Abstract
Three ant species nest obligately in the swollen-thorn domatia of the African ant-plant Vachellia (Acacia) drepanolobium, a model system for the study of ant-defence mutualisms and species coexistence. Here we report on the characteristic fungal communities generated by these ant species in their domatia. First, we describe behavioural differences between the ant species when presented with a cultured fungal isolate in the laboratory. Second, we use DNA metabarcoding to show that each ant species has a distinctive fungal community in its domatia, and that these communities remain characteristic of the ant species over two Kenyan sampling locations separated by 190 km. Third, we find that DNA extracted from female alates of Tetraponera penzigi and Crematogaster nigriceps contained matches for most of the fungal metabarcodes from those ant species' domatia, respectively. Fungal hyphae and other debris are also visible in sections of these alates' infrabuccal pockets. Collectively, our results indicate that domatium fungal communities are associated with the ant species occupying the tree. To the best of our knowledge, this is the first record of such ant-specific fungal community-level differences on the same myrmecophytic host species. These differences may be shaped by ant behaviour in the domatia, and by ants vectoring fungi when they disperse to establish new colonies. The roles of the fungi with respect to the ants and their host plant remain to be determined.
Keywords: metabarcoding, fungi, species interactions, insects, Tetraponera, Vachellia (Acacia) drepanolobium
1. Background
The Vachellia (Acacia) drepanolobium ant-plant system has long served as a model for understanding ant-defence mutualisms and species coexistence [1–3]. Much of the work on this system has focused on direct interactions among ants, host plants and herbivores, and especially on how these interactions differ depending upon the ant species occupying the trees. For example, ant colony competitive ability, colonization ability, host plant pruning and deterrence of herbivores vary among the four ant species that commonly inhabit these trees [3–5]. But many other ‘third-party’ organisms are also part of this system, and often show strong associations with the resident ant species [6]. In this paper, we examine one such set of third-party species: we describe distinctive domatium fungal communities associated with three ant species, show behavioural differences among the ants with respect to fungi, and report molecular evidence suggesting potential transmission of domatium fungi by dispersing ant reproductives. These observations raise the possibility that domatium fungi play important functional roles in the interaction between V. drepanolobium and its associated ants.
Vachellia drepanolobium ant-plants are widespread in the East African tropics and are typically the dominant tree in black cotton savannahs [2]. While as many as 15 ant species have been documented on V. drepanolobium in Kenya [7], three species of plant-ant are found obligately inhabiting domatia throughout the tree's range: Tetraponera penzigi, Crematogaster nigriceps and Crematogaster mimosae. The domatia inhabited by these ants are formed by the hollow, swollen bases of stipular thorns, into which the ants chew entrance holes prior to occupation (figure 1a,b). A fourth ant species, Crematogaster sjostedti, is also common on V. drepanolobium at some locations where the system has been studied, but colonies of this species are free-living, and typically nest in trunk cavities or in the ground at the tree base [8]. Each tree is normally occupied by one colony [8], although sometimes a single colony may extend over several trees, and it is not uncommon for adjacent trees to host different ant species.
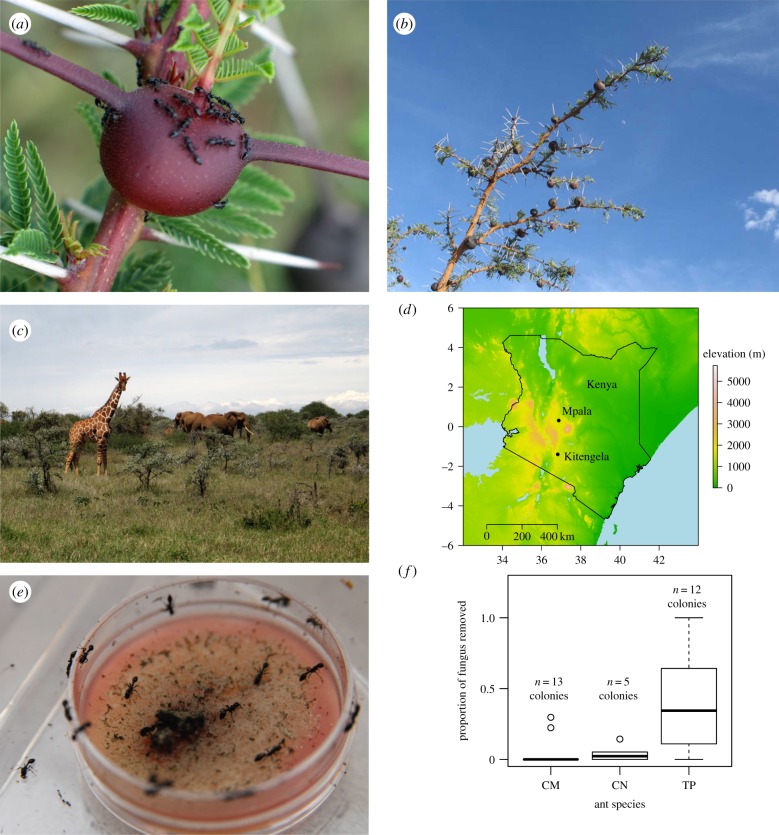
Figure 1.
