Abstract
Diazinon is an organophosphate compound that inhibits the activity of acetylcholinesterase. Standards of the World Health Organization and Environmental Protection Agency for diazinon concentration in water are 0.1 and 9 × 10−6 mg/L, respectively. The aim of this study was the optimization of diazinon biodegradation from aqueous solutions by Saccharomyces cerevisiae using the response surface methodology (RSM). Harvested cells of S. cerevisiae were locally purchased from the Iranian Research Organization for Science and Technology. To obtain the optimum condition for diazinon biodegradation using RSM, input parameters included the initial concentration of diazinon (0.01–10 mg/L), concentration of S. cerevisiae (0.5–5%), pH (4–10), and retention time (1–30 h). The research study had a central composite design where one of the methods was RSM. According to the results, the observed values of the removal efficiency of diazinon were variable in the range of 23–96. The highest removal rate was obtained as 96% under the initial diazinon concentration of 2.5 mg/L, S. cerevisiae concentration of 3.88%, pH of 5.5, and retention time of 22.75 h. The results displayed that the removal efficiency of diazinon had a direct relationship with the concentration of S. cerevisiae and retention time, and an inverse relationship with pH and the initial concentration of diazinon. We can conclude that S. cerevisiae has the ability to remove diazinon with the lowest cost and a high efficiency.
Keywords: Saccharomyces cerevisiae, Diazinon, Biodegradation, RSM
Introduction
The pollution of water resources by pesticides is considered an environmental concern. The use of pesticides in the agricultural sector has increased due to the population growth and increasing demand for agricultural products and foodstuffs (Khazaei et al. 2010; Arjmandi et al. 2010). Several applications of pesticides in agriculture can cause environmental issues such as contamination of water, soil, and the food. Pesticides include organophosphate, carbamate, and pyrethroid, and the organophosphorus compounds are the largest and most diverse pesticides. Because of their effect on a wide range of pests and also their low cost, organophosphate pesticides are used by farmers more than other pesticides (Shayeghi et al. 2008). Different types of organophosphorus compounds inhibit the cholinesterase enzyme and therefore cause a range of toxic effects. These effects include headaches, vomiting, and respiratory tract problems. Depending on the chemical nature of organophosphorus compounds, inhibiting the cholinesterase may be reversible or irreversible (Hashemian et al. 2015). Diazinon is an organophosphate compound with the chemical formula of C12H21N2O3PS (Hunter et al. 1985; WHO 1998). According to the standard of the European Union, the maximum allowable concentration for the total pesticide residue and for each pesticide in drinking water sources is 0.5 μg/L and 0.0001 mg/L, respectively (Cai and Yun 2008). World Health Organization (WHO) and the Environmental Protection Agency (EPA) standards for the value of diazinon in drinking water are 0.1 μg/L and 9 × 10−6 mg/L, respectively (Goodrich et al. 1989). The maximum allowable concentrations of diazinon in drinking water in Australia, Canada, Germany, England and America are 20, 10, 0, and 0.6 μg/L, respectively (Readman et al. 1992). Different methods such as filtration, ozonation, and adsorption using the granular activated carbon are used for the removal of pesticides from aqueous solutions, but these methods have been associated with problems such as high capital and operating costs, saturation of activated carbon, and production of toxic substances (Reynolds et al. 1989; Lai et al. 1995; Jiang and Adams 2006; Ormad et al. 2008). Saccharomyces cerevisiae is a single-cell and non-pathogenic yeast (Cabral et al. 2003; Teixeira et al. 2007). It is considered for the biological removal of diazinon due to advantages such as safety, low cost, simplicity, widespread distribution, rapid growth of cells, and easy cultivation. The main advantage of Saccharomyces cerevisiae is its selectivity in active transport and competitiveness in biological absorption (Goksungur et al. 2005; Peinado and Morenoa 2006; Yarke-Salkhori et al. 2011). Mutagenicity studies indicated