Abstract
Signaling through the canonical NF-κB pathway is critical for the generation and maintenance of mature B-cells and for antigen-dependent B-cell activation. c-REL (rel) and RELA (rela) are the downstream transcriptional activators of the canonical NF-κB pathway. Studies of B-cells derived from constitutional rel knockout mice and chimeric mice repopulated with rela−/− fetal liver cells provided evidence that the subunits can have distinct roles during B-cell development. However, the B-cell-intrinsic functions of c-REL and RELA during B-cell generation and antigen-dependent B-cell activation have not been determined in vivo. To clarify this issue, we crossed mice with conditional rel and rela alleles individually or in combination to mice that express Cre-recombinase in B-cells. We here report that, whereas single deletion of rel or rela did not impair mature B-cell generation and maintenance, their simultaneous deletion led to a dramatic reduction of follicular and marginal zone B-cells. Upon T-cell-dependent immunization, B-cell-specific deletion of the c-REL subunit alone abrogated the formation of germinal centers (GC), whereas rela deletion did not affect GC formation. T-independent responses were strongly impaired in mice with B-cell-specific deletion of rel, and only modestly in mice with RELA-deficient B-cells. Our findings identify differential requirements for the canonical NF-κB subunits c-REL and RELA at distinct stages of mature B-cell development. The subunits are jointly required for the generation of mature B-cells. During antigen-dependent B-cell activation, c-REL is the critical subunit required for the initiation of the GC-reaction and for optimal T-independent antibody responses, with RELA being largely dispensable at this stage.
Keywords: B-cell, B-cell compartment, B-cell activation, follicle, marginal zone, germinal center, immune response, NF-κB transcription factors
INTRODUCTION
Signaling through the nuclear factor-κB (NF-κB) signal transduction pathway is critical for the generation of a normal mature B-cell compartment and for antigen-dependent B-cell activation.1-3 The NF-κB pathway comprises two branches, a canonical (classical) and an alternative (non-canonical, non-classical) pathway, that transmit signals elicited by distinct cell surface receptors, eventually leading to the nuclear translocation of NF-κB transcription factors.4,5 c-REL, RELA and NF-κB1/p50 are the subunits of the canonical pathway, while RELB and NF-κB2/p52 transduce signals of the alternative pathway. In B-cells, activation of the canonical NF-κB pathway predominantly results in the nuclear translocation of c-REL/p50 and RELA/p50 heterodimers and occurs during various stages of antigen-independent and antigen-dependent B-cell development.1-3 Only c-REL and RELA are transcriptionally active as they contain transactivation domains (TAD).4,5 A large body of work has provided insights into the role of the NF-κB pathway in lymphocyte biology and during the antibody response; however, the B-cell-intrinsic functions of RELA and c-REL in the generation and maintenance of mature B-cells and during antigen-dependent B-cell activation remains incompletely defined due to the lack of suitable experimental systems.
Studies in which upstream regulators of the canonical NF-κB pathway were deleted in mouse B-cells have firmly established that activation of this pathway is required for the generation and maintenance of mature B-cells.6-9 However, the roles of the downstream transcription factors c-REL and RELA in this process are less well defined. Germ-line deletion of rela causes embryonic lethality at day 15 of gestation.10,11 Transfer of rela−/− fetal liver cells into irradiated SCID mice indicated that mature B-cells developed in the recipient mice,11,12 although analogous experiments with irradiated Rag-1−/− mice found only a partial reconstitution of splenic B-cells.13 In contrast, mice with germ-line deletion of rel or that lack the TAD of c-REL (relΔTAD/ΔTAD) are viable.14-16 These mice showed normal generation of mature B-cells. Results from experiments performed with chimeric mice repopulated with rel−/−rela−/− fetal liver hematopoietic stem cells suggested a requirement for both RELA and c-REL in this process.17 However, the extent to which c-REL and RELA, either individually or in combination, contribute to mature B-cell development is unclear.
With regard to antigen-dependent B-cell development, the canonical NF-κB pathway shows a biphasic activation pattern during T-cell dependent B-cell activation and the germinal center (GC) reaction where antigen-activated B-cells undergo somatic hypermutation and class switch recombination.18-20 While nuclear translocation of NF-κB is observed rapidly upon B-cell activation.21,22 NF-κB signaling is not activated in the majority of GC B-cells.23,24 However, the canonical NF-κB subunits show nuclear translocation in a small subset of B-cells in the light zone of the GC.24 We recently demonstrated that c-REL and RELA exert distinct functions in these light zone B-cells.25 Thus, conditional deletion of rel in GC B-cells revealed that c-REL is required for the maintenance of the GC reaction, whereas RELA was found to be dispensable at this stage.25 However, rela deletion in GC B-cells impaired plasma cell development.25 While it is clear that c-REL and RELA have specific roles in late B-cell development, the B-cell intrinsic functions of the separate subunits during the antigen-dependent B-cell activation and the initiation of the GC reaction have not been determined in vivo. However, this is a particularly relevant issue in the light of recent studies that demonstrated distinct functions for the same transcription factors in the early initiation of the GC reaction and later in the established GC. For example, c-MYC is required for the formation of the GC reaction26 and in a later developmental stage for GC maintenance.27 Similarly, interferon regulatory factor 4 (IRF4) controls the formation of the GC upon T-dependent immunization28,29 and in the established GC is required for plasma cell differentiation.30,31
Several observations imply critical roles for c-REL and RELA during B-cell activation. A recent study demonstrated that the two subunits follow a different activation pattern in B-cells stimulated with anti-IgM in vitro,21 suggesting that RELA and c-REL may exert unique functions upon antigen encounter also in vivo. RELA-deficient fetal liver-derived B-cells proliferated similarly to wild-type B-cells upon mitogenic stimulation in vitro.12 In contrast, c-REL-deficient B-cells showed defects in activation and proliferation following mitogenic stimulation in vitro,14-16 and rel−/− and relΔTAD/ΔTAD mice displayed an impaired T-dependent B-cell response in vivo. Accordingly, constitutional rel knockout mice were characterized by the appearance of smaller GCs relative to control mice.15,32 Since rel−/− T-cells also have impaired activation and proliferation upon T-cell receptor stimulation in vitro,14 it is unclear to what extent the defective T-dependent B-cell response and GC formation are due to the loss of c-REL function in B-cells.
We here deleted rel and/or rela conditionally in B-cells in order to unequivocally identify the specific, B-cell-autonomous roles of c-REL and RELA in the generation and maintenance of mature B-cells and in T-dependent and T-independent immune responses in vivo. We found that whereas c-REL and RELA were functionally redundant during B-cell generation and maintenance, c-REL deficiency in antigen-activated B-cells dramatically impaired the formation of GCs upon T-dependent immunization and also led to a strong reduction of T-independent responses. Conversely, RELA deficiency in B-cells did not impede GC formation and led to only a modest reduction in serum immunoglobulin (Ig) levels upon T-independent immunizations.
