Abstract
Zmpste24 is a metalloproteinase required for the processing of prelamin A to lamin A, a structural component of the nuclear lamina. Zmpste24 deficiency results in the accumulation of prelamin A within cells, a complete loss of mature lamin A, and misshapen nuclear envelopes. Zmpste24-deficient (Zmpste24–/–) mice exhibit retarded growth, alopecia, micrognathia, dental abnormalities, osteolytic lesions in bones, and osteoporosis, which are phenotypes shared with Hutchinson–Gilford progeria syndrome, a human disease caused by the synthesis of a mutant prelamin A that cannot undergo processing to lamin A. Zmpste24–/– mice also develop muscle weakness. We hypothesized that prelamin A might be toxic and that its accumulation in Zmpste24–/– mice is responsible for all of the disease phenotypes. We further hypothesized that Zmpste24–/– mice with half-normal levels of prelamin A (Zmpste24–/– mice with one Lmna knockout allele) would be subjected to less toxicity and be protected from disease. Thus, we bred and analyzed Zmpste24–/–Lmna+/– mice. As expected, prelamin A levels in Zmpste24–/–Lmna+/– cells were significantly reduced. Zmpste24–/–Lmna+/– mice were entirely normal, lacking all disease phenotypes, and misshapen nuclei were less frequent in Zmpste24–/–Lmna+/– cells than in Zmpste24–/– cells. These data suggest that prelamin A is toxic and that reducing its levels by as little as 50% provides striking protection from disease.
Keywords: Hutchinson–Gilford progeria syndrome, prelamin A
The metalloproteinase Zmpste24 (1, 2), the mammalian orthologue of Ste24p in Saccharomyces cerevisiae (3, 4), is required for the biogenesis of mature lamin A (a key component of the nuclear lamina) from its precursor protein prelamin A (1, 2). In Zmpste24-deficient (Zmpste24–/–) cells, mature lamin A is absent and prelamin A accumulates (1, 2). These perturbations in lamin A metabolism adversely affect the integrity of the nuclear envelope. Many Zmpste24–/– cells contain blebs in the nuclear envelope (2), much like cells lacking Lmna (the gene encoding prelamin A and lamin C) (5). At the whole-animal level, the consequences of Zmpste24 deficiency are striking (1, 2). Zmpste24–/– mice appear normal at birth but soon manifest retarded growth, dental abnormalities, broken bones, osteolytic lesions in bones, and alopecia. They also have muscle weakness, manifested by an abnormal gait and a reduced ability to hang on to an upside-down grid. It seems plausible that all of these phenotypes are due to defective lamin A metabolism, given that muscular dystrophy is caused by missense mutations in LMNA (6) and most of the other phenotypes are observed in mandibuloacral dysplasia and Hutchinson–Gilford progeria syndrome (HGPS), human diseases caused by LMNA mutations (7, 8). On the other hand, it is unclear whether prelamin A is the sole substrate for Zmpste24, and thus it has never been possible to discount the possibility that some of the relatively nonspecific phenotypes in Zmpste24–/– mice are caused by defective processing of an as-yet-unidentified protein substrate (1, 2).
The processing of prelamin A is complex (9, 10). A CAAX motif at the carboxyl terminus (amino acids 661–664) specifies three sequential modifications: farnesylation, endoproteolytic release of the last three amino acids of the protein (i.e., the –AAX), and methylation of the newly exposed carboxyl-terminal farnesylcysteine. Next, 15 aa are clipped from the carboxyl terminus of the protein (amino acids 647–661), releasing mature lamin A (11). Zmpste24 is clearly essential for the production of mature lamin A. By analogy to a-factor processing by Ste24p in yeast (3, 4), we hypothesized that Zmpste24 is capable of carrying out the first protease step (release of the –AAX) and carries out the final endoproteolytic processing step (1). Recent biochemical studies suggest that Zmpste24 indeed can carry out both steps (12).
Interest in Zmpste24 deficiency and prelamin A has increased with the finding that most cases of HGPS are caused by de novo single-nucleotide substitutions in codon 608 of prelamin A (exon 11 of LMNA) (8). These mutations result in an alternatively spliced transcript that yields a mutant prelamin A protein with an in-frame deletion of 50 carboxyl-terminal amino acids (residues 606–656) but retaining the carboxyl-terminal eight amino acids including the CAAX motif. The retention of the CAAX motif, but not the second endoproteolytic cleavage site, means that the mutant prelamin A in HGPS likely would retain a farnesylcysteine methyl ester at the carboxyl terminus. Humans with HGPS have many of the phenotypes observed in the Zmpste24–/– mice, except for muscle weakness (8). Relatively small amounts of the mutant prelamin A are present in HGPS cells (8), yet the disease is severe (8), and the mutant protein adversely affects the function of the nuclear lamina (13). The latter observations led to the suggestion that the mutant prelamin A might have “dominant-negative” effects on nuclear lamina function (13).
