Abstract
Neutrophils are generally the first immune cells recruited during the development of sterile or microbial inflammation. As these cells express many innate immune receptors with the potential to directly recognize microbial or endogenous signals, we set out to assess whether their functions are locally influenced by the signals present at the onset of inflammation. Using a mouse model of peritonitis, we demonstrate that neutrophils elicited in the presence of C-type lectin receptor ligands have an increased ability to produce cytokines, chemokines and lipid mediators in response to subsequent Toll-like receptor stimulation. Importantly, we found that licensing of cytokine production was mediated by paracrine TNFα-TNFR1 signaling rather than direct ligand sensing, suggesting a form of quorum sensing among neutrophils. Mechanistically, licensing was largely imparted by changes in the post-transcriptional regulation of inflammatory cytokines, whereas production of IL-10 was regulated at the transcriptional level. Altogether, our data suggest that neutrophils rapidly adapt their functions to the local inflammatory milieu. These phenotypic changes may promote rapid neutrophil recruitment in the presence of pathogens while limiting inflammation in their absence.
Introduction
Innate immune responses can be initiated by a variety of signals broadly classified as either microbial or endogenous, the latter of which are the main drivers of sterile inflammation(1, 2). In responses induced by either class of ligand, recruitment of neutrophils is typically one of the earliest events. Neutrophils represent the most abundant leukocyte in circulation and are critical for elimination of microbial pathogens (3). However, neutrophil activation can also lead to tissue damage, such as joint and cartilage erosion during rheumatoid arthritis (4). Neutrophil recruitment can be mediated by multiple factors either produced by host cells, such as chemokines (e.g., CXCL1) and lipid mediators, or released by bacteria and mitochondria, such as formylated peptides. Engagement of specific G-protein coupled receptors expressed by neutrophils (CXCR2, BLT1, and members of the formylated peptide receptor (FPR) family) leads to their extravasation and migration to the site of injury or infection (5). Neutrophils also express many innate immune receptors, including Toll-Like Receptors (TLR) and C-type Lectin Receptors (CLR)(6), allowing them to recognize a large number of molecular patterns associated with both tissue damage and microbes locally at sites of inflammation.
Engagement of TLRs, CLRs or FPRs generally leads to neutrophil activation, potentially inducing degranulation, production of reactive oxygen species and lipid mediators, and, in some cases, release of the cell’s DNA to form neutrophil extracellular traps (NETs). Neutrophils can also produce cytokines and chemokines, such as TNFα (7) and IL-10 (8), although the latter seems to be specific for mouse neutrophils (as discussed in (9)). However, cytokine production by neutrophils has been largely understudied, in part because neutrophils are generally poor protein producers (10) and because of the species differences mentioned above. While neutrophils are generally thought to contribute only modestly to cytokine production when compared with macrophages and monocytes, this issue has remained a subject of debate (11, 12).
There is now accumulating evidence that the functional properties of neutrophils, once thought to be fairly stereotyped, can be altered by the local micro-environment during chronic inflammation or within tumors (13). However, it remains unclear how local cues can influence neutrophils during the onset of inflammation. We reasoned that different inflammatory milieus may modify the capacity of neutrophils to produce anti-microbial compounds and pro- and anti-inflammatory cytokines. Such a mechanism could maximize elimination of microbes while limiting tissue damage during sterile inflammation.
To address these questions, we compared the functional properties of murine neutrophils recruited to different types of peritoneal inflammation. We found that neutrophils recruited by microbial ligands displayed increased cytokine production compared to naïve neutrophils or neutrophils recruited during sterile inflammation. This difference was mediated by a paracrine signal that we identified as TNFα. Altogether, our results provide evidence for early phenotypic changes in neutrophils and open new avenues to study the regulation of inflammatory responses through the perspective of the most abundant cells present at the onset of inflammation.
Material and methods
Mice and peritonitis model
C57BL/6 WT, C57BL/6.SJL (CD45.1) and TNRF single or double-deficient mice (Tnfrsf1a−/−, Tnfrsf1b−/− and Tnfrsf1a−/− x Tnfrsf1b−/−) were obtained from Jackson laboratories. Fcer1g−/− deficient mice (14) were a kind gift from Jessica Hammerman. All mice were bred in our colony at University of California, Berkeley, and used at 6–12 weeks of age. All experiments were approved by the local Animal Care and Use Committee (Animal Use Protocol R298-0816BR). Bone marrow chimeras were generated by irradiating mice in an X-ray irradiator (900 rads split across 2 doses, 5 hours apart). The irradiated mice were reconstituted by injection of 5×106 donor bone marrow cells i.v. immediately after the second irradiation.
Peritonitis was induced by intra-peritoneal injection of 100μL of a 10% w/v sonicated solution of uric acid (15) (Sigma), 500μg of zymosan or 100μg TDB (both from Invivogen) in 200μL PBS. After 16 or 42 hours, peritoneal cells were collected by lavage with 5mL PBS 2% FCS.
Bone marrow neutrophil harvest
Bone marrow was harvested by flushing the femur and tibia of mice with complete RPMI. Cells were passed through a 70μm strainer and RBC were lysed with ACK (Gibco).
For neutrophil purification, total bone marrow cells were underlayed with a solution of 68% isotonic Percoll (i.e. 90% Percoll + 10% 10x PBS) and 79% isotonic Percoll, then spun for 30 minutes at 750g without brake. Neutrophils were collected at the 68/79 interface. Neutrophil purity was >90% as tested by flow cytometry after Ly6G-staining.
Flow cytometry and intracellular cytokine staining
Peritoneal cells, total bone marrow cells or purified neutrophils were plated in complete RPMI. For ex vivo experiments, cells were directly restimulated with TLR ligands (Pam3CSK4, 500ng/mL, LPS, 500ng/mL, ultrapure flagellin, 1μg/mL, R848, 1μM, CpG-B ODN 1668, 1μM, all from Invivogen; Sa23, 4μM, from IDT DNA). After 30 minutes, brefeldin (GolgiPlug, BD Biosciences) was added to cells before incubation for another 5h30.
For in vitro experiments, cells were first stimulated as indicated (zymosan, 5μg/mL, TDB, 1μg/mL, both from Invivogen, recombinant TNFα aa84-235 at the indicated concentrations, from R&D, or supernatants) for 12–16h, before restimulation as described above.
At the end of the restimulation, cells were washed once in PBS, stained with anti-Ly6G, Ly6C, CD11b and CD62L antibodies in the presence of FcBlock. Cells were then fixed and permeabilized (Cytofix/perm kit, BD Biosciences) then stained for TNFα.
For signaling assays, cells were immediately fixed with 1.6% PFA at room temperature after restimulation, then washed and resuspended in ice-cold methanol. After 3h of incubation at −20°C, cells were stained with FcBlock, stained with anti-p-Erk, p-p38 and IκBα for 1h at room temperature.
For measurements of de novo protein synthesis, cells were incubated for 1h with 50μM O-propargyl-puromycin (Jena Bioscience) after overnight stimulation with TNFα as described above. When indicated, cyclohexamide (Sigma, 10μg/mL) was added 30 minutes before incubation with OP-puromycin. At the end of the incubation, cells were fixed and permeabilized as described above, then stained with Click-IT reaction kit and Alexa488-azide (Invitrogen) according to manufacturer’s instructions. After 2 washes, cells were stained with antibodies as described above.
A complete list of antibodies, fluorophores, clones and providers is available in supplemental table 1. All cells were analyzed on an LSR II or LSR Fortessa (BD Biosciences), sorting was performed on a MoFlo (Beckman Coulter) where indicated, and data was analyzed with FlowJo (TreeStar, v9.9.3). Absolute cell counts were obtained by adding 10μL of CountBright counting beads (Invitrogen) to each sample.