RESULTS
Combined c-REL and RELA deficiency leads to a severe reduction in splenic B-cells
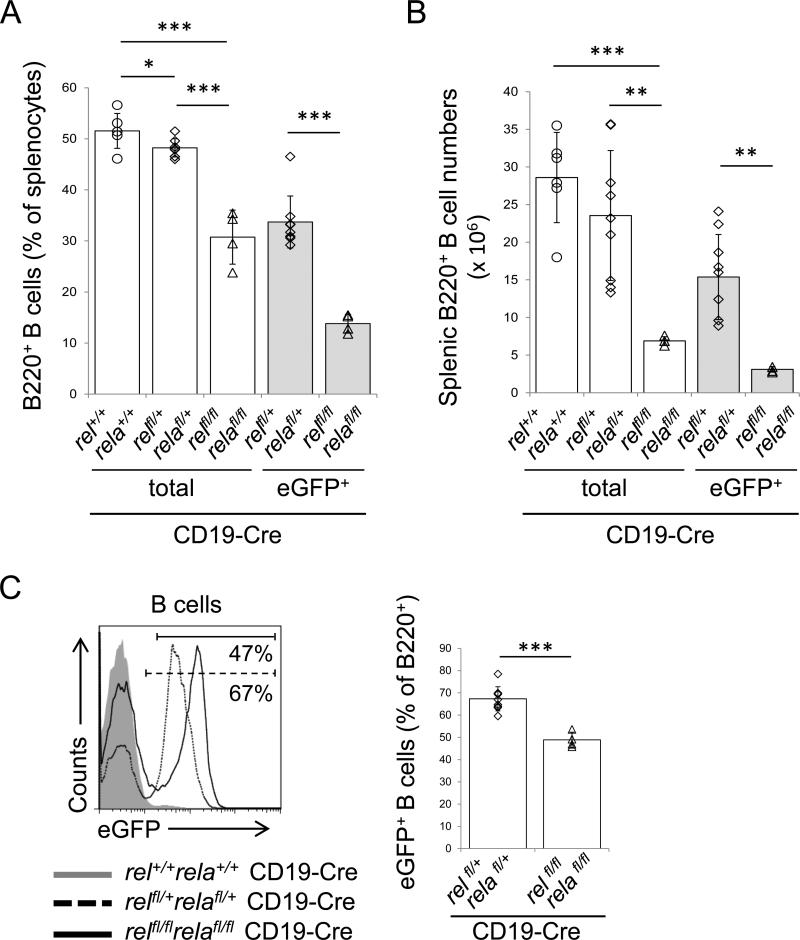
The study of chimeric mice repopulated with rel−/−rela−/− fetal liver hematopoietic stem cells implicated a role for both c-REL and RELA in the generation of mature B-cells.17 To determine the extent of the B-cell-intrinsic requirement of c-REL and RELA in the generation and maintenance of mature B-cells in vivo, we crossed mice with conditional rel and rela alleles25 to CD19-Cre mice that express Cre-recombinase in B-cells33 to jointly ablate the canonical NF-κB subunits in B-cells. We observed a marked reduction in the fraction and cell number of splenic B-cells in relfl/flrelafl/flCD19-Cre mice vs. relfl/+relafl/+CD19-Cre and CD19-Cre control mice (Fig. 1A,B). B220+ cells comprised ~31% of total splenocytes in relfl/flrelafl/flCD19-Cre mice vs. ~52% and ~48% in relfl/+relafl/+CD19-Cre and CD19-Cre control mice, respectively, and, in total cell numbers, ~7×106 B220+ cells were present in relfl/flrelafl/flCD19-Cre mice compared to ~24×106 and ~29×106 B220+ cells in the control mice. In the conditional mice, rel and rela deletion is concomitantly linked to the expression of an eGFP gene,25 which allows the tracking of the rel/rela-deleted B-cells in the tissues. Analysis for eGFP expression among B-cells of relfl/flrelafl/flCD19-Cre mice revealed distinct eGFP+ and eGFP− peaks of equal proportions (~49% eGFP+ vs. ~51% eGFP− B-cells; Fig. 1C); in contrast, ~67% of B-cells of relfl/+relafl/+CD19-Cre mice were eGFP+ (Fig. 1C). This indicates that eGFP+ B-cells double-deficient for c-REL and RELA were outcompeted by eGFP− B-cells that escaped Cre-deletion. Rel/rela-deleted eGFP+ B-cells therefore represented only ~14% of total splenocytes in relfl/flrelafl/flCD19-Cre mice compared to ~52% observed in the CD19-Cre control mice (total cell numbers, ~3×106 rel/rela-deleted eGFP+ B-cells were present in relfl/flrelafl/flCD19-Cre mice vs. ~29×106 B-cells in the control mice) (Fig. 1A,B).
Figure 1. Combined c-REL and RELA deficiency leads to a severe reduction in splenic B-cells.
(A) Percentage and (B) number of splenic B-cells in relfl/flrelafl/flCD19-Cre and the corresponding heterozygous and CD19-Cre control mice as determined by flow cytometry. (C) Flow cytometry of eGFP expression in splenic B-cells of the indicated genotypes. The numbers below the gate indicate the percentage of eGFP+ B-cells among of B220+ B-cells of relfl/flrelafl/flCD19-Cre and relfl/+relafl/+CD19-Cre mice (left). Summary of the frequency of eGFP+ cells among the corresponding B-cell subsets (right). (A-C) Data are cumulative from independent experiments (n=4-9 per group), with each symbol representing a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student's t test (*, P<0.05; **, P<0.01; ***, P<0.001).
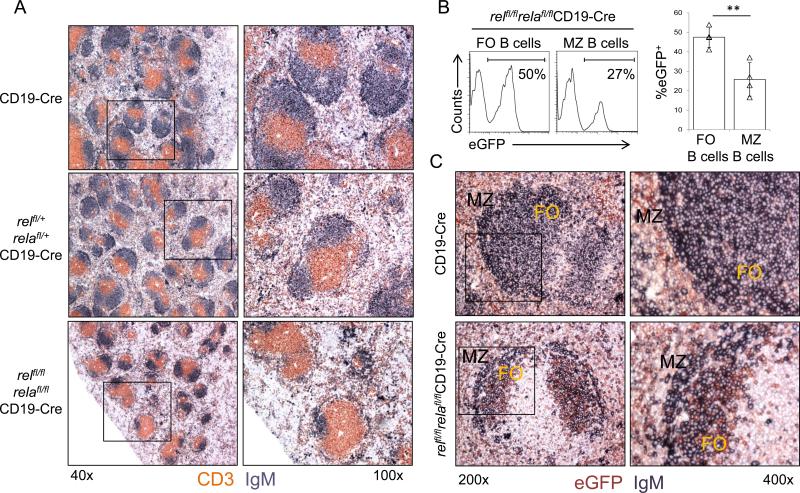
Immunohistochemistry (IHC) analysis of spleen sections for IgM and CD3 revealed that, in line with the severely reduced B-cell fraction observed by flow cytometry, relfl/flrelafl/flCD19-Cre mice had fewer B-cell follicles within the splenic white pulp relative to relfl/+relafl/+CD19-Cre and CD19-Cre control mice (Fig. 2A). The B-cell follicles in relfl/flrelafl/flCD19-Cre mice were also more heterogeneous in size compared to the controls. Together, the simultaneous, B-cell-specific deletion of rel and rela leads to a severe reduction in the number of splenic B-cells that is reflected by abnormalities in the architecture of the white pulp.
Figure 2. relfl/flrelafl/flCD19-Cre mice display fewer B-cell follicles within the splenic white pulp compared to controls and are characterized by counter selection against rel/rela-deleted MZ B-cells.
(A) Spleen sections from mice of the indicated genotypes were analyzed via IHC for the expression of CD3 and IgM. One representative mouse of three per group is shown. Original magnification ×40 (left) and ×100 (right). (B) The fractions of eGFP+ cells among splenic follicular (FO; CD23+CD21int) and marginal zone (MZ; CD21hiCD23−) B-cells in relfl/flrelafl/flCD19-Cre mice were determined by flow cytometry. Numbers below gates indicate the percentage of eGFP+ B-cells among the indicated B-cell subsets (left). Data are cumulative from independent experiments (n=4 per group), with each symbol representing a mouse, showing the frequency of eGFP+ cells among the corresponding B-cell subsets (right). Each symbol represents a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student's t test (**, P<0.01). (C) Spleen sections from mice of the indicated genotypes were analyzed by IHC for the expression of eGFP and IgM. FO, follicular area; MZ, marginal zone area. One representative mouse of three per group is shown. Original magnification ×200 (left) and ×400 (right).
