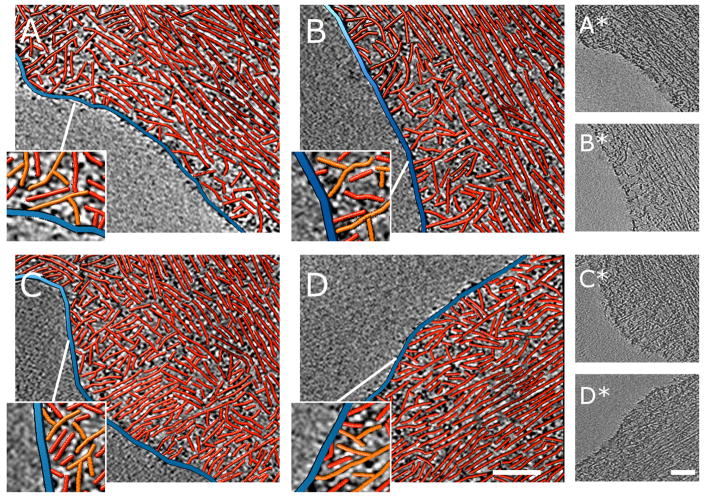
Figure 3.
Nanometer-scale characterization of actin filament assemblies at the cell peripheries of wild-type fibroblasts. Each example shows a 3.8-nm thick slice through a three-dimensional cyro-tomogram in grayscale with a paired three-dimensional rendering of red actin filaments and blue plasma membrane traced in the boxed areas of the reconstructions. The orientation of the slicing direction was adjusted so the long axis of the bundles is parallel to the plane of the slices. Only traced filaments contained in a thin slab (up to 4 slices) are shown to simplify visual appearance. In wild-type cells a narrow lamellipodial region of the cell periphery shows a dendritic network between the cell membrane (blue) and actin bundles running parallel to the cell membrane. The enlarged regions show branched actin filaments in orange. Bars are 160 nm. Raw tomogram slices of the traced regions are shown to the right.
Figure 4.
Nanometer-scale characterization cell peripheries of ARPC3−/− fibroblasts. Each example shows a 3.8-nm thick slice through a three-dimensional cyro-tomogram in grayscale with a paired three-dimensional rendering of red actin filaments and blue plasma membrane traced in the boxed areas of the reconstructions. The periphery of ARPC3−/− fibroblasts shows a characteristic arrangement of actin filament bundles parallel to the edge and closely approaching the membrane. Bars are 160 nm. Raw tomogram slices of the traced regions are shown to the right.
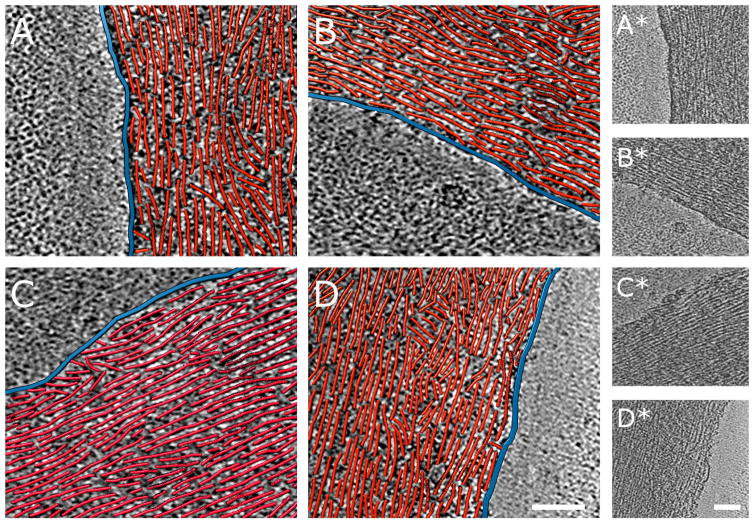
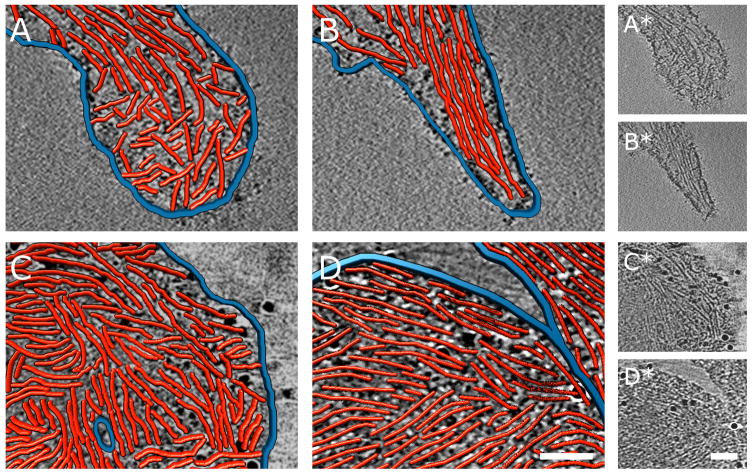
Figure 5. Nanometer-scale comparison of the tips of FLPs of ARPC3−/− fibroblasts and of filopodia of their wild-type isogenic control cells. Each example shows a 3.8 nm thick slice through a three-dimensional cyro-tomogram in grayscale with a paired three-dimensional rendering of red actin filaments and blue plasma membrane traced in boxed areas of the reconstructions. Bars are 200 nm. Raw tomogram slices of the traced regions are shown to the right.
A. Wild-type filopodium tip with ‘terminal cone’ geometry.
B. Wild-type filopodium tip with parallel filaments. Figure S3 shows complementary information.
D, E. Bulbous ARPC3−/− FLP tips with densely packed actin filaments. Occasionally, an FLP tip interacts with the shaft of another FLP as in (E).