(a) T. penzigi workers on domatium. (b) V. drepanolobium sapling. (c) Resident ant colonies defend host plants against damage by large mammalian herbivores. (d) Kitengela and Mpala field sites are located approximately 190 km apart. (e,f) T. penzigi ants (TP) removed significantly more fungus than either C. mimosae ants (CM) or C. nigriceps ants (CN). Removal of agar growth medium for Phoma fungal isolates was negligible.
The four ant species engage in a protection mutualism with their host plants, defending against large mammalian herbivores in exchange for housing (figure 1c). However, the exchange varies among the ant species [1]. The ants differ markedly in the extent to which they patrol and deter herbivores [5,7], but they also exert different direct and indirect effects on their hosts. Workers of all three Crematogaster species harvest secretions from host plant extrafloral nectaries, while workers of T. penzigi destroy them [2,9]. Workers of C. nigriceps prune axillary buds, stimulating terminal growth but eliminating flowering [2]. Workers of C. mimosae and C. sjostedti tend phloem-feeding scale insects [2,7], presumably imposing a cost that partly offsets the protection those ants offer against mammalian herbivores. These and other differences are reflected in the host plant, as trees occupied by different ant species often appear distinctly different in the field: in addition to differences in typical host plant size among the ants [2,7], T. penzigi-occupied trees often appear weak and spindly, C. nigriceps trees dense and thorny, and C. mimosae trees leafy and broad-canopied [3,7]. Demographic modelling indicates positive synergistic effects of these multiple ant species on host performance over time [9], suggesting that plants obtain different kinds of benefits from each ant species.
An unresolved question in the V. drepanolobium system is how T. penzigi can afford to forfeit extrafloral nectar by destroying its host's extrafloral nectaries. Destruction of nectaries by T. penzigi is thought to make host plants less attractive to other ant species that rely on the nectar for food, making takeovers less likely and facilitating ant species coexistence [2]. But the diet of T. penzigi is otherwise unknown: workers do not tend Hemiptera; few ants are normally found outside of the domatia; and unlike the three Crematogaster species, workers of T. penzigi are not known to prey on insects [2,7,10]. Some authors have suggested that T. penzigi might feed on pollen and fungal spores [10,11], but this has not been verified.
In this paper, we explore potential ant–fungal relationships with three lines of evidence. First, we test the hypothesis that the three obligate domatium-dwelling ant species show different behavioural interactions with fungi using a behavioural assay with a fungus isolated from an ant-occupied domatium. Second, we hypothesize that these behavioural or other differences among the ants might alter fungal communities inside the domatia, and explore this hypothesis using multiplexed amplicon sequencing or ‘metabarcoding’ [12]. Third, we hypothesize that dispersing ant reproductives (alates) may vector fungi when colonizing new trees by carrying fungal hyphae or spores in their infrabuccal pockets, thereby contributing to differences among the domatium fungal communities, and examine this hypothesis with additional metabarcoding.
2. Material and methods
(a). Recruitment experiment
Following previous anecdotal reports of potential fungus-tending behaviour in T. penzigi colonies [13], we used a behavioural assay in the laboratory to test the hypothesis that workers of T. penzigi show different behaviours towards fungi than workers of either C. mimosae or C. nigriceps. We presented each of 30 ant colonies collected from Kitengela in Kenya (figure 1d and the electronic supplementary material, S1) with a Petri dish containing a culture of Phoma sp. (Ascomycota: Dothideomycetes: Pleosporales) on potato dextrose agar media, plus a control containing only media. We imaged the plates after 8 h, and determined the proportion of the dish contents that had been removed (figure 1e) by using Photoshop to quantify disturbed and undisturbed areas of the plate.
We selected the Phoma isolate for our assay from approximately 60 fungal cultures that we isolated from ant-occupied V. drepanolobium domatia because it grew readily and appeared common: it was obtained 19 times among our cultures, from two C. mimosae and three T. penzigi colonies. The particular Phoma isolate that we selected was derived from a C. mimosae-occupied domatium.
(b). Domatium fungal metabarcoding
After finding that the three obligate domatium-dwelling ants differed in their behaviour towards our fungal isolates in the laboratory, we hypothesized that fungal communities in ant-occupied domatia in the field would differ among the three ant species. As our preliminary observations and recruitment experiment left open the possibility that any ant–fungus association might involve multiple fungal taxa, we used DNA metabarcoding [12] to characterize entire domatium fungal communities for each of the ant species. To do this, we sampled the contents of 56 ant-occupied domatia from different V. drepanolobium trees at Kitengela and Mpala in Kenya, approximately 190 km apart (figure 1d and the electronic supplementary material, S1). Twenty of these trees were occupied by C. mimosae, 17 by C. nigriceps and 19 by T. penzigi (see the electronic supplementary material, S2 for breakdown by site). Each domatium's contents were typical for the occupying ant species: old Vachellia leaflets for C. nigriceps, carton lamellae for C. mimosae and loose fibrous particles for T. penzigi [7]. The fourth ant species associated with V. drepanolobium at Mpala, C. sjostedti, was not sampled because it does not nest obligately in the domatia [8] and is not found at Kitengela.