Counterselection against rel/rela-deleted marginal zone (MZ) B-cells
To investigate whether B-cell-specific ablation of c-REL or RELA individually or in combination affects the development of mature B-cell subsets in the spleen, we performed flow cytometry analysis for CD23 and CD21 expression on splenocytes from relfl/flCD19-Cre, relafl/flCD19-Cre and relfl/flrelafl/flCD19-Cre and littermate controls to determine the fractions of FO B-cells (B220+CD23+CD21int) and marginal zone (MZ) B-cells (B220+CD23−CD21hi). With regard to the single conditional knockouts, the results revealed only minor (c-REL cohort) or no significant (RELA cohort) differences in the fractions of FO and MZ B-cell subsets (Suppl. Fig. 1), or in the fractions of IgM+IgDhi vs. IgMhiIgDlo B-cells (data not shown), among relfl/flCD19-Cre or relafl/flCD19-Cre mice and the corresponding heterozygous and wild-type controls. In accordance, analysis of H&E stained sections showed normal splenic architecture in both mouse models (data not shown). These results are in line with earlier observations based on the analysis of constitutional knockout mice that c-REL and RELA have redundant functions in the generation and maintenance of mature B-cells.11,12,14-16
In contrast to the single conditional knockouts, relfl/flrelafl/flCD19-Cre mice displayed a reduced fraction of FO B-cells compared to relfl/+relafl/+CD19-Cre and CD19-Cre mice (~43% vs. ~77%), with no significance increase in the fraction of MZ B-cells (Suppl. Fig. 2). However, analysis of eGFP+ vs. eGFP− cells in the MZ B-cell compartment of relfl/flrelafl/flCD19-Cre mice revealed a strong counterselection against rel/rela-deleted MZ B-cells, which surpassed that observed in the FO B-cell compartment (Fig. 2B). Specifically, only ~26% of MZ B-cells in these mice were eGFP+ (and thus rel/rela-deleted) compared with ~48% of FO B-cells. In accordance with the flow cytometry results, IHC analysis of splenic sections revealed that the majority of B-cells in the MZ of relfl/flrelafl/flCD19-Cre mice were eGFP-negative, in contrast to B-cells in the follicular area that mostly stained for eGFP (Fig. 2C). This severe counterselection against rel/rela-deleted MZ B-cells is consistent with the published observation that deletion of upstream regulators of the canonical NF-κB pathway causes impaired development and/or persistence of MZ B-cells.1,2,9
Mature B-cells require c-REL and RELA for their survival
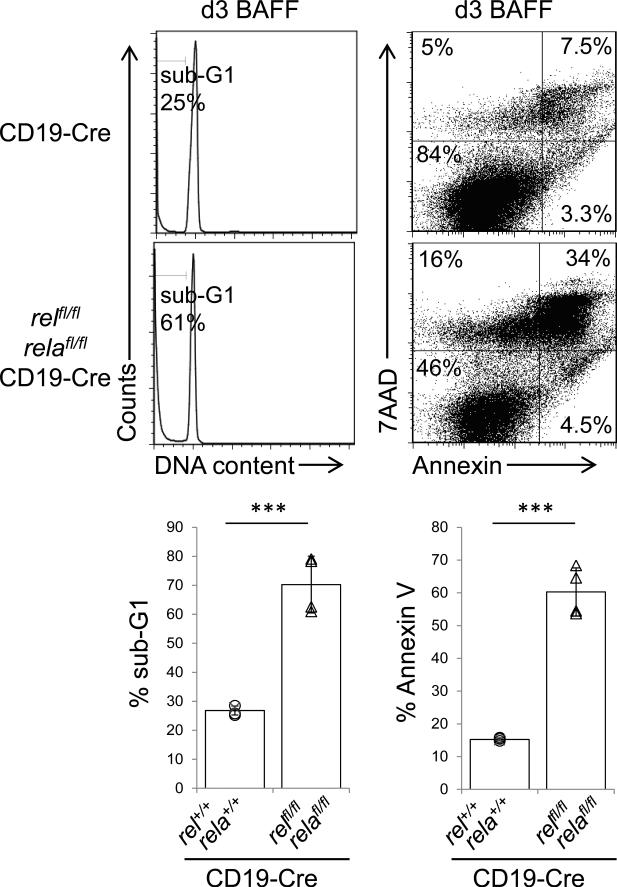
To determine whether c-REL/RELA deficiency in mature B-cells impairs survival in vitro, we stimulated B-cells from relfl/flrelafl/flCD19-Cre or CD19-Cre control mice with B-cell activating factor (BAFF), which is required for the survival of mature B-cells,34 for three days. While BAFF is a strong activator of the alternative NF-κB pathway, it partly signals also through the canonical pathway,35,36 and it was recently shown that c-REL and RELA indeed undergo nuclear translocation in B-cells following BAFF stimulation.37 The results showed significantly enhanced cell death in the cultures of c-REL/RELA-deficient vs. control B-cells in response to BAFF stimulation (~70% vs. ~27% by propidium iodide (PI) staining and ~ 60 % vs. ~15% by annexin V/7AAD staining) (Fig. 3). These findings provide additional evidence that the canonical NF-κB pathway transmits signals derived from BAFF-mediated activation35-37 and may explain in part the importance of canonical NF-κB signaling for mature B-cell maintenance.6,7
Figure 3. Combined c-REL and RELA deficiency impairs BAFF-mediated cell survival.
(left) Flow cytometric analysis of BAFF-stimulated purified B-cells from relfl/flrelafl/flCD19-Cre and CD19-Cre mice at d3 for DNA content by propidium iodide (PI) staining and summary of the corresponding percentage sub-G1 (bottom), and (right) for apoptotic/dead cells by annexin V/7AAD staining and summary of the corresponding percentage of annexin V/7AAD+ cells (bottom). Data are cumulative from independent experiments (n=4 per group), with each symbol representing a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student's t test (***, P<0.001).
Combined c-REL and RELA deficiency does not impair B-cell maturation in the bone marrow
In CD19-Cre mice, the Cre recombinase is expressed from the pre-B cell stage in the bone marrow (BM) on. Therefore, we were able to determine whether the reduction in mature B-cells in the spleen may be the result of a defect during B-cell maturation in the BM of relfl/flrelafl/flCD19-Cre versus relfl/+relafl/+CD19-Cre and CD19-Cre mice by flow cytometric analysis for pro-B (CD93hiB220intIgM−), pre-B (CD93hiB220intIgM+) and mature (CD93loB220+) B-cells. We observed that both the fraction and cell number of immature B-cells did not differ among the genotypes (Suppl. Fig. 3). However, in accordance with the observed reduction of mature B-cells in the periphery, mature B-cells were significantly reduced in the BM of relfl/flrelafl/flCD19-Cre mice compared to the controls (~6% compared to ~14% and ~11% in the control mice; and in total cell numbers ~0.6×106 compared to ~1.7×106 and ~1.5×106 in the controls; Suppl. Fig. 3). These results demonstrate that c-REL and RELA are redundant during the generation of B-cells in the BM in a B-cell intrinsic fashion, extending previous findings on chimeric mice repopulated with rel−/−rela−/− fetal liver hematopoietic stem cells that concluded that the combined absence of c-REL and RELA did not perturb the development of B-cell progenitors in the BM.17
