Abstract
Adaptive immune cells, such as T cells, integrate information from their extracellular environment through complex signaling networks with exquisite sensitivity in order to direct decisions on proliferation, apoptosis, and cytokine production. These signaling networks are reliant on the interplay between finely tuned secondary messengers, such as Ca2+ and H2O2. Frequency response analysis, originally developed in control engineering, is a tool used for discerning complex networks. This analytical technique has been shown to be useful for understanding biological systems and facilitates identification of the dominant behaviour of the system. We probed intracellular Ca2+ dynamics in the frequency domain to investigate the complex relationship between two second messenger signaling molecules, H2O2 and Ca2+, during T cell activation with single cell resolution. Single-cell analysis provides a unique platform for interrogating and monitoring cellular processes of interest. We utilized a previously developed microfluidic device to monitor individual T cells through time while applying a dynamic input to reveal a natural frequency of the system at approximately 2.78 mHz stimulation. Although our network was much larger with more unknown connections than previous applications, we are able to derive features from our data, observe forced oscillations associated with specific amplitudes and frequencies of stimuli, and arrive at conclusions about potential transfer function fits as well as the underlying population dynamics.
Introduction
As part of the adaptive immune response, T cell lymphocytes function to recognize and respond to pathogens present in the body. Due to their central role in immunity, T cell dysfunction has been implicated in numerous diseased conditions, such as autoimmune disorders 1, tumour immunity 2, and allergic reactions 3. T cell activation induces rapid proliferation and a change in intracellular signalling cascades to alter gene expression and ultimately cytokine release 4. More specifically, when an antigen-presenting cell (APC) engages the T cell receptor (TCR), a cascade of activated kinases drive intracellular signalling through protein modification 5, 6, ultimately enabling the release of intracellular stores of calcium, which drive a multitude of signalling events. Calcium signalling involves oscillations, thought to be the result of the stochastic distribution of IP3R within the membrane and the result of calcium influx from external sources 7, 8. This signalling profile illustrates the ability for calcium to produce complex signals as opposed to molecules that produce binary state switches 9. Different frequencies have been shown with varying levels of stimulation 10 and have an effect on the activation of downstream transcription factors, such as NFAT and NF-κB 6, 11.
The increased signalling capacity during T cell activation is also associated with an increase in glucose metabolism and subsequent burst of reactive oxygen species (ROS) from NADPH oxidases 12 and the mitochondria 4. ROS, such as hydrogen peroxide (H2O2), are produced within the cell and act as secondary messengers in numerous cellular processes through protein thiol oxidation, such as reversible phosphatase inactivation and protein localization13. Alteration of T cell ROS production and regulation in signalling has been reported in immunological diseases such as systemic lupus erythematosus and rheumatoid arthritis 14, 15. Aberrant T cell signalling has also been associated with tumour cell immunity. However, it is often difficult to measure intracellular ROS and many techniques only allow for population averages through time. There is known cross talk between calcium and H2O2 during T cell activation; ROS, including H2O2, is able to activate calcium release channels 16–18 and increase the channel activity of two ER membrane channels, IP3R and RyR 19–23, while NADPH oxidases Duox1/224 and many mitochondrial proteins (e.g. VDAC) are calcium sensitive 25. The relationships between these signalling molecules are difficult to analyse due to the fast, dynamic kinetics and subcellular localization. We seek to better understand these connections in the context of frequency encoding, looking to answer the question of whether dynamic stimulation with H2O2 is able to affect Ca2+ signalling in the frequency domain within T cell lymphocytes. Furthermore, we seek to determine which frequencies of input oscillatory conditions of H2O2 elicit the maximum Ca2+ response.
Current approaches are not sufficient to analyse the signalling network in question because of the complexity and lack of understanding of all molecular mechanisms. Novel computational models are needed to overcome these limitations and enable an investigation of T cell signalling in a more complete and systematic fashion. Control-based computational methods have been developed for discerning complex, interconnected networks of signalling molecules that are difficult to interrogate with bulk measurements 26. Ultimately, these techniques, borrowed from control engineering, can help identify and model only dominant interactions within the network by characterizing the behaviour of a system from its responses to well-characterized inputs. This is accomplished by applying known stimuli to cells and measuring the resultant gain and phase shift of the output signal. This behaviour can be modelled with a transfer function, reducing the parameters necessary to describe a biological system. We present here an approach to interrogating calcium dynamics with dynamic H2O2 input to elucidate characteristics of the signalling network.
A secondary challenge in studying non-adherent immune cell behaviour is a constraint in live cell imaging. T cells develop in the thymus and reside as suspension cells in the blood. As suspension cells, they have been historically difficult to analyse dynamically at a single-cell level because they often float out of the focal place during fluorescent microscopy imaging. There are techniques, such as flow cytometry, that enable single-cell analysis, but these are end-point assays that cannot monitor a single-cell through time. Appropriate experimental techniques require the ability to trap individual T cells and maintain their location while delivering precisely tuned stimulation profiles.
Advances in microfluidics have enabled more advantageous methods for interrogating T cells, such that hundreds of single T cells can be loaded into a single device and monitored dynamically with fluorescent molecules 27–32, providing insight into the underlying signalling networks with commonly available fluorescent probes and markers. Recent microfluidic devices have also enabled delivery of robust, time-varying chemical signals, in contrast to conventional experimental techniques which measure the response of cells to a single perturbation of step increase or bolus of stimulus 33, 34. The enhanced experimental capability can be combined with frequency response analysis such that the underlying complex signalling networks can be discerned more easily. As calcium signalling is an almost immediate response to T cell stimulation, occurring within seconds of stimulation, it is an appropriate molecular 35 candidate for this analysis technique. Prior examples of this approach include biological applications in the osmotic stress response 36 and the galactose response pathway 37 in S. cerevisiae. More recently, the utility of microfluidic delivery of periodic chemical stimuli to interrogate calcium regulation has been demonstrated in GPCR activated HEK293 cells 38.