that diazinon does not have negative effects on the metabolic system of S. cerevisiae (Mohn 1973; Fahrig 1974), and only at concentrations greater than 50,000 ppm can it have negligible toxic effects on S. cerevisiae (Bianchi et al. 1994). Sadeghi et al. (2014) used S. cerevisiae for the biological removal of carmoisine and Reactive Black 5 dyes (Sadeghi et al. 2014). Fabrizio et al. (1983) demonstrated that this type of yeast can biologically remove the folpet and metalaxyl fungicides and the pyrethroid and deltametrin insecticides (Fabrizio et al. 1983). Also, Tijana and Rada (2015) reduced the chlorpyrifos-methyl pesticide with this method (Tijana and Rada 2015), and Cabras and Angioni (2000) and George et al. (2014) carried out a similar study (Cabras and Angioni 2000; George et al. 2014). The technique of experimental design is a useful tool that uses statistical models to optimize the interaction between different parameters. Using the response surface methodology (RSM), we can study the interaction between two or more parameters (Yi et al. 2012). The present study was conducted with the aim of optimizing diazinon biodegradation from aqueous solutions by Saccharomyces cerevisiae using RSM.
Materials and methods
Chemicals
All chemicals used in the experiments were reagent grade. All solutions were prepared with distilled water. Diazinon was obtained from Sigma-Aldrich.
Microorganism
The harvested cells of S. cerevisiae [Persian-type culture collection (PTCC):5052] were locally purchased from the Iranian Research Organization for Science and Technology, Tehran, Iran. In this experiment, yeast was prepared at the concentration of 5%. For this purpose, 5 g of yeast was suspended in 100 mL of toxic substance solution.
Preparation of reaction mixtures
In this study, 100 cc of reaction mixture at the concentration of 0.01–10 mg/L of diazinon and 0.5–5% of S. cerevisiae, with the pH of 4–10 and retention times of 1–30 h were prepared. The experiments were performed at the fixed agitation speed of 120 rpm and room temperature (28 ± 2 °C). Figure 1 shows the reaction mixture with the volume of 100 mL at different pH concentration and reaction times.
Fig. 1.

Biodegradation of diazinon using wet cells of S. cerevisiae at different pH, concentration and reaction times
Analytical methods
Ten microliter of sample was taken from each Erlenmeyer flask at definite time intervals. Samples were centrifuged at 4000 rpm for 10 min to remove the medium. The diazinon concentration was measured using isocratic reverse-phase HPLC (model of KNAUER, Germany) and the EZ-chrome software with a UV–Vis absorbance detector at the wavelength of 269 nm in column C-18. The operation was conducted at room temperature with a mobile phase containing methanol: water (50:50) with the column size 4.6 × 150 mm, the flow rate of 0.8 mL/min, and the maximum pressure of 40 MPa (APhA et al. 1998).
Experimental design and statistical analysis
The experiment was designed based on RSM. As shown in Table 1, four parameters were selected for this study, including the initial concentration of diazinon (mg/L), reaction time (h), concentration of S. cerevisiae (%), and pH which was high, low, and medium for the three levels coded +1, 0, and −1, respectively. Moreover, assisting points coded +α and −α were considered for model validation based on the models.
Table 1.
Experimental ranges and levels of independent parameters according to RSM design
| Parameters | Symbol | Levels | ||||
|---|---|---|---|---|---|---|
| −α | −1 | 0 | +1 | +α | ||
| Concentrations of diazinon (mg/L) | A | 0.01 | 2.5 | 5 | 7.5 | 10 |
| Reaction time (h) | B | 1 | 8.25 | 15.5 | 22.75 | 30 |
| Concentration of S. cerevisiae (%) | C | 0.5 | 1.63 | 2.75 | 3.88 | 5 |
| pH | D | 4 | 5.5 | 7 | 8.5 | 10 |
In this study, 30 runs were performed using RSM with 16 real points, 8 pivot points, and 6 central focal point. The quadratic model for the variables is presented below:
| 1 |
where Y, β0, βi, βii, βij and xi or xj are the predicted response, the constant coefficient, regression coefficients for linear effects, quadratic coefficients, interaction coefficients, and the coded values of the parameters, respectively. The fit of the models was evaluated by determining the coefficients (R2) and adjusted R2 (R2adj) (Soumasree et al. 2012).