Mature B-cells are characterized by the expression of low levels of the CD24 (heat stable antigen, HSA) cell surface antigen.38 Consistent with the strong reduction of mature B-cells in the spleen of relfl/flrelafl/flCD19-Cre mice, these mice were characterized by a small fraction of B220+CD21intCD24lo FO B-cells compared to the control mice (~23% compared to ~70% and ~58% in the control mice; Suppl. Fig. 4). Instead, most splenic B220+ cells in relfl/flrelafl/flCD19-Cre mice were CD24hi immature B-cells (~48% compared to ~16% and ~26% in the control mice; Suppl. Fig. 4), a population that comprises transitional B-cells which are the precursors of mature B-cells. Transitional (T) B-cells are further distinguished into recent immigrants from the BM (transitional 1; T1) that differentiate into T2 and T3 B-cells.39
Combined c-REL and RELA deficiency leads to a developmental block at the T1 stage
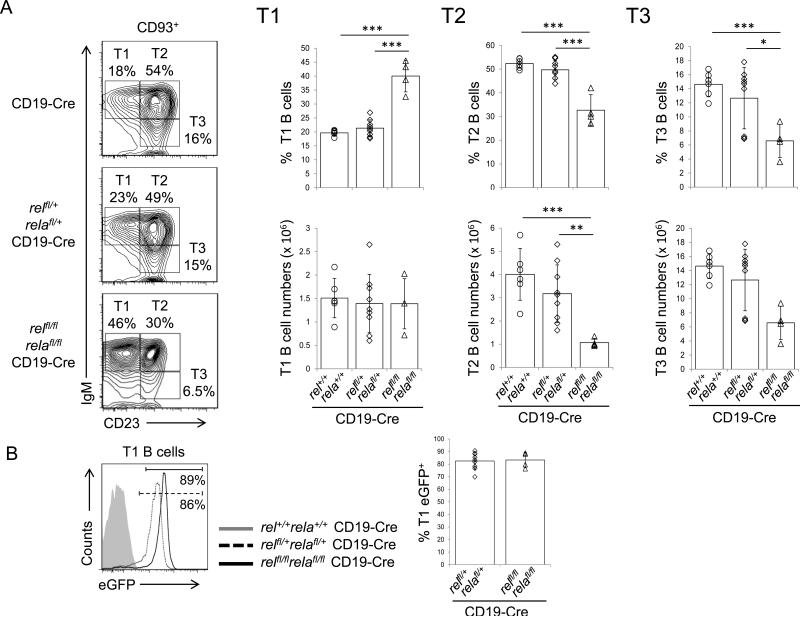
To determine whether the canonical NF-κB subunits are functionally required during the B-cell transitional phase (T1 to T3) development, we analyzed splenic B-cells from relfl/flrelafl/flCD19-Cre and control mice for their expression of CD93+IgMhiCD23− (T1), CD93+IgMhiCD23+ (T2), and CD93+IgMloCD23+ (T3) B-cells.39 We found a significantly increased fraction of T1 B-cells with a concomitant reduction in the populations of T2 and T3 B-cells in relfl/flrelafl/flCD19-Cre mice compared to the CD19-Cre littermate controls (~40% vs. ~20% T1 B-cells, ~33% vs. ~52% T2 B-cells, and ~7% vs. ~15% T3 B-cells) (Fig. 4A). Of note, in contrast to what we observed for the MZ B-cells (Fig. 2B), there was no counterselection of rel/rela-deleted eGFP+ T1 cells in relfl/flrelafl/flCD19-Cre vs. relfl/+relafl/+CD19-Cre mice (~83% were eGFP+ in both genotypes; Fig. 4B). To further characterize the block in the T1 to T2 transition, we determined the T1/T2 correlation as described by Derudder et al.9 which in normal mice is positive as T2 cells arise from T1 cells.39 As expected, we observed a positive T1/T2 correlation for the CD19-Cre and relfl/+relafl/+CD19-Cre control mice, which however was not observed for the relfl/flrelafl/flCD19-Cre mice (Suppl. Fig. 5A). Also, since CD23—which is used along with other markers to identify transitional B-cells—is a potential NF-κB target gene,40 we used CD93 (AA4.1), which is expressed at lower levels on T2 cells,41,42 as an independent marker to confirm the identity of the T1 and T2 B-cell subsets.9 We found that relfl/flrelafl/flCD19-Cre mice harbor a significantly reduced fraction of CD93lo cells among T2 cells compared to the control mice (Suppl. Fig. 5B). In addition, we observed that T1 B-cells occurred at equal numbers across all genotypes (~1.4×106, ~1.4×106 and ~1.5×106 in relfl/flrelafl/flCD19-Cre, relfl/+relafl/+CD19-Cre and CD19-Cre mice, respectively, Fig. 4A), indicating normal generation up to the T1 stage in relfl/flrelafl/flCD19-Cre mice. Together, these findings reveal the importance of a c-REL/RELA-controlled biological program at this B-cell developmental stage, in agreement with the results from studies in which the deletion of upstream components of the canonical NF-κB pathway caused a block in the T1 stage of B-cell development.8,42 The underlying mechanism for the T1 to T2 block upon combined c-REL and RELA-deficiency in B-cells remains to be determined. Since T1 cells do not proliferate,39 the T1 to T2 block is unlikely to be associated with cell cycle control. Interestingly, the recent finding that a bcl2-transgene was unable to rescue the T1 to T2 block observed in NEMO-deficient mice9 (NEMO is an upstream regulator of the canonical pathway ultimately resulting in c-REL and RELA nuclear translocation) suggests that activation of the canonical NF-κB pathway may be required for a developmental transition rather than cell survival.
Figure 4. Combined c-REL and RELA deficiency leads to a developmental block at the T1 stage.
(A) IgM and CD23 expression of CD93+ (AA4.1+) splenic B-cells from mice of the indicated genotypes were analyzed by flow cytometry. Numbers beside gates indicate the percentage of T1 (CD93+IgMhiCD23−), T2 (CD93+IgMhiCD23+), and T3 (CD93+IgMloCD23+) B-cells (left). Summary of the frequencies of T1-T3 B-cells (right). Data are cumulative from independent experiments (n=4-9 per group), with each symbol representing a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student's t test (*, P<0.05; **, P<0.01; ***, P<0.001). (B) The fractions of eGFP+ cells among splenic T1 B-cells in relfl/flrelafl/flCD19-Cre and relfl/+relafl/+CD19-Cre mice were determined by flow cytometry. Numbers below gates indicate the percentage of eGFP+ B-cells among the indicated B-cell subsets (left). Data are cumulative from independent experiments (n=4-9 per group), with each symbol representing a mouse, showing the frequency of eGFP+ cells among the corresponding B-cell subsets (right).
c-REL is required for the formation of GCs in a B-cell intrinsic fashion
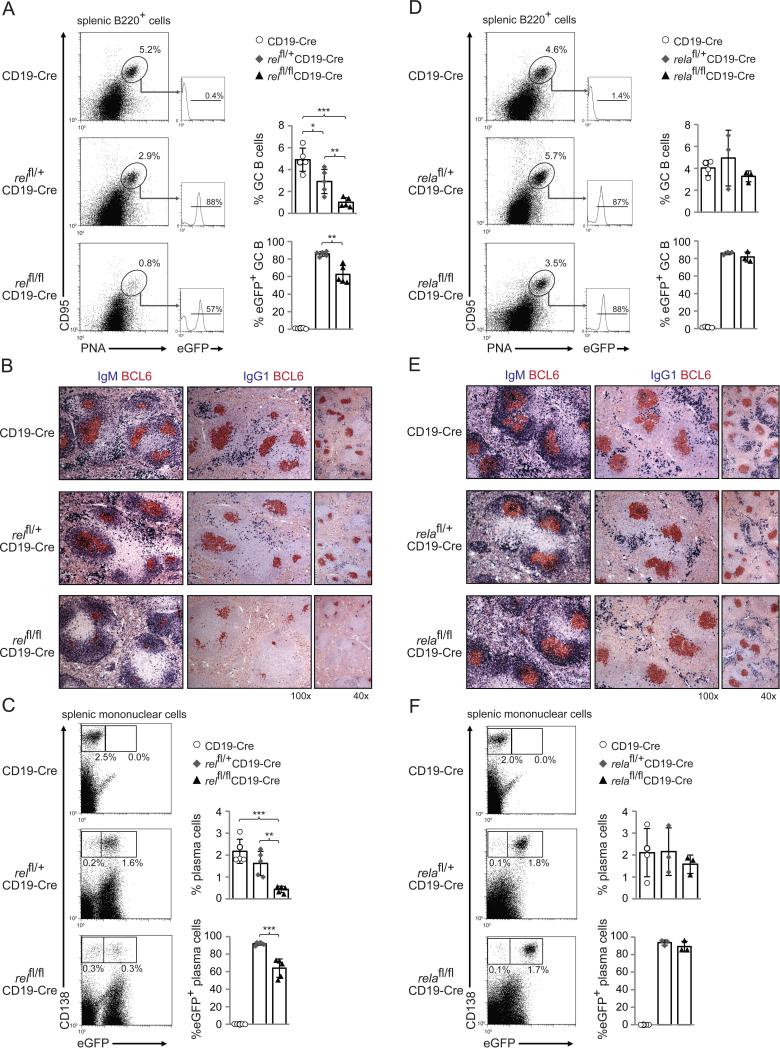
To determine the B-cell-intrinsic role of c-REL in the formation of GCs upon T-dependent B-cell activation, relfl/flCD19-Cre, relfl/+CD19-Cre and CD19-Cre control mice were immunized with the T-dependent antigen sheep red blood cells (SRBC) to induce GC formation, and analyzed 5 days later, i.e. when the initiation phase of the GC reaction has been completed and the early GC has formed.43,44 The results showed a marked reduction of CD95hiPNAhi (Fig. 5A) and BCL6+ (Fig. 5B) GC B-cells in the spleens of relfl/flCD19-Cre mice, with a concurrent reduction in rel-deleted (eGFP+) GC B-cells compared to relfl/+CD19-Cre mice (Fig. 5A). Similarly, the generation of total and eGFP+ splenic plasma cells was strongly impaired at this time-point in relfl/flCD19-Cre compared to relfl/+CD19-Cre and CD19-Cre control mice (Fig. 5C). These findings indicate that c-REL is required for the formation of GCs and for the extrafollicular plasma cell response upon T-dependent immunization.