In this study, we utilized a custom microfluidic platform and frequency response analysis to investigate cross-communication between two secondary messenger molecules in Jurkat T cells, an immortal T cell lymphocyte line. We probed the dynamics between H2O2 and Ca2+ by varying the extracellular H2O2 environment of the cell and recording the intracellular cytoplasmic Ca2+ response to varying frequencies. Cells in any given experiment received a single frequency of stimulation and cells were combined across conditions for an experimental Bode Plot, which provides insight into the filtering properties of cytoplasmic Ca2+ signalling in response to H2O2.
Results
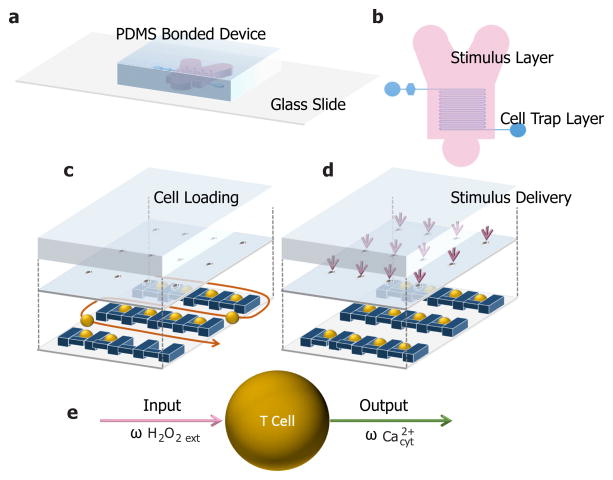
Experimental device enables dynamic input with precision
A previously characterized microfluidic device was used for the trapping and subsequent fluorescent imaging of suspension Jurkat T cells 33. The two-layer design of this device enables fast, robust switching of fluids while cells are maintained in a low-shear stress environment for the duration of the experiment. As can be seen in Figure 1a–d, this device consists of cell trap layer, capable of trapping and holding cells for subsequent analysis, connected to a large stimulus delivery layer, capable of fast fluid switching, via small pores 33. We imaged individual T cells through time with precise, uniform control of the cellular environment. With this custom device, we stimulated Jurkat T cells with an oscillatory input of H2O2 and measured the gain, phase, and dominant frequencies of the output signal (Figure 1e). Here, we were interested in probing the dynamics between calcium and H2O2 by varying the extracellular H2O2 environment of the cell and recording the intracellular cytoplasmic calcium response to varying frequencies. In this device, cells received a single frequency of stimulation; data were then combined from all conditions for a Bode Plot, which provides insight into the filter dynamics of cytoplasmic calcium signalling in response to H2O2. To complement this enabling experimental pipeline, we developed computational analysis techniques to gain an unprecedented view of calcium signalling in Jurkat T cells.
Figure 1.
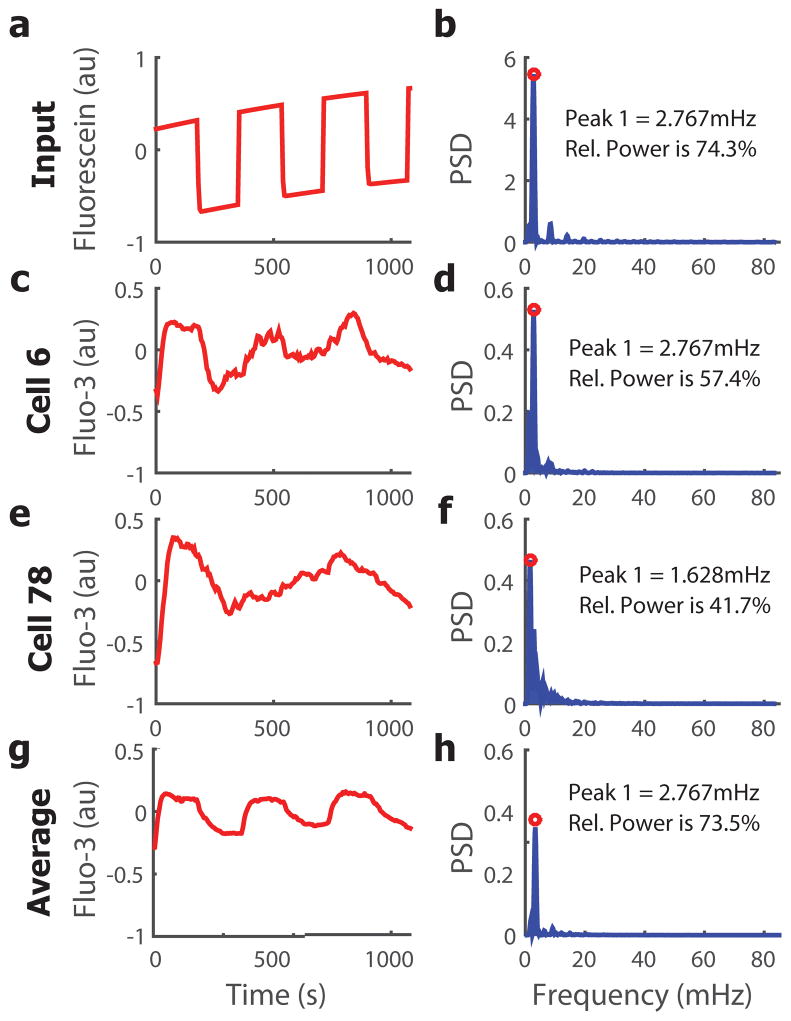
We first characterized the stimulus delivery using alternating buffer and buffer carrying a dissolved fluorophore to characterize the frequencies delivered in the input (Figure 2a,b). Data indicated that the cells should receive a consistent waveform with the designated frequency for all subsequent conditions. The resulting single-cell calcium traces were recorded and heterogeneity was observed in individual responses (such as two cells shown in Figure 2c–f). Traces were analysed for dominant frequencies, as defined by exhibiting an area under the curve greater than a designated threshold in the power spectral density (PSD) (Figure 2d,f). Interestingly, for given input frequencies, the average trace corresponded to the single dominant frequency of oscillatory input (Figure 2g,h). Some cells were forced to oscillate similarly to the population average (Figure 2c,d) while others exhibit alternative frequency responses (Figure 2e,f), illustrating the necessity to use single-cell analysis in this approach.
