Abstract
Membrane fusion induced by herpes simplex virus (HSV) requires the action of four viral membrane glycoproteins (gB, gD, gH, and gL) and the binding of gD to one of its receptors, such as the herpesvirus entry mediator or nectin-1. The related animal herpesvirus, pseudorabies virus (PRV), encodes a homologous set of glycoproteins and its gD can also use nectin-1 as an entry receptor. We show here that PRV gD, when coexpressed with HSV gB, gH, and gL, cannot substitute for HSV gD in inducing fusion with target cells expressing nectin-1. Chimeric gD molecules composed of HSV and PRV sequences can substitute, provided the first 285 aa are from HSV gD. Because the first 261 aa were sufficient for receptor binding, this suggested that amino acids 262–285 contain a region required for cell fusion but not for receptor binding. Deletions from amino acids 250–299 failed to identify a specific subregion critical for cell fusion, except possibly for amino acids 250–255, which also influenced receptor binding. Instead, presence of a flexible stalk between the membrane and receptor-binding domain appears to be required, perhaps to enable conformational changes in gD on receptor binding and subsequent interactions of undefined regions of gD with the other glycoproteins required for membrane fusion.
Enveloped viruses of humans and animals invade cells by inducing fusion between the viral envelope and a cell membrane. Viral envelope glycoproteins initiate and mediate this fusion. In some cases, a single viral glycoprotein can mediate binding of virus to the cell surface and fusion with a cell membrane. In other cases, two viral glycoproteins or subunits of a single translation product are required for binding and fusion (reviewed in ref. 1). In the case of herpes simplex virus (HSV), four distinct glycoproteins (gB, gD, gH, and gL) are required for membrane fusion, whereas the initial attachment of virus to cell can be mediated by gB or gC binding to cell surface heparan sulfate (reviewed in refs. 2 and 3). The initiation of membrane fusion requires the interaction of gD with one of its receptors. These include the herpesvirus entry mediator (HVEM); nectin-1 and nectin-2, cell adhesion molecules in the Ig superfamily; and specific sites in heparan sulfate generated by particular 3-O-sulfotransferases (reviewed in ref. 4).
It remains unclear why HSV, and herpesviruses in general, require multiple envelope glycoproteins to induce membrane fusion. It seems unlikely that gD is an actual fusogen. All known viral fusogens must be anchored to the viral envelope as a transmembrane protein, whereas a glycosylphosphatidylinositol-linked gD ectodomain is functional for cell fusion (5) and soluble forms of the gD ectodomain can complement the entry defect of a gD-negative HSV (6). It has been proposed that interactions of gD with one of its receptors causes conformational changes in gD that enable it to activate the fusogenic activity of gB, a homooligomer, and/or gH-gL, a heterodimer (6–8).
HSV-1 gD is a 369-residue type 1 membrane glycoprotein. Its 316-aa ectodomain has three N-glycosylation sites and six cysteine residues forming three disulfide bonds (9, 10). X-ray structures have been reported for a truncated form of the HSV-1 gD ectodomain (first 285 aa), crystallized alone and in complex with HVEM (7). The structures revealed an Ig-like domain with N-terminal and C-terminal extensions. The N terminus was extended and partially disordered when gD was crystallized alone, whereas it formed an ordered hairpin in the gD–HVEM complex. All contacts with HVEM were within the hairpin (first 32 aa). However, studies with soluble truncated forms of gD have shown that the first 240–250 aa are required for stable binding to HVEM or nectin-1 (11).
Pseudorabies virus (PRV) is a porcine herpesvirus related to HSV-1 and its gB, gD, gH, and gL homologs have been well characterized (12). PRV and HSV-1 gDs (gD-P and gD-H1, respectively) exhibit only 30% amino acid identity. However, the six cysteine residues in the ectodomain are spatially conserved, implying that a common structure is retained. Consistent with the existence of shared structural features, gD-P binds to nectin-1 (13) and can use nectin-1 as an entry receptor (14). However, gD-P cannot substitute for gD-H1 in inducing cell fusion when it is coexpressed with HSV-1 gB, gH, and gL (this study). This finding raised the possibility that gD-P lacks determinants critical for putative interactions with the HSV-1 glycoproteins that participate in membrane fusion.
The purpose of this study was to identify regions of gD-H1 that are required for cell fusion, but not for receptor binding, and to determine whether the sequences of these regions influence fusion activity. The approach was in part to generate chimeric forms of gD, in which structurally conserved regions were switched between gD-H1 and gD-P, and to test these chimeras for binding to nectin-1 and HVEM and for cell fusion activity after coexpression with HSV-1 gB, gH, and gL. The results showed that a region of gD-H1 encompassing amino acids 262–285 was required for cell fusion but not for receptor binding. Amino acid deletions covering this region, and amino acid substitutions, failed to identify a specific sequence necessary for cell fusion. We propose that a Pro-rich stalk between the membrane and receptor-binding domain of gD-H1 is necessary for HSV-1-induced cell fusion, probably to permit interactions between N-terminal and C-terminal regions of the gD-H1 ectodomain and conformational changes in the receptor-binding domain, but not for specific interactions with the other HSV-1 glycoproteins.
