Abstract
Mammalian cells have evolved a common DNA damage response (DDR) that sustains cellular function, maintains genomic integrity, and suppresses malignant transformation. In pre-B cells, DNA double strand breaks (DSBs) induced at Igκ loci by the Rag1/Rag2 (RAG) endonuclease engage this DDR to modulate transcription of genes that regulate lymphocyte-specific processes. We previously reported that RAG DSBs induced at one Igκ allele signal through the ATM kinase to feedback inhibit RAG expression and RAG cleavage of the other Igκ allele. Here, we show that DSBs induced by ionizing radiation, etoposide, or bleomycin suppress Rag1 and Rag2 mRNA levels in primary pre-B cells, pro-B cells, and pro-T cells, indicating that inhibition of Rag1 and Rag2 expression is a prevalent DSB response among immature lymphocytes. DSBs induced in pre-B cells signal rapid transcriptional repression of Rag1 and Rag2, causing downregulation of both Rag1 and Rag2 mRNA but only Rag1 protein. This transcriptional inhibition requires the ATM kinase and the NF-κB essential modulator protein, implicating a role for ATM-mediated activation of canonical NF-κB transcription factors. Finally, we demonstrate that DSBs induced in pre-B cells by etoposide or bleomycin inhibit recombination of Igκ loci and a chromosomally integrated substrate. Our data indicate that immature lymphocytes exploit a common DDR signaling pathway to limit DSBs at multiple genomic locations within developmental stages wherein monoallelic antigen receptor locus recombination is enforced. We discuss the implications of our findings for mechanisms that orchestrate the differentiation of mono-specific lymphocytes while suppressing oncogenic antigen receptor locus translocations.
Introduction
DSBs are unavoidable, common, and hazardous genomic lesions. They are induced by endogenous factors including cellular metabolites, gene transcription, and DNA replication, and by exogenous factors including ionizing radiation (IR) and genotoxic drugs. DSBs can impair cellular function, induce apoptosis, or create genomic alterations that cause cancer if they are not repaired or are aberrantly repaired. Mammalian cells have evolved a conserved DDR that coordinates DSB repair with cellular proliferation and survival to maintain cellular function, preserve genomic integrity, and suppress malignant transformation. A central component of this shared DDR is the Ataxia Telangiectasia mutated (ATM) kinase, which is activated by DSBs (1). ATM phosphorylates and activates DNA repair proteins to enhance the kinetics and fidelity of DSB repair (2). ATM also activates intracellular signaling pathways that maintain cellular survival and halt DNA synthesis as cells attempt to repair DSBs, and promote apoptosis if DSBs are not repaired (1). For example, ATM phosphorylates the NF-κB essential modulator (Nemo) protein to activate NF-κB-mediated transcription of anti-apoptotic genes (3). In parallel, ATM-dependent phosphorylation of the Tp53 protein activates transcription of cell cycle checkpoint and pro-apoptotic genes (1). The importance of the conserved mammalian DDR is highlighted by the increased predisposition of humans and mice lacking ATM protein to oncogenic genomic instability (4–8).
Despite their inherent danger, DSBs are essential for mammalian biology. The programmed induction of DSBs by tissue-specific proteins is necessary to establish the genetic diversity that drives evolution and provides adaptive immunity (9, 10). A paradigm for this concept is the assembly of Ag receptor (AgR) gene variable region exons via recombination of variable (V), diversity (D), and joining (J) gene segments in developing B and T cells (11). The lymphocyte-specific RAG endonuclease catalyzes V(D)J recombination in G1 phase cells by cleaving at recombination signal sequences (RSSs) that flank all V, D, and J segments (12). The resolution of RAG DSBs by non-homologous end-joining (NHEJ) factors generates V(D)J coding joins and RSS signal joins (2). These V(D)J exons and downstream constant region exons comprise assembled AgR genes. The large number of possible V(D)J joining events and the inherent imprecision of coding join formation cooperate to establish AgR gene diversity. The essential role for RAG DSBs in establishing immunity is emphasized by mutations of RAG1, RAG2, RSSs, or NHEJ factors that impair lymphocyte development, restrict AgR gene repertoires, and cause fatal severe combined immunodeficiency (13–15).
RAG DSBs and the DDR cooperate to promote and police AgR gene assembly and differentiation of lymphocytes in the bone marrow (B lymphocytes) and thymus (T lymphocytes). The assembly of IgH or TCRβ genes in pro-B or pro-T cells, respectively, generates pre-BCR or pre-TCR complexes that signal cellular proliferation and differentiation into pre-B or pre-T cells (16). Similarly, assembly of IgL (Igκ or Igλ) or TCRα genes in pre-B or pre-T cells, respectively, forms BCR and αβ TCR AgRs that signal cellular differentiation or apoptosis (16). Autoreactive AgRs induce receptor editing through further IgL recombination or apoptosis; while innocuous AgRs halt Rag1 and Rag2 transcription and drive differentiation of conventional B and αβ T cells (17). In pro-T cells, the assembly of TCRγ and TCRδ genes also occurs and when successful produces γδ TCRs that terminate Rag1 and Rag2 transcription and signal differentiation of γδ T cells (18). RAG cleavage of AgR loci activates the ATM kinase (19), which phosphorylates DSB repair proteins to prevent degradation and enhance joining of RAG-liberated DNA ends (2). ATM also activates Tp53 to trigger the G1/S cell cycle checkpoint, inhibiting cells with RAG DSBs from entering S phase where DNA replication-associated DSBs increase the probability for AgR locus translocations (20, 21) At least in pre-B cells, RAG DSBs signal through both ATM-dependent and ATM-independent pathways to activate a broad multifunctional genetic program that includes genes known to regulate lymphocyte differentiation and lymphocyte-specific processes (22). One of these pathways involves ATM-dependent Nemo phosphorylation, which induces NF-κB-mediated transcription of the anti-apoptotic Pim2 gene to prolong the survival of pre-B cells recombining IgL genes (22, 23).
Recent work from our lab and the Sleckman lab has revealed that lymphocyte-specific RAG DSBs cooperate with common DDR signaling pathways to transiently feedback inhibit AgR gene assembly. We demonstrated that RAG cleavage of one Igκ allele in primary pre-B cells signals via the ATM kinase to suppress RAG cleavage of the other allele (24). ATM also downregulates the levels of Rag1 and Rag2 mRNA and Rag1 protein, as well as mRNA and protein levels of Gadd45α (24), which had been shown to signal Rag1 and Rag2 transcription in a pre-B cell line (25). However, the asynchronous induction of RAG DSBs at Igκ alleles precluded investigating the kinetics or mechanism of RAG downregulation or a potential mechanistic link between suppression of Gadd45α protein and repression of Rag1 and Rag2 mRNA. We proposed that RAG DSB-induced feedback inhibition of RAG expression limits simultaneous RAG cleavage of distinct AgR alleles, helping to enforce mono-allelic assembly and expression (allelic exclusion) of Igκ, IgH, and TCRβ genes and to suppress oncogenic IgH, TCRβ, and TCRδ translocations during proliferation of pro-B and pro-T cells (24, 26). Consistent with this idea, we showed that mice lacking ATM develop higher than normal frequencies of lymphocytes with bi-allelic expression and/or translocations of Igκ, IgH, or TCRβ loci (26). The Sleckman lab found that RAG DSBs induced at one Igκ allele signal via ATM to activate expression of the hematopoietic-specific SpiC transcriptional repressor (27). SpiC inhibits RAG cleavage of the other Igκ allele by suppressing transcription-linked accessibility of Jκ gene segments to the RAG endonuclease (27). This body of work established the concept of DSB-induced feedback inhibition of Igκ recombination by RAG cleavage and demonstrated that this regulation involves ATM-dependent DDR signaling pathways that repress RAG expression and Igκ recombination potential.
Since DSBs induced in pre-B cells by ionizing radiation or etoposide can activate expression of some lymphocyte-specific genes induced by RAG DSBs (22, 28), we hypothesized that any type of DSB can signal inhibition of RAG expression and V(D)J recombination. In this study, we demonstrate that DSBs induced in primary lymphocytes by IR or etoposide signal through ATM and Nemo to transcriptionally repress Rag1 expression and inhibit RAG cleavage of Igκ loci and RSSs integrated at another genomic location. While we were preparing our manuscript, the Guikema lab reported that DSBs induced in immortalized and transformed human and mouse pre-B cell lines signal via ATM and the FOXO1 transcription factor to inhibit RAG expression and activity (29). We discuss how our study corroborates the central findings of the Guikema lab and presents novel data that provide further insights into the mechanisms and the scope by which lymphocytes direct DSB-induced feedback inhibition of V(D)J recombination. We also outline the implications of both studies to mechanisms that orchestrate the differentiation of mono-specific lymphocytes while suppressing oncogenic AgR locus translocations.
Materials and Methods
Mice
All mice used within this study were housed, bred, and used under pathogen-free conditions at the Children's Hospital of Philadelphia (CHOP). All experiments were performed using 4–6 week old mice using both male and female mice. Experimental mice were euthanized by exposure to CO2 followed by cervical dislocation. Animal husbandry and experiments were performed in accordance with national guidelines and regulations and approved by the CHOP Institutional Animal Care and Use Committee. The EμBCL2 (30), Rag1D708A (31), Mb1Cre+Atmflox/flox (24), VavCre+p53flox/flox(32), Mb1Cre+Nemoflox/flox (33) mice were on a mixed 129S1/SvImJ and C57BL/6 background, while Rag1−/− (34) and Rag2−/− (35) mice were on the C57BL/6 background. The wild-type control mice were of the appropriate background.
Irradiation
All mice were subject to irradiation using an XRAD320 X-ray irradiator (Precision X-Ray) and a dose rate of 0.74 Gy/min. Primary cells and cell lines were subject to irradiation using a Gammacell 1000 Cs-137 irradiator (Nordion Inc) and a dose rate of 1.8 Gy/min.
qRT-PCR quantification of Rag1, Rag2, and p21 mRNAs
Non-irradiated and irradiated cells were harvested at indicated time points and immediately lysed in Trizol (Life Technologies) and processed according to the manufacturer's instructions. Isolated RNA was treated with DNase (Promega) according to manufacturer directions, primed with random nonamers (New England Biolabs, NEB), and reverse transcribed with M-MuLV (NEB) to generate cDNA. The cDNAs were then used as template for qRT-PCR reactions were performed with SYBR green mastermix (Applied Biosystems) and run on an Applied Biosystems 7500 Fast machine. Values were normalized to housekeeping genes as indicated in the text and fold change was determined by the ΔΔCT method. The primers used for qRT-PCR reactions are listed in Supplementary Table 1. When pre-B cells were subject to irradiation, non-irradiated pre-B cells also were transported to the irradiator, but not exposed to ionizing radiation along with irradiated samples. The irradiated and non-irradiated pre-B cells were each placed back into culture until they were harvested. Since pre-B cells were transported to and from the irradiator at room temperature, the basal levels of Rag1, Rag2, and p21 mRNAs were lower at 1 hour after the irradiation time point than at 4 hours after the irradiation time point. To inactivate the ATM kinase, the KU55933 ATM kinase inhibitor was added to media at a concentration of 15 µM To block new protein synthesis, the cycloheximide ribosome inhibitor was added to media at a concentration of 10 µg/mL.
Primary pre-B cell cultures
Primary bone marrow (BM) cells were harvested by flushing BM from all leg bones of at least four mice of the appropriate genotype for each experiment. These BM cells were cultured for 3–5 days in RPMI supplemented with 10% FBS, 10 mM HEPES, 1 × non-essential amino acids, 1 mM L-glutamine, 1 mM sodium pyruvate, 100 U/mL pen-strep, 30 µM β-mercaptoethanol and 50 ng/mL IL-7. Cells were plated at a density of 5 million cells per mL of media. Each day, cells were harvested and put back into culture in fresh media at a density of 5 million cells per mL. To induce G1 arrest and activate transcription of Rag1 and Rag2 by IL7 withdrawal, cells were pelleted by centrifugation, re-suspended in the same media lacking IL7 at a density of 5 million cells per mL, and treated as described for each experiment.