Bacterial killing and reactive oxygen species production
To determine bacterial killing, 105 purified neutrophils were incubated with 103 serum-opsonized bacteria (E.coli, K12 strain) for 1h at 37°C in RPMI (complete w/o Pen/Strep)(16). Cultures were then plated on agar plates and the killing efficiency was calculated as the ratio of CFU to the CFU obtained in the absence of neutrophils.
Reactive oxygen species production was measured by culturing 105 purified neutrophils in HBSS in the presence of 50μM luminol and 4U/mL HRP (Sigma). Luminescence was measured on a L384 luminometer every 30s, for 10 minutes before and 20 minutes after the addition of 1μM fMLP (Sigma).
Supernatant production and protein purification, cytokine measurements
Bone marrow cells or RAW.264 macrophages were stimulated in the presence of curdlan (100ug/mL, from Invivogen) for 12–16 hours. Supernatants were harvested after centrifugation for 10 min at 4000g, and further filtered through a 0.22μm syringe filter.
Supernatants were either used directly or concentrated and exchanged into 20mM Tris pH 8 using 30K ultrafiltration membranes (Millipore). Samples were then loaded on a HiTrap Q HP column (GE) using an NGS chromatography system (Biorad) and eluted over 40mL of a 0–400mM NaCl gradient, with 1 fraction collected each mL.
2D gel electrophoresis of the concentrated samples or fractions, spot picking and mass spectrometry identification of candidates was performed by Applied Biomics.
Measurements of cytokine levels in supernatants were performed using a custom-ordered Bio-Plex assay (Biorad) according to manufacturer’s instructions.
Quantitative RT-PCR
Neutrophils were lysed in RNAzol (MRC) and RNA was purified according to manufacturer’s instructions and concentrated using on a column (RNA Clean and Concentrator, Zymo Research). cDNA was prepared with iScript cDNA synthesis kit (Biorad), and quantitative PCR was performed with SYBR Green Master mix (Biorad) on a StepOnePlus thermocycler (Applied Biosystems). Primers were obtained from PrimerBank (17) and synthesized by IDT (see supplemental table 2).
Lipid mediator production assay
Neutrophils were harvested and purified from bone marrow as described above, and plated at 5×106 cells/mL in cRPMI with 10ng/mL TNFα. After 12h, cells were restimulated for 30 min with Pam3CSK4 or fMLP. Eicosanoids in neutrophils and secreted into the culture medium were quantified via liquid chromatography-tandem mass spectrometry (LC-MS/MS) as described before (18–20). In brief, cold methanol was added to cultured neutrophils in media to stabilize lipid mediators. Deuterated internal standards (PGE2-d4, LTB4-d4, 15-HETE-d8, LXA4-d5, DHA-d5, AA-d8) were added to samples to calculate extraction recovery. LC/MS/MS system consisted of an Agilent 1200 Series HPLC, Kinetex C18 minibore column (Phenomenex, Torrance, CA, USA), and AB Sciex QTRAP 3200 mass spectrometer. Analysis was carried out in negative ion mode, and lipid mediators quantitated using scheduled multiple reaction monitoring (MRM) mode using specific and established transition ions.
Statistical analysis
Statistical significance was assessed using either a two-tailed t-test for unpaired data points (comparison of two groups), or in the case of multiple comparisons, an ANOVA followed by Tukey’s post test. All tests were performed using Prism (GraphPad, v6). *, ** and *** indicate p-values lower than 0.05, 0.01 and 0.001, respectively.
Results
Microbial signals can license neutrophils for increased cytokine production
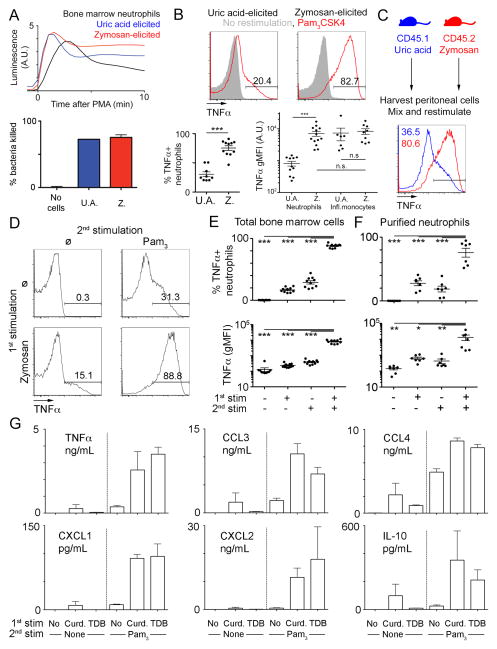
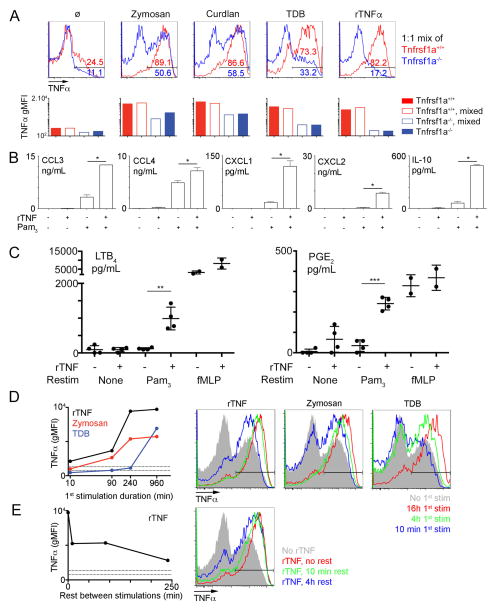
To investigate how different inflammatory conditions may influence neutrophil functions, we harvested neutrophils from the peritoneal cavity of mice injected with uric acid or zymosan to mimic sterile or microbial inflammation, respectively. As expected, neutrophils recruited to the peritoneal cavity by either stimuli displayed signs of priming, as evidenced by increased reactive oxygen (ROS) production (Fig. 1A, top). We also observed no difference in bacterial killing when neutrophils elicited with uric acid or zymosan were co-incubated with serum-opsonized bacteria (Fig. 1A, bottom). Next, we assessed neutrophil cytokine production by intracellular TNFα staining following restimulation with the synthetic TLR2 ligand Pam3CSK4. Surprisingly, only neutrophils harvested from mice injected with zymosan produced significant amounts of TNFα, while neutrophils harvested from mice injected with uric acid produced only low amounts of this cytokine (Fig. 1B). We also examined TNFα production by inflammatory monocytes restimulated in the same well. Interestingly, we did not observe any changes in levels of TNFα produced by inflammatory monocytes, but neutrophils elicited in the presence of zymosan produced similar levels of TNFα as inflammatory monocytes (Fig. 1B). The observed increase in neutrophil cytokine production could be cell-intrinsic or mediated by differences occurring during the restimulation, such as different cell populations present in the peritoneal lavage. To distinguish between these possibilities, we injected congenically distinct mice with uric acid or zymosan and then mixed and restimulated the harvested cells with TLR2 ligands (Fig. 1C). In these conditions, neutrophils harvested from mice injected with zymosan clearly produced higher levels of TNFα than cells from mice injected with uric acid, demonstrating that the increase in cytokine production is intrinsic to neutrophils. Given that neutrophils recruited to the peritoneum in either condition were capable of efficient ROS production and bacterial killing, a phenotypical change referred to as ‘priming’, we decided to refer to the increase in cytokine production observed only in zymosan elicited neutrophils as ‘licensing’, to clearly distinguish the two phenomena.
Figure 1. Neutrophil cytokine production is licensed locally by microbial ligands.