Materials and Methods
Cells and Virus. CHO-K1 cells, CHO cells expressing human nectin-1 (14), and CHO cells expressing human HVEM (15) were grown in Ham's F12 medium supplemented with 10% FBS and Geneticin (400 μg/ml). HSV-1(KOS)gD6 expresses β-galactosidase from an insertion that replaces the gD gene (16). This virus was propagated on complementing VD60 cells, a Vero cell line inducible for gD-H1 expression (17). Vero cells and VD60 cells were grown in DMEM plus 10% FBS.
Plasmids Expressing Chimeric or Mutant gDs. Plasmid pPEP99 expresses HSV-1(KOS) gD from its ORF cloned into pCAGGS (18). Plasmid pAZPD was constructed by cloning into pCAGGS the PRV(Becker) gD ORF amplified by PCR, using a PRV bacterial artificial chromosome as template (19) and primers PRVD1 (5′-GGAATTCCGCCCCAGGTTCCCATACACTCA-3′) and PRVD2 (5′-ACATGCATGCATGTTCATCGACGCCGGTACTGC-3′). These oligonucleotides match sequences upstream and downstream of the gD-P ORF and have added EcoRI and SphI restriction sites (bold). The sequence of the cloned PCR product was similar to that of PRV(Rice) gD (GenBank accession no. VGBE50) with only one mismatch that affected coding (Y231 instead of H231). Sequence of the PRV(Becker) gD gene has not been deposited in GenBank. The gD chimeric ORFs were created by PCR. Briefly, for each chimera the gD-H1 and gD-P sequences were amplified in separate reactions by using pPEP99 and pAZPD, respectively, as templates. One of the primers used in gD-H1 or gD-P sequence amplification contained nucleotides (18–20 bases) on its 5′ end that matched the PRV or HSV-1 sequence, respectively. The amplification products were purified and combined in a final PCR using only the outside primers GDCH1 (5′-GGTTGTTGTGCTGTCTCATC-3′) and GDCH6 (5′-GATCTGCTAGCTCGAGGCAT-3′), which match sequences in pCAGGS upstream and downstream of EcoRI and SphI sites in the polylinker. These products were then purified, digested with EcoRI and SphI, and cloned into the corresponding sites of pCAGGS. Plasmids were sequenced to ensure against unintended mutations. Table 2, which is published as supporting information on the PNAS web site, indicates which amino acids from gD-H1 and gD-P were present in each chimera.
Plasmid pCJ9 expressing gD-H1Δ290–299 was constructed by excising the gD ORF from pHC240 (20), which has amino acids 290–299 deleted and GKIFP inserted at the site of the deletion, at EcoRI and BglII sites and cloning it directly into pCAGGS. pCJ11 containing the gD-H1 ORF with amino acids 250–299 deleted was constructed by amplifying the first 250 aa of the gD-H1 ectodomain, using the primer pair 5′-CACTGCTTACTGGCTTATCG and 5′-GCGGCGTCCTGCGTGTATGGGGC; amino acids 299–369 of gD-H1 were amplified by using the primer pair 5′-GCCCCATACACGCAGGACGCCGC and 5′-TGATCAGCGAGCTCTAGCAT. The resulting PCR products, which contained overlapping overhangs, were combined by using the primers 5′-CACTGCTTACTGGCTTATCG and 5′-TGATCAGCGAGCTCTAGCAT, digested with BglII and EcoRI, and inserted into pCAGGS. The other deletions and amino acid substitutions were constructed with the QuikChange Kit (Stratagene) in a pUC19 vector containing the gD-H1 ORF, then transferred to pCAGGS by using the EcoRI and SphI sites.
Binding of Antibodies and Soluble Receptors to CHO Cells Expressing Viral Glycoproteins. Binding was quantified as described (21, 22). Briefly, CHO-K1 cells in six-well plates were transfected with pCAGGS-based plasmids expressing various forms of gD along with HSV-1 gB, gH, gL, and T7 polymerase for 6 h. The latter plasmids have been described (18). The cells were then detached, replated on 96-well plates, and incubated overnight. Then, the cells were incubated at 37°C for 30 min with polyclonal or monoclonal anti-gD antibodies. Anti-gD-H1 rabbit sera R7 (23) and R45 were used at 1:10,000 dilution. Anti-gD-H1 mAbs 1D3 (24), DL6 (24), DL11 (25), HD1 (26), AP7 (27), and anti gD-P mAbs 6D8MB4 (American Type Culture Collection), and c14-c27 and b51-b11 (provided by T. Mettenleiter, Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany) were used at 1:1,000 dilution. Alternatively, the cells were incubated with soluble HVEM:Fc or nectin-1:Fc (500 ng/ml), hybrids of the receptor ectodomains joined to the Fc domain of rabbit IgG, prepared as described (28). Then the cells were washed and fixed with PBS containing 2% formaldehyde and 0.2% glutaraldehyde. To detect bound antibodies, the fixed cells were incubated with biotinylated anti-rabbit IgG (Sigma) or anti-mouse IgG (Sigma), streptavidin-conjugated horseradish peroxidase (HRP, Amersham Pharmacia) and HRP substrate (BioFx Laboratories, Owings Mills, MD). To detect bound soluble receptors, the fixed cells were incubated sequentially with an HRP-coupled anti-rabbit Fc antibody (Chemicon) and the same HRP substrate. HRP product was quantified at 370 nm in a Victor Wallac spectrophotometer (PerkinElmer).