Figure 5. Impaired GC formation in mice with deletion of rel, but not rela, in B-cells.
relfl/flCD19-Cre, relfl/+CD19-Cre and CD19-Cre littermates (A-C), and relafl/flCD19-Cre, relafl/+CD19-Cre and CD19-Cre littermates (D-F) were immunized with SRBC and analyzed 5 days later. (A,D) CD95, PNA and eGFP expression by splenic B-cells were analyzed by flow cytometry. Numbers above gates indicate the percentage of CD95hiPNAhi (dot plots) or eGFP+CD95hiPNAhi (histograms) GC B-cells. (B,E) Spleen sections from mice of the corresponding genotypes were analyzed for the expression of BCL6 and IgG1 or IgM; IgG1 stainings were counterstained with haematoxylin. One representative mouse out of 3 per group is shown. (C,F) CD138 and eGFP expression by splenic mononuclear cells from mice of the corresponding genotypes were analyzed by flow cytometry. Numbers below gates indicate the percentage of CD138hieGFP+ or CD138hieGFP− cells. (A,C,D,F) Data are cumulative from independent experiments (n=3-5 per group), with each symbol representing a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student's t test (*, P<0.05; **, P<0.01; ***, P<0.001).
RELA is dispensable for the formation of GCs
The embryonic lethality of rela−/− mice10 practically impeded the study of RELA's B-cell-intrinsic function in antibody responses in vivo. To determine the role of RELA in T-dependent B-cell responses, we immunized relafl/flCD19-Cre, relafl/+CD19-Cre and CD19-Cre control mice with SRBC. In contrast to what we observed upon rel deletion, relafl/flCD19-Cre mice mounted a GC response similar to control mice 5 days after SRBC-immunization (Fig. 5D,E). Accordingly, the fraction of eGFP+ GC B-cells was similar in relafl/flCD19-Cre and relafl/+CD19-Cre mice (Fig. 5D). Also, relafl/flCD19-Cre mice generated equal amounts of splenic plasma cells compared to relafl/+CD19-Cre and CD19-Cre control mice (Fig. 5F), and showed similar fractions of eGFP-expressing plasma cells compared to relafl/+CD19-Cre mice (Fig. 5F). Thus, RELA seems to be dispensable for both the formation of GCs and the generation of extrafollicular plasma cells during the T-dependent immune response.
Mice with B-cell-specific deletion of c-REL show strong impairment of T-independent type-II and type-I antibody responses
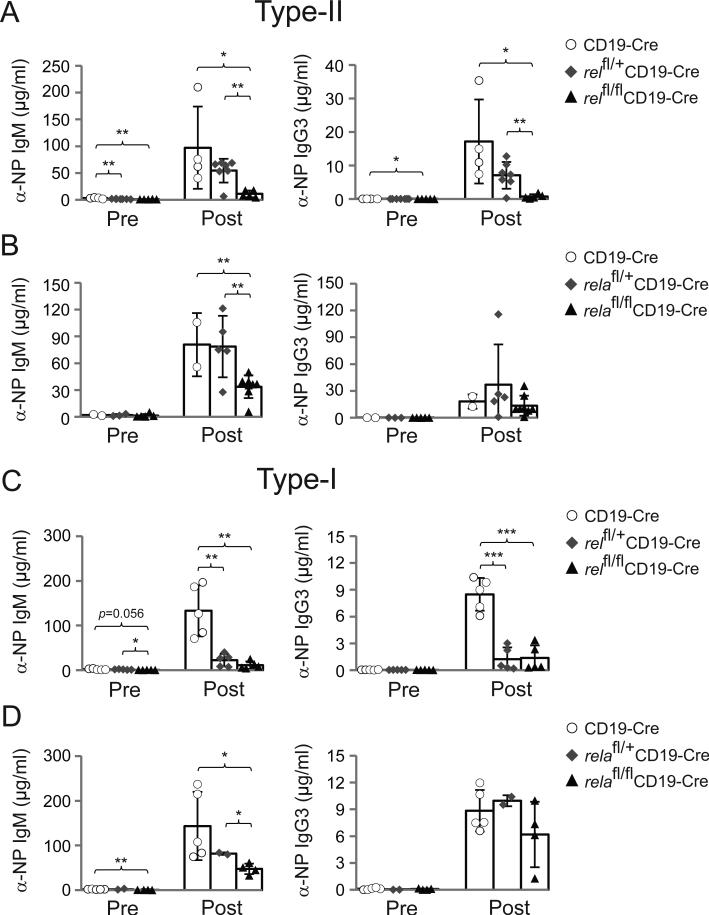
To investigate the roles of c-REL and RELA in T-independent type-II responses against polysaccharides, relfl/flCD19-Cre or relafl/flCD19-Cre and the corresponding heterozygous and CD19-Cre control mice were immunized with the hapten 4-hydroxy-3-nitrophenyl acetyl (NP) conjugated to aminoethyl carboxymethyl (AECM)-Ficoll and assessed for NP-specific IgM and IgG3 serum titers 7 days later. This analysis showed a strong reduction (5 to 10-fold) in both NP-specific IgM and IgG3 serum titers in relfl/flCD19-Cre mice compared to relfl/+CD19-Cre and CD19-Cre mice (Fig. 6A). relafl/flCD19-Cre mice showed a less severe (~2-fold) reduction in NP-specific IgM titers, and no significant changes in the corresponding IgG3 titers (Fig. 6B).
Figure 6. Mice with B-cell-specific deletion of c-REL show strong impairment of T-independent type-II and type-I antigen responses.
(A) α-NP IgM (left) and α-NP IgG3 (right) serum response of relfl/flCD19-Cre, relfl/+CD19-Cre and CD19-Cre mice to NP-ficoll immunization before (Pre), and 7d after immunization (Post) with NP-ficoll (left). (B) α-NP IgM (left) and α-NP IgG3 (right) serum response of relafl/flCD19-Cre, relafl/+CD19-Cre and CD19-Cre mice to NP-ficoll immunization before (Pre), and 7d after immunization with NP-ficoll (left). (C) α-NP IgM (left) and α-NP IgG3 (right) serum response of relfl/flCD19-Cre, relfl/+CD19-Cre and CD19-Cre mice to NP-LPS immunization before (Pre), and 7d after immunization with NP-LPS (left). (D) α-NP IgM (left) and α-NP IgG3 (right) serum response of relafl/flCD19-Cre, relafl/+CD19-Cre and CD19-Cre mice to NP-LPS immunization before (Pre), and 7d after immunization with NP-LPS (left). (A-D) Data are cumulative from independent experiments (n=2-7 per group), with each symbol representing a mouse. Data are shown as mean ± standard deviation. Statistical significance was determined by Student's t test (*, P<0.05; **, P<0.01; ***, P<0.001).
Type-I T-independent B-cell responses are characterized by the co-activation of antigen-specific B-cells through Toll-like receptors (TLRs) such as TLR4, the receptor for LPS. We immunized relfl/flCD19-Cre or relafl/flCD19-Cre and the corresponding heterozygous and CD19-Cre control mice with NP-LPS and analyzed for NP-specific IgM and IgG3 serum titers 7 days later. NP-specific IgM and IgG3 serum titers in relfl/flCD19-Cre and relfl/+CD19-Cre mice were strongly reduced (8 to 12-fold) compared to CD19-Cre mice (Fig. 6C), implying that the previously observed reduction in NP-specific IgG3 serum titers upon NP-LPS immunization in rel−/− mice14 is a B-cell intrinsic effect. In contrast, relafl/flCD19-Cre mice showed only a 3-fold reduction in serum IgM levels compared to controls, but no significant changes in IgG3 levels were observed among the different genotypes (Fig. 6D). Together, the results indicate a more prominent role for c-REL in T-independent type-II and type-I antibody responses in comparison to RELA.