A. Representative luminescence after PMA stimulation in the presence of luminol (upper panel) of neutrophils purified from the peritoneal cavity of mice injected with uric acid (blue) or zymosan (red), or as a control, from the bone marrow of uninjected mice (black). Average bacterial killing by neutrophils purified as above, or in the absence of any neutrophils (lower panel).
B. Representative flow cytometry plots gated on neutrophils in the peritoneal exudate of mice injected with uric acid or zymosan and left unstimulated (grey) or restimulated with Pam3CSK4 (red). Percentage of TNF-positive neutrophils are indicated in the lower left panel. Geometric mean fluorescence intensities (gMFI) for TNFα of neutrophils or inflammatory monocytes in the peritoneal exudate of mice injected with uric acid or zymosan, after Pam3CSK4 restimulation, are indicated in the lower right panel.
C. Representative flow cytometry plot of a mixed culture of cells harvested from mice injected with uric acid (blue) and zymosan (red) after Pam3CSK4 restimulation.
D–F. Representative flow cytometry plots (D) and percentage of TNF-positive neutrophils or gMFI for TNFα (E, F) from neutrophils gated out of total bone marrow cells (D–E) or Percoll-purified neutrophils (F) after stimulation with zymosan or not (1st stim, + and − respectively), followed by restimulation with Pam3CSK4 or not (2nd stim, + and − respectively).
G. Supernatants from purified neutrophils (as in F) stimulated with media alone, Curdlan or TDB and restimulated with or without Pam3CSK4 were analyzed by Luminex for the indicated cytokines.
Data were representative of 2 (A, C, G), 6 (B) or 10 (D) experiments. Dots plots (B, E, F) are pooled from 6–10 mice from 6 experiments. *, p<0.05;**, p<0.01;***, p<0.001
As neutrophils are among the first immune cells to infiltrate the peritoneum during local inflammation, we hypothesized that they directly sense signals delivered by zymosan in the peritoneum, resulting in an increase in their ability to produce cytokines. To test our hypothesis, we used bone marrow as a source of naïve neutrophils and pre-treated bone marrow cells with zymosan before restimulation with Pam3CSK4. Similar to our in vivo experiments, we observed a striking increase in the TNFα production by neutrophils pre-treated with zymosan before stimulation with Pam3CSK4 (Fig. 1D–E).
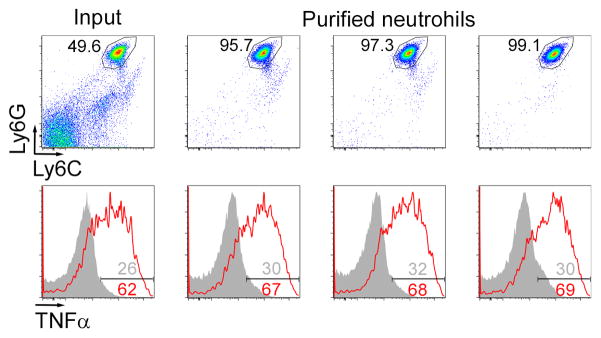
We next performed experiments with purified bone marrow neutrophils and observed similar results, suggesting that neutrophils can acquire the capacity to produce cytokines directly, in the absence of accessory cells (Fig. 1F) Of note, as Percoll purified neutrophils are 90–95% pure, we also sorted neutrophils by FACS to 97–99% purity and restimulated them as above (Fig. 2). Although we did observe slightly lower levels of cytokine production, perhaps due to a decrease in viability after sorting, licensing was similar in total bone marrow (input) and highly purified cells. Using purified cells also allowed us to directly measure cytokines secreted by neutrophils in the supernatant. Strikingly, induction of the anti-inflammatory cytokine IL-10 and several chemokines (CCL3-4, CXCL1-2) produced by neutrophils followed a pattern similar to TNFα (Fig. 1G). Moreover, other CLR ligands, such as curdlan or the synthetic Mincle/MCL ligand TDB (Trehalose Di-Behenate), also strongly increased secretion of cytokines in response to TLR2 ligands (Fig. 1G, top left).
Figure 2. Licensing of highly purified neutrophils.
Representative flow cytometry plots of total bone marrow (input) or FACS-purified neutrophils showing purity (top) and intracellular TNFα levels (bottom) after Pam3CSK4 restimulation (red) or no stimulation (grey). Numbers indicate the percentage of neutrophils (top) or percentage of TNFα-positive cells (bottom). Data are representative of 2 independent experiments. Neutrophils were sorted on a MoFlo (Beckman Coulter) either on the basic of FSC-SSC in the absence of antibodies (columns 1 and 2) or on the basis of Ly6G expression (column 3).
Altogether, these experiments suggest that the ability of neutrophils to produce cytokines is modulated during certain types of inflammation. In particular, the low cytokine production exhibited by naïve neutrophils can be greatly increased by microbial signals. We propose to call this process neutrophil licensing to distinguish it from neutrophil priming, the rapid differentiation process that neutrophils undergo when recruited by either microbial or sterile inflammatory signals.
Licensing is mediated by a paracrine signal
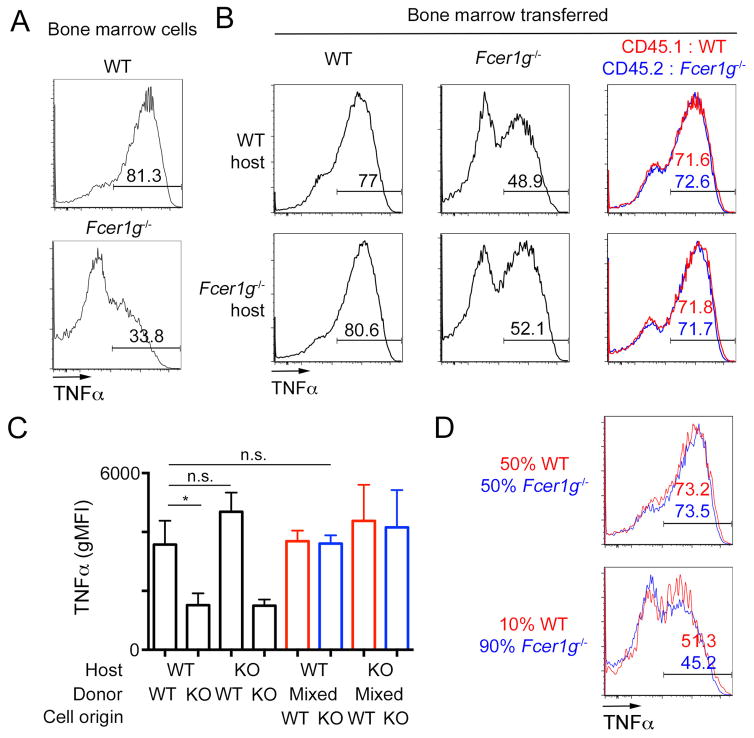
Next, we sought to determine the nature of the signal that licenses neutrophils for cytokine production. Our experiments with purified neutrophils argued for direct sensing of ligands by neutrophils (Fig. 1F), but these results did not rule out the possible contribution of other cells in vivo. The synthetic CLR ligand TDB is recognized by Mincle and MCL, and both receptors depend on the adaptor protein FcεRIγ for signaling as well as cell surface expression (21). In agreement with these previous studies, we observed TDB-induced neutrophil licensing in wild-type (WT) but not Fcer1g−/− mice (Fig. 3A). To determine if hematopoietic cells are responsible for ligand sensing and neutrophil licensing in vivo, we generated bone-marrow chimeras with wild type mice and mice deficient for Fcer1g. In chimeras generated with Fcer1g−/− bone marrow, TDB injection in the peritoneum failed to induce an increase in the TNFα production by the recruited neutrophils (Fig. 3B–C, left). The requirement for Fcer1g was restricted to hematopoietic cells, consistent with a model in which direct sensing of the ligand by neutrophils is sufficient for licensing. However, in mixed bone marrow chimeras receiving both WT and Fcer1g−/− cells, injection of TDB led to a similar increase in neutrophil cytokine production in both WT and Fcer1g−/− cells (Fig. 3B–C, right), even though the latter population cannot directly recognize TDB. This result raised the possibility that neutrophil licensing can also occur via paracrine signaling. Indeed, Fcer1g−/− neutrophils could be licensed when co-cultured with WT cells in vitro (Fig. 3D). Interestingly, at low ratios of WT/Fcer1g−/− cells, we noticed a reduction in the TDB-induced increase in TNFα production in both WT and Fcer1g−/− neutrophils (Fig. 3D). This result suggests that even though WT cells can directly sense TDB, the paracrine signal is necessary for efficient licensing, and that this signal becomes limiting with low numbers of WT cells.