Figure 2.
We also extracted the gain and phase for each individual cell and combined them for further, population-based analyses (figure 2d,h). This analysis pipeline allowed us to view individual calcium traces as signals in the frequency domain, ultimately providing additional information about the response of cytoplasmic calcium to the experimental perturbation that cannot be captured with dose-response, bulk measurements.
Single-cell analysis reveals response to oscillatory stimulation
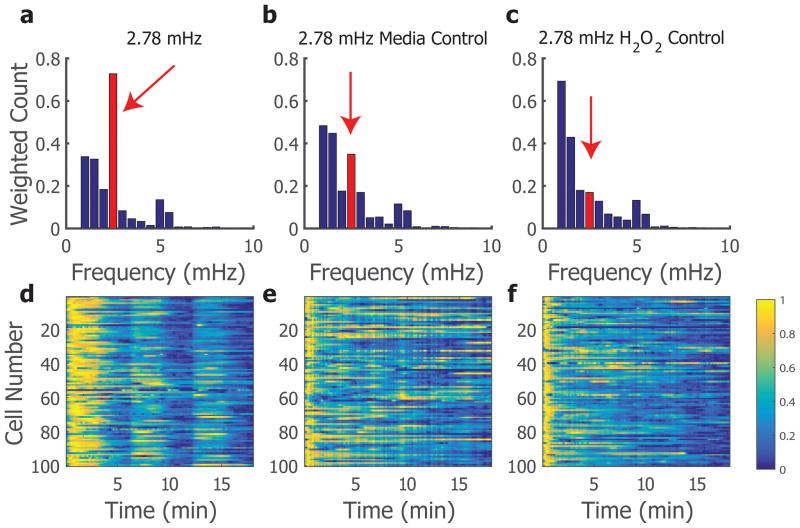
Once single-cell calcium traces were analysed, they were compiled to provide insight to the response of cells to frequencies of interest. For cells receiving a frequency of 2.78 mHz (corresponding to a period of 6 minutes) we observed diverse dynamic responses resulting in a range of dominant frequencies to the stimulation, thus highlighting the emphasis to look at a single-cell response as opposed to the population average of these experiments (Figure 3a,d). This experimental condition was compared to two controls for mechanical switching of fluid flow: (1) a media control where the cellular microenvironment switched at the same frequency but both solutions contained standard RPMI cell culture media, and (2) a H2O2 control where the two solutions were both 25 μM H2O2 supplemented RPMI media. We observed a clear oscillatory behavior of many cells to an oscillating H2O2 stimulation, more so than in the control conditions (Figure 3). It is evident from these single-cell traces that cells can oscillate under specific driving frequencies, in particular the oscillating H2O2 condition as compared to the two controls (Figure 3d–f).
Figure 3.
To quantify cell behaviour, we used spectral analysis to calculate 39 the dominant frequencies present in cell signals when driven with a frequency of 2.78 mHz, and found a peak in dominant frequencies of the treated cells corresponding to the driving frequency. Both the media and H2O2 controls showed a reduced peak in dominant frequency at the entrainment value (Figure 3a–c), indicating the experimental application of oscillatory H2O2 drives this calcium response. This population-based visualization shows that although the population is heterogeneous in responses, there is an appreciable shift to correspond with the driving frequency of H2O2 stimulation, in at least a subset of the population.
Input amplitude modulates cellular oscillation for a given frequency
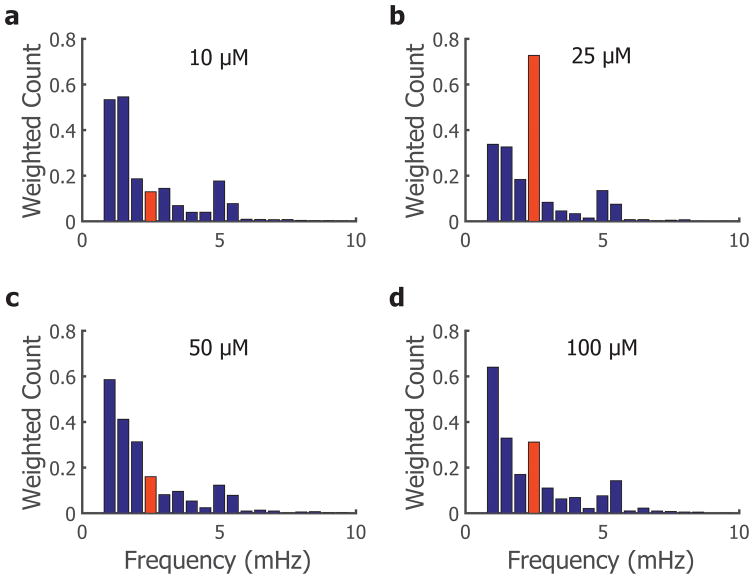
Experiments were repeated for different concentrations of H2O2 (10, 25, 50, and 100 μM) to test the dependence of the forced oscillation effect on concentration of the stimuli. From the dominant frequency histograms, we observed that the forced oscillation is dependent on input amplitude (Figure 4a–d). These experiments were all driven at the same frequency, as shown before, of 2.78 mHz and only the 25 μM condition exhibits a large peak in output signal at this frequency (Figure 4b). This suggests that 25 μM is ideal for eliciting the calcium signal: concentrations lower than 25 μM H2O2 were not able to elicit the robust response; and similarly, those concentrations tested above 25 μM H2O2 also had diminished responsiveness at the driving frequency, potentially due to cytotoxic effects of high concentrations of ROS. Thus, all subsequent experiments were performed with 25 μM H2O2.
Figure 4.