Western blot analysis of Rag1, Rag2, and actin proteins
Pre-B cells were washed with PBS and re-suspended in ice cold lysis buffer (20 mM Tris pH 7.4, 20 mM glycerol phosphate, 10 mM sodium orthovanadatem, 10% glycerol, 0.5 mM EDTA, 0.5 mM MgCl2, 200 mM NaCl, and 0.2% Triton-X) and then sonicated at intervals of 30 seconds on and 30 seconds off for 8 minutes at 4°C. The sonicated cells were incubated on ice for 10 minutes and then centrifuged to remove insoluble material. Laemmli buffer was added and then samples were boiled for 5 minutes. Lysates from equivalent numbers of pre-B cells were loaded and run on NuPage tris-glycine gels (Life Technologies). Electrophoresed proteins were transferred to Immobilon-FL PVDF membrane (EMD Millipore). Membranes were blocked with Odyssey blocking buffer (Li-Cor) according to manufacturer’s instructions. Antibodies used are: anti-Rag1 or anti-Rag2 monoclonal antibody (36); anti-actin antibody (Sigma AV40173); anti-Gadd45α (Santa Cruz sc-797). Blots were washed and incubated with appropriate IRDye800 secondary antibodies (LiCor) according to manufacturer’s instructions. Following washing, blots were scanned on an Odyssey infrared scanner (Li-Cor).
Click-It analysis of Rag1 and Rag2 mRNA turnover and Rag1 and Rag2 transcription
These assays were conducting using Click-It® Nascent RNA Capture kits (Life Technologies). For mRNA turnover assays, ethynyl uridine (EU) was added to medium of IL7 withdrawn pre-B cell cultures at a final concentration of 0.2 mM for the final 16 hours of culture time. Pre-B cells were washed, placed into media lacking EU, and then split into pools that were immediately irradiated or left non-irradiated. Cell pools were collected for RNA isolation immediately before EU removal or at indicated times after EU removal or EU removal and irradiation. For transcriptional assays, pre-B cells were grown in media lacking EU. Cultures were split into pools that were either irradiated or left non-irradiated. Immediately after irradiation of some pools, EU was added to the media of all pools at a final concentration of 0.5 mM. After the indicated times, cells were collected for RNA isolation. For both assays, RNA was isolated using Trizol (Life Technologies) according to the manufacturer's instructions. Click chemistry and streptavidin pull down of EU-labeled RNA were performed according to the Click-It Nascent RNA Capture kit’s instructions. Pulled-down RNA was reverse transcribed and analyzed by qRT-PCR as described above. The loading controls for mRNA turnover and transcription experiments were 18S RNA and Hprt mRNA, respectively.
Abelson pro-B cell cultures
The EμBCL2 (A2) (37) and Artemis−/−EμBCL2 (Art2.1) (38) Abelson transformed and immortalized pro-B cell lines were previously described. We made the EμBCL2-pINV (39) Abelson pro-B cell line using the same procedures that we previously used to generate cell lines with recombination substrates (40). Abelsons pro-B cells were cultured in RPMI media supplemented with 10% FBS, 100 U/mL pen-strep, and 30 µM β-mercaptoethanol To induce G1 cell cycle arrest, differentiation into pre-B cells, expression of Rag1 and Rag2 mRNA, and V(D)J recombination, 5 µM of STI571 was added to the culture media. To induce DSBs by genotoxic drugs, etoposide was added to the media at a concentration of 10 µg/mL or bleomyin was added at a concentration of 5 µM.
Southern blot analysis
Southern blotting was performed as described (38). Briefly, 15–20 µg of genomic DNA was digested with Sac1-HF (NEB) and EcoR1-HF (NEB). Southern blot membranes were probed with a 3’Jκ probe and then a 5'Hprt probe as a loading control. The intensities of bands were quantified using ImageJ software (NIH). For Artemis−/−EμBLC2 cells, the percentage of Jκ cleavage at each time point was calculated by dividing the total intensities of Jκ coding end bands by the combined intensities of the germline Jκ band and Jκ coding end bands (40). For EμBLC2 cells, the percentage of Jκ cleavage at each time point was calculated by dividing the intensity of the germline Jκ band by the intensity of the 5'Hprt control band.
Flow cytometry
For analysis of GFP expression in EμBCL2-pINV cells, cells were washed in FACS buffer (PBS with 3% FCS and 0.25 mM EDTA) and then stained with PE-conjugated anti-human CD4 antibody (BD Pharmigen, clone RPA-T4) to stain cells containing the substrate. Flow cytometry was conducted using an LSR-II flow cytometer (BD Biosciences) and data was analyzed using FlowJo 10.
Statistical analyses
All statistics were performed using GraphPad Prism 5 or Prism 7 software.
Results
The suppression of Rag1 and Rag2 mRNA in response to ionizing radiation is a prevalent response of immature lymphocytes
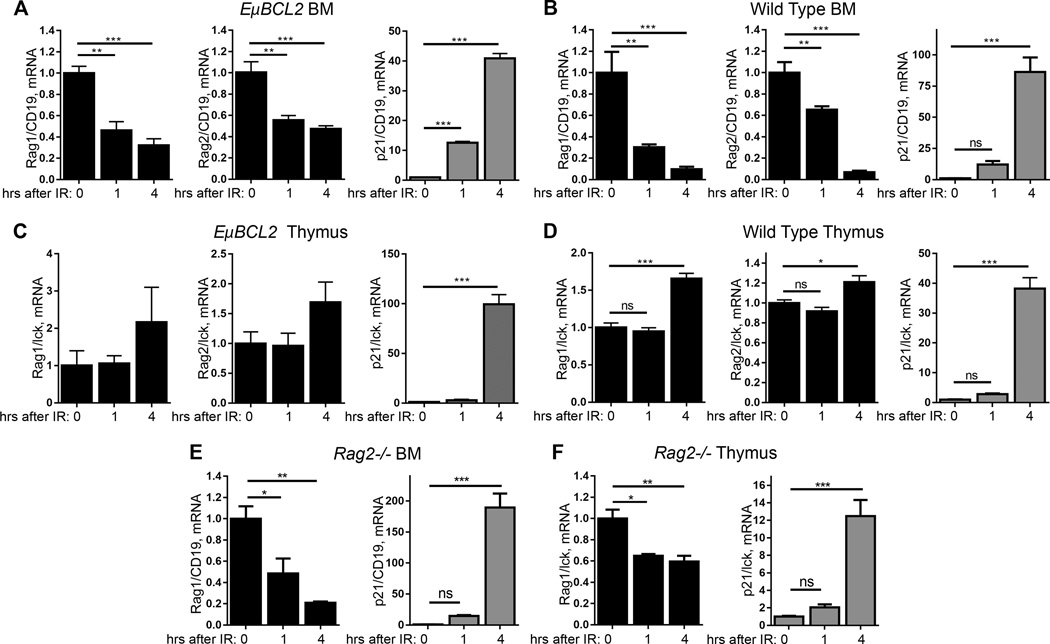
We first sought to determine the ability of DSBs induced by ionizing radiation to suppress Rag1 and Rag2 expression in developing B and T lymphocytes. Since we discovered that the DDR of immature lymphocytes frequently is abnormal in immortalized cell lines and in primary cells cultured ex vivo (41), we conducted initial experiments by exposing live mice to IR. To limit apoptosis of immature lymphocytes, we utilized EμBCL2 mice with transgenic expression of the anti-apoptotic BCL2 protein in all B and T lineage cells. We exposed these mice to IR and harvested cells from their thymuses and bone marrow (BM) 1 or 4 hours later. We conducted qRT-PCR on RNA isolated from these cells or corresponding cells of non-irradiated EμBCL2 mice. We quantified levels of Rag1 and Rag2 mRNA relative to levels of the T lineage-specific Lck mRNA for thymus or the B lineage-specific Cd19 mRNA for BM. To control for the induction of DSBs, we quantified mRNA levels of the p21 cyclin dependent inhibitor gene whose transcription is activated by ATM and Tp53 following DSBs (1). We detected lower levels of Rag1 and Rag2 mRNAs and higher levels of p21 mRNA in BM from irradiated EμBCL2 mice as compared to BM from non-irradiated EμBCL2 mice (Fig. 1A). The levels of Rag1 mRNA were reduced by more than 50% at 1 and 4 hours after exposure to 10 Gy of ionizing radiation, while levels of Rag2 mRNA were reduced by ~50% at both times (Fig. 1A), indicating that immature B lymphocytes rapidly inhibit Rag1 and Rag2 expression in response to IR. In contrast, we observed no decrease in the level of Rag1 or Rag2 mRNA in thymuses from irradiated EμBCL2 mice relative to thymuses from non-irradiated EμBCL2 mice, despite induction of DSBs as reflected by elevated p21 mRNA levels (Fig. 1B). Indeed, the levels of Rag1 and Rag2 mRNA each were increased by ~40% at 4 hours after exposure to 10 Gy of ionizing radiation (Fig. 1B). We corroborated that IR rapidly downregulates Rag1 and Rag2 mRNA in BM but not thymuses of wild-type mice that lack the pro-survival EμBCL2 transgene (Fig. 1C, D). Collectively, these data suggest that DSBs induced by ionizing radiation inhibit Rag1 and Rag2 expression in developing B lymphocytes but not developing T lymphocytes. However, considering that BM and thymuses each contain a much greater number of pre-lymphocytes than pro-lymphocytes, this experimental approach likely is not sensitive enough to determine whether or not pro-lymphocytes downregulate Rag1 and Rag2 expression in response to DSBs.
FIGURE 1.
Immature B and T lymphocytes suppress Rag1 and Rag2 expression in response to ionizing radiation. (A) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in BM from non-irradiated EμBCL2 mice or EμBCL2 mice at indicated times after exposure to 10 Gy IR. Data are from one experiment with 3–4 mice for each timepoint. (B) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in thymuses from non-irradiated EμBCL2 mice or EμBCL2 mice at indicated times after exposure to 10 Gy IR. Data are from three independent experiments including a total of 3–6 mice per timepoint. (C) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in BM from non-irradiated wild-type mice at indicated times after exposure to 10 Gy IR. Data are from two independent experiments including a total of 4–5 mice per timepoint. (D) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in thymus from non-irradiated wild-type mice or wild-type mice at indicated times after exposure to 10 Gy IR. Data are from two independent experiments including a total of 4–5 mice per timepoint. (E) qRT-PCR quantification of Rag1 and p21 mRNA in BM from non-irradiated Rag2−/− mice or Rag2−/− mice at indicated times after exposure to 10 Gy IR. Data are from one experiment with 3 or 4 mice per timepoint. (F) qRT-PCR quantification of Rag1 and p21 mRNA in thymus from non-irradiated Rag2−/− mice or Rag2−/− mice at indicated times after exposure to 10 Gy IR. Data are from one experiment with 3 or 4 mice per timepoint. (A–F) Data averages are shown with error bars indicating SEM. p-values calculated using Dunnett's post-test after ANOVA. *p<0.05, **p<0.01, ***p<0.001.
We next sought to determine the effects of DSBs on Rag1 and Rag2 expression in primary pro-B and pro-T cells. For this purpose, we turned to Rag1−/− and Rag2−/− mice in which lymphocyte development is blocked at the pro-B and pro-T cell stages (34, 35). The Rag1−/− mice have a drug-resistance gene driven by a constitutive promoter in place Rag1 coding sequences, while the Rag2−/− mice have a loxP site in place of Rag2 coding sequences. We observed decreased levels of Rag1 mRNA in BM from irradiated Rag2−/− mice relative to BM from non-irradiated Rag2−/− mice (Fig. 1E). The levels of Rag1 mRNA in Rag2−/− BM were reduced ~75% at 1 hour after 10 Gy IR and ~90% at 4 hours after 10 Gy IR. Notably, we detected lower levels of Rag1 mRNA in thymuses from irradiated Rag2−/− mice as compared to thymuses from non-irradiated Rag2−/− mice (Fig. 1F). The levels of Rag1 mRNA in Rag2−/− thymuses were reduced ~40% at 1 and 4 hours after 10 Gy IR. Consistent with these findings, we observed that IR downregulated Rag2 mRNA in BM and thymus of Rag1−/− mice to a similar extent as Rag1 mRNA in Rag2−/− mice. (Supplemental Fig. 1). The results of our experiments with Rag1−/− and Rag2−/− mice suggest that DSBs downregulate Rag1 and Rag2 expression in primary pro-B and pro-T cells.