Results
In this study, the effect of parameters such as concentration of diazinon, concentration of S. cerevisiae, reaction time, and pH were studied on the removal efficiency of diazinon. The summary of the results of the study are presented in Table 2.
Table 2.
Experimental design and response values at different runs of diazinon removal
| Run order | Parameters | The removal efficiency of diazinon (%) | |||
|---|---|---|---|---|---|
| A | B | C | D | ||
| 1 | 5 | 15.5 | 5 | 7 | 58.34 |
| 2 | 10 | 15.5 | 2.75 | 7 | 27 |
| 3 | 2.5 | 22.75 | 1.63 | 8.5 | 47 |
| 4 | 5 | 15.5 | 0.5 | 7 | 34 |
| 5 | 2.5 | 8.25 | 1.63 | 5.5 | 5 |
| 6 | 7.5 | 22.75 | 1.63 | 5.5 | 45 |
| 7 | 5 | 15.5 | 2.75 | 4 | 60 |
| 8 | 7.5 | 22.75 | 1.63 | 8.5 | 32 |
| 9 | 5 | 15.5 | 2.75 | 10 | 27 |
| 10 | 5 | 15.5 | 2.75 | 7 | 43 |
| 11 | 5 | 15.5 | 2.75 | 7 | 44 |
| 12 | 2.5 | 22.75 | 3.88 | 8.5 | 65 |
| 13 | 5 | 15.5 | 2.75 | 7 | 27 |
| 14 | 7.5 | 22.75 | 3.88 | 5.5 | 48 |
| 15 | 2.5 | 8.25 | 3.88 | 8.5 | 44 |
| 16 | 5 | 15.5 | 2.75 | 7 | 38.02 |
| 17 | 5 | 15.5 | 2.75 | 7 | 43.86 |
| 18 | 5 | 30 | 2.75 | 7 | 85 |
| 19 | 2.5 | 22.75 | 3.88 | 5.5 | 96 |
| 20 | 2.5 | 8.25 | 3.88 | 5.5 | 58.93 |
| 21 | 7.5 | 8.25 | 3.88 | 5.5 | 33 |
| 22 | 7.5 | 8.25 | 1.63 | 5.5 | 26 |
| 23 | 5 | 15.5 | 2.75 | 7 | 35 |
| 24 | 7.5 | 22.75 | 3.88 | 8.5 | 40 |
| 25 | 2.5 | 8.25 | 1.63 | 8.5 | 32 |
| 26 | 7.5 | 8.25 | 3.88 | 8.5 | 30 |
| 27 | 5 | 1 | 2.75 | 7 | 25 |
| 28 | 7.5 | 8.25 | 1.63 | 8.5 | 23 |
| 29 | 0.01 | 15.5 | 2.75 | 7 | 87 |
| 30 | 2.5 | 22.75 | 1.63 | 5.5 | 65 |
Table 3 shows the regression results of quadratic model for the removal efficiency of diazinon; Fig. 2 demonstrates the correlations between the actual and predicted removal of diazinon.
Table 3.
ANOVA of the quadratic model for the removal efficiency of diazinon
| Source | Sum of squares | Degree of freedom | Mean square | F-value | P value |
|---|---|---|---|---|---|
| Model | 9889.93 | 14 | 706.42 | 14.45 | <0.0001 |
| Residual | 733.30 | 15 | 48.89 | ||
| Lack of fit | 509.34 | 10 | 50.93 | 1.14 | 0.4715 |
| Pure error | 223.96 | 5 | 44.79 | ||
| Total | 10623.23 | 29 | |||
| R2 = 9310 Adj R2 = 0.8665 Pred R2 = 0.6935 Adeq precision = 14.84 | |||||
Fig. 2.