Previous cryo-tomography studies near leading edges of fibroblasts cells were focused on the focal adhesion machinery (Patla et al., 2010; Elad et al., 2013) where actin filaments are organized in parallel to the long axis of the adhesion site but no quantitative analysis of actin network properties other than angular spread were provided. The actin network properties of small numbers of filopodia (four in total) and stress fibers (eight in total) from cryo-tomograms of epithelial PtK2 cells were analyzed in detail for comparison with Listeria comet tails (Jasnin et al., 2013). The average length of the filaments in filopodia and stress fibers in that study was around, 200–300 nm, about twice the average length measured in our study. It is possible that this is a consequence of comparing different cell types (fibroblasts versus epithelial cells) or the fact that the tomograms were recorded in different regions of the cells. In fact, filaments at the leading edge are expected to be very short owing to the fact that they must be able to exert force on the membrane and tend to buckle when they become too long (Mogilner and Oster, 1996).
While the average width of lamellipodia in moving fibroblasts is about 1.5 μm, in spreading fibroblasts the expected width is well below 1μm (Giannone et al., 2003). We observe that spreading wild-type cells typically assemble dendritic networks of actin filaments in a narrow lamellipodial zone <0.5 μm wide between the plasma membrane and actin filaments running parallel to the membrane (Fig. 3), corresponding to the lamella region. Most actin filaments in the mutant ARPC3−/− fibroblasts are organized in thick bundles either within FLPs or at the periphery of the cell where they very closely approach the membrane (Fig. 4). Measurements of the spacing between parallel filaments within the bundles yield a minimum distance of about 16 nm for both genotypes (Figs. 2C–D). The localization of fascin by correlative fluorescence microscopy (Fig. 1) in that region suggests that the crosslinkers responsible for the minimal distance correspond to fascin. Interfilament distances for actin filaments crosslinked by fascin vary widely in the literature from 8–9 nm from 2D analysis of negatively stained samples (Ishikawa et al., 2003), through 11–12 nm for cryo-tomography analysis of 3D reconstituted bundles (Yang et al., 2013) to about 15–16 nm in the fascin-based drosophila bristles (Tilney et al., 1995). We used our established technique of lipid monolayers (Volkmann et al., 2001) to reconstitute two-dimensional arrays of actin filaments crosslinked by fascin (Fig. 2F), yielding an average interfilament distance of 15 nm, similar to that in Drosophila bristles and that within the bundles investigated in this study. Very few regions in the bundles observed here display the type of paracrystalline order with regular cross-linker bands that occur in our in vitro reconstituted actin-fascin bundles, possibly due to the existence of additional actin-binding proteins in these assemblies in cells, as studied here.