The induction of DSBs in pre-B cells downregulates levels of Rag1 and Rag2 mRNAs
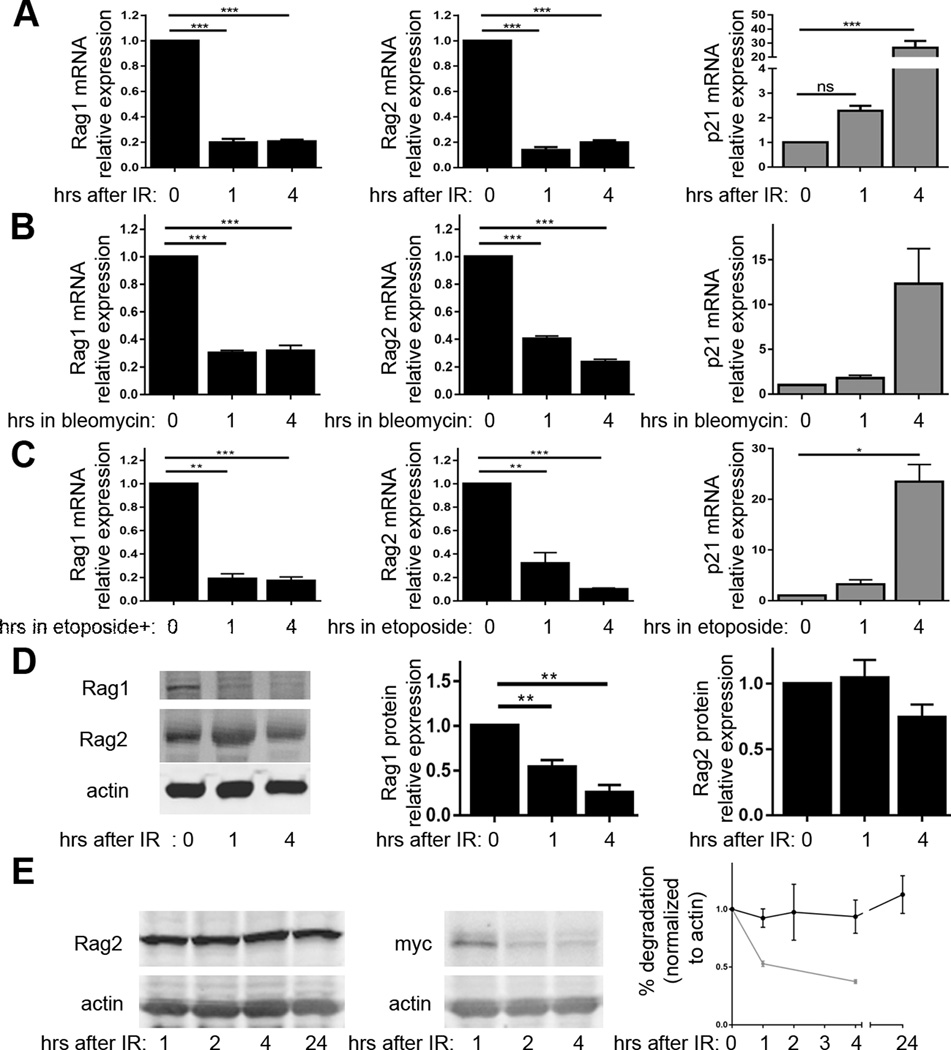
To validate that DSBs induced by ionizing radiation in primary pre-B cells downregulate Rag1 and Rag2 expression, we turned to the ex vivo culture system wherein we discovered that RAG DSBs inhibit Rag1 and Rag2 expression (24). The culture of primary mouse BM in media containing IL7 cytokine selectively promotes survival and proliferation of pre-B cells, such that within a few days over 95% of cells in culture are pre-B cells(42). IL7 also suppresses Rag1 and Rag2 expression (43). Following removal of IL7, pre-B cells arrest in G1 phase and activate Rag1 and Rag2 transcription (42). Expression of the EμBCL2 transgene enables pre-B cells to survive and express Rag1 and Rag2 for several days after IL7 removal (42). We cultured pre-B cells from EμBCL2 mice, removed IL7 for 48 hours to arrest cells in G1 phase and induce Rag1 and Rag2 transcription, exposed cells to 4 Gy IR, and then isolated RNA at 1 or 4 hours following irradiation. In parallel, we harvested RNA from non-irradiated pre-B cells. We used qRT-PCR to quantify Rag1 and Rag2 mRNA levels relative to actin mRNA levels. We also quantified p21 mRNA to confirm the induction of DSBs. We detected lower levels of Rag1 and Rag2 mRNA and higher levels of p21 mRNA in irradiated EμBCL2 pre-B cells as compared to non-irradiated EμBCL2 pre-B cells (Fig. 2A). The levels of Rag1 and Rag2 mRNA each were reduced ~80% at 1 and 4 hours following exposure to ionizing radiation (Fig. 2A), indicating that pre-B cells rapidly inhibit Rag1 and Rag2 expression in response to IR.
FIGURE 2.
DSBs induced in pre-B cells suppress downregulate Rag1 and Rag2 mRNA and Rag1 protein. (A) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in non-irradiated EμBCL2 pre-B cells or irradiated EμBCL2 pre-B cells at indicated times after exposure to 4 Gy IR. Data are from 11 independent experiments. (B) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in EμBCL2 pre-B cells or EμBCL2 pre-B cells at indicated times after addition of 5 µM bleomycin to culture media. Data are from 3 independent experiments. (C) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in EμBCL2 pre-B cells or EμBCL2 pre-B cells at indicated times after addition of 10 mg/mL etoposide to culture media. Data are from 3 independent experiments. (D) Representative western blot analysis and graphs depicting quantification of Rag1 and Rag2 protein in non-irradiated EμBCL2 pre-B cells or irradiated EμBCL2 pre-B cells at indicated times after exposure to 4 Gy IR. Data are from 8 independent experiments. (E) Representative western blot analysis and graphs depicting quantification of Rag2 and c-Myc protein in cycloheximide-treated non-irradiated EμBCL2 pre-B cells or irradiated EμBCL2 pre-B cells at indicated times after exposure to 4 Gy IR. Data are from 3 independent experiments. (A–E) Data are normalized to 1.0 for non-irradiated cells within each experiment. For irradiated cells, data averages are shown with error bars indicating SEM. p-values calculated using Dunnett's post-test after ANOVA. **p<0.01, ***p<0.001.
While our data suggest that DSBs induced by ionizing radiation inhibit Rag1 and Rag2 expression, they cannot rule out that other types of cellular damage caused by IR promote this response. To address this issue, instead of exposing pre-B cells to IR, we exposed cells to drugs that predominantly induce DSBs. Bleomycin is a genotoxic drug that induces DSBs throughout the cell cycle, but also damages RNA (44). We observed lower levels of Rag1 and Rag2 mRNAs and higher levels of p21 mRNA in EμBCL2 pre-B cells treated with bleoymcin relative to EμBCL2 pre-B cells treated with only vehicle (Fig. 2B). The levels of Rag1 and Rag2 mRNA each were reduced ~60–80% at 1 and 4 hours with treatment with bleomycin (Fig. 2B), indicating that DSBs induced in pre-B cells rapidly inhibit Rag1 and Rag2 expression. Etoposide is a genotoxic drug whose only demonstrated biological effect is that it inhibits re-ligation of DNA by type II topoisomerase enzymes, causing accumulation of DSBs (45). Although etoposide causes DSBs mainly in S phase cells (45), this genotoxic agent does induce DSBs in G1 arrested pre-B cells (46). We detected lower levels of Rag1 and Rag2 mRNAs and higher levels of p21 mRNA in EμBCL2 pre-B cells treated with etoposide relative to EμBCL2 pre-B cells treated with only vehicle (Fig. 2C). The levels of Rag1 and Rag2 mRNA each were reduced ~80% at 1 and 4 hours with treatment with etoposide (Fig. 2C), indicating that DSBs induced in pre-B cells rapidly inhibit Rag1 and Rag2 expression. Considering that ionizing radiation, bleomycin, etoposide, and RAG cleavage of Igκ loci (24) each suppress Rag1 and Rag2 mRNA levels, we conclude that any type of DSB activates DSB-induced feedback inhibition of Rag1 and Rag2 expression.
DSBs induced in pre-B cells inhibit expression of Rag1 protein but not Rag2 protein
For DSB-induced feedback inhibition of Rag1 and Rag2 expression to suppress V(D)J recombination, Rag1 and/or Rag2 protein levels must be diminished as a result of decreased Rag1 and Rag2 mRNA. Previous studies reported that the Rag1 and Rag2 proteins each have a half-life of ~10–12 minutes (29, 47, 48). Considering these reports and our observed kinetics and extents of Rag1 and Rag2 mRNA loss in response to IR, one would predict appreciable decreased levels of Rag1 and Rag2 protein at 1 hour after irradiation. To confirm this prediction, we conducted western blot analysis of IL7 withdrawn primary pre-B cells at 1 and 4 hours after their exposure to 4 Gy of IR. In parallel, we assayed Rag1 and Rag2 protein in non-irradiated pre-B cells. We also conducted western blot analysis for actin protein and quantified Rag1 and Rag2 protein levels relative to actin protein. We observed reduced levels of Rag1 protein in irradiated EμBCL2 pre-B cells as compared to non-irradiated EμBCL2 pre-B cells (Fig. 2D). Consistent with our prediction, Rag1 protein was reduced ~50% at 1 hour after 4 Gy IR and ~75% at 4 hours after 4 Gy IR. In marked contrast, we detected no change in Rag2 protein levels after irradiation of EμBCL2 pre-B cells (Fig. 2D), which is not consistent with our prediction. Regardless, as both Rag1 and Rag2 are required for RAG endonuclease activity (49), the downregulation of Rag1 protein levels in response to the induction of DSBs could be sufficient to inhibit V(D)J recombination.
Endogenous Rag2 protein in primary pre-B cells has an immeasurably long half-life
We found the retention of Rag2 protein following the loss of Rag2 mRNA interesting, particularly because it was not predicted from the published half-life of Rag2 protein. The possible explanations of our finding are that the basal half-life of Rag2 protein in primary pre-B cells is much longer than ~45 minutes and/or DSBs induced in primary pre-B cells increase the half-life of Rag2 protein. The Rag2 protein is expressed in G1 but not in subsequent cell cycle phases due to its phosphorylation and consequent degradation upon S phase entry (48, 50, 51), supporting the possibility that Rag2 protein might be very stable in G1 phase cells. The published half-life of Rag2 protein is only 10–12 minutes, however this value was obtained by measuring asynchronously cycling non-lymphoid cell lines with ectopic expression of Rag2 (48, 51). Therefore, we investigated the basal half-life of Rag2 protein in G1-arrested primary pre-B cells. We conducted western blot analysis of IL7 withdrawn primary pre-B cells at increasing times after addition of cycloheximide to block new protein synthesis. As a positive control for inhibition of protein synthesis, we also conducted western blot analysis of the c-Myc protein, which has a half-life of less than one hour (52). Strikingly, we detected no loss of Rag2 protein at any time point after cycloheximide addition, despite rapid loss of c-Myc protein (Fig. 2E). In this context, we could not calculate the half-life of Rag2 protein because we detected no loss of Rag2 protein levels at 24 hours after cycloheximide (Fig. 2E), following which pre-B cells begin to die from lack of new protein synthesis. Since we could not detect loss of Rag2 protein in non-irradiated cells, we were not able to ascertain the potential effect of DSBs on Rag2 protein stability.