Cellular oscillation is dependent on input frequency
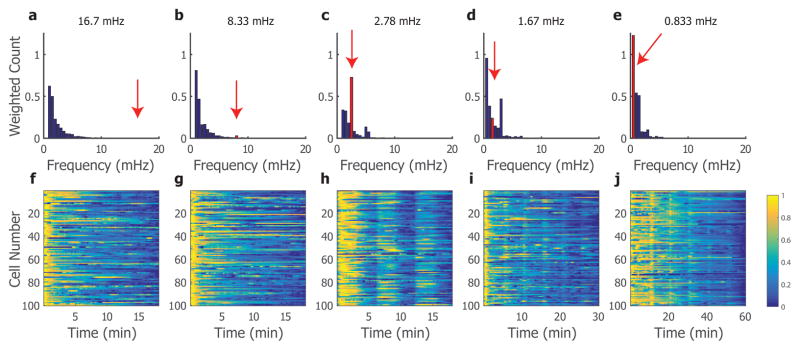
While keeping the concentration of H2O2 constant at 25 μM H2O2, the input frequency was varied in different experiments between 16.7 mHz (1 minute period) and 0.83 mHz (20 minute period). The cells typically responded to at least the initial input of H2O2 signal, but not all frequencies elicited the same response seen with 2.78 mHz (Figure 5f–j). For instance, at the higher frequencies of 16.7 and 8.3 mHz, cells did not respond to later inputs of H2O2 but instead appear to slowly dampen intracellular calcium signalling (Figure 5f–g). This was corroborated by the histograms of dominant frequencies, which show very low or no response in the frequency domain at the driving frequency, as denoted with the green arrow (Figure 5a,b). This could potentially be the result of fatigue or refractory nature of the system, such that the calcium signalling mechanistically cannot recover and respond at this rate. For experiments done at lower frequencies, the cells appeared to recover between oscillations and exhibit an increase in calcium signalling when the environment is altered, although this increase was not sustained through the duration of H2O2 application (Figure 5i,j). These higher frequencies elevated the H2O2 in the cellular environment for longer periods, potentially negatively impacting the cells’ signaling or exhausting the calcium signalling. The cellular response heatmaps also reflect the population heterogeneity; single cells often have varying responses to the same input signal and this platform best captures those differences. The majority of cells responded to the 2.78 mHz frequency with the greatest response.
Figure 5.
Second order system captures cell behaviour
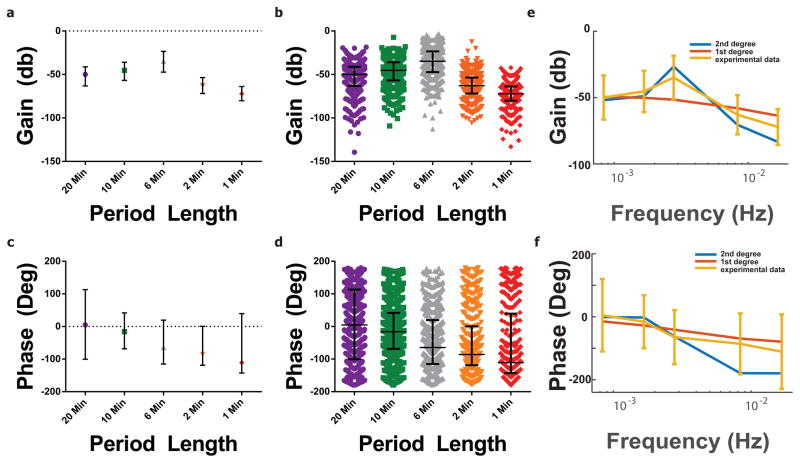
To test whether the complex Ca-ROS signalling dynamics can be captured by simpler models, we next examined the gain and the phase behaviour of individual cells in frequency space. The gain and phase of individual cells was compiled across at least 3 independent experiments for each frequency (Figure 6a–d). A definitive peak in the gain response for the 6 minute period emerges in contrast to all others (Figure 6a,b). It should also be noted that there is a drop in phase as the frequency increases, concomitant with an increase in variability. More specifically, there is a noticeable decline in gain and phase past the 2.78 mHz experimental condition, indicating a decrease in the cells’ ability to respond to the oscillatory input to periods lower than 6 minutes. This observation is in line with previous reports of biological systems behaving as low-pass filters 40. As cells must filter out numerous environmental cues and assimilate them to mount a response 41–43; it would require great energy expenditure to respond to every fluctuation in the environment. As such, these cells appear to filter out signals above and below the experimental condition of 2.78 mHz, but are slightly responsive with the 10- and 20-minute period conditions. We further plotted these experimental conditions in histograms to capture the population heterogeneity across gain and phase values (Supplemental Figure 4). Our visualization also provides an unparalleled view of single-cell responses within a population of genetically similar Jurkat T cells, with a surprisingly large variation in magnitude.
Figure 6.
We next sought to determine a transfer function that would enhance our understanding of the underlying biological network while providing a model for future hypothesis driven experimentation. The experimental data suggests a resonant frequency at approximately 2.78 mHz. This data trend aligns with an overdamped, second order transfer function of the form:
| (1) |
where K is the system gain, ωn is the natural frequency in rad/s, and ζ is the damping factor (parameter values provided in Table 1). We compared this fitting to that of a first order system with transfer function:
| (2) |
where τc is the time constant and K is the system gain. As can be seen in Figure 7. τc can be converted to the cutoff frequency of the resultant low pass filter and describes the frequency at which the filter begins attenuating signal (Supplemental Figure 5). As this is the only parameter that can be altered, we are unable to reflect the increase in gain around the 2.78 mHz. The first order system does not recapitulate the natural frequency observed increase in gain at 2.78 mHz and we chose to move forward with the second order transfer function.
Table 1.
Optimized Parameter Values for Transfer Function Fits.
| Parameter | Optimized Parameter Values | Bounds | ||
|---|---|---|---|---|
| Population | Top 25% | Bottom 25% | ||
| K | 7.09e-7 | 7.80e-6 | 1.53e-7 | [1e-8, 1e-3] |
| ζ | 0.0220 | 0.104 | 0.00240 | [1e-5, 1] |
| ωn | 0.0177 rad/s | 0.0168 rad/s | 0.0190 rad/s | [0.015, 0.019] rad/s |
| Error | 3.7640 | 2.5980 | 3.4388 | |
| Poles | −0.0004+/− 0.0177i | −0.0017+/− 0.0167i | −0.000045+/− 0.0190i | |
| Stable? | Yes | Yes | Yes | |
Figure 7.