The packing density in FLPs of ARPC3−/− fibroblasts is significantly increased compared to wild-type filopodia
Quantitative measurements using filament-tracing software confirmed the visual impression that actin filaments are more densely packed in FLPs of ARPC3−/− cells than in filopodia of wild-type cells (Table 1). Actin filaments occupy 25% of the total volume in FLPs and 19% in filopodia, a difference of high statistical significance. In addition to this higher actin-filament packing density in FLPs, the morphology of the FLP tips also differs from wild-type filopodia. Filopodia often show the ‘terminal cone’ geometry previously described for Dictyostelium filopodia (Medalia et al., 2007) with short filaments at various angles touching the inside of the membrane (Fig. 5A). In other filopodia, parallel actin filaments emanate from the tip into the shaft (Figs 5B). In contrast, the tips of FLPs tend to form large bulbs that are densely packed with actin filaments (Fig. 5C–D). Many filaments in these bulbs are parallel to the membrane, forming a circular pattern around the circumference of the bulb.
Table 1.
Actin filament packing densities in different bundle types
| vol | N | avg dis | p(fil) | p(FLP) | |
|---|---|---|---|---|---|
| Wt Filopdia | 19±6% | 74 | 20 nm | ≪0.0001 | |
| Mut FLPs | 25±5% | 175 | 18 nm | ≪0.0001 | |
| Wt peripheral | 15±4% | 156 | 22 nm | ≪0.0001 | ≪0.0001 |
| Mut peripheral | 24±6% | 81 | 18 nm | ≪0.0001 | 0.0863 |
vol: volume percentage occupied by actin filaments, N: number of bundle regions (extracted from 198 mutant and 92 wild-type cells) contributing to statistics, avg dis: average distance between filaments; p(.): statistical t-test p-values if packing density is compared with that in filopodia, p(fil), or in FLPs, p(FLP).
ARPC3−/− fibroblasts have a reduced variety of actin network types
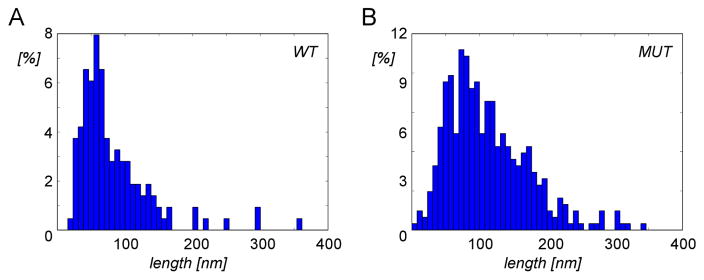
In addition to bundles inside filopodia or FLPs, both wild-type and ARPC3−/− cells also have bundles of actin at the cell periphery that run parallel to the plasma membrane (supplementary movies). The density of filaments in the peripheral bundles of wild-type cells was significantly lower than in bundles in filopodia of wild-type cells (Table 1), indicating different types of actin networks in these two locations. In contrast, the packing densities of filaments in the peripheral bundles and FLPs of ARPC3−/− fibroblasts were statistically indistinguishable. Thus, wild-type cells have at least two types of actin filament bundles in different locations, while the packing of bundles inside FLPs and in bundles near the cell periphery is the same for ARPC3−/− fibroblasts. Quantitation of individual filament lengths in the cryo-tomograms showed that the length distributions of mutant and wild-type cells differ significantly with the ARPC3−/− fibroblasts distribution shifted towards longer filaments (Fig. 6).