Cell Fusion Assay. The cell fusion assay was performed as described (29) by using plasmids described here and in ref. 18. The effector cells were CHO-K1 cells transfected with pCAGGS-based plasmids expressing various forms of gD, HSV-1 gB, gH, and gL and pCAGT7 expressing the T7 RNA polymerase. The target cells were CHO-HVEM or CHO-nectin-1 cells transfected with pT7EMCLuc, encoding the firefly luciferase gene under control of the T7 promoter. The effector and target cells were mixed in 1:1 ratio in 96-well plates, incubated for 18 h, and then lysed for the quantitation of luciferase activity as a measure of cell fusion.
Complementation of gD-Negative Virions for Viral Entry. As described (30), Vero cells in six-well plates were transfected with a plasmid expressing WT or chimeric forms of gD and then infected with HSV-1(KOS)gD6 (20 plaque-forming units per cell). After 2 h the virus inocula were removed and unpenetrated virus were inactivated by treatment of the cells with citrate buffer (pH 3.0) for 1 min. The cells were then incubated in DMEM-1% FBS for 24 h, harvested, and lysed to prepare virus stocks. Samples of the complemented viruses were added in triplicate to CHO-HVEM or CHO-nectin-1 cells in 96-well plates and incubated for 24 h. The cells were then washed, permeabilized, and incubated with O-nitrophenyl-β-d-galactopyranoside (Sigma) as described (15). The β-galactosidase reaction product was monitored at 405 nm to quantify viral entry.
Results
Construction and Cell Surface Expression of HSV-1/PRV gD Chimeras. Use of the structural coordinates for gD-H1 (7) and the amino acid sequence for gD-P permitted structural alignment of part of the PRV sequence (amino acids 28–282) to the HSV-1 sequence (amino acids 44–285) (Fig. 1). Amino acid sequences of the N and C termini of the two proteins are even more divergent than those of the structurally related midsections. In particular, the N terminus of gD-P is shorter and distinct from that of gD-H1, perhaps explaining why HVEM is not an entry receptor for gD-P (15). Moreover, the membrane-proximal region of the gD-H1 ectodomain is shorter than that for gD-P.
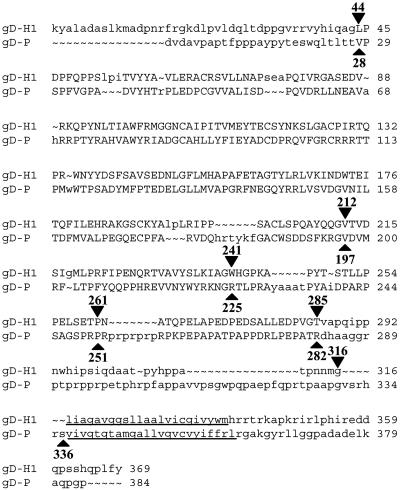
Fig. 1.
Structure-based sequence alignment of gD-H1 and gD-P. For both sequences the first amino acid after signal peptidase cleavage is number 1. Uppercase letters indicate regions of structural homology as determined by the program cn3d (National Center for Biotechnology, Bethesda). Inputs to the program were structural coordinates of gD-H1 deposited in the Protein Data Bank (33) for 1JMA (7) and the amino acid sequence for gD-P. The underlined regions are amino acids predicted to span the membrane. Arrowheads indicate amino acid residues just before the switch points for construction of the chimeras.
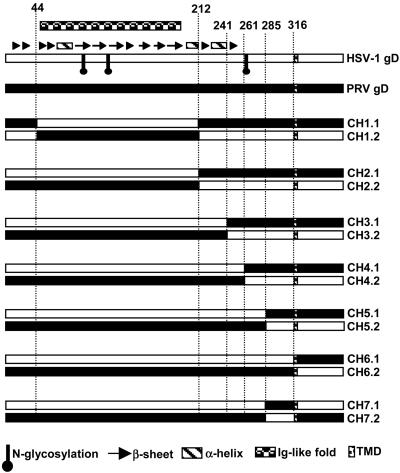
We used the alignment results of Fig. 1 to construct the chimeric gD molecules examined in this study, so that prominent structural domains remained intact (donated entirely by one or the other parental gD) in the chimeras (Fig. 2). The first pair of chimeras (CH1.1 and CH1.2) was constructed by interchanging, between gD-H1 and gD-P, the region containing all six cysteine residues involved in stabilization of the conserved Ig-like fold. Chimeric sets 2, 3, and 4 had switch points just downstream of the Ig fold, a long α-helix in gD-H1, or after amino acid 261 in gD-H1. Switch points for chimeric sets 5, 6, and 7 were the last amino acids in the structurally aligned region (285 for HSV-1 and 282 for PRV) and amino acids just before the membrane span (316 for HSV-1 and 336 for PRV).
Fig. 2.
Stick diagrams of gD-H1, gD-P, and chimeric forms of gD. Sequences from gD-H1 are represented by empty boxes, and sequences from gD-P are represented by filled boxes. Secondary structural features of gD-H1 and positions of N-linked glycans are indicated above and below the gD-H1 line, according to the legend at the bottom. Names of the chimeras are given on the right.