Figure 3. Neutrophil licensing is mediated by a paracrine signal.
A. Representative flow cytometry plots gated on neutrophils from the indicated mice after in vitro stimulation of bone marrow cells with TDB followed by Pam3CSK4.
B–C. Representative flow cytometry plots gated on neutrophils harvested from chimeras generated by transfer of CD45.1 WT cells (red), CD45.2 Fcer1g−/− cells (FcRγ-deficient, blue) or a 1:1 mix of those cells on WT or Fcer1g−/− hosts. Mice were injected with TDB, and peritoneal exudates were restimulated with Pam3CSK4. C. Quantification of the gMFI in the samples described above
D. Congenically marked cells from the indicated mice were mixed at a 1:1 or 1:9 ratio before restimulation as above. Representative overlays of CD45.1 WT cells (red) and CD45.2 Fcer1g−/− neutrophils (blue) are shown. Data presented are representative of 3 independent experiments with 3 mice per group.
Identification of TNFα as a paracrine licensing factor
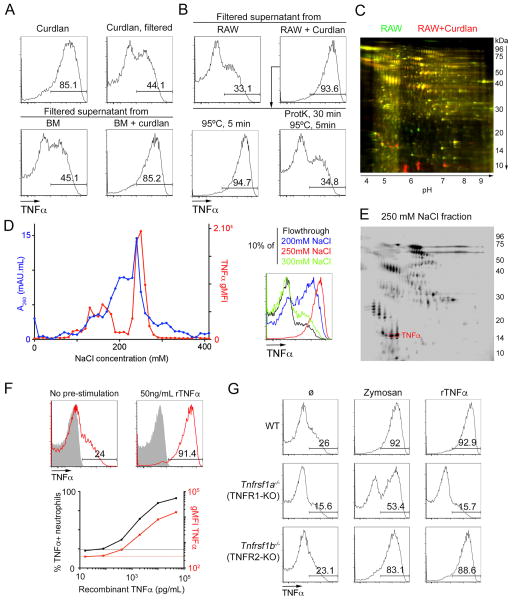
Paracrine activation can be mediated either by soluble factors or by cell-cell interactions. If a soluble signal is sufficient for neutrophil licensing, we reasoned that supernatant from activated cells should be able to license naïve neutrophils even in the absence of the original microbial ligand. To test this hypothesis, we took advantage of the fact that the curdlan, another Dectin-1 agonist, is composed of large particles, which can easily be removed from solution by filtration. Indeed, while curdlan can directly induce neutrophil licensing, passage through a 0.22μm filter abrogated this effect (Fig. 4A, top). However, when we treated bone marrow cells with curdlan, collected and filtered the supernatant of those cells, and transferred the supernatant to naïve cells, we observed robust neutrophil licensing, demonstrating the presence of a soluble licensing factor (Fig. 4A, bottom). Next, we tested whether other cell lines might be able to produce a soluble factor with the same properties in order to generate enough material for purification and identification. The macrophage cell line RAW 264.7 also produced significant levels of a licensing factor after treatment with curdlan (Fig. 4B). Treatment with proteinase K completely abrogated the activity of the supernatant, strongly suggesting that the soluble factor is a protein (Fig. 4B).
Figure 4. Identification of TNFα as the licensing factor.
A–B. Representative flow cytometry plots of bone marrow neutrophils incubated with curdlan, 0.22um-filtered curdlan (A, top) or 0.22um-filtered supernatants of bone marrow cells stimulated with curdlan or not (A, bottom) or 0.22um-filtered supernatants of RAW 264.7 cells stimulated with curdlan or not (B, top). Supernatants of RAW 264.7 cells stimulated with curdlan were also treated with proteinase K and incubated for 5 min at 95°C before incubation with neutrophils (B, bottom). Data are representative of three independent experiments.
C. 2D gel electrophoresis of total supernatants from RAW 264.7 cells (green) or RAW 264.7 cells stimulated with curdlan (red).
D. Supernatants from RAW 264.7 cells stimulated with curdlan were fractionated by anion exchange chromatography, and naïve neutrophils were incubated with fractions, indicated by the NaCl at which the fraction eluted. Total protein in the fraction (in A260.mL, in blue) and gMFI for TNFα in the neutrophils incubated with that fraction and restimulated with Pam (left panel). Representative plots of neutrophils incubated with a 1:10 dilution of the flow-through (=0mM NaCl) or the indicated fractions and restimulated with Pam3CSK4 (right panel).
E. 2D gel electrophoresis of proteins present in the 250mM NaCl fraction obtained as in D.F. (Top) Representative flow cytometry plots of neutrophils gated from bone marrow cells left untreated or incubated with 50ng/mL recombinant TNFα, then restimulated with Pam3CSK4 (red) or not (solid grey).
(Bottom) Percentage of TNFα+ neutrophils and gMFI for TNFα in neutrophils pretreated with the indicated concentrations of TNFα, then restimulated with Pam3CSK4. Dotted lines represent these values for neutrophils in the absence of recombinant TNFα.
G. Representative flow cytometry plots gated on neutrophils from the indicated mice after in vitro stimulation of bone marrow cells with zymosan or 50ng/mL recombinant TNFα or in the absence of pre-stimulation (ø), followed by restimulation with Pam3CSK4. Data are representative of 3 independent experiments.
To identify potential candidates, we used two parallel approaches. First, we performed 2D-gel electrophoresis on supernatants from untreated RAW 264.7 cells and cells treated with curdlan, to identify proteins secreted specifically in the presence of curdlan (Fig. 4C). We also performed anion exchange chromatography on the supernatant and tracked the licensing activity across 40 fractions. Most of the licensing activity eluted in a single peak around 250mM NaCl (Fig. 4D). We then included that fraction in a 2D-gel electrophoresis assay together with the original active and control supernatants, to detect proteins that are specifically enriched after purification by anion exchange chromatography (Fig. 4E). We then selected spots corresponding to proteins induced by curdlan and enriched in the fraction eluted at 250mM NaCl for identification by mass spectrometry (Fig. 4E).
TNFα itself was present in the candidate proteins identified by purification, so we tested whether recombinant TNFα can directly induce neutrophil licensing. Bone marrow neutrophils incubated with recombinant TNFα before restimulation produced significantly higher levels of TNFα after restimulation with Pam3CSK4 (Fig. 4F, top). This effect was dose dependent and robust at concentrations above 10ng/mL (Fig. 4F, bottom). Importantly, treatment with TNFα alone was not sufficient for cytokine production by neutrophils, but required subsequent activation by a TLR2 ligand (Fig. 4F).