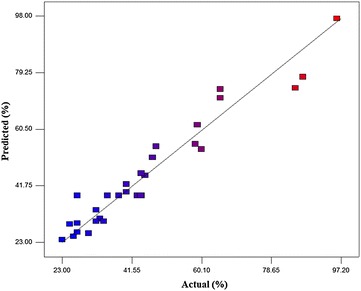
Actual and predicted removal of diazinon
The quadratic model is shown in Eq. 2 in terms of coded factors of the removal efficiency of diazinon (Y):
| 2 |
As can be seen in the equation, each model has a fixed part and a variable part. On the basis of Eq. (2), the removal efficiency of has have been 38.48% that is affected by different factors. The main effects of A, B, C, and D have the coefficients of −12.12, +11.29, +6.4, and −6.87, respectively. The main effect belongs to the variable which is shown with the coefficient of +11.29. The highest interaction effect belongs to the AC with the coefficient of 3.43, and the highest square effects of the factors belong to the A2 with the coefficient of +3.78. The effects of the main variables (between −1 and +1 levels) on the removal efficiency of diazinon are demonstrated in Fig. 3. With regard to Fig. 3, it is necessary to note that to describe the effect of one factor on a response, other variables are fixed at the zero level. For example, when the variable of reaction time or B increases from level −1 (1 h) to +1 (30 h), the other three variables of the concentrations of diazinon or A (5 mg/L), concentration of S. cerevisiae or C (2.75%), and pH or D (7) are at the zero level. The optimized values for pH, concentrations of diazinon, concentration of S. cerevisiae and reaction time were found to be 5.51, 2.52 mg/L, 3.84% and 22.6 h, respectively. At these conditions, the predicated diazinon removal percentage was more than 96.1% with desirability 1. Conduction of similar experiments at specified optimum conditions reveal the high repeat ability of method for prediction of real removal percentage with relative deviation less than 2%.
Fig. 3.
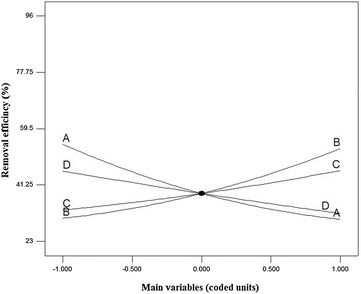
Response surface (perturbation) showing the effect of the main variables (A, B, C and D) on the removal efficiency of diazinon
Discussion
According to the results presented in Table 2, the maximum and minimum removal efficiency of diazinon were 96 and 23%, respectively. Results showed that the observed values of the removal efficiency of diazinon were in the range of 23–96 which was not different from the values predicted by the model (RSM). The findings of this study (using Eq. 2) showed that this treatment process was able to remove 96% of diazinon from aquatic environments. This process underwent the main, interaction, and square effects. Each of these effects have coefficients with positive or negative signs that indicate positive or negative effects are on removal efficiency. The best model to fit the experimental data with independent variables was the quadratic model. ANOVA was used to determine the significance of the model (P values < 0.05). Overall, results showed that this process was significance (P values < 0.0001). Results presented in Table 3 showed that R2, justified R2, and adequacy precision were 0.931, 0.8665, and 14.842, respectively. Figure 3 indicated that with increasing the parameters from −1 level (2.5 mg/L) to +1 level (7.5 mg/L), the removal efficiency of diazinon reduced significantly. There is an inverse relationship between the removal efficiency of diazinon and its initial concentration, so that with increasing the diazinon concentration from −1 level to +1 level, its removal efficiency decreased 24.25% while the reaction time, pH, and concentration of yeast were at the zero level (P value < 0.0001). This is because with increasing the concentration of diazinon, adsorption sites on yeast walls are saturated and decrease the absorption capacity (Mahmoud 2016). Mahmoud (2016) confirmed that with the use of S. cerevisiae, the removal efficiency of reactive