DSBs induced in pre-B cells signal rapid transcriptional repression of Rag1 and Rag2
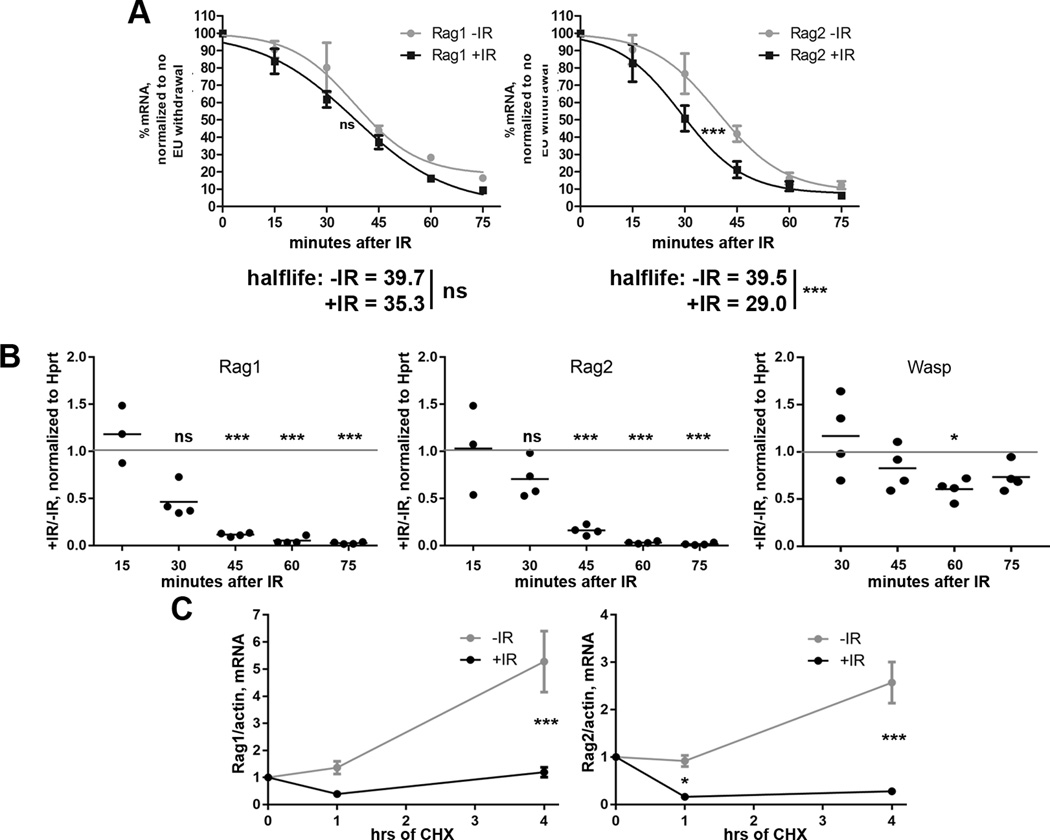
We next sought to elucidate mechanisms by which DSBs cause downregulation of Rag1 and Rag2 mRNA. This loss of mRNA could be due to increased turnover of Rag1 and Rag2 mRNAs and/or repression of Rag1 and Rag2 transcription. We utilized Click-iT mRNA capture technology to investigate these possibilities. Click-iT® mRNA capture incorporates ethylene uridine (EU) into RNAs during transcription; EU-labeled RNAs are conjugated with biotin, isolated by streptavidin beads, and quantified by qRT-PCR. To measure turnover of Rag1 and Rag2 mRNAs in irradiated and non-irradiated primary pre-B cells, we added EU to the media of EμBCL2 pre-B cell cultures for the last 16 hours of IL7 withdrawal to fully label existing mRNAs. We washed out EU immediately before exposure of cells to 4 Gy IR to block further EU incorporation so that we could measure the amounts of EU-labeled Rag1 and Rag2 mRNA remaining over time in irradiated and non-irradiated cells. We found that the Rag1 and Rag2 mRNAs each have a half-life of ~40 minutes (Fig. 3A), similar to published results using nuclear run-on assays on primary pre-B cells (53). At all times assayed, we detected no difference in the levels of labeled Rag1 mRNA between irradiated and non-irradiated cells (Fig. 3A), indicating that pre-B cells do not increase turnover of Rag1 mRNA in response to DSBs. In contrast, we observed lower levels of labeled Rag2 mRNA at 30 and 45 minutes following irradiation (Fig. 3A), reflecting that DSBs induced in pre-B cells decrease the half-life of Rag2 mRNA from ~40 minutes to ~30 minutes. To investigate Rag1 and Rag2 transcription, we added EU to IL7-withdrawn EμBCL2 pre-B cells immediately after exposure to 4 Gy IR. We also added EU to non-irradiated IL7-withdrawn EμBCL2 pre-B cells. Over time, we measured the amounts of newly synthesized, EU-labeled Rag1 and Rag2 mRNA, and the loading control Hprt mRNAs. In this experiment, the balance of transcription and mRNA turnover determines the amount of a labeled mRNA. By 45 minutes after irradiation, we observed near complete loss of labeled Rag1 and Rag2 mRNAs in irradiated cells relative to non-irradiated cells (Fig. 3B). Considering the ~30–40 minute turnover of Rag1 and Rag2 mRNAs in irradiated pre-B cells (Fig. 3A), the kinetics and extents by which EU-labeled Rag1 and Rag2 mRNAs are lost in irradiated cells suggests that DSBs cause an immediate halt of Rag1 and Rag2 transcription.
FIGURE 3.
DSBs induced in primary pre-B cells immediately repress Rag1 and Rag2 transcription. (A) qRT-PCR quantification of EU-labeled Rag1 and Rag2 mRNA levels relative to EU-labeled 18S RNA levels in non-irradiated EμBCL2 pre-B cells or irradiated EμBCL2 pre-B cells at indicated times after EU washout and/or exposure to 4 Gy of IR. Data are from 4 independent experiments. Data averages are shown with error bars the SEM. Prism 5 was used to calculate best-fit curves, half-lives and p-values. ***p<0.001. (B) qRT-PCR quantification of EU-labeled Rag1, Rag2, and Wasp mRNA levels relative to EU-labeled Hprt mRNA levels in non-irradiated EμBCL2 pre-B cells and irradiated EμBCL2 pre-B cells at indicated times after addition of EU and exposure to 4 Gy of IR. Data are presented as the ratio of relative levels of each mRNA in irradiated cells compared to non-irradiated cells. The dotted line represents a value of 1, which would indicate that IR had no effect on the transcription rate of a gene. Data are from 4 independent experiments. p-values for whether the average ratio at each time point is different from 1.0 was determined using one-tailed T test with Bonferroni's correction for multiple testing. ***p<0.001. (C) qRT-PCR quantification of Rag1 and Rag2 mRNA in cycloheximide-treated non-irradiated EμBCL2 pre-B cells or irradiated EμBCL2 pre-B cells at indicated times after exposure to 4 Gy IR. Data are from 3 independent experiments. Data averages are shown with error bars the SEM. p-values were calculated by T test with Bonferroni's correction for multiple testing. ***p<0.001.
To investigate whether DDR activation in immature lymphocytes leads to repression of genes non-essential to the genotoxic stress response, we also measured the amount of newly synthesized, EU-labeled Wasp mRNA since Wasp has no documented role in the DDR and irradiation of primary pre-B cells has no effect on steady-state mRNA levels (data not shown). Our analysis reveals that irradiation of primary pre-B cells has minimal effect on the synthesis of Wasp mRNA (Fig. 3B). Thus, we conclude that Rag1 and Rag2 transcription is specifically targeted by the DDR, and not simply part of a generalized inhibition of transcription in the presence of widespread DNA damage.
Consistent with the rapid timeframe of Rag1and Rag2 transcriptional suppression, inhibition of new protein synthesis by cycloheximide treatment does not prevent the loss of Rag1 and Rag2 mRNA levels following irradiation of EμBCL2 pre-B cells (Fig. 3C). This finding suggests that DSBs suppress Rag1 and Rag2 transcription through factors constitutively expressed in primary pre-B cells. Collectively, these data are consistent with repression of Rag1 and Rag2 transcription being the predominant mechanism by which pre-B cells signal DSB-induced loss of Rag1 and Rag2 expression.
DSB-induced downregulation of Rag1 and Rag2 expression requires the ATM kinase
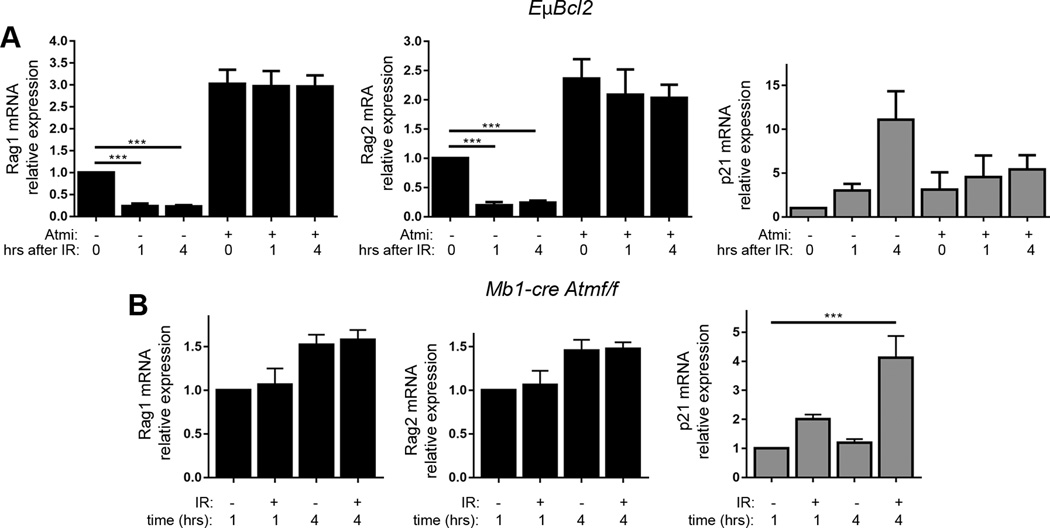
We next launched a line of research to elucidate the factors by which DSBs induced in primary pre-B cells suppress Rag1 and Rag2 transcription. We previously reported that RAG DSBs induced in pre-B cells signal via ATM to downregulate Rag1 and Rag2 expression (24). Therefore, we investigated the role of ATM through chemical inhibition of the ATM kinase or genetic inactivation of the ATM protein. First, we added the ATM kinase inhibitor KU55933 or vehicle only at the same time that we removed IL7 from EμBCL2 pre-B cell cultures; 48 hours later we exposed cells to 4 Gy IR or left them untreated, and then harvested cells for qRT-PCR quantification of Rag1 and Rag2 mRNA. We also quantified p21 mRNA as a control for both induction of DSBs and inactivation of the ATM kinase, since genetic deletion of ATM impairs but does not block the ability of DSBs to induce p21 mRNA levels (54). We detected ~2–3-fold higher levels of Rag1 and Rag2 mRNAs in non-irradiated cells treated with KU55933 versus vehicle only (Fig. 4A), likely reflecting ATM-dependent suppression of Rag1 and Rag2 transcription from DSBs induced by RAG during Igκ recombination and by other transcription and cellular metabolites. Notably, the addition of KU55933 also blocked downregulation of Rag1 and Rag2 mRNA following irradiation (Fig. 4A). Next, we analyzed primary pre-B cells cultured from Mb1Cre+Atmflox/flox mice with B lineage-specific conditional deletion of Atm initiating at the pro-B cell stage (24). Unirradiated Mb1Cre+Atmflox/flox IL-7 withdrawn pre-B cells expressed ~2-fold higher levels of Rag1 and Rag2 mRNAs compared to EμBCL2 pre-B cells (data not shown), possibly reflecting ATM-dependent inhibition of Rag1 and Rag2 expression from DSBs. Moreover, irradiation of Mb1Cre+Atmflox/flox pre-B cells caused no change in Rag1 and Rag mRNA levels relative to non-irradiated Mb1Cre+Atmflox/flox pre-B cells (Fig. 4B). The retention of Rag1 and Rag2 mRNA from 1–4 hours after irradiation of pre-B cells that lack ATM indicates that IR-induced DSBs signal through ATM to downregulate Rag1 and Rag2 expression.
FIGURE 4.