TNFα can signal through two related receptors, TNFR1 and TNFR2 (encoded by Tnfrsf1a and Tnfrsf1b, respectively). Deletion of Tnfrsf1a completely abrogated the licensing induced by TNF, whereas deletion of Tnfrsf1b had little to no effect, demonstrating the critical role of TNFR1 in licensing of neutrophils (Fig. 4G). Of note, deletion of Tnfrsf1a had a significant but only partial effect on the cytokine production of neutrophils treated with zymosan (Fig. 4G, middle column), suggesting that some microbial ligands may, directly or through another soluble factor, increase cytokine production to some extent, but that the bulk of this increase is mediated by paracrine TNFR1 signaling.
Altogether, our results suggest that TNFα, through a TNFR1-dependent signaling pathway, mediates an increase in the ability of neutrophils to produce TNFα itself in response to microbial ligands.
TNFα treatment induces long-term changes in neutrophil cytokine and lipid mediator production
To address definitively whether TNFα can directly license neutrophils we examined cytokine production of WT and TNFR1-deficient cells in mixed cultures stimulated with CLR ligands and Pam3CSK4. Under these conditions, we did not observe any rescue of TNFα production by Tnfrsf1a−/− cells when cultured with WT cells (Fig. 5A). These results contrast with what we observed in mixed cultures of WT and Fcer1g−/− cells (Fig. 3D) and confirm that TNFα directly modifies neutrophil cytokine production, rather than causing the induction of another downstream paracrine signal.
Figure 5. Consequences and kinetics of TNFα-mediated licensing.
A. Top. Representative flow cytometry plots gated on neutrophils from a mixed culture of congenically marked WT (red) and TNFR1-KO (blue) bone marrow cells. Cells were stimulated with the indicated ligands, followed by restimulation with Pam3CSK4.
Bottom. Geometric mean fluorescence intensity for TNFα in a mixed culture (empty columns) or individual cultures (full boxes) of WT (red) and TNFR1-KO (blue) neutrophils treated as above.
B. Supernatants from purified neutrophils, stimulated with 10 ng/mL recombinant TNFα or not and restimulated with Pam3CSK4 or not, were analyzed by Luminex for the indicated cytokines.
Data for A, B are representative of 4 independent experiments.
C. Purified neutrophils, stimulated with 10 ng/mL recombinant TNFα or not and restimulated Pam3CSK4, fMLP or not, were lysed in methanol for analysis of lipid mediator production by LC-MS. Data are representative of two independent experiments performed in duplicate.
D. Left. Geometric MFI for TNFα in neutrophils gated from bone marrow cells and treated with the indicated ligands (recombinant TNFα : black, zymosan : red, TDB : blue) for 10min to 16h. All cells were then restimulated with Pam3CSK4 for 6 hours. Right. Representative flow cytometry plots of neutrophils treated with the indicated ligands (recombinant TNFα : left, zymosan : center, TDB : right) or left untreated (solid grey). Colors indicate the duration of treatment before Pam3CSK4 restimulation.
E. Geometric TNFα MFI and representative plots in neutrophils gated from bone marrow cells treated with recombinant TNFα for 12h, then washed and rested for 10 min to 4h, before restimulation with Pam3CSK4. 0 min (=no rest) positive controls were kept in the presence of rTNFα during restimulation. Data for D, E are representative of two independent experiments with duplicates at each timepoint.
Next, we tested whether the effect of TNFα-TNFR1 signaling on TNFα production could be extended to other cytokines produced by neutrophils. Pre-stimulation of neutrophils with TNFα was sufficient to increase production of CXCL1-2, IL-10, and to some extent CCL3-4 in purified neutrophils treated with Pam3CSK4 (Fig. 5B). Again, TNFα stimulation itself did not induce any cytokine production on its own but enhanced TLR-induced production.
A primary function of neutrophils is to generate eicosanoids such as leukotrienes and prostaglandins, which initiate and amplify acute inflammation, adaptive immune responses, vascular leakage and pain (22). Formation of eicosanoids by neutrophils is tightly regulated and usually triggered by G-protein coupled receptors such as FPR and C5AR. To assess whether treatment with TNFα could also modulate eicosanoid production, we measured lipid mediator release in neutrophils treated with Pam3CSK4 for 30 minutes, after a 12h incubation with or without TNFα. As expected, TNFα or TLR stimulation alone did not induce significant production of leukotriene B4 (LTB4) and prostaglandin E2 (PGE2); however we observed a synergistic effect of these two stimuli that induced significant levels of LTB4 and PGE2 release (Fig. 5C). As a control, we also treated neutrophils with fMLP, and, as expected, observed robust lipid mediator production independently of TNF. These results suggest that TNF treatment directly modulates the outcome of TLR signaling in neutrophils, namely licensing TLRs to trigger the release of LTB4 and PGE2, which are potent mediators of neutrophil and effector T cell recruitment.
Finally, we assessed the kinetics of the effects of TNFα as well as of other ligands. While TNFα has been reported to prime reactive oxygen species production in minutes, we observed that the increase of cytokine production was maximal only after hours of incubation with TNFα or zymosan (Fig. 5D). In the case of the ligand TDB, increased cytokine production required over 8 hours of stimulation, likely reflecting the fact that licensing occurs entirely indirectly in this case and first requires accumulation of sufficient amounts of TNFα in the culture media. This result suggests that TNFα has long term effects on neutrophil phenotype (i.e., licensing), in addition to its short term effect on ROS production (i.e., priming), the latter of which is mediated by phosphorylation of the NADPH oxidase (23).
We also tested whether the continuous presence of TNFα was required to maintain licensing. To this end, cells were washed and cultured in fresh media for increasing amounts of time before restimulation. We still observed a strong increase in cytokine production in neutrophils that had been treated with TNFα and subsequently washed and cultured in fresh media although the licensing effect decreased as the rest duration increased (Fig. 5E). Of note, the prolonged licensing observed in vitro is consistent with what we observed in ex vivo neutrophils, in which increased cytokine production is maintained even after the cells are taken from the inflammatory milieu of the peritoneum (Fig. 1C). These results argue that TNFα can durably modify neutrophil activity.
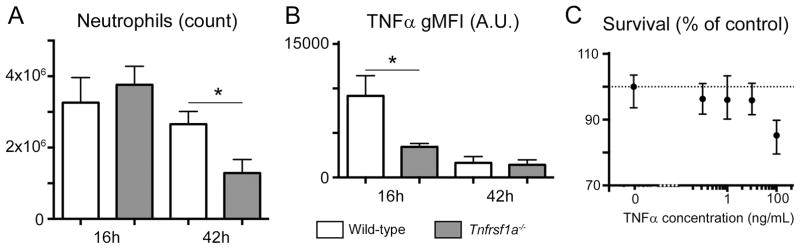
Finally, we assessed the role of TNFR1-mediated licensing in a model of zymosan induced peritonitis in vivo. As described above, injection of zymosan leads to neutrophil accumulation in the peritoneum at 16 hours, and to an increase in their capacity to produce TNFα (Fig. 1B). In TNFR1-deficient mice, we observed a comparable recruitment of neutrophils (Fig. 6A), but, in agreement with our in vitro results, the recruited neutrophils displayed a lower production of TNFα upon restimulation (Fig. 6B). Interestingly, 42 hours after zymosan injection, i.e. during the resolution of inflammation, the number of neutrophils present in wild type mice was significantly higher than in Tnfrsf1a−/− mice (Fig. 6A). Altogether, these experiments argue that neutrophil licensing, likely through increased cytokine or lipid mediator production, contributes to the maintenance of inflammation through increased neutrophil recruitment. Of note, it is also possible that the different inflammatory conditions in WT and Tnfrsf1a−/− mice alter the half-life of neutrophils, which would also influence their numbers, but we did not observe any positive effect of TNFα treatment on neutrophil survival in vitro (Fig. 6C), leading us to favor the hypothesis of an increased recruitment.