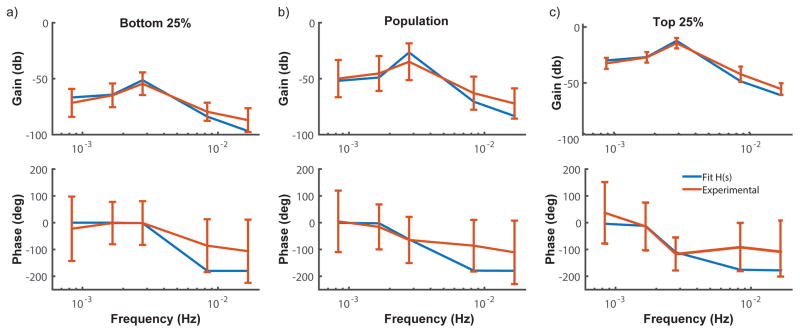
When fit to the median of the experimental conditions across various frequencies, our resultant second order transfer function aligned well (Figure 6e,f). We then subdivided the population of cells into quartiles based on the gain response and again fit the data to a second order band pass filter to compare the parameters between subpopulations (Figure 7b and Figure 7c). For the top 25% of cells, a much more damped system resulted and the range of phase values decreased when compared to both the total population and to the bottom quartile (Figure 7b). Interestingly, this result suggests that the cells with highest gain values have less variable phase response, implying cells are more in phase with one another than when visualizing the entire population. This is not true for the bottom quartile population, as the phase variation appears to be more similar to the overall population of cells (Figure 7c). Although cells in a given experiment were sampled for a single input frequency and not across the entire frequency domain, this subpopulation analysis indicates that different cells within the population have distinct and separate filtering capabilities.
Discussion
Cells continually respond to dynamic environmental conditions through intracellular Ca2+ signalling. Yet questions remain unanswered about how cells are able to use this secondary messenger to elicit a wide range of context dependent responses. Recent reports have indicated that the answer lies in better understanding the diverse spatiotemporal dynamics giving rise to infinite patterns of Ca2+ responses 44, 45. Ca2+ signalling is believed to increase information transmission by reducing extrinsic noise factors on the signal to noise ratio for intracellular signalling cascades 46. However, current technological approaches limit the delivery of environmental cues and subsequent analysis of single-cell behaviour for testing these theories. In this work, we integrated microfluidic and computational technologies to overcome this technical barrier and to gain a better understanding of Ca2+ signalling in Jurkat T cell lymphocytes by applying frequency response analysis.
Our experimental design varied extracellular H2O2, a reactive oxygen species that has been shown to be dynamically induced in T cell activation and plays a role in signal transduction. Our advanced view of the interplay between calcium and H2O2 delineates differences between individual cells in response to the same, robustly controlled, environmental signal. Such a view provides novel insight both for intracellular signalling dynamics as well as for differences between cells in a relatively clonal population.
To examine the interplay between these two signalling molecules, we applied oscillatory H2O2 input at varying frequencies and amplitudes and compiled a Bode Plot of the results to obtain the estimated resonant frequency and damping coefficient of this second order system. We found an oscillatory H2O2 input was able to force strong oscillations in the calcium dynamics in cells unlike either constant media or H2O2 controls (Figure 3). Furthermore, we sampled different amplitudes, or concentrations, of the input H2O2 signal and found an optimal concentration of 25 μM H2O2 (Figure 4). This aligns with previous literature suggesting different concentrations of Ca2+ stimulation can result in oscillatory or refractory signalling 38; the findings further suggest the 25 μM H2O2 concentration is within the oscillatory regime of stimulation for our sampled system. With this concentration held constant, a frequency sweep was performed for 5 different frequencies and results showed a damping of signals both above and below the optimal frequency of 2.78 mHz (Figure 5). Identification of a resonant frequency provides experimental conditions by which Jurkat T cells may be rapidly assessed for activation potential. Physiologically, this indicates a frequency at which information may be encoded for specific downstream signalling. As Ca2+ is a ubiquitous signalling molecule capable of moderating multiple downstream effects, signalling systems may be finely tuned to specific frequencies and we suggest this frequency warrants further investigation to understand underlying biochemical signals.
To our knowledge, this is the first application of a sinusoidal input to Jurkat T cells, but our findings can be related to previous reports of T cell transcriptional encoding with Ca2+ spikes at varying frequencies. Observed Ca2+ frequencies vary widely based on cell type and stimulation, with values ranging from 100 Hz in excitable cells to 0.01 mHz for non-excitable cells 47. Our experimental system falls within this reported range, with the natural frequency estimated to be at approximately 2.78 mHz for non-excitable T cell system.
In general, cells must decode a wide range of dynamic stimuli, usually by sensing molecules that can appropriately modify their behaviour 48. On a molecular level, Ca2+ binds to phosphatases and kinases to affect downstream targets, and these on-off kinetics are most likely responsible for decoding a range of possible frequencies. With oscillations below appropriate frequencies, the signal cannot integrate to mount a response. Ca2+ binds to many effectors with high cooperatively and high dissociation, again supporting the idea that a range of moderate frequencies, corresponding to these kinetics, encodes more information than constant Ca2+ signals or high frequency signals 44. Multiple proteins involved in T cell activation are thought to be appropriate for this signalling, including PLC-γ 49, 50, PKCβ 51 and the mitochondrial Ca2+ uniporter 52.