In this study, we assessed the importance of prelamin A accumulation for the development of disease phenotypes in Zmpste24–/– mice (1). We hypothesized that prelamin A is toxic to Zmpste24–/– cells and tissues, accounting directly or indirectly for all of the disease phenotypes, and we further hypothesized that the disease phenotypes would be ameliorated by reduced expression of the molecule. In this study, we tested whether half-normal amounts of prelamin A expression (heterozygosity for Lmna deficiency) would ameliorate disease phenotypes in Zmpste24–/– mice.
Materials and Methods
Mice. Zmpste24+/– mice (1, 14) were mated with Lmna+/– mice (5), and offspring from intercrosses of Zmpste24+/–Lmna+/– mice were analyzed. Lmna genotyping was performed by Southern blotting (5). Zmpste24 genotyping was performed by PCR with oligonucleotides 5′-TCACATGGAGTGAATGCTCTG-3′ and 5′-AGTGAACACCAGGCCAGTTT-3′; the mutant PCR product is 240 bp, and the wild-type product is 320 bp. All mice had a mixed genetic background (≈90% C57BL/6 and ≈10% 129/SvJae). The mice were fed a chow diet and housed in a virus-free barrier facility with a 12-h/12-h light/dark cycle. Mice were weighed weekly; their ability to hang on to an upside-down grid was assessed as described in ref. 1.
Cultured Embryonic Fibroblasts and Immunofluorescence Microscopy. Primary mouse embryonic fibroblasts (MEFs) were isolated from 13.5-d-postcoitus embryos of intercrosses of Zmpste24+/– Lmna+/– mice as described in ref. 15. For immunocytochemistry experiments, primary MEFs (passages 3–10, matched for different genotypes, not immortalized) were grown on coverslips and fixed with 3% paraformaldehyde, permeabilized with 0.2% Triton X-100, and incubated with a mouse LAP2 monoclonal antibody (1:200 dilution; BD Biosciences), a rabbit polyclonal antibody to emerin, or a goat polyclonal to lamin B (1:300 and 1:200 dilutions, respectively; Santa Cruz Biotechnology). The slides then were incubated with rabbit anti-goat IgG-Cy3, goat anti-rabbit IgG-Cy3, or rabbit anti-mouse IgG-Cy3 (1:1,000 dilution; Zymed). DNA was detected with SYTOX Green (Molecular Probes). Cell imaging was performed by confocal microscopy with an Olympus microscope and a Bio-Rad Laser2000 system. Nuclear shape abnormalities were scored by two independent observers blinded to genotype.
Western Blot Analyses. Urea-soluble MEF extracts were prepared as described in ref. 16. Briefly, adherent cells in T175 flasks were washed with PBS and scraped into ice-cold PBS containing the Roche EDTA-free Protease Inhibitor Mixture. The cell pellets were resuspended in an urea-containing buffer (8.6 M urea/10 mM Tris, pH 8/0.2% 2-mercaptoethanol/10 μM EDTA/1.0 mM NaF/1.0 mM PMSF/protease inhibitor mixture), sonicated, and centrifuged (10,000 × g for 5 min) to sediment insoluble materials. An aliquot of the supernatant was separated on 4–12% gradient polyacrylamide [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane gels with the NuPage System (Invitrogen). Equivalent amounts of protein were loaded onto the gels, as judged by a protein assay from Bio-Rad. The size-separated proteins were then electrophoretically transferred to nitrocellulose membranes for Western blotting. The antibody dilutions were 1:400 anti-lamin A/C mouse IgM (sc-7293), 1:400 anti-lamin A (carboxyl terminus) goat IgG (sc-6214), 1:1,000 anti-actin goat IgG (sc-1616), 1:6,000 horseradish peroxidase-labeled anti-goat IgG (sc-2020), and 1:4,000 horseradish peroxidase-labeled anti-mouse IgM (sc-2064) (all from Santa Cruz Biotechnology). Antibody binding was detected with the ECL Plus chemiluminescence system (Amersham Pharmacia) with subsequent exposure to x-ray film.