Figure 6. TNFR1-mediated licensing prolongs zymosan-induced inflammation.
A.B. Absolute counts (A) and TNFα gMFI (B) for neutrophils present in the peritoneal exudates of WT or TNFR1-deficient mice injected with zymosan for 16 or 42h. Data were obtained in two independent experiments with 3 mice/group. *, P<0.05
C. Relative survival of bone-marrow neutrophils cultured for 24h in the presence of the indicated concentrations of recombinant TNFα. Survival is calculated as a percentage of the number of live neutrophils harvested in media alone.
Mechanistic changes induced by neutrophil licensing
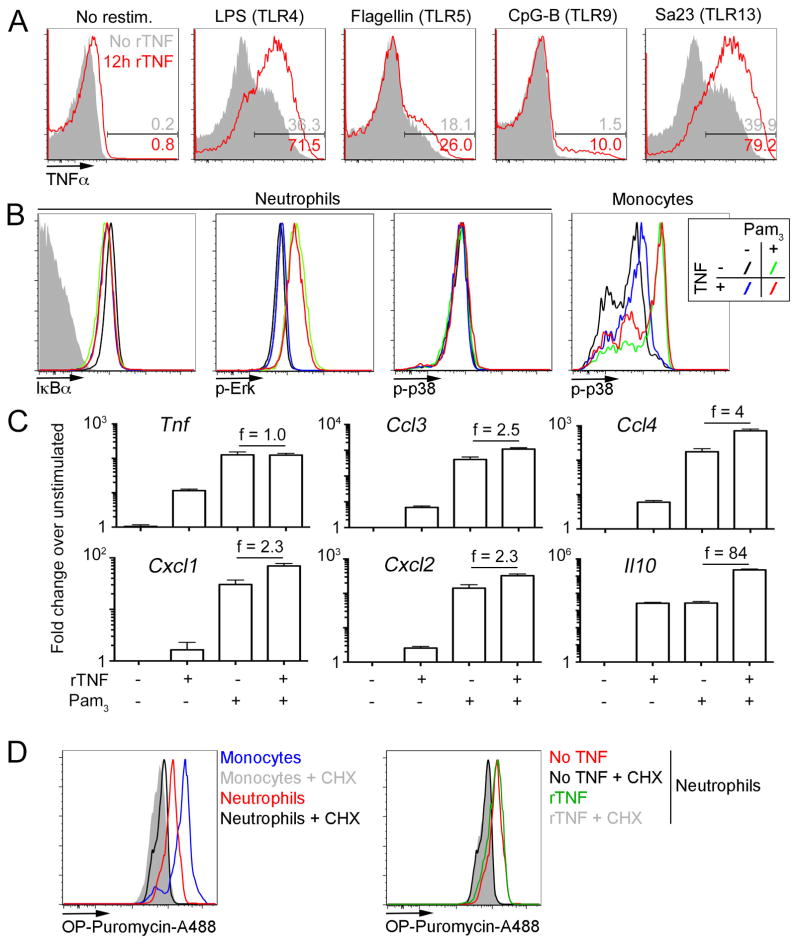
In the experiments presented so far, we relied on TLR2 ligands to stimulate neutrophils and measure cytokine production. To determine whether increased cytokine production is a general feature of neutrophil licensing, we tested ligands for other TLRs. While the magnitude of responses to TLR ligands varied drastically, TNFα pre-treatment increased cytokine production for all TLRs tested (Fig. 7A). In particular, TLR4 and TLR13 ligands yielded similar cytokine production patterns as TLR2. Of note, increased cytokine production in response to other TLR ligands was also observed with pre-treatments other than TNFα, such as zymosan and TDB (data not shown).
Figure 7. Mechanistic changes induced by neutrophil licensing.
A. Representative flow cytometry plots of neutrophils treated with recombinant TNFα (red) of left untreated (solid grey) for 12 hours before restimulation with the indicated ligands for TLR receptors. Numbers display the percentage of TNFα positive neutrophils.
B. Representative flow cytometry plots of neutrophils (or for the rightmost plot, inflammatory monocytes) gated from bone marrow cells left untreated (black, green) or incubated with 10ng/mL recombinant TNFα (blue, red), then restimulated Pam3CSK4 (green, red) or not (black, blue), depicting levels of IkBα, phospho-Erk and phospho-p38.
C. qPCR analysis of purified neutrophils left untreated or incubated with 10 ng/mL recombinant TNFα, then restimulated with Pam3CSK4 or not, for the indicated genes. Values plotted represent fold change over untreated neutrophils (no TNF, no Pam3CSK4). Numbers represent the fold change between unlicensed and licensed neutrophils after Pam3CSK4 restimulation.
D. Left. Bone marrow cells were cultured in the presence of OP-puromycin with or without cyclohexamide (CHX). Cells were then stained to label the OP group with Alexa 488 and gated on monocytes (grey, blue) or neutrophils (black, red). Right. Neutrophils were treated as in left after treatment with TNFα (grey, green) or in the absence of TNFα (black, red).
Data are representative of 3 (A, B, D) or 4 (C) independent experiments.
Engagement of TLRs leads to the activation of IRAK family members downstream of MyD88 or TRIF, and ultimately to the activation of the NFkB and MAPK pathways. Therefore, we measured the degradation of IkBα and phosphorylation of Erk and p38 in neutrophils after TLR2 stimulation to examine whether TNFα pre-treatment affected downstream signaling. We detected robust phosphorylation of Erk following TLR2 stimulation but did not see any effect of pre-treatment with TNFα on this response (Fig. 7B). Similarly, degradation of IkBα in response to TLR2 was not affected by pre-treatment. Interestingly, we did not observe phosphorylation of p38 in neutrophils, even though we could detect it reliably in inflammatory monocytes (Fig. 7B, right). Altogether, we found no evidence that TNFα-mediated licensing of neutrophils modifies the signaling directly downstream of TLR activation.
Since signaling is not affected by licensing, we reasoned that increased cytokine production could be due to either epigenetic changes leading to increased mRNA production, or post-transcriptional regulation of cytokines. To discriminate between these possibilities, we measured the levels of messenger RNA for cytokines affected by licensing. In the case of TNFα, CCL3-4 and CXCL1-2, stimulation with Pam3CSK4 alone was sufficient to induce a maximal or near maximal upregulation of the respective mRNA. On the other hand, IL-10 mRNA was strongly induced by TNFα alone, and we observed a synergistic effect with additional TLR stimulation (Fig. 7C). Altogether, these results suggest that cytokine production was largely regulated at the post-transcriptional level. It is noteworthy that IL-10, the main anti-inflammatory cytokine produced by neutrophils, did not fit this pattern and is strongly upregulated at the transcriptional level, suggesting that the kinetics of pro- and anti-inflammatory gene induction in neutrophils may differ.
In the case of pro-inflammatory cytokines, it remained unclear how increased cytokine production is achieved when mRNA levels are largely unchanged following licensing. Neutrophils have been reported to be poor protein producers, so we tested whether licensing changes this aspect of neutrophil function. To that end, we measured incorporation of an analog of puromycin in nascent polypeptides. First, we confirmed that protein synthesis in neutrophils occurs at much lower levels than in monocytes (Fig. 7D, left). Indeed, using cyclohexamide as a background control, we only observed a low level of puromycin incorporation in neutrophils, compared to inflammatory monocytes cultured in the same conditions. These results are in agreement with a recent study of protein synthesis across bone marrow cells (10). Treatment with TNFα did not noticeably increase the incorporation of puromycin in neutrophils (Fig. 7D, right), suggesting that changes in global translation levels are unlikely to account for the difference in cytokine production we observed. Thus, our data suggest that the increased protein production we observed in licensed neutrophils may be due to selective translation or import of pro-inflammatory cytokines, and a selective transcriptional up-regulation of the anti-inflammatory cytokine IL-10. Taken together, these experiments argue that while naïve neutrophils are competent for TLR signaling and cytokine mRNA induction, efficient secretion of cytokines requires an additional licensing step mediated by TNFR1 signaling.