Each of the chimeras and parental gDs was coexpressed in CHO cells with HSV-1 gB, gH, gL, and T7 polymerase (so that expression of the various forms of gD could be assessed under the conditions used for cell fusion assays). The live intact cells were incubated with a panel of anti-gD antibodies to determine whether the chimeras were expressed on cell surfaces at detectable levels. Two rabbit antisera (R7 and R45) and five mAbs specific for gD-H1 and three mAbs specific for gD-P were used. Binding of these antibodies to cells expressing the chimeras was quantified in comparison with binding to cells expressing the appropriate parental gD and expressed as a percentage of this control binding (Table 1). The results showed that all of the chimeras, except CH1.1 and CH1.2, could be detected on cell surfaces at levels comparable to those of the WT proteins, based on findings that at least one of the antibodies bound at levels from 60% to 100% of control binding (bold results in Table 1). All of the chimeras were recognized by the polyclonal anti-HSV antibodies, above the background binding noted for gD-P (6%). Binding was lowest for the chimeras with the least HSV-1 contribution (CH5.2, CH6.2, and CH7.2), as expected, but also low for CH1.1, CH1.2, and CH2.1. Two of the HSV-specific mAbs recognize linear epitopes, amino acids 272–279 for DL6 and amino acids 11–19 for 1D3 (24). All chimeras containing these sequences, except CH1.2, bound these antibodies at levels 60–100% of the positive controls. The remainder of the HSV-specific and PRV-specific mAbs recognize conformational epitopes. The HSV-specific mAbs HD1 and DL11 and all three PRV-specific mAbs showed significant binding only to the chimeras for which the first 241 aa (at least) were derived from HSV-1 or PRV, respectively. HSV-specific mAb AP7 bound only to CH6.1, the only chimera containing the entire gD-H1 ectodomain, consistent with evidence that amino acids contributing to the AP7 epitope are at both the N terminus (positions 25 and 27) and the C terminus (positions 290–300) (20, 27, 31). Thus, all of the chimeras except for CH1.1 and CH1.2 retained appropriate gD-H1 epitopes based on previous mapping results.
Table 1. Binding of various antibodies to HSV-1/PRV chimeric forms of gD.
| Binding to various forms of gD of these anti-gD antibodies
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Form of gD | R7* | R45* | DL6† | HD1† | AP7† | DL11† | 1D3† | c14-c27‡ | b51-b11‡ | 6D8MB4‡ |
| gD-H1 | 100§ | 100 | 100 | 100 | 100 | 100 | 100 | 0 | 0 | 0 |
| gD-P | 6.3 | 6.5 | 3.8 | 0 | 0 | 4 | 2.5 | 100§ | 100 | 100 |
| CH1.1 | 43.5 | 38 | 6 | 0 | 3.2 | 6 | 0 | 0 | 0 | 0 |
| CH1.2 | 30 | 21 | 21 | 0 | 0 | 3 | 7.6 | 0 | 0 | 0 |
| CH2.1 | 32 | 45.4 | 4 | 0 | 0 | 7 | 100 | 0 | 0 | 0 |
| CH2.2 | 78 | 39 | 61.5 | 0 | 1 | 4 | 0 | 0 | 0 | 0 |
| CH3.1 | 50 | 61.4 | 4 | 102 | 2 | 89 | 96 | 0 | 0 | 0 |
| CH3.2 | 57 | 35 | 68 | 0 | 0 | 4.5 | 0 | 62 | 46.3 | 59 |
| CH4.1 | 62 | 69 | 7 | 106 | 0 | 98 | 82 | 0 | 0 | 0 |
| CH4.2 | 79 | 65 | 87 | 0 | 3 | 6 | 5 | 89 | 80 | 95 |
| CH5.1 | 84.5 | 97.6 | 62 | 50 | 3 | 101 | 92 | 0 | 0 | 2.5 |
| CH5.2 | 59 | 40.6 | 0 | 0 | 0 | 0 | 0 | 79 | 74 | 84 |
| CH6.1 | 65 | 68 | 93 | 90 | 72.3 | 79 | 80 | 0 | 0 | 0 |
| CH6.2 | 32 | 26 | 4 | 0 | 0 | 0 | 0 | 65 | 69 | 65 |
| CH7.1 | 86 | 98.3 | 94.5 | 98 | 0 | 103 | 103 | 0 | 0 | 0 |
| CH7.2 | 25 | 21 | 0 | 0 | 0 | 2.3 | 0 | 44 | 79.6 | 61.5 |
Bold type indicates the highest level of antibody binding for each chimera, as a percentage of binding to gD-H1 or gD-P.
Polyclonal rabbit antisera specific for gD-H1.
mAbs specific for gD-H1.
mAbs specific for gD-P.
Binding to the cognate antigen (gD-H1 or gD-P) is set at 100% for each antibody. Binding to each chimera is expressed as a percentage of binding to gD-H1 or gD-P.