A downstream step in this signalling pathway is transcription of appropriate genes, and one would assume this is maximally achieved within the range of natural frequencies, hence the necessity of appropriately timed Ca2+ oscillations. It has been shown in previous studies, for multiple genes, that maximal expression is tuned to particular Ca2+ signalling characteristics. 11, 44, 53–55. In our approach we conclude H2O2 signalling modulates Ca2+ oscillatory kinetics within an effective range necessary for downstream transcription factor activation. Our findings implicate connections between Ca2+ and H2O2 that tune frequency behaviour of Ca2+ signalling, thus modulating Ca2+ signalling in a less explored domain and demonstrating the necessity to better characterize the dynamic regulation. This also establishes the ability of our approach to investigate the connection between a plethora of chemical cues to better understand the unaltered underlying chemical signalling.
Our investigation of Ca2+ kinetics is the first to compare the Ca2+ response in Jurkat as a Bode plot across frequencies. By optimal fitting of our cellular responses to a 2nd order transfer function, (Figure 6), we have generated a simple model representation of complex behaviour with 3 parameters: ωn, ζ, and K. The benefit of such a reduced system while retaining the pertinent responses in the frequency domain is the utility for in silico perturbation of the interactions between Ca2+ and H2O2. The transfer function can be compared with subsets of mechanistic models to determine dominant feedback controls of the large system through model reduction. Furthermore, in future work the specific topological features of the Ca2+ and H2O2 regulatory networks could be mapped to the control systems-derived parameters discovered here. The molecular contributors, for example, to the damping coefficient ζ would exert significant control over cellular information processing by sustaining dynamic cues into frequency encoded transcriptional responses or curbing the signal. More importantly to near term applications of this work, however, is that the transfer function will be capable of predicting cellular responses to any arbitrary input function 34 that may be delivered to the cells, such as a pre-experiment on-chip assessment of highly active cells.
Lastly, all experimental results showed great population heterogeneity, with a wide range of responses that could not be easily captured with any other conventional experimental platform such as flow cytometry. These single-cell differences could be the result of stochastic differences in gene expression, growth phase of the cells, or epigenetic alterations. Maintaining a plethora of responses at the population level would be advantageous for mounting an effective immune response 56 by being more or less responsive (gain) to a given input and filtering dynamic inputs with more or less stringency. Future experiments may attempt to control or visualize more of these levels of variability for a more complete understanding of the underlying mechanisms.
Conclusions
Oscillating cytoplasmic calcium spikes have been shown to encode a variety of transcription factors in a complex way 11. The ability to extract more of this encoded information using frequency response analysis will shed light on potential dominant feedback connections and new therapeutic targets. We report a widely applicable platform of technology advancements that enable complex and intelligent perturbation of Ca2+ signalling. This includes a microfluidic device enabling uniform, dynamic stimulation of suspension cells, and a set of analysis techniques to gain single-cell resolution. Together, this approach can be utilized for different cell types and signalling molecules of interest. We demonstrated this applicability on Jurkat T cells by probing Ca2+ dynamics with H2O2 and found novel biological insight that Ca2+ signals exhibit a natural frequency and our results align with literature reports of maximal downstream transcription. We highlight the potential for different cells within the population to have distinct and separate filtering capabilities, an unprecedented understanding of the natural responses of Jurkat T cells to frequency-based stimulation.
Experimental Methods
Microfluidic Device Fabrication
Devices were moulded with polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning) and two master wafers as previously described 33. Both layers were moulded with a 10:1 mixture of PDMS pre-polymer to cross linker. The first layer of PDMS was spun on the master with cell traps to a level of ~10–12 μm so the pore structures that connect the two layers were above the PDMS and thus created holes for fluid to flow through. This layer was baked at 70°C for approximately 15 minutes, until partially cured. The second layer was poured on the stimulus chamber wafer to a height of ~2–3 mm and also partially cured at 70°C for 20 minutes. Once both layers were partially cured, the stimulus chamber layer was cut, aligned with the cell trap layer, and thermally bonded for an additional 40 minutes at 70°C. Once cured, holes were punched with a 19-gauge needle and the two-layer PDMS device was plasma bonded onto a clean glass slide.
Cell Culture and Treatments
Experiments were performed on the Jurkat E6-1 human acute T cell lymphoma cell line (American Type Culture Collection) grown in conditions as described before 34. Briefly, the cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 without Phenol Red (Lonza) and with L-glutamine (Sigma-Aldrich), supplemented with 10 mM HEPES buffer (Corning), 1 mM sodium pyruvate (Cellgro), 50 units mL-1 penicillin-streptomycin (Cellgro), 1× MEM nonessential amino acids (Cellgro), and 10% fetal bovine serum (Sigma-Aldrich).
To visualize cytoplasmic calcium in response to varying experimental conditions, the cells were loaded with 5 μM Fluo-3 AM, cell permeant (Life Technologies) and 0.05% w/v Pluronic F-127 (Sigma-Aldrich) for 30 minutes at 37°C. Cells were subsequently washed with PBS and resuspended in complete RPMI media without Phenol Red. Cells were loaded into the device at a density of 1 × 106 cells/mL.
Device Operation
Devices were primed with 2% BSA in PBS to prevent unintentional cell-adhesion and non-specific binding. Once all bubbles were removed, the top, stimulus chamber, was connected to two pressurized reservoirs of fluid at 1 psi. Specifically, these reservoirs contained either complete media or complete media with the addition of 10, 25, 50, or 100 μM H2O2. Cells were loaded into the device via gravity driven flow as previously described 33. Once cells were loaded, the different solutions were delivered at alternating frequencies, as defined with user input to custom MATLAB (MathWorks™) scripts, which controlled off-chip pinch solenoid valves.
Image Acquisition
The device was positioned on the automatic stage of an inverted Nikon Eclipse Ti inverted fluorescent microscope and time-lapse microscopy was automated using Elements Software (Nikon). Cells were imaged at 10× magnification at a single position on the stage that encompassed cells loaded into the traps. Images were acquired using a FITC filter (Omega XF22) with exposure time of 800 ms. The sampling rate was every 6 s for a duration of 18–60 minutes, depending on the oscillatory input condition.