Discussion
In the present study, we have focused on the ability of neutrophils to produce cytokines during different inflammatory conditions. We found that different inflammatory milieus enable neutrophils to produce high concentrations of cytokines, chemokines, and lipid mediators in response to TLR ligands, and propose to describe this phenomenon as ‘licensing’. It is important to note that the priming of neutrophil functions by several factors, including TNFα and integrin crosslinking, increases ROS production and enhances bacterial killing (23, 24). However, priming does not necessarily increase cytokine production and is mechanistically distinct from licensing, as we discuss below. Indeed, after licensing, we observed a ~10-fold increase in cytokine production, raising TNFα production of neutrophils to levels equivalent to that of inflammatory monocytes stimulated similarly. Even more strikingly, licensed neutrophils could produce eicosanoids in response to TLR stimulation, even though this activity is normally gated by GPCRs.
We also demonstrated that licensing is mediated by TNFα in a paracrine manner. Importantly, production of TNFα is increased in licensed neutrophils, raising the possibility that neutrophil cytokine production is regulated by a feed-forward loop. In agreement with that observation, we observed that neutrophils stimulated ex vivo either all robustly produced TNFα in the presence of microbial ligands, or all produced low amounts of this cytokine (Fig. 1B). This unimodality is consistent with the hypersensitivity associated with feed-forward loops, and disappeared in the absence of paracrine signaling in TNFR1-deficient cells (Fig. 4G). Our results thus suggest a form of quorum sensing in neutrophils, where recognition of microbial ligands will increase local concentrations of TNFα and license other neutrophils migrating to this site for increased cytokine production as long as TLR ligands are present.
Consistent with this idea, we also observed licensing of the production of chemokines and lipid mediators involved in neutrophil recruitment (CXCL1-2, LTB4). Altogether, the results presented here point to a model for escalation of inflammation, where the local licensing of neutrophils allows them to recruit and activate additional neutrophils and other immune cells. Importantly, since activation of this positive feedback loop requires both local licensing signals and the persistent presence of ligands for TLRs, the elimination of microbes and decrease in the availability of TLR ligands would lead to a termination of the cytokine production. There is, however, evidence for activation of TLRs by endogenous ligands, that may mediate a continuous neutrophil recruitment and cytokine production in the context of inflammatory diseases such as rheumatoid arthritis (4). It is interesting to note that TNFα is a critical mediator of this disease, and our results suggest that its role on neutrophils may be involved in the persistence of inflammation. In the context of normal responses, the fact that cytokine production by neutrophils requires licensing over the course of a few hours may also be important to limit inflammation, as our results would predict that neutrophils will produce very few cytokines in the context of a transient presence of TLR ligands, for example after a wound infected by an easily eliminated commensal. Conversely, anti-TNFα therapies are also associated with increased risks of infection, and our results suggest that defects in neutrophil licensing or accumulation may play a part in these side-effects (25).
Somewhat expectedly, licensed neutrophils also produce increased levels of the regulatory cytokine IL-10, which may concomitantly limit inflammation. Interestingly, IL-10 was mostly regulated at the transcriptional level, while pro-inflammatory cytokines were mostly regulated at the post-transcriptional level. This discrepancy may lead to differences in the kinetics of expression of these proteins both in vivo and in vitro, and may underlie the differences observed in previous reports on neutrophil cytokine production (8, 26). In particular, Zhang et al. measured neutrophil cytokine production 24h post-stimulation, and found the response to be dominated by IL-10, suggesting that anti-inflammatory cytokine production is more sustained than pro-inflammatory cytokine production. It is also important to note that human neutrophils differ from their murine counterparts, in particular with regards to IL-10 production, and it will be interesting to compare the effects of licensing in human cells.
In this study, we demonstrate that TNFα is a critical regulator of neutrophil protein and eicosanoid synthesis. It is interesting that TNFα has long been used as a priming agent for neutrophil studies (24). In this setting, TNFα treatment leads to a rapid but transient increase in the ability of neutrophils to produce ROS through phosphorylation of the NADPH oxidase and other proteins (23). However, we did not observe any difference in ROS production by neutrophils elicited with uric acid or zymosan, even though only the latter are able to produce large amounts of cytokines. This observation is consistent with the idea that priming can be induced by integrin cross-linking (27), such as when cells are migrating to the peritoneum. Altogether, our data suggest that priming of reactive oxygen species production for bacterial elimination and licensing of cytokine production are distinct, but not mutually exclusive, phenomena. Specifically, we hypothesize that priming will occur rapidly as neutrophils migrate to most inflammatory sites, while licensing will be restricted to sites of prolonged inflammation, for example in the presence of a pathogen.
Mechanistically, our results suggest that TNFα directly increases the ability of neutrophils to produce pro-inflammatory cytokines, independently of transcriptional regulation. Previous reports have shown that priming of neutrophils, including with TNFα, can induce a transient transcriptional upregulation of cytokine genes (28), but this effect was only modest at later time points in our hands. While further studies will be necessary to fully understand the mechanisms involved, it is interesting to note that translation in mature neutrophils is largely affected by the loss of nucleoli (29), defects in mRNA splicing (30) and overall lower translational ability (10). Our results would seem to suggest that TNFα can reverse some of these hallmarks of neutrophil differentiation, and allow them to produce cytokines at levels consistent with those seen in monocytes. However, since we did not observe global changes in translation in a puromycin based assay, we favor the hypothesis that there is a selective increase in translation or secretion of cytokines. It is also possible that translation elongation is regulated by licensing, as this would not be revealed by puromycin labeling.
In summary, our work shows that neutrophil cytokine production is locally regulated, and that this regulation is unexpectedly mediated by neutrophils themselves. While the study of cytokine production by neutrophils has generally been limited to cells activated in vitro, our results argue for a re-assessment of the role of neutrophils in the early shaping of the inflammatory micro-environment. These studies may pave the way to the development of new therapies in the context of inflammatory diseases.
Supplementary Material
Acknowledgments
This project was supported by the NIH (AI063302, AI095587, AI104914, AI072429 to G.M.B, EY011108, EY026082 to K.G). J.D. is supported by a Long-Term Fellowship from the Human Frontier Science Program (LT-000081/2013-L). G.M.B. is a John P. Stock Faculty Fellow and the recipient of a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award.
We thank members of the Barton lab, Gabrielle Reiner, Hélène Beuneu and Béatrice Bréart for helpful comments on the manuscript, Julie Choe and Jeanette Tenthorey for technical help with protein purification, and Hector Nolla, Alma Valeros and Kartoosh Heydari for assistance with flow cytometry.
J.D. and G.M.B. designed research and wrote the manuscript. J.D. performed and analyzed experiments, R.B. designed and performed initial experiments, J.W. and K.G. designed and performed experiments related to lipid measurements.
Footnotes
The authors have no conflicting financial interests.