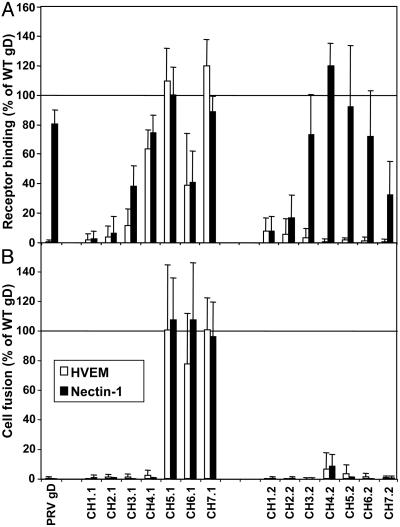
Binding of Soluble Receptors to the Chimeric gDs. Each of the chimeras and parental gDs was coexpressed in CHO cells with HSV-1 gB, gH, gL, and T7 polymerase, and the cells were then divided into two samples, one for cell fusion as described below, and the other for assessment of receptor binding. Live intact cells were incubated with soluble forms of human HVEM and nectin-1 (HVEM:Fc and nectin-1:Fc), and binding was quantified by use of an Fc detection system. Fig. 3A presents the binding results expressed as a percentage of binding to gD-H1. As expected, nectin-1:Fc, but not HVEM:Fc, bound to gD-P at levels 80% of the gD-H1 control level. Consistent with results obtained by using soluble truncated forms of gD-H1 (11), only chimeras having at least the first 241 aa exclusively from gD-H1 or gD-P (series 3–7) bound detectable levels of nectin-1:Fc. Thus, proper conformation of the nectin-1 receptor-binding domain requires, from either gD-H1 or gD-P, at least regions encompassing the Ig fold and two α-helices downstream of this fold (Fig. 2). Nectin-1:Fc binding to the chimeras containing HSV-1 sequence from the N terminus up to or through amino acid 241 was not entirely equivalent to its binding to the comparable set containing PRV sequences from the N terminus. CH3.1 and CH4.1 bound this receptor less efficiently than did CH3.2 and CH4.2, and CH7.2 bound less efficiently than did CH7.1. These results must reflect subtle differences between gD-H1 and gD-P in effects of the sequence switches at various positions on integrity of the nectin-1 binding domain. Also CH6.1, containing the entire ectodomain from HSV-1, bound nectin-1:Fc less efficiently than did gD-H1, indicating that the transmembrane and tail sequences from PRV somehow reduced binding.
Fig. 3.
Activities of the gD chimeras in receptor binding (A) and cell fusion (B). CHO cells were cotransfected with plasmids expressing one of the parental gDs or gD chimeras, HSV-1 gB, gH, gL, and T7 polymerase. Negative control cells were similarly prepared except that the gD-expressing plasmid was replaced with empty vector. The cells were then detached and divided. (A) Samples of each cell population were plated in triplicate in 96-well plates and then incubated with HVEM:Fc or nectin-1:Fc. After washing and fixation of the cells, binding of the soluble receptors was quantified by use of an Fc detection system. (B) Replicate samples of each cell population were mixed with target cells (HVEM-expressing or nectin-1-expressing CHO cells transfected with a plasmid carrying the luciferase gene under control of the T7 promoter) and plated in triplicate in 96-well plates. After 18 h lucerifase was quantified as a measure of cell fusion. Values for the negative controls were subtracted from each experimental value, and the results are expressed as a percentage of binding to, or cell fusion activity with, gD-H1. For receptor binding, negative control values ranged from 0.10 to 0.34 for HVEM and 0.09 to 0.32 for nectin-1, whereas uncorrected values for gD-H1 ranged from 0.31 to 1.1 for HVEM and 0.20 to 0.76 for nectin-1. For cell fusion, negative control values ranged from 602 to 14,865 for HVEM and 25 to 10,991 for nectin-1, whereas uncorrected values for gD-H1 ranged from 5,671 to 101,782 and 24,069 to 125,970. The means and SD for three independent experiments are shown.
Binding of HVEM:Fc to the chimeras containing HSV-1 sequences at the N terminus was comparable to the nectin-1:Fc binding (somewhat lower for CH3.1 and CH4.1) whereas no binding to the equivalent PRV set of chimeras was observed, as expected. Consistent with previous results (11), at least the first 261 aa of gD-H1 must be present in the chimeras for significant binding to HVEM:Fc, despite the fact that all known contacts for HVEM on gD-H1 are in the N-terminal 32 aa (7). Mutational analysis of gD (32) also showed that downstream regions are required for binding to HVEM, either for additional as yet undiscovered contact sites or to provide support for proper conformation of the N terminus.
CH1.1, CH1.2, CH2.1, and CH2.2 failed to bind either nectin-1:Fc or HVEM:Fc, possibly because in part of lower levels of expression on the cell surface, at least for CH1.1 and CH1.2. The results obtained with both sets of chimeras indicate, however, that the Ig fold alone from either gD-H1 or gD-P, with or without the N terminus, is insufficient to provide the proper interfaces for receptor binding.
Cell Fusion and Viral Entry Activities of the Chimeric gDs. For cell fusion, effector cells expressing the viral glycoproteins and T7 polymerase, prepared as described in the preceding section, were mixed with target CHO cells stably expressing either HVEM or nectin-1 and transfected with a plasmid encoding luciferase under control of the T7 promoter. After 18 h, luciferase activity was quantified as a measure of cell fusion. The cell fusion activities are presented in Fig. 3B and expressed as a percentage of activity observed with gD-H1. Note that gD-P, coexpressed with HSV-1 gB, gH, and gL, cannot substitute for gD-H1 in inducing cell fusion, whereas it can induce cell fusion when coexpressed with the PRV homologs (data not shown).
Fusion with cells expressing either HVEM or nectin-1 was observed only with CH5.1, CH6.1, and CH7.1, indicating that at least the first 285 aa of gD-H1 are necessary for this activity. Interestingly, the fusion activity observed was comparable to that of gD-H1 even for CH6.1, which exhibited reduced binding to both receptors. Clearly, binding to receptors is not sufficient for induction of cell fusion because other chimeras could bind one or both of the receptors but failed to induce cell fusion. Rather, gD-H1 sequences not required for receptor binding are necessary for cell fusion activity (amino acids 262–285).