Microcomputed Tomography (μCT) Scans. Wild-type, Zmpste24–/–, and Zmpste24–/–Lmna+/– mice were examined (n = 2 of each genotype, 28–30 weeks of age) by compact cone-beam type tomography (MicroCT 40 scanner, Scanco Medical, Bassersdorf, Switzerland) (1). Whole-body scans were performed in the axial plane mounted in a cylindrical sample holder with a current of 0.16 mA and a voltage of 70 kV at an isotropic voxel size of 20.5 μm for the skulls and 30.7 μm for the remainder of the skeleton. The system was calibrated with a hydroxyapatite phantom of known density. μCT images were constructed in 1,024 × 1,024-pixel matrices with a standard convolution-back projection procedure with a Shepp and Logan filter. Three-dimensional images of whole bones reconstructed from individual μCT slices were used to qualitatively evaluate whole-bone structure and morphology. A constrained three-dimensional Gaussian filter was used to partially suppress noise. Bone tissue was segmented from marrow and soft tissue with a thresholding procedure. All samples were binarized with the same parameters for the filter width, filter support, and threshold.
Histology. After decalcification and paraffin embedding, the rib cages of mice (three per group) were transversely sectioned and stained with hematoxylin/eosin for histological evaluation with a light microscope.
Results
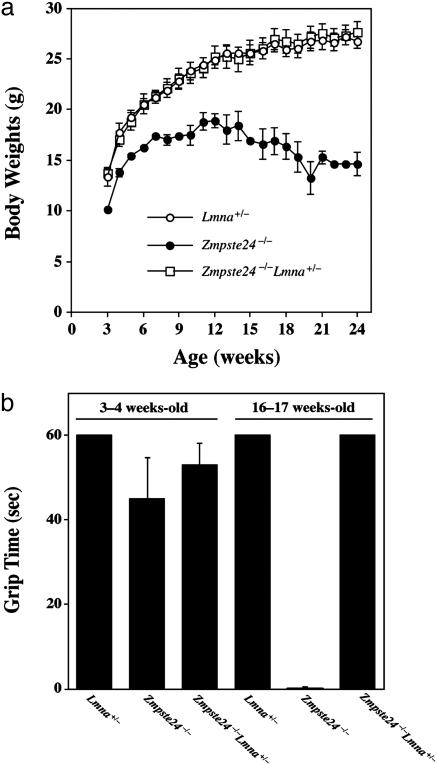
We compared groups of male and female Zmpste24–/– and Zmpste24–/–Lmna+/– mice produced from intercrosses of Zmpste24+/–Lmna+/– mice. Zmpste24–/–Lmna+/– mice grew as well as wild-type littermates and grew far better than Zmpste24–/– littermate controls (Fig. 1a). Moreover, muscle strength, judged by the ability to hang onto an upside-down grid (1), was similar in wild-type mice and Zmpste24–/–Lmna+/– mice but was markedly impaired in Zmpste24–/– littermate controls (Fig. 1b). Homozygous Lmna knockout mice (Lmna–/–) exhibited severely retarded growth and muscle weakness, and they died by 4–6 weeks of age, as originally described by Sullivan et al. (5). We did not detect any differences among the phenotypes of Lmna–/–, Zmpste24+/–Lmna–/–, and Zmpste24–/–Lmna–/– mice. We saw no abnormalities in Zmpste24+/–Lmna+/– mice.
Fig. 1.
Growth rates and grip strength in mice. (a) Female Zmpste24–/– Lmna+/– mice (n = 6) exhibit normal growth, indistinguishable from wild-type mice (n = 5). Similar results were obtained with male mice (data not shown). (b) Muscle strength in 3- to 4-week old and 16- to 17-week-old female wild-type (Lmna+/–, n = 7), Zmpste24–/– (n = 5), and Zmpste24–/–Lmna+/– mice (n = 9), as judged by the length of time that they were able to hang on to an upside-down grid (1) (grip time; P < 0.001). Similar results were observed for male mice. Differences became statistically significant at 8 weeks of age.