ATM and Nemo are required for DSBs induced in pre-B cells to downregulate Rag1 and Rag2 expression. (A) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in non-irradiated EμBCL2 pre-B cells or irradiated EμBCL2 pre-B cells at indicated times after exposure to 4 Gy of IR. Cells were treated with 15 μM of the KU55933 ATM kinase inhibitor or vehicle (DMSO) for 48 hours prior to irradiation or harvesting of non-irradiated cells. Data are from 8 independent experiments. Data are normalized to 1.0 for non-irradiated, vehicle-treated cells within each experiment. For other conditions, data averages are shown with error bars indicating SEM. p-values were calculated by Dunnett's post-test after ANOVA. ***p<0.001. (B) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in non-irradiated Mb1Cre+Atmflox/flox pre-B cells or irradiated Mb1Cre+Atmflox/flox pre-B cells at indicated times after exposure to 4 Gy IR. Data are from 3 independent experiments. Data are normalized to 1.0 for non-irradiated cells harvested one hour following IR exposure of irradiated samples. For other samples, data averages are shown with error bars indicating SEM. p-values were calculated by Dunnett's post-test after ANOVA. ***p<0.001.
DSB-induced Rag1 and Rag2 suppression does not depend on suppression of Gadd45α expression
The intracellular signaling pathways through which ATM represses gene transcription remain incompletely characterized. We previously reported that RAG DSBs induced in primary pre-B cells signal via ATM to decrease levels of Gadd45α mRNA and protein (24). Since Gadd45α had been shown to signal Foxo1-dependent transcription of Rag1 and Rag2 in an immortalized mouse pre-B cell line (25), our findings supported a model wherein DSBs suppress Rag1 and Rag2 transcription through ATM-mediated downregulation of Gadd45α mRNA. To investigate this model, we studied the effects of DSBs induced by IR in primary pre-B cells on Gadd45α mRNA levels. Although we found that Gadd45α mRNA levels are decreased following irradiation of EμBCL2 pre-B cells (Supplemental Fig. 2A, B), the slow kinetics and limited extent of this response suggest that downregulation of Gadd45α mRNA is not a driving force in suppressing Rag1 and Rag2 expression (compare Fig. 3B and Supplemental Fig. 2B). Consistent with this notion, we observed that ectopic retroviral expression of Gadd45α in a mouse pre-B cell line does not prevent DSB-induced inhibition of Rag1 and Rag2 expression (Supplemental Fig. 2C). However, it is important to note that while our data argue against a role for downregulation of Gadd45α expression in DSB-induced inhibition of Rag1 and Rag2 transcription in response to DSBs, our experiments do not address the possibility for inactivation of Gadd45α protein activity by post-translational modifications or other mechanisms.
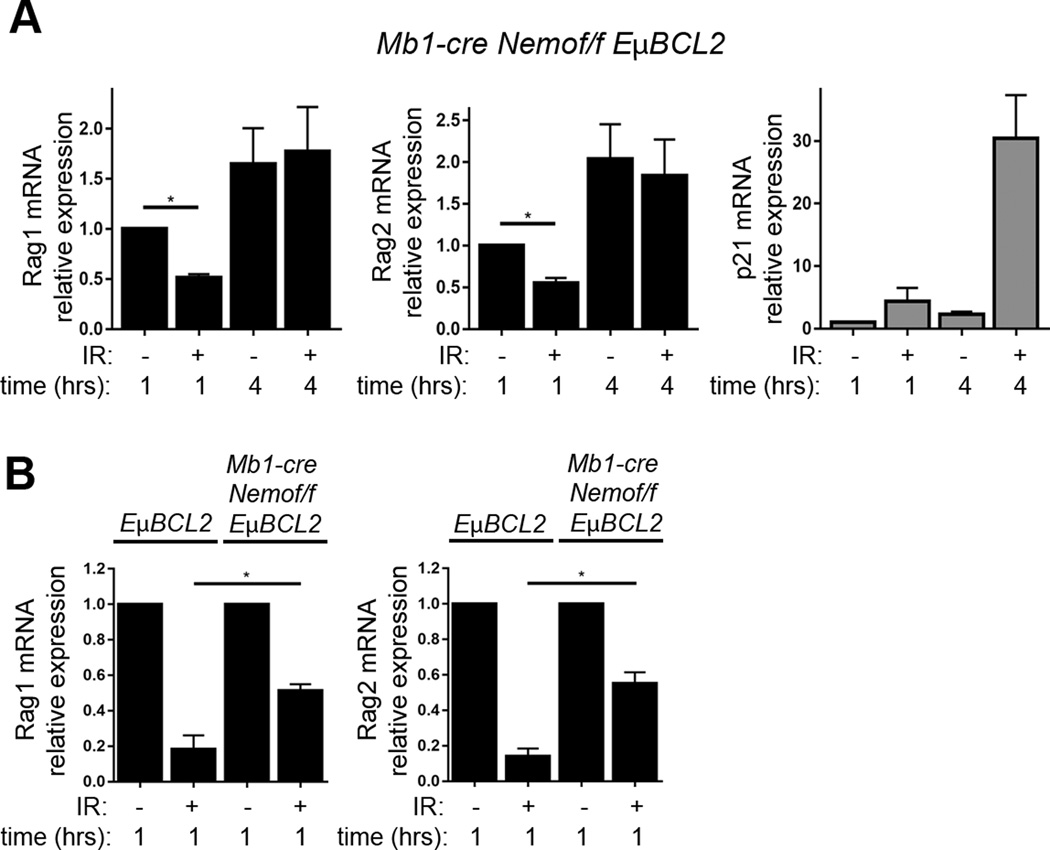
DSB-induced downregulation of Rag1 and Rag2 expression requires the NF-κB essential modulator protein
In pre-B cells, ATM activates transcription of target genes through Tp53-mediated or Nemo-mediated intracellular signaling pathways (1, 22). Thus, we next investigated the roles of Tp53 and Nemo in suppressing Rag1 and Rag2 expression following the induction of DSBs in primary pre-B cells. We observed normal downregulation of Rag1 and Rag2 mRNAs following irradiation of primary pre-B cells cultured from VavCre+Tp53flox/flox mice with hematopoietic lineage-specific deletion of Tp53 (Supplemental Fig. 2E)(32). In contrast, we detected impairment of this DSB response in pre-B cells from Mb1Cre+Nemoflox/floxEμBCL2 mice with B lineage-specific Nemo deletion and ectopic BCL2 expression to enhance survival of Nemo-deficient pre-B cells (Fig. 5A)(33). Although the Rag1 and Rag2 mRNA levels at 4 hours following IR were equivalent to unirradiated cells, the levels at 1 hour after IR were decreased as compared to unirradiated cells (Fig. 5A). However, this downregulation of Rag1 and Rag2 mRNA in Mb1Cre+Nemoflox/floxEμBCL2 pre-B cells at 1 hour following IR is to a lesser extent than in EμBCL2 pre-B cells (Fig. 5B). Our data indicate that Nemo is required for normal DSB-induced feedback inhibition of Rag1 and Rag2 expression in pre-B cells, with the absence of Nemo resulting in slower kinetics by which pre-B cells downregulate Rag1 and Rag2 expression upon irradiation. This finding suggests a two-phase regulation of Rag1 and Rag2 transcription in response to DSBs: an early initiation phase that is partially dependent on Nemo and a later maintenance phase that requires Nemo expression.
FIGURE 5.
Nemo is required for pre-B cells to normally downregulate Rag1 and Rag2 expression in response to DSBs. (A) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in non-irradiated Mb1Cre+Nemoflox/floxEμBCL2 pre-B cells or irradiated Mb1Cre+Nemoflox/floxEμBCL2 pre-B cells at indicated times after exposure to 4 Gy IR. Data are from 3 independent experiments. p-values were calculated by Dunnett's post-test after ANOVA. *p<0.05. (B) qRT-PCR quantification of Rag1 and Rag2 mRNA in non-irradiated or irradiated pre-B cells from EμBCL2 or Mb1Cre+Nemoflox/floxEμBCL2 mice at 1 hour after exposure to 4 Gy IR. Data are from 3 independent experiments. p-value was calculated by T test. *p<0.05. (A–B) Data are normalized to 1.0 for non-irradiated cells harvested one hour following IR exposure of irradiated samples. For other samples, data averages are shown with error bars indicating SEM.
DSBs induced in pre-B cells by genotoxic drugs inhibit V(D)J recombination of Igκ loci and an integrated substrate
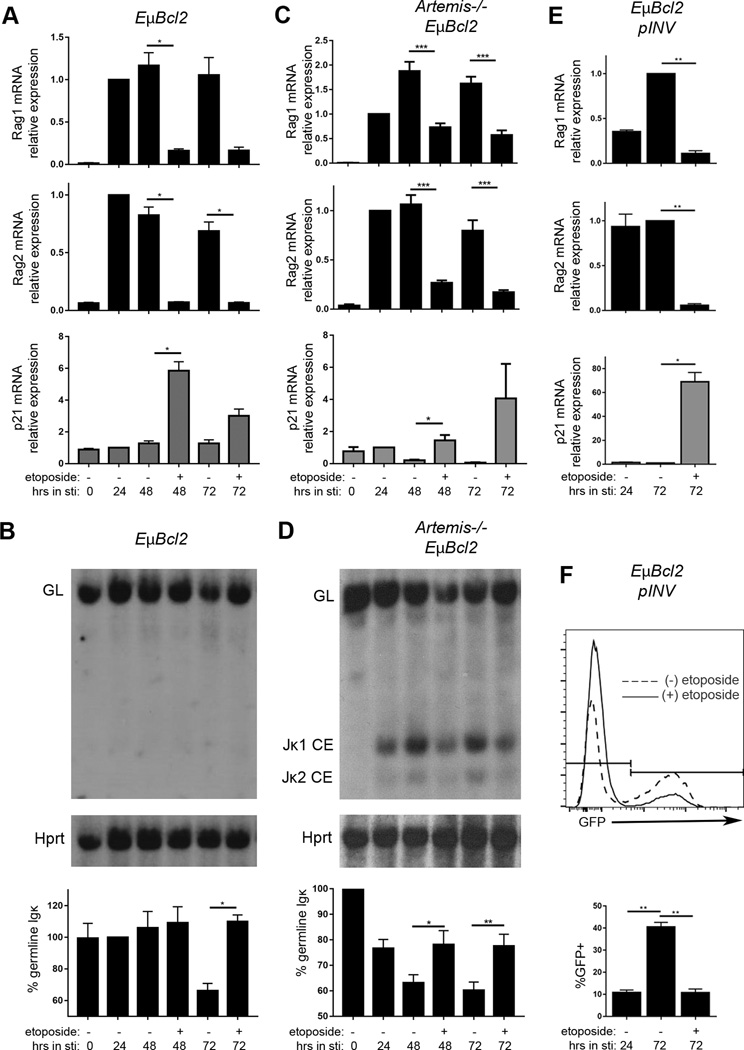
After demonstrating that DSBs induced by IR or genotoxic drugs inhibit Rag1 and Rag2 expression, we sought to test our hypothesis that these types of DSBs inhibit V(D)J recombination. For this purpose, we etoposide and bleomycin since these drugs are active for days in cell culture, which is the time frame over which can activate and monitor V(D)J recombination in primary pre-B cells or pre-B cell lines. We first attempted to study the effect of etoposide-induced DSBs on Igκ recombination in primary EμBCL2 pre-B cells; unfortunately, all of the etoposide doses that we tested caused substantial apoptosis. To circumvent this obstacle, we turned to the Abelson transformed (abl) pro-B cell lines that we had previously used to identify RAG DSB-induced feedback inhibition of Igκ recombination and to study additional cellular responses to RAG DSBs (38, 40). In normal media, Abl pro-B cells proliferate and only sporadically express Rag1 and Rag2 and recombine Igκ loci (55). Addition of the STI571 Abelson kinase inhibitor causes G1 cell cycle arrest and differentiation of pre-B cells that express Rag1 and Rag2 and recombine Igκ loci (55). Expression of the EμBCL2 transgene enables Abl cells to survive RAG DSBs and transcribe Rag1 and Rag2 for several days (37). Moreover, the addition of etoposide to STI571-treated, G1-arrested Abl cells induces DSBs (46). We found that addition of etoposide at 24 hours after STI571 addition had minimal effects on G1 arrest and survival of Abl cells over the next 48 hours. Consistent with our data from primary EμBCL2 pre-B cells (Fig. 2B), the addition of etoposide to EμBCL2 Abl cells caused downregulation of Rag1 and Rag2 mRNA and upregulation of p21 mRNA (Fig. 6A). The small size of the Jκ cluster enables the use of Southern blot analysis to monitor Igκ recombination. In EμBCL2 Abl cells, we measure Igκ recombination by quantifying decreased intensity of the germline Jκ band relative to the intensity of control band from a non-rearranging locus. Our Southern blot analysis revealed that the addition of etoposide 24 hours after activating Igκ recombination in EμBCL2 Abl cells inhibited further Igκ recombination over the next 48 hours (Fig. 6B). We conducted the same experiment with EμBCL2 pre-B cell lines lacking the Artemis NHEJ factor, which is required for coding join formation (56). The addition of etoposide to Artemis−/−EμBCL2 Abl cells caused downregulation of Rag1 and Rag2 mRNA and upregulation of p21 mRNA (Fig. 6C). In Artemis−/−EμBCL2 Abl cells, we measure RAG cleavage of Igκ loci by quantifying the relative intensity of germline Jκ bands and Jκ coding end bands (57). We found that addition of etoposide 24 hours after activating RAG cleavage of Igκ loci in Artemis−/−EμBCL2 Abl cells blocked further RAG cleavage of Igκ loci over the next 48 hours (Fig. 6D). We repeated this same experiment by substituting bleomycin for etoposide. We observed that the addition of bleomycin 24 hours after activating RAG cleavage of endogenous Igκ loci in Artemis−/−EμBCL2 Abl cells also blocked further RAG cleavage of Igκ loci over the next 48 hours (Supplemental Figure 3). These data demonstrate that DSB induced by etoposide or bleomycin in pre-B cells activates DSB-induced feedback inhibition of RAG cleavage to initiate Igκ recombination.