Frequency analysis for single-cell calcium traces
The device was positioned on the automatic stage of an inverted Nikon Eclipse Ti inverted fluorescent microscope and time-lapse microscopy was automated using Elements Software (Nikon). Cells were imaged at 10× magnification at a single position on the stage that encompassed cells loaded into the traps. Images were acquired using a FITC filter (Omega XF22) with exposure time of 800 ms. The sampling rate was every 6 s for a duration of 18–60 minutes, depending on the oscillatory input condition.
Frequency analysis for single-cell calcium traces
As experimental design becomes increasingly complex and capable of collecting large quantities of single-cell data, analysis is becoming dependent on automated techniques to identify cells and collect metrics of interest over time. Many techniques available are unable to discern anomalous or unrelated features that are similar to those of interest. To combat this, our research first utilizes a manual approach for identifying cells of interest, and then builds on a combination of Relative Difference Filtering and Clustering (RDFC), which has shown to be useful for numerous systems with a predictable pattern of signal. Collected images were analysed using custom MATLAB® scripts (MathWorks™). First, cells were automatically identified in one of the initial images using a previously described relative difference filtering and clustering (RDFC) approach (Zhao et al., In Review). Once the mask was created, it was applied to the entire image sequence and the average fluorescent intensity of the cells was calculated and the local background was subtracted, based on a small area to the top left of each identified cell. Cell traces were discarded if the average fluorescent intensity was negative for any value in the measured time points, as this indicated the background subtraction being above the mean fluorescent intensity, suggesting the identified cell may have squeezed through the trap and was no longer present. For the cells that remained in the trap through the duration of the experiment, the average intensity was normalized via a linear transformation such that each cell’s signal varies from 0 to 1. This step helped to reduce variation in initial loading of cell dye.
Once the normalized fluorescent intensity was calculated for each cell, it was then analysed via a modified spectral analysis GUI, as originally developed by Uhlén in 200439. This GUI takes the single-cell signals and first fits a second order polynomial to each individually and subtracts this from the signal to remove artefacts from the experimental conditions 39, such as photo-bleaching of the cytoplasmic calcium indicator, Fluo-3. The Fourier transform is then taken of the signal to identify dominant frequencies in the signals. For each cell, the power spectral density is normalized to a total area of 1 and the area under the curve for each frequency is calculated to determine these dominant frequencies. It is then compiled into histograms with each identified frequency being multiplied by the relative power of the frequency and combined with the frequency information from all other cells in the population and plotted in a weighted histogram. This histogram is then normalized such that the total area under the curve is equal to 1 with bins of width 0.5 mHz.
Transfer function model fitting
Parameters to the 2nd order system function were fit to the median of the single-cell data at each frequency by using the same approach as for the ROS Ca2+ model. These parameters include K, the system gain, ζ, the damping coefficient, and ωn, the systems natural frequency in rad/s. The parameters are allowed to vary between given bounds while evaluating the genetic algorithm (ga) in the MATLAB® Optimization Toolbox™.
Supplementary Material
T cells contribute to adaptive immunity by integrating complex extracellular information and directing cellular behavior with exquisite sensitivity. We present innovation with a microfluidic platform capable of applying robust, oscillatory stimuli of hydrogen peroxide to Jurkat T cells while simultaneously imaging the dynamic calcium response. With this platform, we present insight of the interaction between calcium and reactive oxygen species during T cell activation, with identification of a natural frequency of the system and demonstration of heterogeneity in resulting calcium signal frequency between individual cells. This integration of a microfluidic device with live cell imaging enables the application of control theory for probing and analyzing T cell signaling in an unprecedented manner.
Acknowledgments
The authors would like to thank Margaret Gran and Linda Kippner for their invaluable assistance with cell culture. We also wish to thank Luye He for his technological assistance, and financial support to AK provided through NSF Graduate Research Fellowship Grant DGE-1148903, P.E.O. Scholar Award, and NIH Training Grant 32GM105490, and the Love Family Foundation to HL. This work was supported by NIH R01AI088023 to MLK and HL.
Footnotes
Electronic Supplementary Information (ESI) available: [details of any supplementary information available should be included here]. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.DiPaolo RJ, Brinster C, Davidson TS, Andersson J, Glass D, Shevach EM. Journal of immunology. 2007;179:4685–4693. doi: 10.4049/jimmunol.179.7.4685. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Yamane H, Paul WE. Annual review of immunology. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu J, Paul WE. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, Bryce PJ, Chandel NS. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feske S, Skolnik EY, Prakriya M. Nature Reviews Immunology. 2012;12:532–547. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis RS. Biochem Soc T. 2003;31:925–929. doi: 10.1042/bst0310925. [DOI] [PubMed] [Google Scholar]
- 7.Skupin A, Kettenmann H, Falcke M. PLoS computational biology. 2010:6. doi: 10.1371/journal.pcbi.1000870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skupin A, Kettenmann H, Winkler U, Wartenberg M, Sauer H, Tovey SC, Taylor CW, Falcke M. Biophysical journal. 2008;94:2404–2411. doi: 10.1529/biophysj.107.119495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis RS. Annual review of immunology. 2001;19:497–521. doi: 10.1146/annurev.immunol.19.1.497. [DOI] [PubMed] [Google Scholar]