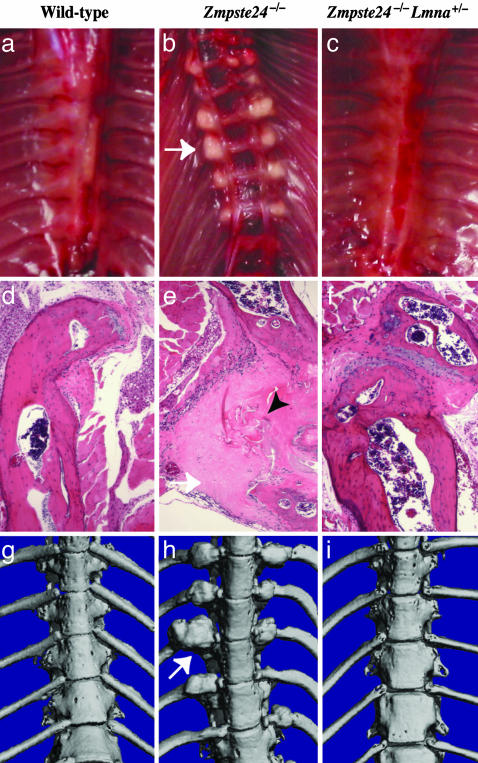
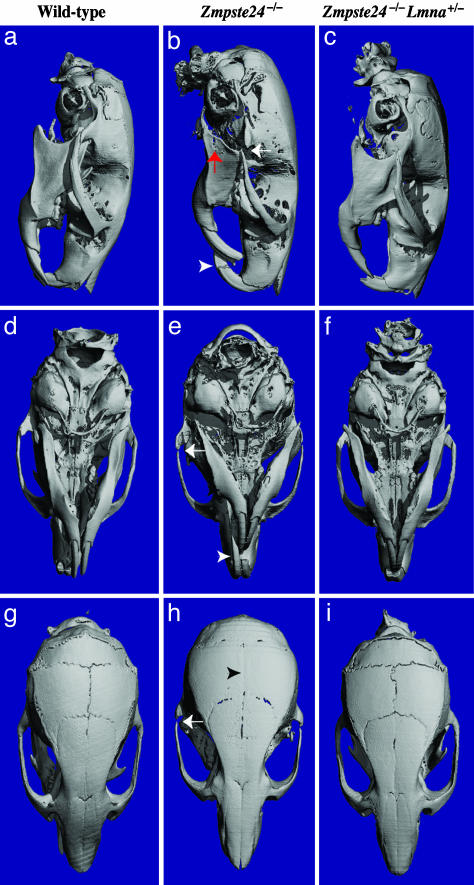
The normal growth and muscle strength in Zmpste24–/– Lmna+/– mice led us to suspect that these mice also might be protected from bone fractures and osteolytic lesions. This suspicion was confirmed. Rib fractures occurred near the costovertebral joints in all Zmpste24–/– mice but were absent in wild-type or Zmpste24–/–Lmna+/– mice (Fig. 2). Also, Zmpste24–/– mice exhibited a variety of skull abnormalities, including a small mandible, long incisors, osteolytic lesions in the mandible and posterior portion of the zygomatic arch, and a loss of the zigzag appearance of the cranial sutures (Fig. 3). These abnormalities were absent in wild-type and Zmpste24–/– Lmna+/– mice (Fig. 3).
Fig. 2.
Fractures of ribs near the costovertebral joint occur in Zmpste24–/– mice but not in wild-type or Zmpste24–/–Lmna+/– mice. (a–c) Photographs of the thorax after removal of the heart and lungs, revealing fracture callus in a Zmpste24–/– mouse (b, arrow indicating callus around a rib fracture). (d–f) Hematoxylin/eosin-stained sections of ribs near the costovertebral joint. In the Zmpste24–/– mouse (e), fibrotic fracture callus (white arrow) contains fragments of necrotic bone (black arrowhead). Identical results were observed in three mice of each genotype. (g–i) Surface renderings of μCT scans of the thoracic spine in 28-week-old wild-type, Zmpste24–/–, and Zmpste24–/–Lmna+/– mice. In the Zmpste24–/– mouse (h), fracture callus is indicated by a white arrow.
Fig. 3.
Absence of skull abnormalities in Zmpste24–/–Lmna+/– mice at 20 weeks of age, as judged by surface renderings of μCT scans. (a, d, and g) Wild-type mouse. (b, e, and h) Zmpste24–/– mouse. (c, f, and i) Zmpste24–/–Lmna+/– mouse. Wild-type and Zmpste24–/–Lmna+/– skulls were indistinguishable. Skulls of Zmpste24–/– mice exhibited a smaller mandible, in this case with an osteolytic lesion (red arrow in b); elongated incisors (white arrowhead in b and e); osteolytic lesion in the posterior portion of the zygomatic arch (white arrows in b, e, and h); and loss of the zigzag appearance of the cranial sutures (black arrowhead in H). Identical results were observed in two mice of each genotype.