FIGURE 6.
Etoposide inhibits recombination of endogenous Igκ loci and an integrated substrate. (A) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in EμBCL2 Abl cells untreated or treated with STI571 or STI571 and etoposide for the indicated amounts of time. Data are from 3 independent experiments. (B) Representative Southern analysis and graphical quantification of Igκ recombination in EμBCL2 Abl cells untreated or treated with STI571 or STI571 and etoposide for the indicated amounts of time. Data are from 3 independent experiments. (C) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in Artemis−/−EμBCL2 Abl cells untreated or treated with STI571 or STI571 and etoposide for the indicated amounts of time. Data are from 6 independent experiments. (D) Representative Southern blot analysis and graphical quantification of Jκ cleavage in Artemis−/−EμBCL2 Abl cells untreated or treated with STI571 or STI571 and etoposide for the indicated amounts of time. Data are from 6 independent experiments. (E) qRT-PCR quantification of Rag1, Rag2, and p21 mRNA in EμBCL2-pINV Abl cells treated with STI571 or STI571 and etoposide for the indicated amounts of time. Data are from 3 independent experiments. (F) Representative flow cytometric analysis and graphical quantification of pINV recombination in EμBCL2-pINV Abl cells treated with STI571 or STI571 and etoposide for the indicated amounts of time. Data are from 4 independent experiments. (A–F) Data averages are shown with error bars the SEM. p-values were determined by Dunnett's post-test after ANOVA. *p<0.05, **p<0.01, ***p<0.001.
The Sleckman laboratory recently reported a locus-specific trans-factor for RAG DSB-induced feedback inhibition of Igκ recombination that functions by downregulating transcription-linked accessibility of Jκ gene segments to the RAG endonuclease (27). In this context, the ability of etoposide to inhibit Igκ recombination could be due to downregulation of Rag1 and Rag2 expression and/or Jκ recombinational accessibility. Therefore, we thought it was important to determine whether DSB-induced feedback inhibition in pre-B cells also suppresses recombination of RSSs inserted at another genomic location. For this purpose, we turned to EμBCL2 Abl cells containing one chromosomally integrated copy of the pINV recombination substrate. The pINV substrate contains a constitutive promoter, and a downstream GFP cDNA oriented in the anti-sense reading frame that is flanked by compatible RSSs (37, 58). The orientation of these RSSs is such that V(D)J recombination inverts the GFP cDNA, leading to GFP expression (37, 58). Due to sporadic Rag1 and Rag2 expression during culture, a small fraction (~10%) of EμBCL2-pINV cells expresses GFP before the addition of STI571. The addition of etoposide but not vehicle only 24 hours after STI571 addition to EμBCL2-pINV Abl cells resulted in lower levels of Rag1 and Rag2 mRNA and higher levels of p21 mRNA after an additional 48 hours of culture (Fig. 6E). Reflecting this suppression of Rag1 and Rag2 expression, flow cytometry analysis revealed that etoposide suppressed generation of GFP+ cells (Fig. 6F). These data indicate that DSB-induced feedback inhibition of V(D)J recombination in pre-B cells occurs independent of Igκ-specific trans-factors.
Discussion
Our study demonstrates that immature lymphocytes downregulate Rag1 and Rag2 expression and inhibit V(D)J recombination in response to DNA damage. The suppression of Rag1 and Rag2 mRNA levels following ionizing radiation is a response shared by pro-T, pro-B, and pre-B cells. Where assayed, this response is dependent on the ATM kinase and also triggered by etoposide or bleomycin, providing strong evidence that immature lymphocytes experiencing DSBs inhibit Rag1 and Rag2 expression. Our use of a primary mouse pre-B cell culture system enabled us to elucidate that transcriptional repression, rather than induction of mRNA turn over, is the mechanism by which DSBs downregulate Rag1 and Rag2 expression. This inhibition is immediate, not requiring protein synthesis, and causes rapid loss in protein levels of Rag1 but not Rag2. Since both Rag1 and Rag2 protein are required for RAG endonuclease activity (12, 49), DSBs induced in pre-B cell lines by etoposide inhibit recombination of endogenous Igκ loci and a chromosomally integrated artificial substrate. These data build on our prior work demonstrating that RAG DSBs induced during Igκ recombination in a primary pre-B cell signal through ATM to downregulate levels of Rag1 and Rag2 mRNAs and Rag1 protein and to feedback inhibit additional RAG cleavage of Igκ gene segments (24). Based on our current and prior findings, we conclude that any type of DSB induced anywhere in the genome of a pre-B cell has the potential to signal immediate cessation of Rag1 and Rag2 transcription, leading to rapid downregulation of Rag1 protein expression and thereby V(D)J recombination throughout the nucleus. Considering that immature T cells express an order of magnitude fewer number of Rag1 monomers than Rag2 monomers (59), DSB-induced downregulation of Rag1 protein in immature lymphocytes would be predicted to be a more effective way to reduce RAG activity than downregulation of Rag2 protein. In this context, the retention of Rag2 protein in thymocytes harboring RAG DSBs at TCR loci (60) may result from a long half-life of endogenous Rag2 protein enabling Rag2 to persists following DSB-induced repression of Rag1/Rag2 transcription and resultant loss of Rag1 protein. While RAG induces two DSBs at an Igκ allele, the IR dose and etoposide concentration that we used in our pre-B cell experiments each induce hundreds of DSBs at random genomic locations (61, 62). Considering that two DSBs induced by RAG during Igκ recombination and a larger number of DSBs induced by IR or etoposide cause similar reduction in Rag1 and Rag2 mRNA levels, it will be important to assess whether intrinsic features of the RAG proteins or the location, proximity, of structures of the two DSBs induced by RAG during Igκ recombination amplify DDR signaling.
Our data that both ATM and Nemo are required for ionizing radiation to rapidly suppress Rag1 and Rag2 mRNA levels in primary pre-B cells implicates a role for canonical NF-κB transcription factors. In pre-B cells, DSBs induced by RAG or etoposide signal ATM-dependent and Nemo-dependent nuclear translocation and DNA binding of canonical NF-κB proteins to upregulate or downregulate expression of numerous genes (22). The post-translational modification of Nemo upon DSBs or other stimuli is essential for rapid cytoplasmic-to-nuclear migration of canonical NF-κB proteins, promoting NF-κB-mediated transcriptional activation or repression of target genes (3, 63–65). The canonical NF-κB proteins consist of p50, p65, and c-Rel, which each can function as homodimers or as heterodimers with each other or with additional transcription factors (63–65). Unlike p65 and c-Rel, p50 lacks a transactivation domain and forms homodimers that repress transcription (63, 66, 67). The mechanisms by which DSBs and other stimuli direct the formation of specific NF-κB complexes and target these to particular gene promoters remain enigmatic (63–65). Several studies have investigated the role of NF-κB proteins in regulating Rag1/Rag2 expression in immature B cells, reaching apparently contradictory conclusions. In immortalized and transformed cell lines, the expression of a dominant negative IκBα protein that sequesters canonical NF-κB proteins in the cytoplasm either has no effect or increases sporadic Rag1/Rag2 expression depending on the lines analyzed (68, 69). The inhibition of canonical NF-κB activity in primary pre-B cells by expression of the same dominant negative IκBα protein or by Mb1Cre-mediated deletion of Nemo has no effect on Rag1 and Rag2 mRNA levels, even after IL7 withdrawal (33). However, the expression of a different dominant negative IκBα protein in primary pre-B cells inhibits antibody-stimulated BCR signaling from inducing Rag1/Rag2 transcription (53). ChIP analyses revealed that p50, p65, and c-Rel each bind at several sites along the Rag1/Rag2 locus, including the ERag B cell specific enhancer and the D3 lymphoid specific enhancer, with BCR activation increasing p65 and c-Rel binding and decreasing p50 binding (53). Moreover, pre-B cells from mice with inactivation of the Nfkb1 gene that encodes p50 exhibit higher basal and BCR-stimulated expression of Rag1 and Rag2 (53). Collectively, our data and these prior findings support a model wherein a balance of positive-acting (p65/c-Rel) and negative-acting (p50) canonical NF-κB proteins helps regulate Rag1 and Rag2 transcription in pre-B cells, with increased binding of p50 in response to DSBs repressing Rag1/Rag2 transcription. However, considering that Nemo can function independent of NF-κB proteins (70, 71), DSBs induced in pre-B cells might repress Rag1 and Rag2 expression through other Nemo-mediated signaling pathways. Another alternative possibility is that Nemo-dependent NF-κB activity is necessary for constitutive expression of ATM-dependent signaling factors and/or transcriptional repressor that target the Rag1/Rag2 locus.
While we were preparing this manuscript, the Guikema lab reported that the DDR response regulates Rag1/Rag2 expression in pre-B cells through ATM-FOXO1 signaling (29). They showed that the genotoxic drug NCS caused rapid downmodulation of Rag1 and Rag2 mRNA and Rag1 protein (they did not report on Rag2 protein) in STI571-treated human BCR-ABL+ pre-B cell lines (BV173 and SUP-B15) and mouse v-Abl transformed pro-B cell lines. Consistent with data from primary mouse pre-B cells (47) the half-life of RAG1 in BV173 cells is relative short (<60 minutes). NCS causes a modest decrease of RAG1 half-life in BV173 cells, but does not affect levels of Rag1 protein expressed in a human embryonic kidney cell line, consistent with transcriptional repression being the mechanism by which DNA damage downmodulates RAG1 protein. Inhibition of ATM kinase activity antagonized the ability of NCS to downregulate Rag1 mRNA levels in pre-B cell lines and primary pre-B cells. NCS promotes both the ATM kinase-dependent loss of FOXO1 binding to ERag and the binding of phospho-ATM to ERag. Since FOXO1 and un-activated ATM are frequently in close proximity, the Guikema lab proposes a model wherein DSBs activate ATM to release FOXO1 binding from ERag and thereby repress Rag1/Rag2 transcription. Finally, they showed that inhibition of ATM kinase activity elevated Rag1 mRNA levels in immature B cell lines and increased sporadic recombination of a retroviral substrate in a pre-B cell line, implying that RAG DSB-induced loss of Rag1 protein expression inhibits V(D)J recombination.