To determine whether the chimeras could substitute for gD-H1 in viral entry, a gD-negative HSV-1 strain was passaged once through Vero cells transfected to express one of the parental gDs or chimeras. This process permits incorporation of the expressed gD into progeny virions. These virions were then plated on CHO-HVEM cells or CHO-nectin-1 cells and entry was quantified. As observed for cell fusion activity, only CH5.1, CH6.1, and CH7.1 mediated viral entry, regardless of the receptor. The entry activity observed, however, was less than that observed for gD-H1 (≈25% for CH5.1 and 50% for CH6.1 and CH7.1). Possibly, PRV sequences in these chimeras may have impeded incorporation of the chimeras into the virion envelope.
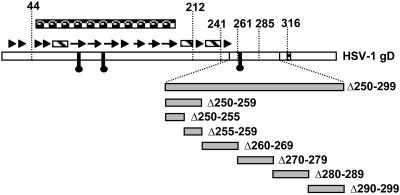
Effects of Deletions and Amino Acid Substitutions Between Amino Acids 250 and 299 in gD-H1 on Receptor Binding and Cell Fusion. A large deletion was introduced into gD-H1 (Δ250–299), and 5- and 10-aa deletions also were introduced, as shown in Fig. 4; Δ290–299 was previously described and its deletion is coupled to an insertion of amino acids GKIFP (20). These deletion mutants were tested for cell surface expression, as described above for the chimeric forms of gD, by using polyclonal rabbit antiserum R7. CHO cells expressing all of the deletion mutants except for the largest (Δ250–299) bound the anti-gD antibodies at levels comparable with those observed for WT gD-H1; binding to gDΔ250–299 was 25% of the positive control values, probably because of loss of epitopes and reduced cell surface expression.
Fig. 4.
Schematic representation of gD-H1 deletion mutants produced for this study. Structural features of gD-H1 are indicated as described in Fig. 2.
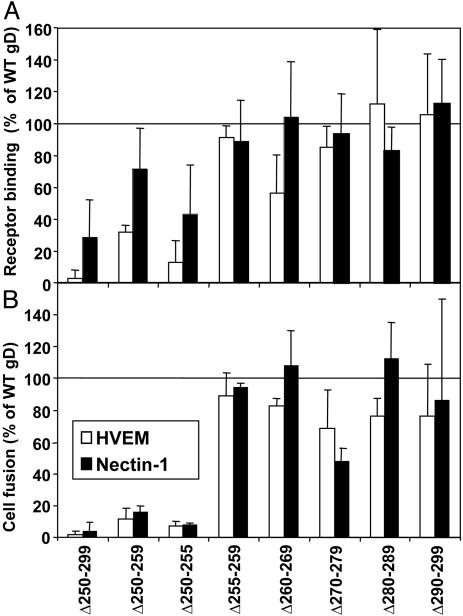
The deletion mutants also were tested for receptor binding and cell fusion activity as described above for the chimeras. The results presented in Fig. 5 show that receptor binding and cell fusion activity were largely abolished for the largest deletion (Δ250–299) except that binding to nectin-1:Fc was at 30% of positive control values. Reduced cell surface expression of this mutant may account in part, but probably not totally, for the loss of cell fusion activity. Mutant Δ250–259 exhibited more severely reduced cell fusion activity than receptor-binding activity, particularly for nectin-1. Two 5-aa deletions were generated to cover this region. Mutant Δ250–255 was severely defective for cell fusion activity but also exhibited reduced receptor binding activity, whereas mutant Δ255–259 was indistinguishable from WT gD in both activities. Ala-scanning mutations were introduced into gD-H1 across the region from amino acids 250–255. None of these point mutants differed from WT gD in cell surface expression, receptor binding, or cell fusion activity (results not shown), indicating that the presence of this region, but not the actual amino acid sequence, is critical for cell fusion and, to a lesser extent, for receptor binding.
Fig. 5.
Activities of gD-H1 deletion mutants in receptor binding (A) and cell fusion (B). The experiments were done and data are presented as described in the legend to Fig. 3, except that the gD-H1 deletion mutants were used instead of gD chimeras.
The other deletion mutants tested exhibited receptor-binding and cell fusion activities indistinguishable from those of WT gD except that Δ270–279 had somewhat reduced cell fusion activity, particularly with nectin-1. Results with the chimeric gDs indicated that the region of gD-H1 from amino acids 262–285 was essential for cell fusion activity, but not for receptor binding, and could not be replaced with the corresponding PRV sequence. However, deletions of 10 aa through this region in gD-H1 had only partial or no effect on cell fusion activity. Thus, no specific amino acid sequence required for cell fusion activity could be identified.
Discussion
Binding of HSV gD to an entry receptor probably mediates conformational changes in gD, which could enable subsequent interactions with gB and/or gH-gL, to induce the membrane-fusing activity required for cell fusion and viral entry. This hypothesis has led to a search for specific domains in gD that could engage in these interactions. We and others (6) have identified a region in gD-H1 (amino acids 262–285) that appears to be required for cell fusion and viral entry but not for receptor binding. We propose, for reasons given below and in contrast to conclusions of the other study (6), that there is probably no specific interaction domain for gB or gH-gL within this region, but that properties of this region (amino acid composition, spacing of particular amino acids such as Pro, and total length) may be necessary for gD to assume conformations critical for fusion activity. It seems likely that domains for functional interaction with gB and/or gH-gL are present in gD but they probably lie elsewhere, perhaps overlapping the receptor-binding regions.