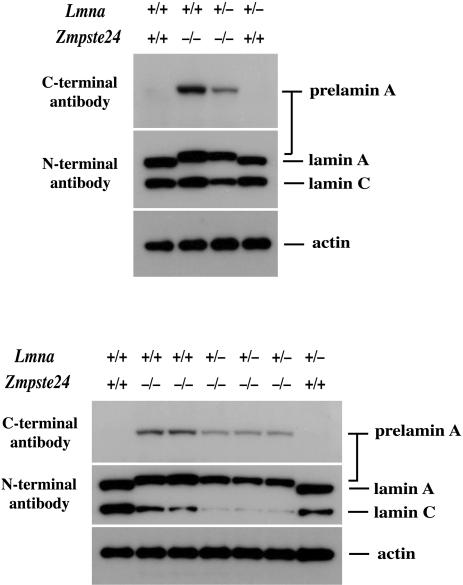
We cultured MEFs from multiple Zmpste24–/–, Zmpste24–/– Lmna+/–, and wild-type embryos and examined cell extracts by Western blots with an antibody recognizing the carboxyl terminus of prelamin A and an antibody recognizing the amino terminus of the molecule (i.e., a lamin A/C antibody). Prelamin A was absent in wild-type cells but was easily detectable in Zmpste24–/– and Zmpste24–/–Lmna+/– cells (Fig. 4). As expected, however, the levels of prelamin A and lamin C in the Zmpste24–/–Lmna+/– cells were significantly reduced compared with the Zmpste24–/– cells, a result that was confirmed by densitometry (Fig. 4).
Fig. 4.
Western blots of extracts from wild-type, Zmpste24–/–, and Zmpste24–/–Lmna+/– MEFs with a carboxyl (C)-terminal prelamin A antibody and an amino (N)-terminal lamin A/C antibody. Analyses from two independent experiments, with cells prepared from two different sets of embryos, are shown. Protein loading was assessed with an antibody against β-actin. Densitometry showed a 58.4 ± 4.1% reduction in prelamin A and a 78 ± 4.9% decrease in lamin C in the Zmpste24–/–Lmna+/– cells (n = 4) relative to the Zmpste24–/– cells.
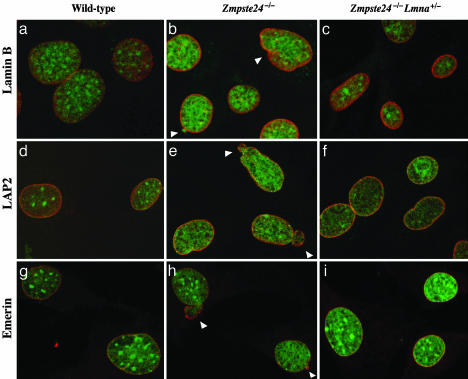
Prelamin A accumulation in Zmpste24–/– cells can be detected at the nuclear envelope by immunofluorescence (2, 13), and the nuclear envelopes of Zmpste24–/– cells frequently contain blebs. We suspected that diminished amounts of prelamin A in Zmpste24–/–Lmna+/– MEFs might result in a reduced frequency of cells with misshapen nuclei. Indeed, this was the case (Fig. 5). In three independent experiments with different fibroblast cell lines, the Zmpste24–/–Lmna+/– MEFs had significantly fewer nuclear envelope blebs than Zmpste24–/– cells (Table 1).
Fig. 5.
Analysis of nuclear shape in wild-type, Zmpste24–/–, and Zmpste24–/–Lmna+/– MEFs by laser-scanning fluorescence microscopy. The nuclear envelope was visualized with antibodies to lamin B (a–c), LAP2 (d–f), or emerin (g–i) (red), and DNA was visualized with SYTOX Green staining (green). White arrowheads indicate blebs.
Table 1. Analysis of nuclear shape in MEFs.
| Nuclear shape
|
||||
|---|---|---|---|---|
| Normal | Blebs | Folds | Irregular borders | |
| Experiment 1 | ||||
| Wild type | 273 (78.0)* | 49 (14.0) | 6 (1.7) | 22 (6.3) |
| Zmpste24-/- | 208 (59.4) | 92 (26.3)† | 27 (7.7) | 23 (6.6) |
| Zmpste24-/-Lmna+/- | 266 (76.0) | 63 (18.0)†† | 5 (1.4) | 16 (4.6) |
| Experiment 2 | ||||
| Wild type | 287 (82.0) | 43 (12.3) | 7 (2.0) | 13 (3.7) |
| Zmpste24-/- | 235 (67.0) | 86 (24.6)† | 10 (2.8) | 19 (2.6) |
| Zmpste24-/-Lmna+/- | 289 (82.6) | 40 (11.4)†† | 4 (1.1) | 17 (4.9) |
| Experiment 3 | ||||
| Wild type | 189 (94.5) | 10 (5) | 0 (0) | 1 (0.5) |
| Zmpste24-/- | 151 (75.5) | 45 (22.5)† | 1 (0.5) | 3 (1.5) |
| Zmpste24-/-Lmna+/- | 174 (87.0) | 20 (10.0)†† | 0 (0) | 6 (3.0) |
Numbers in parentheses are percentages.