Discussion
The B-cell intrinsic roles of the canonical NF-κB transcription factors c-REL and RELA in mature B-cell development and activation are incompletely understood. Our studies revealed differential roles of these subunits during distinct B-cell developmental stages. We found that combined, but not individual ablation of c-REL and RELA strongly impaired the generation of mature B-cells, indicating redundancy of the canonical subunits during this developmental stage. Conversely, the subunits were not functionally redundant during antigen-activation of B-cells, as c-REL deficiency alone strongly impaired the formation of GCs and T-independent antibody responses, whereas RELA was dispensable for the initiation of the GC reaction and only modestly impacted T-independent antibody responses.
The observation that the B-cell-specific combined deletion of rel and rela strongly impaired B-cell development is in keeping with previous publications that demonstrated a crucial role for canonical NF-κB signaling in the generation and maintenance of mature B-cells.6-9 A recent study by Derudder et al. has more precisely dissected the roles of the canonical pathway in these processes by deleting the upstream regulators ikk2 or nemo in different B-cell developmental stages.9 Thus, activation through this pathway is required at the T1 stage of development, and—for those cells that have overcome this developmental block—also later in MZ B-cells for maintenance and in FO B-cells for long-term persistence.9 In agreement with these findings, rel/rela-deletion led to a block in the T1 to T2 transition in the spleen. Moreover, the spleens of relfl/flrelafl/flCD19-Cre mice were characterized by fewer and smaller B-cell follicles (composed predominantly of FO B-cells) and a dramatic counterselection against c-REL/RELA-deficient MZ B-cells (Fig. 2). Of note, Derudder et al. report that a bcl2-transgene rescued FO, but not MZ B-cells in mice with B-cell-specific deletion of nemo9. It therefore appears that, whereas c-REL and RELA may contribute to the maintenance of FO B-cells by upregulating BCL2 expression, in MZ B-cells, c-REL and RELA may be required for the establishment of a biological program beyond the control of cell survival.
The generation of a normal mature B-cell compartment requires activation through the alternative NF-κB pathway in addition to the canonical pathway (reviewed in ref.45). In accordance, we recently showed that the functional abolishment of the alternative pathway in B-cells via combined deletion of the downstream transcription factors RELB and NF-κB2 strongly impaired the generation and maintenance of mature B-cells.46 How do the B-cell phenotypes observed upon inactivation of the canonical vs. the alternative pathway compare qualitatively and quantitatively? By using a similar experimental strategy to conditionally delete the downstream transcription factors of the separate NF-κB pathways, we were able to directly compare the consequences of their inactivation on mature B-cell development. Three observations are evident from this comparison: First, the effects of the ablation of the separate pathways on the size and composition of the mature B-cell compartment were virtually the same (Fig. 1&2A and ref.46). In addition, relb/nfkb2-deleted and rel/rela-deleted MZ B-cells were strongly counterselected, comprising only ~25% of MZ B-cells in mice of both genotypes (Fig. 2B,C and ref.46). These findings suggest that there is some level of complementation among the canonical and the alternative NF-κB pathways during the generation and maintenance of mature B-cells. Second, our observation that relb/nfkb2-deleted46 and rel/rela-deleted B-cells show the same predisposition to undergo apoptosis when cultured with BAFF provides further evidence for the functional requirements of both NF-κB pathways in BAFF signaling.8,37 Third, whereas c-REL and RELA were functionally redundant during the generation and maintenance of mature B-cells, single ablation of RELB or NF-κB2 in B-cells did impair B-cell development—albeit to a lesser extent than the combined deletion.46
In contrast to its redundant role during B-cell generation, c-REL was uniquely required for the formation of GCs upon antigen-activation in the T-dependent immune response. c-REL is known to be crucial for normal B-cell activation in vitro, as c-REL-deficient B-cells showed impaired proliferation in response to several mitogenic stimuli,14-16 a finding which is supported by the observation that BCR-stimulation led to a fast nuclear translocation of c-REL during B-cell activation.21 Our previous observation that CD40+IgM-stimulated c-REL-deficient B-cells in vitro showed defects in the establishment of a metabolic program that precedes proliferation25 suggests that also upon antigen-activation in vivo, c-REL may be required for cell growth and optimal proliferation.
A recent publication demonstrated that inhibition of IKK-induced proteolysis of p105, the precursor of p50, in murine B-cells impaired the antigen-induced formation of both GCs and extrafollicular plasmablasts47 similar to what we here described for rel deletion in B-cells. It is therefore possible that the observed phenotype in the p105 mutant mice is due to the inability of these mice to process p105 which thereby prevents the formation and thus nuclear translocation of c-REL/p50 heterodimers. Conversely, the loss of p105 (which acts as an inhibitory κB protein for c-REL and RELA) in nfkb1−/− mice may lead to enhanced c-REL activity in B-cells, which might contribute to the increased formation of spontaneous GCs in aging mice lacking NF-κB1.48
c-REL shows a biphasic activation pattern during T-dependent B-cell activation and the GC response. c-REL undergoes rapid nuclear translocation upon B-cell activation,21,22 and, while there is no active NF-κB signaling in the majority of GC B-cells,23,24 nuclear translocation of c-REL is detectable within a small subset of light zone B-cells24 and we have previously shown that it is functionally required for the maintenance of the GC reaction.25 Together with our present observations, this indicates that c-REL is required at two stages of the GC reaction, first during the initial antigen-activation phase and later in the fully established GC, presumably during the selection of high-affinity B-cells. Thus, our study adds c-REL to a growing list of transcriptional regulators that have critical functions during the formation of GCs and also in a later stage of GC development which include IRF4 and c-MYC.26-29 Similar to rel ablation in all B-cells (as opposed to GC-specific ablation), B-cell-specific deletion of irf4 dramatically impaired GC formation upon T-dependent immunization,28,29 and c-MYC was found to be required for the initial expansion of GC dark zone cells within the follicle.26 Later, in the established GC, c-REL and c-MYC are required for the maintenance of the GC reaction,25,27 and IRF4 is essential for optimal class switch recombination and plasma cell differentiation.30,31 It will be interesting to determine in future studies to what extent these transcription factors crosstalk among each other49,50 in the different GC B-cell developmental stages.
By deleting rela specifically in GC B-cells, we have recently demonstrated that RELA is required for the generation of GC-derived plasma cells, which was reflected by a dramatic reduction in the serum levels of NP-specific IgG1 antibodies (>10-fold).25 While we here observed that relafl/flCD19-Cre mice showed only a ~2-fold reduction in NP-specific antibodies upon T-independent immunization, we also found that these mice generated equal fractions of plasma cells early in the extrafollicular response upon T-dependent immunization. These results suggest that RELA deficiency does not affect all types of B-cell activation that lead to the generation of antibody-secreting cells to the same extent. One possible explanation is that c-REL may partly compensate for RELA in the T-dependent extrafollicular and T-independent antibody response, but not during the GC response. It will be interesting to identify the specific transcription factor networks involved in the generation of antigen-secreting cells in the different arms of the humoral immune response.
Integrating the results of our present and published25 studies from the conditional deletion of rel and/or rela in all B-cells and GC B-cells, the following picture emerges: c-REL and RELA are jointly required during the generation of the mature B-cell compartment and appear to have non-redundant and entirely distinct functions during antigen-dependent B-cell development. c-REL was found to be the critical subunit for the formation and maintenance of GCs, whereas RELA was dispensable for these processes but instead crucial for the differentiation or physiology of GC-derived plasma cells.25 The canonical NF-κB signaling pathway can be aberrantly activated in B-cell malignancies51,52 and in diseases with chronic B-cell activation.53,54 The differential requirements of c-REL and RELA in B-cell activation and differentiation may be exploited for the development of more specific and thus less toxic therapies aimed at inhibiting pathogenic NF-κB signaling in malignant or chronically activated B-cells at the level of NF-κB subunits, the feasibility of which has recently been demonstrated for a small molecule inhibitor of c-REL.55 Our findings provide additional examples for the diverse roles of separate NF-κB subunits that may be relevant for disease therapies.56
MATERIALS AND METHODS
Mice
Conditional rel, rela and CD19-Cre mice have been described.25,33 All mice were on a C57BL/6 background, male or female, with an age between 2 and 4 months. Mice were housed and treated in compliance with the US Department of Health and Human Services Guide for the Care and Use of Laboratory Animals and according to the guidelines of the Institute of Comparative Medicine at Columbia University. The animal protocol was approved by Columbia University's IACUC. To minimize the number of animals for ethical reasons, experiments used a number of mice per group required to provide the power to detect a two-fold difference between groups. Each experiment was performed multiple times. Littermates were randomly assigned to experimental groups according to genotype. Experiments were not performed in a blinded fashion. No animals were excluded from analysis.