Structural coordinates for regions of the gD-H1 ectodomain downstream of amino acid 259, in gD cocrystallized with HVEM, and both upstream of amino acid 14 and downstream of amino acid 255, in gD crystallized alone, have not been determined even though the truncated form of gD used comprised amino acids 1–285 (7). Two lines of evidence suggest that regions in gD downstream of amino acid 285 may be engaged in interactions with upstream regions of gD. First, soluble forms of gD truncated after amino acids 250, 260, or 285 bind to HVEM and nectin-1 with affinities ≈100-fold higher than that characteristic of gD truncated at amino acid 306 (11), suggesting that a region between amino acids 285 and 306 influences conformation of the upstream receptor-binding domains. Second, results obtained with the mAb AP7 demonstrate that amino acids substitutions at positions 25 and 27 in the N terminus and deletions encompassing amino acids 290–300 in the C terminus can destroy its epitope, but not those of other conformation-dependent mAbs, such as HD1 and DL11 (20, 27, 31). Consistent with these findings, our results showed that the only chimera displaying an AP7 epitope was CH6.1, which has its entire ectodomain from gD-H1. Thus, both the extreme N-terminal and C-terminal regions of the gD ectodomain determine the conformation of the AP7 epitope and may even physically interact to form or influence conformation of this epitope. If so, regions of gD between amino acids 250 and 285 must, at least under certain conditions, assume a conformation enabling the N terminus to interact with the membrane-proximal region of the ectodomain. The sequences of gD-H1 and gD-P downstream of amino acids 241 and 225, respectively, extending to the membrane spans, are very Pro-rich (Fig. 1). The PRV sequence in this region is longer and has more Pro residues, many spaced differently from those in the membrane-proximal region of gD-H1. Although Pro residues introduce constraints on conformation, they are often abundant in flexible regions of a protein, perhaps because their presence precludes certain types of secondary structure.
Our finding that replacement of gD-H1 sequences downstream of amino acid 261 with PRV sequences (CH4.1) failed to permit cell fusion, whereas replacement downstream of amino acid 285 permitted cell fusion (CH5.1), suggests the possibility that a specific HSV-1 sequence between amino acids 262 and 285 is required for cell fusion. Deletions in gD-H1 across this region had little effect on cell fusion activity (Fig. 5), except in the case of Δ270–279. Because this deletion reduced cell fusion activity no more than 50%, it seems likely that there is no specific sequence within the region from amino acids 262–285 that is absolutely essential for fusion activity. An alternative explanation is that this region permits N-terminal/C-terminal interactions within gD that are critical for fusion activity and/or permits receptor-dependent conformational changes within the receptor-binding domains. The former possibility, if true, implies that PRV sequences downstream of amino acid 282 can substitute for gD-H1 sequences downstream of amino acid 285, to mediate these N-terminal/C-terminal interactions, because CH5.1 and CH7.1 both are active in inducing cell fusion. Considering cell fusion activity in proportion to receptor-binding activity (Fig. 3), it may be that CH6.1, containing the entire gD-H1 ectodomain, is even more active in cell fusion than CH5.1 and CH7.1.
Our results also defined a region in gD-H1 (amino acids 250–255) that influences binding to both HVEM and nectin-1 and, to a larger extent, cell fusion activity with both receptors (Fig. 5). This finding appears to be at variance with previous results that soluble forms of gD truncated after amino acid 250 can bind to both receptors with equivalent or higher affinity than does gD truncated after amino acids 260, 285, or 306. The explanation very likely lies in the fact that the deletion mutant tested here was a membrane-bound nontruncated form of gD, constrained by being anchored in a membrane and retaining sequences at the C terminus of the ectodomain that could influence conformation of upstream regions. Ala-scanning mutations at each of the positions from amino acids 250–255 revealed that the precise amino acid sequence of this region is not important for its function in receptor binding or cell fusion.
The results presented here and elsewhere (6) identify a domain in gD-H1 that can be targeted to prevent HSV-induced membrane fusion, independently of blocking receptor binding. Agents that interfere with the function of this domain can be expected to block viral entry and virus-induced cell fusion.
Supplementary Material
Acknowledgments
We thank G. Cohen and R. Eisenberg (University of Pennsylvania, Philadelphia) and T. Mettenleiter for antibodies, G. Smith (Northwestern University) for the PRV bacterial artificial chromosome, and N. Susmarski (Northwestern University) for excellent technical assistance. This work was supported by U.S. Public Health Service Grants CA-21776 and AI-36293.
Author contributions: A.Z., C.R.J., and P.G.S. designed research; A.Z. and C.R.J. performed research; A.Z. and C.R.J. contributed new reagents/analytic tools; A.Z., C.R.J., and P.G.S. analyzed data; and A.Z. and P.G.S. wrote the paper.
Abbreviations: gB, glycoprotein B; gD, glycoprotein D; gH, glycoprotein H; gL, glycoprotein L; HSV, herpes simplex virus; gD-H1, HSV-1 gD; PRV, pseudorabies virus; gD-P, PRV gD; HVEM, herpesvirus entry mediator; HRP, horseradish peroxidase.