P < 0.01 vs. Zmpste24-/- and wild-type nuclei.
Not significantly different from wild-type cells (P = 0.51).
Discussion
Zmpste24–/– mice accumulate prelamin A in cells and tissues and exhibit a host of severe but relatively nonspecific phenotypes, including retarded growth, alopecia, bone abnormalities, and muscle weakness (1). In the current study, we tested the hypothesis that these Zmpste24 phenotypes were due to defective prelamin A processing rather than defective processing of another as-yet-unidentified Zmpste24 protein substrate (1). We also wanted to test the hypothesis that prelamin A has a dose-dependent deleterious effect on the integrity of the nuclear lamina and causes dose-dependent pathological phenotypes in mice. To approach these issues, we compared Zmpste24–/–Lmna+/– mice with Zmpste24–/– littermate control mice. The results of this comparison were striking and intriguing: The Zmpste24–/–Lmna+/– mice were protected from all of the pathological consequences of Zmpste24 deficiency at the whole-animal level, and Zmpste24–/–Lmna+/– MEFs exhibited a significantly reduced frequency of misshapen nuclei. Our data strongly suggest that all of the phenotypes in Zmpste24–/– mice are due to defective lamin A biogenesis. Moreover, reducing the “dose” of prelamin A had an astonishingly beneficial effect on whole-animal pathological phenotypes and on nuclear shape.
The fact that a single Lmna knockout allele caused such profound salutary effects is particularly remarkable because Zmpste24–/–Lmna+/– cells also had reduced expression of lamin C. Indeed, one could persuasively argue that any mutation that reduced lamin C production would worsen nuclear shape and disease phenotypes in Zmpste24-deficient mice rather than improve them. After all, heterozygosity for lamin A/C deficiency in humans (a nonsense mutation after five amino acids) (6) causes Emery–Dreifuss muscular dystrophy, and complete lamin A/C deficiency in mice yields slow growth, muscle weakness, and striking defects in nuclear shape (5) as well as defective mechanical properties of the nuclear envelope (17). So why should heterozygous lamin A/C deficiency in Zmpste24–/– mice ameliorate rather than worsen disease phenotypes? We would contend that the most parsimonious explanation is that a full dose of prelamin A synthesis is injurious to cells but that a “half dose” yields steady-state prelamin A levels that fall below a threshold for toxicity. Is this “threshold” hypothesis plausible? We believe so, in part because of observations made in “progeroid” mice with a gene-targeted L530P mutation (18). In that mouse model, heterozygotes were phenotypically normal, whereas homozygotes exhibited phenotypes that closely resembled those in Zmpste24–/– mice. This concept of wild-type prelamin A as a toxic or injurious molecule is in line with the hypothesis that the mutant prelamin A in HGPS plays a dominant-negative function (13).
Prelamin A in Zmpste24–/– cells is targeted to the nuclear envelope (2), which strongly suggests that it is farnesylated (9, 10, 19). Nonfarnesylated prelamin A is targeted to the nucleoplasm but fails to reach the nuclear envelope, although prenylation can direct it to the nuclear envelope (10, 19). We suspect that farnesyl-prelamin A present at the nuclear lamina in Zmpste24–/– cells lies at the root of the nuclear envelope blebs. If so, one could reasonably hypothesize that inhibitors of prelamin A prenylation (e.g., farnesyltransferase inhibitors) might ameliorate the nuclear envelope shape abnormalities in Zmpste24–/– cells (and might even mitigate the pathological phenotypes in adult Zmpste24–/– mice). This hypothesis needs to be tested. However, the experiment is complex, and the results are not easy to predict, given that the blockade of farnesylation would affect dozens of key cellular proteins, including two structural components of the nuclear lamina, lamin B1 and lamin B2.