Our work corroborates central findings of the Guikema lab and presents additional data that provide substantial insights into potential mechanisms by which immature lymphocytes direct DSB-induced feedback inhibition of V(D)J recombination. Both studies show that DSBs induced by a genotoxic drug in pre-B cells signal via ATM to rapidly downregulate Rag1 and Rag2 expression, but we also show that this DSB response occurs independent of new protein synthesis and involves transcriptional repression of the Rag1/Rag2 locus. In addition, we demonstrate that expression of the Nemo protein is required for normal DSB-induced feedback inhibition of Rag1 and Rag2 expression in pre-B cells, with the absence of Nemo possibly resulting in slower kinetics by which pre-B cells suppress Rag1 and Rag2 expression in response to DSBs. When considered with work of the Guikema lab, our data is consistent with a model wherein ATM-FOXO1 and ATM-Nemo signaling cooperate to rapidly initiate Rag1/Rag2 transcriptional repression, while ATM-Nemo signaling maintains this response. In this context, the release of FOXO1 might facilitate NF-κB binding and/or vice versa; while NF-κB binding may more stably inhibit Rag1/Rag2 expression, such as by persisting longer after cessation of ATM kinase activity following DSB repair. As highlighted by our prior (24) and current Gadd45α data, a potential mechanistic link between release of FOXO1 and inhibition of Rag1/Rag2 transcription should be assessed to distinguish a cause-effect relationship versus another unlinked ATM-dependent correlation. Considering the complexities of ATM-FOXO1 signaling found by the Guikema lab (29), and the unresolved questions over the roles of Nemo and NF-κB proteins in Rag1/Rag2 transcription, major obstacles exist for testing these models. Further characterization of the molecular requirements and associated events of DSB-induced inhibition of Rag1/Rag2 expression should open paths forward.
Our findings that irradiation causes downregulation of Rag1 and Rag2 mRNA levels in pro-B cells, pre-B cells, and pro-T cells, but not in total thymocytes, have important implications for mechanisms by which immature lymphocytes regulate V(D)J recombination. One explanation for this difference between pro-T cells and total thymocytes is that pre-T cells (CD4+CD8+ thymocytes) assembling TCRα genes are not able to downregulate Rag1 and Rag2 expression in response to DSBs. In this scenario, our data may reflect the existence of developmental stage-specific mechanisms by which immature T cells regulate Rag1/Rag2 transcription in response to DSBs. Such putative mechanisms could involve differential expression or activity of DDR factors that suppress Rag1/Rag2 transcription or dominant roles of cis-regulatory elements that drive transcription of Rag1 and Rag2 in pre-T cells (72). An inability of pre-T cells to inhibit Rag1 and Rag2 expression in response to DSBs would be unique among the types of immature B and T cells that assemble AgR genes. Mirroring this, TCRα gene assembly in pre-T cells is not regulated to enforce allelic exclusion, whereas recombination of the only or predominant AgR genes assembled in pro-B cells (IgH), pre-B cells (Igκ), and pre-T cells (TCRβ) is controlled to occur on one allele at a time (73). However, an important caveat to note is that our data cannot rule out that pre-T cells experiencing DSBs from 10 Gy of radiation do not suppress Rag1 and Rag2 transcription because they are directly induced to die. We previously reported that mice lacking ATM develop greater frequencies of mature lymphocytes expressing Igκ, IgH, or TCRβ genes from both alleles (24, 26). These findings provided evidence for RAG DSB-induced feedback inhibition of IgH, Igκ, and TCRβ recombination (24, 26, 57). Yet, as we noted, the slower kinetics of coding join formation in cells lacking ATM could cause this increased allelic inclusion, rather than feedback inhibition of V(D)J recombination through transcriptional repression of Rag1/Rag2 (26). Our current data show that DSBs induced in pro-B, pre-B, and pro-T cells indeed suppress Rag1 and Rag2 expression, providing additional support for our model wherein the responses of these immature lymphocytes to RAG DSBs is one essential mechanism for enforcing AgR allelic exclusion (26). The additional essential mechanisms likely include inefficient Vβ and VH RSSs (58), Ccnd3-mediated cell cycle progression (26), unequal accessibility of Ig/TCR alleles (74–76), and segregation of Ig/TCR alleles from Rag2 protein (60).
Although our data suggest that ATM-mediated downregulation of Rag1 protein expression is a mechanism by which DSBs inhibit V(D)J recombination, we have not investigated potential direct effects of ATM on suppressing AgR gene assembly. For example, ATM phosphorylation of Rag1 and/or Rag2 protein might inactivate RAG endonuclease activity prior to the reduction of Rag1 protein levels. Both Rag1 and Rag2 contain SQ/TQ motifs that could be phosphorylated by ATM and/or the related DNA-PK protein kinase. In fact, DNA-PK can phosphorylate Rag2 on Ser265 in vitro (77). While mutation of all of these motifs to the non-phosphorylatable AQ motif does not impair RAG endonuclease activity or V(D)J recombination in another way (39, 78), the role for phosphorylation of these SQ/TQ motifs in inhibiting RAG cleavage activity has not been reported. Considering that the AMP-activated protein kinase directly phosphorylates Rag1 to enhance the endonuclease activity of RAG1/RAG2 complexes (79), it is conceivable that ATM phosphorylation of Rag1 and/or Rag2 directly suppresses the ability of RAG to bind and/or cleave DNA. Moreover, the ability of ATM to signal nuclear export of a fraction of nuclear Rag2 protein in response to DSBs (46) opens the possibility that ATM might regulate the subcellular location of Rag1 protein. Finally, the finding of the Guikema lab (29) that the genotoxic drug neocarzinostatin increases the degradation of Rag1 protein suggests that ATM also might regulate the stability of Rag1. Clearly, studies are needed to investigate the potential role of ATM-dependent post-translational modifications of the Rag1 and Rag2 proteins in DSB-induced inhibition of V(D)J recombination.
The conservation of DSB-induced feedback inhibition of Rag1 and Rag2 expression among different types of immature lymphocytes and between humans and mice implies that this process is important. As we have outlined previously, it is possible that transiently suppressing RAG-mediated DNA cleavage in response to RAG DSBs induced during V(D)J recombination is critical to suppress autoimmunity from allelic inclusion and leukemia/lymphoma from oncogenic AgR locus translocations (24, 26, 73). The ability of non-RAG DSBs to inhibit RAG-mediated cleavage of an AgR locus could be a byproduct of evolutionary selection for mechanisms that enable RAG DSBs to direct mono-allelic recombination of Igκ, IgH, and TCRβ loci. In pre-T cells, RAG (and non-RAG) DSBs might signal post-translational inhibition of RAG activity and/or repression of TCRα recombination potential. Alternatively, unique requirements for TCRα recombination may have selected for mechanisms that prevent DSB-induced inhibition of V(D)J recombination. Cells that assemble AgR genes via RAG DSB intermediates also experience DSBs from other endogenous factors such as transcription and cellular metabolism. While one DSB can cause a translocation, a second simultaneous DSB increases translocation frequencies by several orders of magnitude (80). Therefore, regardless of whether the ability of immature lymphocytes to downregulate RAG activity in response to non-RAG DSBs is fortuitous, this cellular response likely helps suppress oncogenic AgR locus translocations.
Supplementary Material
Acknowledgments
This research was supported by the T32 AR007442 Training Program in Rheumatic Diseases of the University of Pennsylvania and NRSA grant F31 CA183551 (M.R.F), the GM007229 T32 Training Program in Cell and Molecular Biology of the University of Pennsylvania (A.R.-R.), NIH grant R37 AI32524 (D.G.S.), and the Department of Pathology and Laboratory Medicine of the Children's Hospital of Philadelphia and NIH R01 grant AI112621 (C.H.B).
Footnotes
C.H.B. is a consultant for Regeneron Pharmaceuticals. None of the other authors have potential conflicts of interest.
References
- 1.Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 2.Helmink BA, Sleckman BP. The Response to and Repair of RAG-Mediated DNA Double-Strand Breaks. Annu. Rev. Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCool KW, Miyamoto S. DNA damage-dependent NF-κB activation: NEMO turns nuclear signaling inside out: DNA damage-dependent NF-κB activation. Immunol. Rev. 2012;246:311–326. doi: 10.1111/j.1600-065X.2012.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, Wynshaw-Boris Anthony. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- 5.Taylor AM, Metcalfe JA, Thick J, Mak YF. Leukemia and lymphoma in ataxia telangiectasia. Blood. 1996;87:423–438. [PubMed] [Google Scholar]
- 6.Elson A, Wang Y, Daugherty CJ, Morton CC, Zhou F, Campos-Torres J, Leder P. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Ashley T, Brainerd EE, Bronson RT, Meyn MS, Baltimore D. Targeted disruption of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- 8.Borghesani PR, Alt FW, Bottaro A, Davidson L, Aksoy S, Rathbun GA, Roberts TM, Swat W, Segal RA, Gu Y. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc. Natl. Acad. Sci. U. S. A. 2000;97:3336–3341. doi: 10.1073/pnas.050584897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alt FW, Zhang F-L, Meng Y, Guo C, Schwer B. Mechanisms of Programmed DNA Lesions and Genomic Instability in the Immune System. Cell. 2013;152:417–429. doi: 10.1016/j.cell.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 12.Schatz DG, Swanson PC. V(D)J Recombination: Mechanisms of Initiation. Annu. Rev. Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 13.Notarangelo LD, Kim M-S, Walter JE, Lee YN. Human RAG mutations: biochemistry and clinical implications. Nat. Rev. Immunol. 2016;16:234–246. doi: 10.1038/nri.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodbine L, Gennery AR, Jeggo PA. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair. 2014;16:84–96. doi: 10.1016/j.dnarep.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Feeney AJ, Atkinson MJ, Cowan MJ, Escuro G, Lugo G. A defective Vkappa A2 allele in Navajos which may play a role in increased susceptibility to haemophilus influenzae type b disease. J. Clin. Invest. 1996;97:2277–2282. doi: 10.1172/JCI118669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V (D) J recombination. Cell. 2002;109:S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 17.Nemazee D. Receptor editing in lymphocyte development and central tolerance. Nat. Rev. Immunol. 2006;6:728–740. doi: 10.1038/nri1939. [DOI] [PubMed] [Google Scholar]
- 18.Xiong N, Raulet DH. Development and selection of gammadelta T cells. Immunol. Rev. 2007;215:15–31. doi: 10.1111/j.1600-065X.2006.00478.x. [DOI] [PubMed] [Google Scholar]
- 19.Perkins EJ. Sensing of intermediates in V(D)J recombination by ATM. Genes Dev. 2002;16:159–164. doi: 10.1101/gad.956902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dujka ME, Puebla-Osorio N, Tavana O, Sang M, Zhu C. ATM and p53 are essential in the cell-cycle containment of DNA breaks during V(D)J recombination in vivo. Oncogene. 2010;29:957–965. doi: 10.1038/onc.2009.394. [DOI] [PubMed] [Google Scholar]