References
- 1.Earp, L. J., Delon, S. E., Park, H. E. & White, J. M. (2004) Curr. Top. Microbiol. Immunol. 285, 25–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spear, P. G. (1993) Semin. Virol. 4, 167–180. [Google Scholar]
- 3.Spear, P. G. (2004) Cell. Microbiol. 6, 401–410. [DOI] [PubMed] [Google Scholar]
- 4.Spear, P. G., Eisenberg, R. J. & Cohen, G. H. (2000) Virology 275, 1–8. [DOI] [PubMed] [Google Scholar]
- 5.Jones, N. A. & Geraghty, R. J. (2004) Virology 324, 213–228. [DOI] [PubMed] [Google Scholar]
- 6.Cocchi, F., Fusco, D., Menotti, L., Gianni, T., Eisenberg, R. J., Cohen, G. H. & Campadelli-Fiume, G. (2004) Proc. Natl. Acad. Sci. USA 101, 7445–7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carfi, A., Willis, S. H., Whitbeck, J. C., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Wiley, D. C. (2001) Mol. Cell 8, 169–179. [DOI] [PubMed] [Google Scholar]
- 8.Spear, P. G. & Longnecker, R. (2003) J. Virol. 77, 10179–10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watson, R. J., Weis, J. H., Salstrom, S. S. & Enquist, L. W. (1982) Science 218, 381–383. [DOI] [PubMed] [Google Scholar]
- 10.Long, D., Wilcox, W. C., Abrams, W. R., Cohen, G. H. & Eisenberg, R. J. (1992) J. Virol. 66, 6668–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whitbeck, J. C., Muggeridge, M. I., Rux, A. H., Hou, W., Krummenacher, C., Lou, H., van Geelen, A., Eisenberg, R. J. & Cohen, G. H. (1999) J. Virol. 73, 9879–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mettenleiter, T. C. (2000) Vet. Res. 31, 99–115. [DOI] [PubMed] [Google Scholar]
- 13.Milne, R. S. B., Connolly, S. A., Krummenacher, C., Eisenberg, R. J. & Cohen, G. H. (2001) Virology 281, 315–328. [DOI] [PubMed] [Google Scholar]
- 14.Geraghty, R. J., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Spear, P. G. (1998) Science 280, 1618–1620. [DOI] [PubMed] [Google Scholar]
- 15.Montgomery, R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427–436. [DOI] [PubMed] [Google Scholar]
- 16.Warner, M. S., Geraghty, R. J., Martinez, W. M., Montgomery, R. I., Whitbeck, J. C., Xu, R., Eisenberg, R. J., Cohen, G. H. & Spear, P. G. (1998) Virology 246, 179–189. [DOI] [PubMed] [Google Scholar]
- 17.Ligas, M. W. & Johnson, D. C. (1988) J. Virol. 62, 1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pertel, P., Fridberg, A., Parish, M. L. & Spear, P. G. (2001) Virology 279, 313–324. [DOI] [PubMed] [Google Scholar]
- 19.Smith, G. A. & Enquist, L. W. (2000) Proc. Natl. Acad. Sci. USA 97, 4873–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang, H.-Y., Cohen, G. H. & Eisenberg, R. J. (1994) J. Virol. 68, 2529–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geraghty, R. J., Jogger, C. R. & Spear, P. G. (2000) Virology 268, 147–158. [DOI] [PubMed] [Google Scholar]
- 22.Geraghty, R. J., Fridberg, A., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Spear, P. G. (2001) Virology 285, 366–375. [DOI] [PubMed] [Google Scholar]
- 23.Isola, V. J., Eisenberg, R. J., Siebert, G. R., Heilman, C. J., Wilcox, W. C. & Cohen, G. H. (1989) J. Virol. 63, 2325–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenberg, R. J., Long, D., Ponce de Leon, M., Matthews, J. T., Spear, P. G., Gibson, M. G., Lasky, L. A., Berman, P., Golub, E. & Cohen, G. H. (1985) J. Virol. 53, 634–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long, D., Madara, T. J., Ponce de Leon, M., Cohen, G. H., Montgomery, P. C. & Eisenberg, R. J. (1984) Infect. Immun. 37, 761–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira, L., Klassen, T. & Baringer, J. R. (1980) Infect. Immun. 29, 724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minson, A. C., Hodgman, T. C., Digard, P., Hancock, D. C., Bell, S. E. & Buckmaster, E. A. (1986) J. Gen. Virol. 67, 1001–1013. [DOI] [PubMed] [Google Scholar]
- 28.Jogger, C. R., Montgomery, R. I. & Spear, P. G. (2004) Virology 318, 318–326. [DOI] [PubMed] [Google Scholar]
- 29.Zago, A. & Spear, P. G. (2003) J. Virol. 77, 9695–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manoj, S., Jogger, C. R., Myscofski, D., Yoon, M. & Spear, P. G. (2004) Proc. Natl. Acad. Sci. USA 101, 12414–12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dean, H. J., Terhune, S., Shieh, M.-T., Susmarski, N. & Spear, P. G. (1994) Virology 199, 67–80. [DOI] [PubMed] [Google Scholar]
- 32.Yoon, M. & Spear, P. G. (2004) Proc. Natl. Acad. Sci. USA 101, 17252–17257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. M., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.