Although humans with HGPS synthesize only small amounts of a mutant prelamin A, the disease is merciless (8), and the nuclear shape abnormalities are striking and severe (13). There is little doubt that the mutant prelamin A is a major contributor to these phenotypes (13). At this point, we do not know whether the mutant prelamin A in HGPS has more deleterious effects on the nuclear lamina, and more toxicity to tissues, than the “wild-type” prelamin A in the setting of Zmpste24 deficiency. However, those answers likely will emerge over the next few years from studies with genetically modified mice. In any case, we believe that the current studies with Zmpste24–/–Lmna+/– mice are relevant to HGPS and should provide a small measure of hope to those interested in finding a cure for the disease, simply because they suggest that a 50% reduction in mutant prelamin A levels (or perhaps a 50% reduction in farnesyl-mutant prelamin A) could completely cure the disease. These studies also suggest that therapeutic strategies that would reduce levels of both lamin C and prelamin A may not necessarily be doomed to failure.
We would not be surprised if some cases of progeria in humans were found to be caused by homozygous deficiency of ZMPSTE24. Indeed, one group identified two mutant ZMPSTE24 alleles in DNA from a deceased patient with a progeroid disorder (mandibuloacral dysplasia), although it is unclear whether one of those two mutant alleles was truly dysfunctional, and there was no evidence that prelamin A accumulated in cells (20). Another group found that a mutant ZMPSTE24 allele was fairly common in patients with restrictive dermopathy (21), but some heterozygotes for the same allele have been reported to be normal (20, 21). Fibroblasts from several of the restrictive dermopathy patients with a mutant ZMPSTE24 allele had no lamin A but preserved expression of lamin C. It is extremely difficult to make sense of these human findings in the context of the Zmpste24-deficient mice, but the human findings are certainly important and intriguing and ultimately promise to further illuminate the biochemistry of prelamin A processing and lamin A function in mammals.
Normally, lamin A biogenesis is incredibly efficient, so efficient that prelamin A is essentially undetectable in normal cells. If prelamin A accumulation were indeed injurious to mammalian cells, as suggested by the current studies, then efficient machinery for removing prelamin A would make “evolutionary sense.” Ultimately, however, investigators in this field will need to understand the “physiologic rationale” for the existence of lamin A (in addition to lamin C) and the rationale for prelamin A processing. The latter goal certainly will require defining the functional properties that distinguish prelamin A from mature lamin A. Clearly, several structural differences could explain differences in their functional properties. However, we suspect that the absence of the carboxyl-terminal farnesylcysteine methyl ester in lamin A could turn out to be an important factor. Farnesyl-prelamin A has been shown to bind to the nuclear protein Narf (22), and it is conceivable that the interaction between them could have functional consequences. Also, perhaps the farnesylcysteine methyl ester of prelamin A occupies farnesylcysteine binding sites in the nuclear envelope of Zmpste24–/– cells, interfering with the function or metabolism of farnesyl-lamin B1 or B2. Functional interactions between the A- and B-type lamins almost certainly occur (19, 23), and we hypothesize that these could be relevant to disease pathogenesis. Along these lines, it is intriguing that a lamin B1 knockout in mice causes misshapen nuclei and premature senescence in MEFs (24), like the L530P “progeria mutation” in Lmna (18). Finally, the fact that lamin B1 deficiency causes bone abnormalities may not be a coincidence (24).
Acknowledgments
We thank J. Fish for histology and S. Ordway and G. Howard for criticisms of the manuscript. This work was supported in part by National Institutes of Health Grants AR050200 and HL076839 and grants from the Progeria Research Foundation (to S.G.Y.) and the Swedish Cancer Society (to M.O.B.).
Author contributions: L.G.F., J.K.N., S.H.Y., and S.G.Y. designed research; L.G.F., J.K.N., M.M., N.C., A.B., S.M., K.R., and M.O.B. performed research; C.L.S. and T.S. contributed new reagents/analytic tools; and S.G.Y. wrote the paper.
Abbreviations: HGPS, Hutchinson-Gilford progeria syndrome; μCT, microcomputed tomography; MEF, mouse embryonic fibroblast.