Figure 6. Distributions of actin filament lengths within bundles in wild-type and ARPC3−/− fibroblasts.
A–B. Normalized length distribution of actin filaments extracted from 30 randomly selected regions in wild-type (A) and in ARPC3−/− (B) fibroblasts. Both Mann-Whitney and Kolmogorov-Smirnov tests indicate a highly significant difference between the two length distributions (p ≪ 0.0001).
CONCLUSIONS
Most studies agree that Arp2/3 complex is essential for lamellipodia formation, but cells lacking Arp2/3 complex can migrate at similar rates as wild-type cells (Suraneni et al., 2012; Suraneni et al., 2015). Evidence is mounting that many actin assemblies in cells are controlled by collaborative actions of more than one type of nucleator (Blanchoin and Michelot, 2012). For example, in addition to its confirmed role in lamellipodia formation, Arp2/3 complex has also been implicated in contributing to the formation and regulation of filopodia (Beli et al., 2008; Sarmiento et al., 2008; Korobova and Svitkina, 2008).
Consistent with a regulatory role of Arp2/3 complex in processes guiding filopodia length and frequency, many of our observations indicate that cells lacking functional Arp2/3 complex fail to regulate actin filament growth. Our quantitative analysis at the single filament scale revealed that the massive filopodia-like protrusions (FLPs) of ARPC3−/− fibroblasts are more densely packed and contain longer actin filaments than wild-type filopodia. In addition, the fact that only one bundle type exists in ARPC3−/− mutants indicates that these cells lack the location-dependent fine-tuning of actin-bundle morphology. Instead, ARPC3−/− fibroblasts assemble massive FLPs with densely packed, long actin filaments that are orders of magnitude larger than wild-type filopodia, many with tips so densely packed with actin filaments that they curl up to form distinctive bulbs.
Our data suggest that the role of Arp2/3 complex in cell motility contains, in addition to generating dendritic actin networks at the leading edge and filopodia regulation, an element that is responsible for the fine-tuning of actin-bundle morphology at the cell periphery as well as in protrusions. Our study demonstrates the power of high-throughput analysis of filament network properties at the single filament level in large numbers of cryo-tomograms, allowing the detection of even subtle effects through the use of statistical analysis.
Supplementary Material
Acknowledgments
This work was supported by NIH program project grants P01 GM066311 (TDP, RL) and P01 GM098412 (DH, NV). NIH grant S10 OD012372 (DH) and P01 GM098412-S1 (DH) funded the purchase of the Titan Krios TEM and Falcon II direct detection imaging device. DH is grateful for time provided on the Titan Krios at the Centre for Bioimaging Science at the National University of Singapore (Dr. P. Matsudaira), and on the Polara (FEI company) of the Cryo-Electron Microscopy Facility at the Broad Center, Caltech (Dr. G. Jensen). Special thanks to Drs. Songye Chen, Caltech and Myint Aung, National University of Singapore.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agulleiro JI, Fernandez JJ. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27:582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- Beli P, Mascheroni D, Xu D, Innocenti M. WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat Cell Biol. 2008;10:849–857. doi: 10.1038/ncb1745. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 2000;404:1007–111. doi: 10.1038/35010008. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Michelot A. Actin cytoskeleton: a team effort during actin assembly. Curr Biol. 2012;22:R643–R645. doi: 10.1016/j.cub.2012.07.026. [DOI] [PubMed] [Google Scholar]