dye decreased with increasing the concentration of its pollutant (Mahmoud 2016). Studies show that numerous bacteria are involved in the degradation of diazinon (Kristina et al. 2008). The hydrolase enzyme in bacteria plays a major role in the biodegradation of diazinon. It can biodegrade alkyl and aryl bonds of P-O in organophosphates (Gunner and Zuckerman 1968; Kristina et al. 2008). Abasalt et al. (2014) showed that Pseudomonas plecoglossicida can use diazinon as a carbon and phosphorus source and biodegrade it. Mahiudddin et al. (2014) concluded that Pseudomonas peli, Burkholderia caryophylli and Brevundimonas diminuta can completely biodegrade diazinon in the concentration of 20 mg/L (Mahiudddin et al. 2014). According to Fig. 3, there is a direct relationship between the removal efficiency of diazinon and reaction time, so that with increasing the reaction time from −1 level to +1 level, removal efficiency increased 43.39% while the initial concentrations of diazinon, pH, and the concentration of yeast were at the zero level (P value < 0.0001). This positive effect (Eq. 2) has been shown by the coefficient of +11.29 for parameter B. Mariusz et al. (2009) indicated that Serratia and Pseudomonas can successfully biodegrade diazinon (at the concentration of 50 mg/L for the period of 42 days) by 83 and 87%, respectively (Mariusz 2009). Sadeghi et al. (2014) showed that S. cerevisiae can biodegrade Carmoisine and Reactive Black 5 Dyes within 24 h. Therefore, the maximum removal of carmoisine and Reactive Black 5 Dyes was 85 and 53%, respectively (Sadeghi et al. 2014). Bumpus et al. (1993) reported the ability of Phanerochaete chrysosporium to degrade 27.5% of chlorpyrifos during the 18-day incubation (Bumpus et al. 1993). The results demonstrated a direct relationship between the removal efficiency of diazinon and the concentration of S. cerevisiae, so that the removal efficiency of diazinon (while the initial concentration of diazinon, pH, and reaction time were at the zero level) increased by 27.8% over time from −1 level to +1 level (P value < 0.05). Zaharia et al. (2013) indicated that a 5% concentration of S. cerevisiae can significantly biodegrade the DINOCAP and DNOC pesticides. The findings of this study (Fig. 3) suggest that there is an inverse relationship between the removal efficiency of diazinon and pH. Accordingly, with increasing the pH from −1 level to +1 level, the removal efficiency of diazinon decreases by 30% while the reaction time, initial concentration of diazinon, and the concentration of S. cerevisiae were at the zero level (P value < 0.0001). pH is one of the important parameters influencing chemical reactions and biological aqueous solutions. It affects the surface charge of absorbents and the ionization degree of absorbate (Sadeghi et al. 2015). S. cerevisiae has carboxyl, phosphonate, and amine groups which include negative, negative, and positive charges, respectively. Amine groups are mainly found in proteins and bio-mass and are also more active than other groups in biosorption (Sadeghi et al. 2015). At a low pH, active sites (amino groups) of S. cerevisiae are protonated and the density of positive charge in the absorbent surface increases (Sadeghi et al. 2015). If the adsorption phenomenon is involved alone, a high adsorption must be observed in an alkaline pH, while a high adsorption must observed in an acidic pH (Konstantinos et al. 2012; Sadeghi et al. 2015). Kristina et al. (2008) showed that the removal efficiency of diazinon increases by microorganisms at a low pH (Kristina et al. 2008). Aksu (2003) demonstrated that the efficiency of S. cerevisiae in reactive dye removal increased significantly at a low pH.