References
- 1.Bergo, M. O., Gavino, B., Ross, J., Schmidt, W. K., Hong, C., Kendall, L. V., Mohr, A., Meta, M., Genant, H., Jiang, Y., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 13049–13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pendás, A. M., Zhou, Z., Cadiñanos, J., Freije, J. M. P., Wang, J., Hultenby, K., Astudillo, A., Wernerson, A., Rodríguez, F., Tryggvason, K. & Lopéz-Otín, C. (2002) Nat. Genet. 31, 94–99. [DOI] [PubMed] [Google Scholar]
- 3.Tam, A., Nouvet, F. J., Fujimura-Kamada, K., Slunt, H., Sisodia, S. S. & Michaelis, S. (1998) J. Cell Biol. 142, 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyartchuk, V. L. & Rine, J. (1998) Genetics 150, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sullivan, T., Escalante-Alcalde, D., Bhatt, H., Anver, M., Bhat, N., Nagashima, K., Stewart, C. L. & Burke, B. (1999) J. Cell Biol. 147, 913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonne, G., Di Barletta, M. R., Varnous, S., Bécane, H.-M., Hammouda, E.-H., Merlini, L., Muntoni, F., Greenberg, C. R., Gary, F., Urtizberea, J.-A., et al. (1999) Nat. Genet. 21, 285–288. [DOI] [PubMed] [Google Scholar]
- 7.Novelli, G., Muchir, A., Sangiuolo, F., Helbling-Leclerc, A., D'Apice, M. R., Massart, C., Capon, F., Sbraccia, P., Federici, M., Lauro, R., et al. (2002) Am. J. Hum. Genet. 71, 426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eriksson, M., Brown, W. T., Gordon, L. B., Glynn, M. W., Singer, J., Scott, L., Erdos, M. R., Robbins, C. M., Moses, T. Y., Berglund, P., et al. (2003) Nature 423, 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lutz, R. J., Trujillo, M. A., Denham, K. S., Wenger, L. & Sinensky, M. (1992) Proc. Natl. Acad. Sci. USA 89, 3000–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinensky, M., Fantle, K. & Dalton, M. (1994) Cancer Res. 54, 3229–3232. [PubMed] [Google Scholar]
- 11.Kilic, F., Dalton, M. B., Burrell, S. K., Mayer, J. P., Patterson, S. D. & Sinensky, M. (1997) J. Biol. Chem. 272, 5298–5304. [DOI] [PubMed] [Google Scholar]
- 12.Corrigan, D. P., Kuszczak, D., Rusinol, A. E., Thewke, D. P., Hrycyna, C. A.,Michaelis, S. & Sinensky, M. S. (October 13, 2004) Biochem. J., 10.1042/BJ20041359. [DOI] [PMC free article] [PubMed]
- 13.Goldman, R. D., Shumaker, D. K., Erdos, M. R., Eriksson, M., Goldman, A. E., Gordon, L. B., Gruenbaum, Y., Khuon, S., Mendez, M., Varga, R. & Collins, F. S. (2004) Proc. Natl. Acad. Sci. USA 101, 8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung, G. K., Schmidt, W. K., Bergo, M. O., Gavino, B., Wong, D. H., Tam, A., Ashby, M. N., Michaelis, S. & Young, S. G. (2001) J. Biol. Chem. 276, 29051–29058. [DOI] [PubMed] [Google Scholar]
- 15.Todaro, G. J. & Green, H. (1963) J. Cell Biol. 17, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinert, P., Zackroff, R., Aynardi-Whitman, M. & Goldman, R. D. (1982) Methods Cell Biol. 24, 399–419. [DOI] [PubMed] [Google Scholar]
- 17.Lammerding, J., Schulze, P. C., Takahashi, T., Kozlov, S., Sullivan, T., Kamm, R. D., Stewart, C. L. & Lee, R. T. (2004) J. Clin. Invest. 113, 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mounkes, L. C., Kozlov, S., Hernandez, L., Sullivan, T. & Stewart, C. L. (2003) Nature 423, 298–301. [DOI] [PubMed] [Google Scholar]
- 19.Hennekes, H. & Nigg, E. A. (1994) J. Cell Sci. 107, 1019–1029. [DOI] [PubMed] [Google Scholar]
- 20.Agarwal, A. K., Fryns, J.-P., Auchus, R. J. & Garg, A. (2003) Hum. Mol. Genet. 12, 1995–2001. [DOI] [PubMed] [Google Scholar]
- 21.Navarro, C., Sandre-Giovannoli, A. D., Bernard, R., Bocccaccio, I., Boyer, A., Genevieve, D., Hadj-Raabia, S., Gaudy-Marqueste, C., Smitt, H. S., Vabres, P., et al. (2004) Hum. Mol. Gen. 13, 2493–2503. [DOI] [PubMed] [Google Scholar]
- 22.Barton, R. M. & Worman, H. J. (1999) J. Biol. Chem. 274, 30008–30018. [DOI] [PubMed] [Google Scholar]
- 23.Ye, Q. & Worman, H. J. (1995) Exp. Cell Res. 219, 292–298. [DOI] [PubMed] [Google Scholar]
- 24.Vergnes, L., Peterfy, M., Bergo, M. O., Young, S. G. & Reue, K. (2004) Proc. Natl. Acad. Sci. USA 101, 10428–10433. [DOI] [PMC free article] [PubMed] [Google Scholar]