Immunization
For T-dependent immune responses, mice were immunized i.p. with 1×109 SRBCs in PBS. For T-independent type-I and type-II responses, mice were immunized i.p. with 20 μg NP-LPS or 30 μg of NP-AECM-FICOLL (both Biosearch Technologies), respectively. Peripheral blood and spleens were removed at the indicated time-points for analysis.
B-cell isolation and culture
Single cell suspensions of murine spleen were subjected to hypotonic lysis and ‘untouched’ B cells were purified by magnetic cell separation using the MACS B-cell isolation kit (Miltenyi Biotec). Purified B cells from the respective genotypes were cultured in the presence of 25 ng/ml BAFF (R&D Systems) at a cell density of 1.5 × 106 cells/ml.
Flow cytometry
Spleen cell suspensions or cultured B-cells were stained with the following antibodies as described:25,46 anti-CD138-PE (clone: 281-2); anti-CD95-PE (clone: Jo2); IgM-APC (clone II/41); anti-IgD-PE (clone 11-26c.2a); and anti-CD23-PE (clone B3B4) (all BD Pharmingen); and anti-B220-PerCP (clone: RA3-6B2); anti-CD21-APC (clone: 7E9); anti-CD24 (HSA)-PE (clone: 30-F1), anti-CD93-PE (clone: AA4.1); and anti-CD23-Pacific Blue (clone: B3B4) (all Biolegend); and anti-CD19-CF594 (clone: 1D3) (BD Horizon); and PNA-Biotin (Vector Laboratories) followed by Streptavidin-APC (BD Pharmingen). Annexin V/7-AAD stainings were performed using the APC Annexin V Apoptosis Detection Kit with 7-AAD (Biolegend). For DNA content analysis, cells were lysed and stained with propidium iodide (PI). The cells were analyzed on a FACSCalibur or a LSRII (Becton Dickinson). Transitional B-cells were identified by gating on B220+CD93+ lymphocytes.39 GC B-cells were identified by gating on B220+ lymphocytes. eGFP+ and eGFP− CD138hi plasma cells were identified through the lymphocyte gate. Data were analyzed using FlowJo software.
Enzyme-linked immunosorbent assay (ELISA)
For NP-LPS and NP-Ficoll immunization experiments, 96 well immune-plates (Thermo Fisher Scientific) were coated with NP25-BSA (Biosearch Technologies). Mouse serum samples were incubated for 2h at RT. Standard curves were generated using mouse IgM and IgG3 (Southern Biotech). Bound antibodies were detected by AP-conjugated anti-mouse IgM and IgG3-antibodies (Southern Biotech). Plates were developed with p-nitrophenylphosphate (Southern Biotech) dissolved in substrate buffer.
Histology and immunohistochemistry
Sections of splenic tissue (3 μm) were prepared after overnight fixation in 10% formalin and embedding in paraffin. Sections were stained with H&E for morphologic evaluation. Primary antibodies, rabbit anti-mouse CD3 (clone: SP7; Thermo Fisher Scientific), rabbit anti-GFP (Molecular Probes, Invitrogen), or rabbit anti-mouse BCL6 (clone: N-3; Santa Cruz) or alkaline peroxidase (AP)-conjugated anti-mouse IgM and IgG1-antibodies (Southern Biotech) were applied to tissue sections and incubated overnight at 4°C. Secondary staining with anti-rabbit HRP-labeled polymer (Dako) was performed for BCL6, CD3 and eGFP and developed in aminoethylcarbazole (AEC; Sigma), while AP-conjugated antibodies were developed in nitro blue tetrazolium chloride-5-bromo-4-chloro-3-indolyl phosphate (NBT/BCIP; Roche). Sections stained for BCL6/IgG1 were counterstained with hematoxylin. Images were acquired via a Digital Sight camera mounted to a Nikon Eclipse E600 microscope (Nikon).
Statistical analysis
P values were obtained using unpaired Student's t test.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Laura Pasqualucci for critically reading the manuscript and David Dominguez-Sola for discussion. This work was supported by NCI/NIH grant R01-CA157660 to U.K., a grant from the Stewart Trust Foundation (USA), the HICCC, and through fellowships of the German Research Council (DFG) to N.H. and M.M., and a Cancer Biology Training Program fellowship (NCI/NIH grant 5T32-CA009503-26) to N.S.D.
Abbreviations
- FO
follicular
- MZ
marginal zone
- GC
germinal center
- NP
4-hydroxy-3-nitrophenyl-acetyl
Footnotes
Conflict of Interest Statement: The authors declare no commercial or financial conflict of interest.
REFERENCES
- 1.Gerondakis S, Siebenlist U. Roles of the NF-kappaB pathway in lymphocyte development and function. Cold Spring Harb Perspect Biol. 2010;2:a000182. doi: 10.1101/cshperspect.a000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaileh M, Sen R. NF-kappaB function in B lymphocytes. Immunol Rev. 2012;246:254–271. doi: 10.1111/j.1600-065X.2012.01106.x. [DOI] [PubMed] [Google Scholar]
- 3.Sasaki Y, Iwai K. Roles of the NF-kappaB Pathway in B-Lymphocyte Biology. Curr Top Microbiol Immunol. 2016;393:177–209. doi: 10.1007/82_2015_479. [DOI] [PubMed] [Google Scholar]
- 4.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 5.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 6.Pasparakis M, Schmidt-Supprian M, Rajewsky K. IkappaB kinase signaling is essential for maintenance of mature B cells. J Exp Med. 2002;196:743–752. doi: 10.1084/jem.20020907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li ZW, Omori SA, Labuda T, Karin M, Rickert RC. IKK beta is required for peripheral B cell survival and proliferation. J Immunol. 2003;170:4630–4637. doi: 10.4049/jimmunol.170.9.4630. [DOI] [PubMed] [Google Scholar]
- 8.Sasaki Y, Derudder E, Hobeika E, Pelanda R, Reth M, Rajewsky K, et al. Canonical NF-kappaB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity. 2006;24:729–739. doi: 10.1016/j.immuni.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Derudder E, Herzog S, Labi V, Yasuda T, Kochert K, Janz M, et al. Canonical NF-kappaB signaling is uniquely required for the long-term persistence of functional mature B cells. Proc Natl Acad Sci U S A. 2016;113:5065–5070. doi: 10.1073/pnas.1604529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beg AA, Sha WC, Bronson RT, Ghosh S, Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 11.Doi TS, Takahashi T, Taguchi O, Azuma T, Obata Y. NF-kappa B RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J Exp Med. 1997;185:953–961. doi: 10.1084/jem.185.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horwitz BH, Zelazowski P, Shen Y, Wolcott KM, Scott ML, Baltimore D, et al. The p65 subunit of NF-kappa B is redundant with p50 during B cell proliferative responses, and is required for germline CH transcription and class switching to IgG3. J Immunol. 1999;162:1941–1946. [PubMed] [Google Scholar]
- 13.Prendes M, Zheng Y, Beg AA. Regulation of developing B cell survival by RelA-containing NF-kappa B complexes. J Immunol. 2003;171:3963–3969. doi: 10.4049/jimmunol.171.8.3963. [DOI] [PubMed] [Google Scholar]
- 14.Köntgen F, Grumont RJ, Strasser A, Metcalf D, Li R, Tarlinton D, et al. Mice lacking the c-rel proto oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 15.Carrasco D, Cheng J, Lewin A, Warr G, Yang H, Rizzo C, et al. Multiple hemopoietic defects and lymphoid hyperplasia in mice lacking the transcriptional activation domain of the c-Rel protein. J Exp Med. 1998;187:973–984. doi: 10.1084/jem.187.7.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tumang JR, Owyang A, Andjelic S, Jin Z, Hardy RR, Liou ML, et al. c-Rel is essential for B lymphocyte survival and cell cycle progression. Eur J Immunol. 1998;28:4299–4312. doi: 10.1002/(SICI)1521-4141(199812)28:12<4299::AID-IMMU4299>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 17.Grossmann M, O'Reilly LA, Gugasyan R, Strasser A, Adams JM, Gerondakis S. The anti-apoptotic activities of Rel and RelA required during B-cell maturation involve the regulation of Bcl-2 expression. EMBO J. 2000;19:6351–6360. doi: 10.1093/emboj/19.23.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacLennan IC. Germinal centers. Annu Rev Immunol. 1994;12:117–139. doi: 10.1146/annurev.iy.12.040194.001001. [DOI] [PubMed] [Google Scholar]