No similar study has been published on the use of S. cerevisiae in the removal of diazinon. Results showed that the removal efficiency of diazinon by S. cerevisiae highly depends on pH. The maximum removal rates have been observed at an acidic pH. The removal efficiency of diazinon has a direct relationship with the concentration of S. cerevisiae and reaction time, and an inverse relationship with pH and initial concentration of diazinon. We can conclude that S. cerevisiae has the ability to remove diazinon with the lowest cost and a high efficiency.
Authors’ contributions
MHE and AS conceived and designed the experiments; ZB performed the experiments; AS and MTG made a substantial contribution to the analysis and interpretation of the data presented; ZB wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
The authors wish to thank Prof. Hossein Fallahzadeh from Shahid Sadoughi University of Medical Sciences, Yazd, Iran and also Dr. Ali Asghar Najafpoor from School of Health, Mashhad University of Medical Sciences, Mashhad, Iran for useful discussions.
Competing interests
All authors declare that they have no competing interests.
Funding
This study was financially supported by Grant No: 950402 of the Biotechnology Development Council of the Islamic Republic of Iran.
Abbreviations
- RSM
response surface methodology
- WHO
World Health Organization
- EPA
Environmental Protection Agency
- PTCC
Persian-type culture collection
Contributor Information
Mohammad H. Ehrampoush, Email: Ehram2000@yahoo.com
Abbas Sadeghi, Email: Sadeghia@mums.ac.ir.
Mohammad T. Ghaneian, Email: Ghaneianmt@yahoo.com
Ziaeddin Bonyadi, Phone: +983538209121, Email: zyabonyadi@yahoo.com.
References
- Abasalt B, Ghazal NF, Pouran J, Taghi NF, Saeideh S, Gholamrea K. Cleaning from the inside: biodegradation of organophosphate pesticides by Pseudomonas plecoglossicida. Biotech Health Sci. 2014;1:e19193. [Google Scholar]
- Aksu Z. Reactive dye bioaccumulation by Saccharomyces cerevisiae. Process Biochem. 2003;38:1437–1444. doi: 10.1016/S0032-9592(03)00034-7. [DOI] [Google Scholar]
- APhA, AWWA, WPCF (1998) Standard method for the examination of water and wastewater. Washington, D.C
- Arjmandi A, Tavakol M, Shayeghi M. Determination of organophosphorus insecticide residues in the rice paddies. Int J Environ Sci Technol. 2010;7:175–182. doi: 10.1007/BF03326129. [DOI] [Google Scholar]
- Bianchi L, Zannoli A, Pizzala R, Stivala LA, Chiesara E. Genotoxicity assay of five pesticides and their mixtures in Saccharomyces cerevisiae D7. Mutat Res. 1994;321:203–211. doi: 10.1016/0165-1218(94)90071-X. [DOI] [PubMed] [Google Scholar]
- Bumpus JA, Kakkar SN, Coleman RD. Fungal degradation of organophosphorus insecticides. Appl Biochem Biotechnol. 1993;39(40):715–726. doi: 10.1007/BF02919030. [DOI] [PubMed] [Google Scholar]
- Cabral MG, Viegas CA, Teixeira MC, Sa-Correia I. Toxicity of chlorinated phenoxyacetic acid herbicides in the experimental eukaryotic model Saccharomyces cerevisiae: role of pH and of growth phase and size of the yeast cell population. Chemosphere. 2003;51:47–54. doi: 10.1016/S0045-6535(02)00614-8. [DOI] [PubMed] [Google Scholar]
- Cabras P, Angioni A. Pesticide residues in grapes, wine, and their processing products. J Agric Food Chem. 2000;48:967–973. doi: 10.1021/jf990727a. [DOI] [PubMed] [Google Scholar]