- Block J, Breitsprecher D, Kühn S, Winterhoff M, Kage F, Geffers R, Duwe P, Rohn JL, Baum B, Brakebusch C, Geyer M, Stradal TEB, Faix J, Rottner K. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr Biol. 2012;22:1005–1012. doi: 10.1016/j.cub.2012.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block J, Stradal TEB, Hänisch J, Geffers R, Köstler SA, Urban E, Small JV, Rottner K, Faix J. Filopodia formation induced by active mDia2/Drf3. J Microsc. 2008;231:506–517. doi: 10.1111/j.1365-2818.2008.02063.x. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL. Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science. 2012;336:1164–1168. doi: 10.1126/science.1218062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buades A, Coll B, Morel JM. A non-local algorithm for image denoising. IEEE Comput Soc Conf Comp Vis Pattern Recog. 2005;2:60–65. [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nardo A, Cicchetti G, Falet H, Hartwig JH, Stossel TP, Kwiatkowski DJ. Arp2/3 complex-deficient mouse fibroblasts are viable and have normal leading-edge actin structure and function. Proc Natl Acad Sci U S A. 2005;102:16263–16268. doi: 10.1073/pnas.0508228102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elad N, Volberg T, Patla I, Hirschfeld-Warneken V, Grashoff C, Spatz JP, Fässler R, Geiger B, Medalia O. The role of integrin-linked kinase in the molecular architecture of focal adhesions. J Cell Sci. 2013;126:4099–4107. doi: 10.1242/jcs.120295. [DOI] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Döbereiner HG, Freund Y, Borisy G, Sheetz MP. Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell. 2007;128:561–575. doi: 10.1016/j.cell.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Jiang G, Sutton DH, Critchley DR, Sheetz MP. Talin1 is critical for force-dependent reinforcement of initial integrin-cytoskeleton bonds but not tyrosine kinase activation. J Cell Biol. 2003;163:409–419. doi: 10.1083/jcb.200302001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struc Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Huber F, Käs J, Stuhrmann B. Growing actin networks form lamellipodium and lamellum by self-assembly. Biophys J. 2008;95:5508–5523. doi: 10.1529/biophysj.108.134817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Sakamoto T, Ando T, Higashi-Fujime S, Kohama K. Polarized actin bundles formed by human fascin-1: their sliding and disassembly on myosin II and myosin V in vitro. J Neurochem. 2003;87:676–685. doi: 10.1046/j.1471-4159.2003.02058.x. [DOI] [PubMed] [Google Scholar]
- Jasnin M, Asano S, Gouin E, Hegerl R, Plitzko JM, Villa E, Cossart P, Baumeister W. Three-dimensional architecture of actin filaments in Listeria monocytogenes comet tails. Proc Natl Acad Sci U S A. 2013;110:20521–20526. doi: 10.1073/pnas.1320155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Gautreau A. Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat Rev Mol Cell Biol. 2014;15:577–590. doi: 10.1038/nrm3861. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struc Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–616. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys J. 2006;90:1439–1452. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struc Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Heuser JA, Pollard TD. The interaction of Arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci U S A. 1998;95:6181–6186. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson-Dykstra SM, Higgs HN. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil Cytoskeleton. 2008;65:904–922. doi: 10.1002/cm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460:1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page C, Hanein D, Volkmann N. Accurate membrane tracing in three-dimensional reconstructions from electron cryotomography data. Ultramicroscopy. 2015;155:20–26. doi: 10.1016/j.ultramic.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patla I, Volberg T, Elad N, Hirschfeld-Warneken V, Grashoff C, Fassler R, Spatz JP, Geiger B, Medalia O. Dissecting the molecular architecture of integrin adhesion sites by cryo-electron tomography. Nat Cell Biol. 2010;12:909–915. doi: 10.1038/ncb2095. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Evangelista M, Yang C, Bi E, Zigmond S, Bretscher A, Boone C. Role of formins in actin assembly: nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. The structural basis of actin filament branching by Arp2/3 complex. J Cell Biol. 2008;180:887–895. doi: 10.1083/jcb.200709092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidani M, El-Sibai M, Desmarais V, Holman HA, Kitchen S, Backer JM, Alberts A, Condeelis J. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J Cell Biol. 2008;180:1245–1260. doi: 10.1083/jcb.200708123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Faix J, Resch GP, Linkner J, Wehland J, Small JV, Rottner K, Stradal TEB. Filopodia formation in the absence of functional WAVE- and Arp2/3-complexes. Mol Biol Cell. 2006;17:2581–2591. doi: 10.1091/mbc.E05-11-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser GA, Rahim NA, VanderWaal KE, Gertler FB, Lanier LM. Arp2/3 is a negative regulator of growth cone translocation. Neuron. 2004;43:81–94. doi: 10.1016/j.neuron.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Suraneni P, Fogelson B, Rubinstein B, Noguera P, Volkmann N, Hanein D, Mogilner A, Li R. A mechanism of leading edge protrusion in the absence of Arp2/3 complex. Mol Biol Cell. 2015;26:901–912. doi: 10.1091/mbc.E14-07-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraneni P, Rubinstein B, Unruh JR, Durnin M, Hanein D, Li R. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197:239–251. doi: 10.1083/jcb.201112113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Tilney MS, Guild GM. F actin bundles in Drosophila bristles. I. Two filament cross-links are involved in bundling. J Cell Biol. 1995;130:629–638. doi: 10.1083/jcb.130.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D, Kojima SI, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174:863–875. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinzenz M, Nemethova M, Schur F, Mueller J, Narita A, Urban E, Winkler C, Schmeiser C, Koestler SA, Rottner K, Resch GP, Maeda Y, Small JV. Actin branching in the initiation and maintenance of lamellipodia. J Cell Sci. 2012;125:2775–785. doi: 10.1242/jcs.107623. [DOI] [PubMed] [Google Scholar]
- Volkmann N. An approach to automated particle picking from electron micrographs based on reduced representation templates. J Struc Biol. 2004;145:152–156. doi: 10.1016/j.jsb.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Volkmann N, Amann KJ, Stoilova-McPhie S, Egile C, Winter DC, Hazelwood L, Heuser JE, Li R, Pollard TD, Hanein D. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science. 2001;293:2456–2459. doi: 10.1126/science.1063025. [DOI] [PubMed] [Google Scholar]
- Volkmann N, DeRosier D, Matsudaira P, Hanein D. An atomic model of actin filaments cross-linked by fimbrin and its implications for bundle assembly and function. J Cell Biol. 2001;153:947–956. doi: 10.1083/jcb.153.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann N, Hanein D. Quantitative fitting of atomic models into observed densities derived by electron microscopy. J Struc Biol. 1999;125:176–184. doi: 10.1006/jsbi.1998.4074. [DOI] [PubMed] [Google Scholar]
- Welch MD, Iwamatsu A, Mitchison TJ. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature. 1997;385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- Wu C, Asokan SB, Berginski ME, Haynes EM, Sharpless NE, Griffith JD, Gomez SM, Bear JE. Arp2/3 is critical for lamellipodia and response to extracellular matrix cues but is dispensable for chemotaxis. Cell. 2012;148:973–987. doi: 10.1016/j.cell.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XP, Page C, Volkmann N. Efficient extraction of macromolecular complexes from electron tomograms based on reduced representation templates. Lect Notes Comput Sc. 2015;9256:423–431. doi: 10.1007/978-3-319-23192-1_35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Svitkina T. Visualizing branched actin filaments in lamellipodia by electron tomography. Nat Cell Biol. 2011;13:1012–1013. doi: 10.1038/ncb2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Huang FK, Huang J, Chen S, Jakoncic J, Leo-Macias A, Diaz-Avalos R, Chen L, Zhang JJ, Huang XY. Molecular mechanism of fascin function in filopodial formation. J Biol Chem. 2013;288:274–284. doi: 10.1074/jbc.M112.427971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M. Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol. 2003;13:1820–1823. doi: 10.1016/j.cub.2003.09.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.