- 21.Callén E, Jankovic M, Difilippantonio S, Daniel H-T, Chen JA, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, Sleckman BP, Ried T, Nussenzweig M, Nussenzweig A. ATM Prevents the Persistence and Propagation of Chromosome Breaks in Lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 22.Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins JB, Weaver BK, Mandik-Nayak L, Schreiber RD, Allen PM, May MJ, Paules RS, Bassing CH, Sleckman BP. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bednarski JJ, Nickless A, Bhattacharya D, Amin RH, Schlissel MS, Sleckman BP. RAG-induced DNA double-strand breaks signal through Pim2 to promote pre–B cell survival and limit proliferation. J. Exp. Med. 2012;209:11–17. doi: 10.1084/jem.20112078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinel NC, Lee B-S, Tubbs AT, Bednarski JJ, Schulte E, Yang-Iott KS, Schatz DG, Sleckman BP, Bassing CH. The Ataxia Telangiectasia mutated kinase controls Igκ allelic exclusion by inhibiting secondary Vκ-to-Jκ rearrangements. J. Exp. Med. 2013;210:233–239. doi: 10.1084/jem.20121605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amin RH, Schlissel MS. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 2008;9:613–622. doi: 10.1038/ni.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinel NC, Fisher MR, Yang-Iott KS, Bassing CH. The Ataxia Telangiectasia Mutated and Cyclin D3 Proteins Cooperate To Help Enforce TCR and IgH Allelic Exclusion. J. Immunol. 2014;193:2881–2890. doi: 10.4049/jimmunol.1302201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bednarski JJ, Pandey R, Schulte E, White B-R, Chen LS, Sandoval GJ, Kohyama M, Haldar M, Nickless A, Trott A, Cheng G, Murphy KM, Bassing CH, Payton JE, Sleckman BP. RAG-mediated DNA double-strand breaks activate a cell type-specific checkpoint to inhibit pre–B cell receptor signals. J. Exp. Med. 2016;213:209–223. doi: 10.1084/jem.20151048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Innes CL, Hesse JE, Palii SS, Helmink BA, Holub AJ, Sleckman BP, Paules RS. DNA damage activates a complex transcriptional response in murine lymphocytes that includes both physiological and cancer-predisposition programs. BMC Genomics. 2013;14:163. doi: 10.1186/1471-2164-14-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ochodnicka-Mackovicova K, Bahjat M, Maas C, van der Veen A, Bloedjes TA, de Bruin AM, van Andel H, Schrader CE, Hendriks RW, Verhoeyen E, Bende RJ, van Noesel CJM, Guikema JEJ. The DNA Damage Response Regulates RAG1/2 Expression in Pre-B Cells through ATM-FOXO1 Signaling. J. Immunol. 2016 doi: 10.4049/jimmunol.1501989. [DOI] [PubMed] [Google Scholar]
- 30.Strasser A, Whittingham S, Vaux DL, Bath ML, Adams JM, Cory S, Harris AW. Enforced BCL2 expression in B-lymphoid cells prolongs antibody responses and elicits autoimmune disease. Proc. Natl. Acad. Sci. U. S. A. 1991;88:8661–8665. doi: 10.1073/pnas.88.19.8661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The In Vivo Pattern of Binding of RAG1 RAG2 to Antigen Receptor Loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeMicco A, Yang-Iott K, Bassing CH. Somatic inactivation of Tp53 in hematopoietic stem cells or thymocytes predisposes mice to thymic lymphomas with clonal translocations. Cell Cycle. 2013;12:3307–3316. doi: 10.4161/cc.26299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Derudder E, Cadera EJ, Vahl JC, Wang J, Fox CJ, Zha S, van Loo G, Pasparakis M, Schlissel MS, Schmidt-Supprian M, Rajewsky K. Development of immunoglobulin λ-chain–positive B cells, but not editing of immunoglobulin κ-chain, depends on NF-κB signals. Nat. Immunol. 2009;10:647–654. doi: 10.1038/ni.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 35.Hao Z, Rajewsky K. Homeostasis of Peripheral B Cells in the Absence of B Cell Influx from the Bone Marrow. J. Exp. Med. 2001;194:1151–1164. doi: 10.1084/jem.194.8.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coster G, Gold A, Chen D, Schatz DG, Goldberg M. A Dual Interaction between the DNA Damage Response Protein MDC1 and the RAG1 Subunit of the V(D)J Recombinase. J. Biol. Chem. 2012;287:36488–36498. doi: 10.1074/jbc.M112.402487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bredemeyer AL, Sharma C-Y, Huang GG, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, Sleckman BP. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 38.Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of Dynamic γ-H2AX Domains along Broken DNA Strands Is Distinctly Regulated by ATM and MDC1 and Dependent upon H2AX Densities in Chromatin. Mol. Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gapud EJ, Lee B-S, Mahowald GK, Bassing CH, Sleckman BP. Repair of Chromosomal RAG-Mediated DNA Breaks by Mutant RAG Proteins Lacking Phosphatidylinositol 3-Like Kinase Consensus Phosphorylation Sites. J. Immunol. 2011;187:1826–1834. doi: 10.4049/jimmunol.1101388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin B, Savic V, Juntilla MM, Bredemeyer AL, Yang-Iott KS, Helmink BA, Koretzky GA, Sleckman BP, Bassing CH. Histone H2AX stabilizes broken DNA strands to suppress chromosome breaks and translocations during V(D)J recombination. J. Exp. Med. 2009;206:2625–2639. doi: 10.1084/jem.20091320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeMicco A, Reich T, Arya R, Rivera-Reyes A, Fisher MR, Bassing CH. Lymphocyte Lineage-Specific and Developmental Stage Specific Mechanisms Suppress Cyclin D3 Expression in Response to DNA Double Strand Breaks. Cell Cycle Georget. Tex. 2016 doi: 10.1080/15384101.2016.1198861. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rolink A, Grawunder U, Haasner D, Strasser A, Melchers F. Immature surface Ig+ B cells can continue to rearrange kappa and lambda L chain gene loci. J. Exp. Med. 1993;178:1263–1270. doi: 10.1084/jem.178.4.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Billips LG, Nuñez CA, Bertrand FE, Stankovic AK, Gartland GL, Burrows PD, Cooper MD. Immunoglobulin recombinase gene activity is modulated reciprocally by interleukin 7 and CD19 in B cell progenitors. J. Exp. Med. 1995;182:973–982. doi: 10.1084/jem.182.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hecht SM. Bleomycin: new perspectives on the mechanism of action. J. Nat. Prod. 2000;63:158–168. doi: 10.1021/np990549f. [DOI] [PubMed] [Google Scholar]
- 45.Montecucco A, Zanetta F, Biamonti G. Molecular mechanisms of etoposide. EXCLI J. 2015;14:95–108. doi: 10.17179/excli2015-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodgers W, Byrum JN, Sapkota H, Rahman NS, Cail RC, Zhao S, Schatz DG, Rodgers KK. Spatio-temporal regulation of RAG2 following genotoxic stress. DNA Repair. 2015;27:19–27. doi: 10.1016/j.dnarep.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grawunder U, Schatz DG, Leu TM, Rolink A, Melchers F. The half-life of RAG-1 protein in precursor B cells is increased in the absence of RAG-2 expression. J. Exp. Med. 1996;183:1731–1737. doi: 10.1084/jem.183.4.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin WC, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 49.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 50.Lin W-C, Desiderio S. Cell cycle regulation of V (D) J recombination-activating protein RAG-2. Proc. Natl. Acad. Sci. 1994;91:2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V (D) J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 52.Hann SR, Eisenman RN. Proteins encoded by the human c-myc oncogene: differential expression in neoplastic cells. Mol. Cell. Biol. 1984;4:2486–2497. doi: 10.1128/mcb.4.11.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verkoczy L, Aït-Azzouzene D, Skog P, Märtensson A, Lang J, Duong B, Nemazee D. A Role for Nuclear Factor Kappa B/Rel Transcription Factors in the Regulation of the Recombinase Activator Genes. Immunity. 2005;22:519–531. doi: 10.1016/j.immuni.2005.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Barlow C, Brown KD, Deng CX, Tagle DA, Wynshaw-Boris A. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat. Genet. 1997;17:453–456. doi: 10.1038/ng1297-453. [DOI] [PubMed] [Google Scholar]
- 55.Muljo SA, Schlissel MS. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus‒transformed pre-B cell lines. Nat. Immunol. 2003;4:31–37. doi: 10.1038/ni870. [DOI] [PubMed] [Google Scholar]
- 56.Moshous D, Callebaut I, de Chasseval R, Corneo B, Cavazzana-Calvo M, Le Deist F, Tezcan I, Sanal O, Bertrand Y, Philippe N, Fischer A, de Villartay JP. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 57.Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, Farrar MA, Sleckman BP, Schatz DG, Busslinger M, Bassing CH, Skok JA. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat. Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liang H-E, Hsu L-Y, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y-H, Shetty K, Surleac MD, Petrescu AJ, Schatz DG. Mapping and Quantitation of the Interaction between the Recombination Activating Gene Proteins RAG1 and RAG2. J. Biol. Chem. 2015;290:11802–11817. doi: 10.1074/jbc.M115.638627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan EAW, Teng G, Corbett E, Choudhury KR, Bassing CH, Schatz DG, Krangel MS. Peripheral subnuclear positioning suppresses Tcrb recombination and segregates Tcrb alleles from RAG2. Proc. Natl. Acad. Sci. 2013;110:E4628–E4637. doi: 10.1073/pnas.1310846110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rothkamm K, Löbrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muslimović A, Nyström S, Gao Y, Hammarsten O. Numerical analysis of etoposide induced DNA breaks. PloS One. 2009;4:e5859. doi: 10.1371/journal.pone.0005859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smale ST. Dimer-specific regulatory mechanisms within the NF-κB family of transcription factors. Immunol. Rev. 2012;246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 65.Smale ST. Hierarchies of NF-κB target-gene regulation. Nat. Immunol. 2011;12:689–694. doi: 10.1038/ni.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tong X, Yin L, Washington R, Rosenberg DW, Giardina C. The p50-p50 NF-kappaB complex as a stimulus-specific repressor of gene activation. Mol. Cell. Biochem. 2004;265:171–183. doi: 10.1023/b:mcbi.0000044394.66951.4d. [DOI] [PubMed] [Google Scholar]
- 67.Bohuslav J, Kravchenko VV, Parry GC, Erlich JH, Gerondakis S, Mackman N, Ulevitch RJ. Regulation of an essential innate immune response by the p50 subunit of NF-kappaB. J. Clin. Invest. 1998;102:1645–1652. doi: 10.1172/JCI3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scherer DC, Brockman JA, Bendall HH, Zhang GM, Ballard DW, Oltz EM. Corepression of RelA and c-rel inhibits immunoglobulin kappa gene transcription and rearrangement in precursor B lymphocytes. Immunity. 1996;5:563–574. doi: 10.1016/s1074-7613(00)80271-x. [DOI] [PubMed] [Google Scholar]
- 69.Ochodnicka-Mackovicova K, Bahjat M, Bloedjes TA, Maas C, de Bruin AM, Bende RJ, van Noesel CJM, Guikema JEJ. NF- B and AKT signaling prevent DNA damage in transformed pre-B cells by suppressing RAG1/2 expression and activity. Blood. 2015;126:1324–1335. doi: 10.1182/blood-2015-01-621623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verma UN. Nuclear Role of I B Kinase- /NF- B Essential Modulator (IKK /NEMO) in NF- B-dependent Gene Expression. J. Biol. Chem. 2003;279:3509–3515. doi: 10.1074/jbc.M309300200. [DOI] [PubMed] [Google Scholar]
- 71.Yang Y, Xia F, Hermance N, Mabb A, Simonson S, Morrissey S, Gandhi P, Munson M, Miyamoto S, Kelliher MA. A cytosolic ATM/NEMO/RIP1 complex recruits TAK1 to mediate the NF-kappaB and p38 mitogen-activated protein kinase (MAPK)/MAPK-activated protein 2 responses to DNA damage. Mol. Cell. Biol. 2011;31:2774–2786. doi: 10.1128/MCB.01139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yannoutsos N, Barreto V, Misulovin Z, Gazumyan A, Yu W, Rajewsky N, Peixoto BR, Eisenreich T, Nussenzweig MC. A cis element in the recombination activating gene locus regulates gene expression by counteracting a distant silencer. Nat. Immunol. 2004;5:443–450. doi: 10.1038/ni1053. [DOI] [PubMed] [Google Scholar]
- 73.Brady BL, Steinel NC, Bassing CH. Antigen Receptor Allelic Exclusion: An Update and Reappraisal. J. Immunol. 2010;185:3801–3808. doi: 10.4049/jimmunol.1001158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, Bergman Y. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- 75.Schlimgen RJ, Reddy KL, Singh H, Krangel MS. Initiation of allelic exclusion by stochastic interaction of Tcrb alleles with repressive nuclear compartments. Nat. Immunol. 2008;9:802–809. doi: 10.1038/ni.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Skok JA, Gisler R, Novatchkova M, Farmer D, de Laat W, Busslinger M. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat. Immunol. 2007;8:378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 77.Hah Y-S, Lee JH, Kim DR. DNA-dependent protein kinase mediates V(D)J recombination via RAG2 phosphorylation. J. Biochem. Mol. Biol. 2007;40:432–438. doi: 10.5483/bmbrep.2007.40.3.432. [DOI] [PubMed] [Google Scholar]
- 78.Lin JM, Landree MA, Roth DB. V (D) J recombination catalyzed by mutant RAG proteins lacking consensus DNA-PK phosphorylation sites. Mol. Immunol. 1999;36:1263–1269. doi: 10.1016/s0161-5890(99)00099-1. [DOI] [PubMed] [Google Scholar]
- 79.Um J-H, Brown AL, Singh SK, Chen Y, Gucek B-S, Lee M, Luckey MA, Kim J-H, Park MK, Sleckman BP, Gellert M, Chung JH. Metabolic sensor AMPK directly phosphorylates RAG1 protein and regulates V(D)J recombination. Proc. Natl. Acad. Sci. 2013;110:9873–9878. doi: 10.1073/pnas.1307928110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.