- Cai M, Yun Z (2008) A rapid method for trace analysis of organophosphorus pesticides in drinking water. https://www.agilent.com
- Fabrizio F, Giovanni A, Pietrino D, Paolo C, Marco M, Filippo MP. A preliminary investigation into the effect of Saccharomyces cerevisiae on pesticide concentration during fermentation. Eur J Appl Microbiol Biotechnol. 1983;18:323–325. doi: 10.1007/BF00500500. [DOI] [Google Scholar]
- Fahrig JR (1974) Mutagenicity studies with pesticides, in chemical carcinogenesis essays. IARC scientific publications 10
- George N, Singh CP, Sondhi S, Saini S, Puri N, Gupta N, Gupta N. Biodegradation and analytical methods for detection of organophosphorous pesticide: chlorpyrifos. Int J Pure Appl Sci Technol. 2014;20(2):79–94. [Google Scholar]
- Goksungur Y, Uren S, Guvenc U. Biosorption of cadmium and lead ions by ethanol treated waste baker’s yeast biomass. Bioresour Technol. 2005;96:103–109. doi: 10.1016/j.biortech.2003.04.002. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Lykins BW, Clark RM. Drinking water from agriculturally contaminated groundwater. J Environ Qual. 1989;20:707–717. doi: 10.2134/jeq1991.00472425002000040001x. [DOI] [Google Scholar]
- Gunner HB, Zuckerman BM. Degradation of diazinon by synergistic microbial action. Nature. 1968;217:1183–1184. doi: 10.1038/2171183a0. [DOI] [PubMed] [Google Scholar]
- Hashemian AM, Ebrahimi M, Vafadar E, Salarirad M, Fadavi H. Bilateral acute angle-closure glaucoma following acute poisoning with organophosphate. J Clin Toxicol. 2015;5:1–2. [Google Scholar]
- Hunter RL, Faulkner L, Culver F, Hill J. Draft user manual for the QSAR system. Bozeman: Center for Data Analysis, Montana State University; 1985. [Google Scholar]
- Jiang H, Adams C. Treatability of chloro-s-triazines by conventional drinking water treatment technologies. Water Res. 2006;40:1657–1667. doi: 10.1016/j.watres.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Khazaei H, Korasani N, Talebijahromi KH (2010) Quality survey and health status mazandaran groundwater from use of diazinon insecticide (case study: mahmoudabad city). 12th Environmental Health Conference. Tehran. (Persian)
- Konstantinos VP, Plakas J, Anastasios J. Removal of pesticides from water by NF and RO membranes. Desalination. 2012;287:255–265. doi: 10.1016/j.desal.2011.08.003. [DOI] [Google Scholar]
- Kristina D, Tjas D, Polonca T, David S. Microorganisms trigger chemical degradation of diazinon. Int Biodeterior Biodegrad. 2008;62:293–296. doi: 10.1016/j.ibiod.2008.02.003. [DOI] [Google Scholar]
- Lai MS, Jensen JN, Weber AS. Oxidation of simazine: ozone, ultraviolet, and combined ozone/ultraviolet oxidation. Water Environ Res. 1995;67:340–346. doi: 10.2175/106143095X131565. [DOI] [Google Scholar]
- Mahiudddin M, Fakhruddin ANM, Abdullah AM, Chowdhury MAZ, Rahman MA, Alam MK. Degradation of the orgnophosphorus insecticide diazinon by soil bacterial isolated. Int J Biotechnol. 2014;3:12–23. [Google Scholar]
- Mahmoud MS. Decolorization of certain reactive dye from aqueous solution using Baker’s Yeast (Saccharomyces cerevisiae) strain. HBRC J. 2016;12:88–98. doi: 10.1016/j.hbrcj.2014.07.005. [DOI] [Google Scholar]