- 10.Dolmetsch RE, Lewis RS. J Gen Physiol. 1994;103:365–388. doi: 10.1085/jgp.103.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolmetsch RE, Xu K, Lewis RS. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 12.Kwon J, Shatynski KE, Chen H, Morand S, de Deken X, Miot F, Leto TL, Williams MS. Science signaling. 2010;3:ra59. doi: 10.1126/scisignal.2000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dwivedi G, Kemp ML. Antioxidants & redox signaling. 2012;16:374–380. doi: 10.1089/ars.2011.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths HR. Redox report : communications in free radical research. 2005;10:273–280. doi: 10.1179/135100005X83680. [DOI] [PubMed] [Google Scholar]
- 15.Kesarwani P, Murali AK, Al-Khami AA, Mehrotra S. Antioxidants & redox signaling. 2013;18:1497–1534. doi: 10.1089/ars.2011.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorn GW, Maack C. Journal of Molecular and Cellular Cardiology. 2013;55:42–49. doi: 10.1016/j.yjmcc.2012.07.015. [DOI] [PubMed] [Google Scholar]
- 17.Csordas G, Hajnoczky G. Bba-Bioenergetics. 2009;1787:1352–1362. doi: 10.1016/j.bbabio.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trebak M, Ginnan R, Singer HA, Jourd’heuil D. Antioxidants & redox signaling. 2010;12:657–674. doi: 10.1089/ars.2009.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abramson JJ, Zable AC, Favero TG, Salama G. Journal of Biological Chemistry. 1995;270:29644–29647. doi: 10.1074/jbc.270.50.29644. [DOI] [PubMed] [Google Scholar]
- 20.Bootman MD, Berridge MJ, Taylor CW. The Journal of physiology. 1992;450:163–178. doi: 10.1113/jphysiol.1992.sp019121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bootman MD, Taylor CW, Berridge MJ. Journal of Biological Chemistry. 1992;267:25113–25119. [PubMed] [Google Scholar]
- 22.Oda T, Yang Y, Uchinoumi H, Thomas DD, Chen-Izu Y, Kato T, Yamamoto T, Yano M, Cornea RL, Bers DM. J Mol Cell Cardiol. 2015;85:240–248. doi: 10.1016/j.yjmcc.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav VR, Song T, Joseph L, Mei L, Zheng YM, Wang YX. American journal of physiology Lung cellular and molecular physiology. 2013;304:L143–151. doi: 10.1152/ajplung.00310.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambeth JD, Kawahara T, Diebold B. Free radical biology & medicine. 2007;43:319–331. doi: 10.1016/j.freeradbiomed.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gincel D, Zaid H, Shoshan-Barmatz V. The Biochemical journal. 2001;358:147–155. doi: 10.1042/0264-6021:3580147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toettcher JE, Weiner OD, Lim WA. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobel S, Valero A, Latt J, Renaud P, Lutolf M. Lab on a chip. 2010;10:857–863. doi: 10.1039/b918055a. [DOI] [PubMed] [Google Scholar]
- 28.Chung K, Kim Y, Kanodia JS, Gong E, Shvartsman SY, Lu H. Nature methods. 2011;8:171–176. doi: 10.1038/nmeth.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramji R, Wong VC, Chavali AK, Gearhart LM, Miller-Jensen K. Integr Biol. 2015;7:998–1010. doi: 10.1039/c5ib00094g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Lab on a chip. 2010;10:1391–1400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Nature methods. 2009;6:147–152. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chung K, Rivet CA, Kemp ML, Lu H. Analytical chemistry. 2011;83:7044–7052. doi: 10.1021/ac2011153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chingozha L, Zhan M, Zhu C, Lu H. Analytical chemistry. 2014;86:10138–10147. doi: 10.1021/ac5019843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He LY, Kniss A, San-Miguel A, Rouse T, Kemp ML, Lu H. Lab on a chip. 2015;15:1497–1507. doi: 10.1039/c4lc01070a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mandikova J, Volkova M, Pavek P, Cesnek M, Janeba Z, Kubicek V, Trejtnar F. Toxicology. 2013;311:135–146. doi: 10.1016/j.tox.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 36.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. Science. 2008;319:482–484. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennett MR, Pang WL, Ostroff NA, Baumgartner BL, Nayak S, Tsimring LS, Hasty J. Nature. 2008;454:1119–1122. doi: 10.1038/nature07211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jovic A, Wade SM, Neubig RR, Linderman JJ, Takayama S. Integrative biology : quantitative biosciences from nano to macro. 2013;5:932–939. doi: 10.1039/c3ib40032h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uhlen P. Science’s STKE : signal transduction knowledge environment. 2004:pl15. doi: 10.1126/stke.2582004pl15. [DOI] [PubMed] [Google Scholar]
- 40.Muller J, Kuttler C, Hense BA. Bio Systems. 2008;92:76–81. doi: 10.1016/j.biosystems.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 41.Balazsi G, van Oudenaarden A, Collins JJ. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang HW, Zheng L, Othmer HG. Interface focus. 2012;2:465–486. doi: 10.1098/rsfs.2011.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng L, Chen M, Nie Q. Journal of Mathematical Physics. 2012;53:115616. doi: 10.1063/1.4762825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smedler E, Uhlen P. Biochimica et biophysica acta. 2014;1840:964–969. doi: 10.1016/j.bbagen.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 45.Berridge MJ, Lipp P, Bootman MD. Nature reviews Molecular cell biology. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 46.Selimkhanov J, Taylor B, Yao J, Pilko A, Albeck J, Hoffmann A, Tsimring L, Wollman R. Science. 2014;346:1370–1373. doi: 10.1126/science.1254933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boulware MJ, Marchant JS. Current biology : CB. 2008;18:R769–r776. doi: 10.1016/j.cub.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parekh AB. Trends in biochemical sciences. 2011;36:78–87. doi: 10.1016/j.tibs.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 49.Cifuentes ME, Honkanen L, Rebecchi MJ. The Journal of biological chemistry. 1993;268:11586–11593. [PubMed] [Google Scholar]
- 50.Rebecchi MJ, Eberhardt R, Delaney T, Ali S, Bittman R. The Journal of biological chemistry. 1993;268:1735–1741. [PubMed] [Google Scholar]
- 51.Kohout SC, Corbalan-Garcia S, Torrecillas A, Gomez-Fernandez JC, Falke JJ. Biochemistry. 2002;41:11411–11424. doi: 10.1021/bi026041k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vinogradov A, Scarpa A. The Journal of biological chemistry. 1973;248:5527–5531. [PubMed] [Google Scholar]
- 53.Tomida T, Hirose K, Takizawa A, Shibasaki F, Iino M. Embo Journal. 2003;22:3825–3832. doi: 10.1093/emboj/cdg381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- 55.Shibasaki F, Price ER, Milan D, McKeon F. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 56.Richard M, Yvert G. Frontiers in Genetics. 2014;5:374. doi: 10.3389/fgene.2014.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.