References
- 1.Chen GY, Nunez G. Sterile inflammation: sensing and reacting to damage. Nat Rev Immunol. 2010;10:826–837. doi: 10.1038/nri2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern Recognition Receptors and Inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Nathan C. Neutrophils and immunity: challenges and opportunities. Nat Rev Immunol. 2006;6:173–182. doi: 10.1038/nri1785. [DOI] [PubMed] [Google Scholar]
- 4.Wright HL, Moots RJ, Edwards SW. The multifactorial role of neutrophils in rheumatoid arthritis. Nat Rev Rheumatol. 2014;10:593–601. doi: 10.1038/nrrheum.2014.80. [DOI] [PubMed] [Google Scholar]
- 5.McDonald B, Pittman K, Menezes GB, Hirota SA, Slaba I, Waterhouse CCM, Beck PL, Muruve DA, Kubes P. Intravascular Danger Signals Guide Neutrophils to Sites of Sterile Inflammation. Science. 2010;330:362–366. doi: 10.1126/science.1195491. [DOI] [PubMed] [Google Scholar]
- 6.Thomas CJ, Schroder K. Pattern recognition receptor function in neutrophils. Trends Immunol. 2013;34:317–328. doi: 10.1016/j.it.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Bennouna S, Bliss SK, Curiel TJ, Denkers EY. Cross-Talk in the Innate Immune System: Neutrophils Instruct Recruitment and Activation of Dendritic Cells during Microbial Infection. J Immunol. 2003;171:6052–6058. doi: 10.4049/jimmunol.171.11.6052. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Majlessi L, Deriaud E, Leclerc C, Lo-Man R. Coactivation of Syk Kinase and MyD88 Adaptor Protein Pathways by Bacteria Promotes Regulatory Properties of Neutrophils. Immunity. 2009;31:761–771. doi: 10.1016/j.immuni.2009.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Davey MS, Tamassia N, Rossato M, Bazzoni F, Calzetti F, Bruderek K, Sironi M, Zimmer L, Bottazzi B, Mantovani A, Brandau S, Moser B, Eberl M, Cassatella MA. Failure to detect production of IL-10 by activated human neutrophils. Nat Immunol. 2011;12:1017–1018. doi: 10.1038/ni.2111. [DOI] [PubMed] [Google Scholar]
- 10.Signer RAJ, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassatella MA. Neutrophil-derived proteins: selling cytokines by the pound. Adv Immunol. 1999;73:369–509. doi: 10.1016/s0065-2776(08)60791-9. [DOI] [PubMed] [Google Scholar]
- 12.Tecchio C, Micheletti A, Cassatella MA. Neutrophil-Derived Cytokines: Facts Beyond Expression. Front Immunol. 2014;5:508. doi: 10.3389/fimmu.2014.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridlender ZG. Polarization of tumor-associated neutrophil phenotype by TGF-b: ,“N1” versus “N2” TAN. Cancer Cell. 2009;16:183–194. doi: 10.1016/j.ccr.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SY, Ueda S, Ohno H, Hamano Y, Tanaka M, Shiratori T, Yamazaki T, Arase H, Arase N, Karasawa A, Sato S, Ledermann B, Kondo Y, Okumura K, Ra C, Saito T. Resistance of Fc receptor-deficient mice to fatal glomerulonephritis. J Clin Invest. 1998;102:1229–1238. doi: 10.1172/JCI3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itou T, Collins LV, Thoren FB, Dahlgren C, Karlsson A. Changes in activation states of murine polymorphonuclear leukocytes (PMN) during inflammation: a comparison of bone marrow and peritoneal exudate PMN. Clin Vaccine Immunol. 2006;13:575–583. doi: 10.1128/CVI.13.5.575-583.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clarke TB, Davis KM, Lysenko ES, Zhou AY, Yu Y, Weiser JN. Recognition of peptidoglycan from the microbiota by Nod1 enhances systemic innate immunity. Nat Med. 2010;16:228–231. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012. Nucleic Acids Res. 2012;40:D1144–1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leedom AJ, Sullivan AB, Dong B, Lau D, Gronert K. Endogenous LXA4 circuits are determinants of pathological angiogenesis in response to chronic injury. Am J Pathol. 2010;176:74–84. doi: 10.2353/ajpath.2010.090678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sapieha P, Stahl A, Chen J, Seaward MR, Willett KL, Krah NM, Dennison RJ, Connor KM, Aderman CM, Liclican E, Carughi A, Perelman D, Kanaoka Y, Sangiovanni JP, Gronert K, Smith LEH. 5-Lipoxygenase metabolite 4-HDHA is a mediator of the antiangiogenic effect of ω-3 polyunsaturated fatty acids. Sci Transl Med. 2011;3:69ra12. doi: 10.1126/scitranslmed.3001571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Moltke J, Trinidad NJ, Moayeri M, Kintzer AF, Wang SB, van Rooijen N, Brown CR, Krantz BA, Leppla SH, Gronert K, Vance RE. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 2012;490:107–111. doi: 10.1038/nature11351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyake Y, Toyonaga K, Mori D, Kakuta S, Hoshino Y, Oyamada A, Yamada H, Ono KI, Suyama M, Iwakura Y, Yoshikai Y, Yamasaki S. C-type lectin MCL is an FcRγ-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity. 2013;38:1050–1062. doi: 10.1016/j.immuni.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Dennis EA, Norris PC. Eicosanoid storm in infection and inflammation. Nat Rev Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewas C, Dang PMC, Gougerot-Pocidalo MA, El-Benna J. TNF-alpha induces phosphorylation of p47(phox) in human neutrophils: partial phosphorylation of p47phox is a common event of priming of human neutrophils by TNF-alpha and granulocyte-macrophage colony-stimulating factor. J Immunol. 2003;171:4392–4398. doi: 10.4049/jimmunol.171.8.4392. [DOI] [PubMed] [Google Scholar]
- 24.Berkow RL, Dodson MR. Biochemical mechanisms involved in the priming of neutrophils by tumor necrosis factor. J Leukoc Biol. 1988;44:345–352. doi: 10.1002/jlb.44.5.345. [DOI] [PubMed] [Google Scholar]
- 25.Bongartz T, Sutton AJ, Sweeting MJ, Buchan I, Matteson EL, Montori V. Anti-TNF antibody therapy in rheumatoid arthritis and the risk of serious infections and malignancies: systematic review and meta-analysis of rare harmful effects in randomized controlled trials. JAMA. 2006;295:2275–2285. doi: 10.1001/jama.295.19.2275. [DOI] [PubMed] [Google Scholar]
- 26.Bennouna S, Denkers EY. Microbial Antigen Triggers Rapid Mobilization of TNF-alpha to the Surface of Mouse Neutrophils Transforming Them into Inducers of High-Level Dendritic Cell TNF-a Production. J Immunol. 2005;174:4845–4851. doi: 10.4049/jimmunol.174.8.4845. [DOI] [PubMed] [Google Scholar]
- 27.Liles WC, Ledbetter JA, Waltersdorph AW, Klebanoff SJ. Cross-linking of CD18 primes human neutrophils for activation of the respiratory burst in response to specific stimuli: implications for adhesion-dependent physiological responses in neutrophils. J Leukoc Biol. 1995;58:690–697. doi: 10.1002/jlb.58.6.690. [DOI] [PubMed] [Google Scholar]
- 28.Wright HL, Thomas HB, Moots RJ, Edwards SW. RNA-seq reveals activation of both common and cytokine-specific pathways following neutrophil priming. PLoS One. 2013;8:e58598. doi: 10.1371/journal.pone.0058598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salati S, Bianchi E, Zini R, Tenedini E, Quaglino D, Manfredini R, Ferrari S. Eosinophils, but not neutrophils, exhibit an efficient DNA repair machinery and high nucleolar activity. Haematologica. 2007;92:1311–1318. doi: 10.3324/haematol.11472. [DOI] [PubMed] [Google Scholar]
- 30.Wong JJL, Ritchie W, Ebner OA, Selbach M, Wong JWH, Huang Y, Gao D, Pinello N, Gonzalez M, Baidya K, Thoeng A, Khoo TL, Bailey CG, Holst J, Rasko JEJ. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.