- 19.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996;381:751–758. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 20.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 21.Damdinsuren B, Zhang Y, Khalil A, Wood WH, 3rd, Becker KG, Shlomchik MJ, et al. Single round of antigen receptor signaling programs naive B cells to receive T cell help. Immunity. 2010;32:355–366. doi: 10.1016/j.immuni.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JL, Chiles TC, Sen RJ, Rothstein TL. Inducible nuclear expression of NF-kappa B in primary B cells stimulated through the surface Ig receptor. J Immunol. 1991;146:1685–1691. [PubMed] [Google Scholar]
- 23.Shaffer AL, Rosenwald A, Hurt EM, Giltnane JM, Lam LT, Pickeral OK, et al. Signatures of the immune response. Immunity. 2001;15:375–385. doi: 10.1016/s1074-7613(01)00194-7. [DOI] [PubMed] [Google Scholar]
- 24.Basso K, Klein U, Niu H, Stolovitzky GA, Tu Y, Califano A, et al. Tracking CD40 signaling during germinal center development. Blood. 2004;104:4088–4096. doi: 10.1182/blood-2003-12-4291. [DOI] [PubMed] [Google Scholar]
- 25.Heise N, De Silva NS, Silva K, Carette A, Simonetti G, Pasparakis M, et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-kappaB transcription factor subunits. J Exp Med. 2014;211:2103–2118. doi: 10.1084/jem.20132613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calado DP, Sasaki Y, Godinho SA, Pellerin A, Kochert K, Sleckman BP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13:1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominguez-Sola D, Victora GD, Ying CY, Phan RT, Saito M, Nussenzweig MC, et al. The proto oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012;13:1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochiai K, Maienschein-Cline M, Simonetti G, Chen J, Rosenthal R, Brink R, et al. Transcriptional regulation of germinal center B and plasma cell fates by dynamical control of IRF4. Immunity. 2013;38:918–929. doi: 10.1016/j.immuni.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Willis SN, Good-Jacobson KL, Curtis J, Light A, Tellier J, Shi W, et al. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J Immunol. 2014;192:3200–3206. doi: 10.4049/jimmunol.1303216. [DOI] [PubMed] [Google Scholar]
- 30.Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 31.Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity. 2006;25:225–236. doi: 10.1016/j.immuni.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Pohl T, Gugasyan R, Grumont RJ, Strasser A, Metcalf D, Tarlinton D, et al. The combined absence of NF-kappa B1 and c-Rel reveals that overlapping roles for these transcription factors in the B cell lineage are restricted to the activation and function of mature cells. Proc Natl Acad Sci U S A. 2002;99:4514–4519. doi: 10.1073/pnas.072071599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25:1317–1318. doi: 10.1093/nar/25.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackay F, Browning JL. BAFF: a fundamental survival factor for B cells. Nat Rev Immunol. 2002;2:465–475. doi: 10.1038/nri844. [DOI] [PubMed] [Google Scholar]
- 35.Hatada EN, Do RK, Orlofsky A, Liou HC, Prystowsky M, MacLennan IC, et al. NF-kappa B1 p50 is required for BLyS attenuation of apoptosis but dispensable for processing of NF-kappa B2 p100 to p52 in quiescent mature B cells. J Immunol. 2003;171:761–768. doi: 10.4049/jimmunol.171.2.761. [DOI] [PubMed] [Google Scholar]
- 36.Enzler T, Bonizzi G, Silverman GJ, Otero DC, Widhopf GF, Anzelon-Mills A, et al. Alternative and classical NF-kappa B signaling retain autoreactive B cells in the splenic marginal zone and result in lupus-like disease. Immunity. 2006;25:403–415. doi: 10.1016/j.immuni.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Almaden JV, Liu YC, Yang E, Otero DC, Birnbaum H, Davis-Turak J, et al. B-cell survival and development controlled by the coordination of NF-kappaB family members RelB and cRel. Blood. 2016;127:1276–1286. doi: 10.1182/blood-2014-10-606988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loder F, Mutschler B, Ray RJ, Paige CJ, Sideras P, Torres R, et al. B cell development in the spleen takes place in discrete steps and is determined by the quality of B cell receptor-derived signals. J Exp Med. 1999;190:75–89. doi: 10.1084/jem.190.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Allman D, Lindsley RC, DeMuth W, Rudd K, Shinton SA, Hardy RR. Resolution of three nonproliferative immature splenic B cell subsets reveals multiple selection points during peripheral B cell maturation. J Immunol. 2001;167:6834–6840. doi: 10.4049/jimmunol.167.12.6834. [DOI] [PubMed] [Google Scholar]
- 40.Debnath I, Roundy KM, Weis JJ, Weis JH. Defining in vivo transcription factor complexes of the murine CD21 and CD23 genes. J Immunol. 2007;178:7139–7150. doi: 10.4049/jimmunol.178.11.7139. [DOI] [PubMed] [Google Scholar]
- 41.Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell. 2004;117:787–800. doi: 10.1016/j.cell.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Derudder E, Cadera EJ, Vahl JC, Wang J, Fox CJ, Zha S, et al. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nat Immunol. 2009;10:647–654. doi: 10.1038/ni.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Victora GD. SnapShot: the germinal center reaction. Cell. 2014;159:700–700. e701. doi: 10.1016/j.cell.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 44.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gardam S, Brink R. Non-Canonical NF-kappaB Signaling Initiated by BAFF Influences B Cell Biology at Multiple Junctures. Front Immunol. 2014;4:509. doi: 10.3389/fimmu.2013.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Silva NS, Silva K, Anderson MM, Bhagat G, Klein U. Impairment of Mature B Cell Maintenance upon Combined Deletion of the Alternative NF-kappaB Transcription Factors RELB and NF-kappaB2 in B Cells. J Immunol. 2016;196:2591–2601. doi: 10.4049/jimmunol.1501120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jacque E, Schweighoffer E, Visekruna A, Papoutsopoulou S, Janzen J, Zillwood R, et al. IKK-induced NF-kappaB1 p105 proteolysis is critical for B cell antibody responses to T cell-dependent antigen. J Exp Med. 2014;211:2085–2101. doi: 10.1084/jem.20132019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Valle E, Grigoriadis G, O'Reilly LA, Willis SN, Maxwell MJ, Corcoran LM, et al. NFkappaB1 is essential to prevent the development of multiorgan autoimmunity by limiting IL-6 production in follicular B cells. J Exp Med. 2016;213:621–641. doi: 10.1084/jem.20151182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grumont RJ, Gerondakis S. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor kappaB. J Exp Med. 2000;191:1281–1292. doi: 10.1084/jem.191.8.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grumont RJ, Strasser A, Gerondakis S. B cell growth is controlled by phosphatidylinosotol 3-kinase-dependent induction of Rel/NF-kappaB regulated c-myc transcription. Mol Cell. 2002;10:1283–1294. doi: 10.1016/s1097-2765(02)00779-7. [DOI] [PubMed] [Google Scholar]
- 51.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pasqualucci L, Dalla-Favera R. The genetic landscape of diffuse large B-cell lymphoma. Semin Hematol. 2015;52:67–76. doi: 10.1053/j.seminhematol.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barnes PJ, Karin M. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 54.Brown KD, Claudio E, Siebenlist U. The roles of the classical and alternative nuclear factor-kappaB pathways: potential implications for autoimmunity and rheumatoid arthritis. Arthritis Res Ther. 2008;10:212. doi: 10.1186/ar2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shono Y, Tuckett AZ, Liou HC, Doubrovina E, Derenzini E, Ouk S, et al. Characterization of a c-Rel Inhibitor That Mediates Anticancer Properties in Hematologic Malignancies by Blocking NF-kappaB-Controlled Oxidative Stress Responses. Cancer Res. 2016;76:377–389. doi: 10.1158/0008-5472.CAN-14-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer. 2012;12:121–132. doi: 10.1038/nrc3204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.