- Mariusz C, Marcin W, Zofia PS. Biodegradation of the organophosphorus insecticide diazinon by Serratia sp. and Pseudomonas sp. and their use in bioremediation of contaminated soil. Chemosphere. 2009;76:494–501. doi: 10.1016/j.chemosphere.2009.03.023. [DOI] [PubMed] [Google Scholar]
- Mohn G. 5-Methyltryptophan resistance mutations in Escherichia coil K-12 Mutagenic activity of monofunctional alkylating agents including organophosphorus insecticides. Mutat Res. 1973;20:7–15. doi: 10.1016/0027-5107(73)90092-4. [DOI] [PubMed] [Google Scholar]
- Ormad MP, Miguel N, Claver A, Matesanz JM, Ovelleiro JL. Pesticides removal in the process of drinking water production. Chemosphere. 2008;71:97–106. doi: 10.1016/j.chemosphere.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Peinado RA, Morenoa JJ. Yeast biocapsules: a new immobilization method and their applications. Enzyme Microb Technol. 2006;40:79–84. doi: 10.1016/j.enzmictec.2005.10.040. [DOI] [Google Scholar]
- Readman JW, Liong W, Mee LD, Bartocci J, Nilve G, Rodriguez JA, Gonzalez-Farias F. Persistent organophosphorus pesticides in tropical marine environments. Mar Pollut Bull. 1992;24:398–402. doi: 10.1016/0025-326X(92)90500-6. [DOI] [Google Scholar]
- Reynolds G, Graham N, Perry R, Rice RG. Aqueous ozonation of pesticides: a review, Ozone. Sci Eng. 1989;11:339–382. [Google Scholar]
- Sadeghi A, Kazemi Bazardehi MA, Raffe S, Zarif B. Biotransformation of carmoisine and reactive black 5 dyes using saccharomyces cerevisiae. Health. 2014;6:859–864. doi: 10.4236/health.2014.610108. [DOI] [Google Scholar]
- Sadeghi A, Dolatabadi M, Asadzadeh SN, Jamali BF. Ability of the yeast saccharomyces cerevisiae for biological removal of ciprofloxacin antibiotic in aqueous solution. J North Khorasan Univ. 2015;7:71–79. [Google Scholar]
- Shayeghi M, Khobdel M, Bagheri F, Abtai M. Azynfosmethyl and diazinon residues in Qarasu river and gorganrood in Golestan province. J Pub Heal and Health Res Instit. 2008;6:75–82. [Google Scholar]
- Soumasree C, Atul K, Srabanti B, Susmita D. Application of response surface methodology for methylene blue dye removal from aqueous solution using low cost adsorbent. Chem Eng J. 2012;181–182:289–299. [Google Scholar]
- Teixeira MC, Duque P, Sa-Correreia I. Environmental genomic: mechanistic insights into toxicity of and resistance to the herbicide 2,4-D. Trends Biotechnol. 2007;25:363–370. doi: 10.1016/j.tibtech.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Tijana M, Rada D. Dissipation of chlorpyrifos-methyl by Saccharomyces cerevisiae during wheat fermentation. LWT Food Sci Technol. 2015;61:516–523. doi: 10.1016/j.lwt.2014.12.044. [DOI] [Google Scholar]
- WHO (1998) Environmental Health Criteria, 198—Diazinon. International Programme on Chemical Safety. World Health Organization. http://www.who.int
- Yarke-Salkhori N, Ghaemi N, Nouhi A. Optimizing conditions of lead removal by Saccharomyces cerevisiae in laboratory condition. J Qazvin Univ Med. 2011;15:9. [Google Scholar]
- Yi L, Jintao W, Yian Zh, Aiqin W. Adsorption of methylene blue by kapok fiber treated by sodium chlorite optimized with response surface methodology. Chem Eng J. 2012;184:248–255. doi: 10.1016/j.cej.2012.01.049. [DOI] [Google Scholar]
- Zaharia M, Maftei D, Dumitras-Hutanu CA, Gradinaru R. Biodegradation of pesticides DINOCAP and DNOC by yeast suspensions in a batch system. Rev Chim. 2013;64:388–392. [Google Scholar]