In addition to the 56 domatia from Kenya, we sampled the contents of 13 ant-occupied domatia from different V. drepanolobium trees grown from seed in our greenhouse in Cambridge, MA (electronic supplementary material, S1), to see whether ants might be able to recreate their Kenyan domatium communities even in trees that had never been exposed to the Kenyan environment. We also sampled leaves from six trees at Mpala to represent a potential environmental source of fungal spores in the field. Finally, we tried to amplify fungal DNA from nine domatia from different trees in the greenhouse that had never been occupied by ants (unoccupied domatia are rare in the field), but none of these samples yielded any appreciable PCR product.
We extracted total DNA from each of the domatium and leaf samples, and sent DNA extracts from all suitable samples to Research and Testing Laboratory, Lubbock, TX (RTL) for PCR and multiplexed 454 pyrosequencing using ITS1F and ITS4 primers to target the internally transcribed spacer regions (ITS) from fungi. We processed the raw 454 data using the QIIME bioinformatics pipeline [14], before isolating the ITS1 region from all reads [15] and picking operational taxonomic units (OTUs) using uclust [16] with a similarity threshold of 95%. We assigned putative taxonomic descriptions to OTUs using blast, and examined differences in OTU richness and community composition between sample types using R [17]. Detailed methods are included in the electronic supplementary material, S3.
(c). Alate fungal metabarcoding
After finding that domatium fungal communities varied depending on ant species, we hypothesized that ants might vector fungi in their infrabuccal pockets when dispersing. If so, we would expect fungi present in the infrabuccal pocket of dispersing alates to resemble the fungal community from that ant species' domatia. We therefore performed additional DNA metabarcoding to compare alate infrabuccal pocket fungal communities to domatium fungal communities. We extracted and where possible sequenced DNA from the heads of 22 female C. nigriceps alates and 10 female T. penzigi alates, collected at Kitengela (figure 1d and the electronic supplementary material, S1) as they were leaving their domatia for their mating flights. To minimize any contribution from fungi on the outside of the alates, we surface-sterilized each alate prior to DNA extraction by rinsing for 60 s in 100% ethanol, followed by 60 s in 10% bleach and a final 60 s in ethanol. Any remaining fungal DNA in the head extractions was thus likely to reflect material in the infrabuccal pocket.
We extracted DNA from these alates, and sent DNA extracts from suitable samples to RTL for PCR and multiplexed 454 pyrosequencing using ITS1F and ITS4 primers. As for the domatium and leaf samples, we used QIIME to process the alate 454 data. ITS1 sequence data from alates, domatia and leaves were combined before picking OTUs using uclust with a 95% similarity threshold and assigning putative taxonomic descriptions using BLAST. Differences in OTU richness and community composition were examined using R, focusing particularly on the overlap between the alates and the ant-occupied domatia. Detailed methods are included in the electronic supplementary material, S3.
(d). Alate microscopy
To examine the contents of T. penzigi and C. nigriceps infrabuccal pockets, we collected female alates at Kitengela as they departed from domatia for their mating flights. The alates' heads were fixed in glutaraldehyde and embedded in araldite prior to sectioning with a Leica EM UC6 ultramicrotome. Semi-thin 1 µm sections were stained with methylene blue and thionin and viewed using an Olympus BX-51 microscope.
3. Results
(a). Recruitment experiment
Workers of T. penzigi chewed and removed significantly more mycelium than did workers of C. mimosae or C. nigriceps (figure 1e,f), but fragments of the mycelium were never observed being deposited elsewhere. Ant species was a significant predictor of fungus removal in a generalized least-squares model with recruitment to the fungus plate and colony size included as predictors (ant species: F2,24 = 5.1, p = 0.01; number of ants at fungus: F1,24 = 91.5, p < 0.01; ant colony size: F2,24 = 24.3, p < 0.01). The removal of fungus appeared to involve harvesting of hyphae, as no specialized structures such as gonglydia or reproductive structures were observed on the fungal isolate. All three ant species attended both fungus and control plates; however, removal of agar from the control plates was negligible, so the greater rate of fungus removal by T. penzigi is unlikely to be an artefact of T. penzigi attempting to access the growth medium for nutrition or moisture.
(b). Domatium fungal metabarcoding
We obtained 407 769 sequences across more than 660 OTUs from the 75 domatium and leaf samples. Many OTUs showed low abundance, with just 84 OTUs represented by greater than or equal to 100 sequences each. Sequencing depth varied between 1119 and 21 345 sequences per sample, with a median of 3980 sequences.
OTU richness varied substantially among the samples, but ant species accounted for little of that variation. Rarefaction curves indicated that the leaf samples contained more fungal OTUs than the Kenyan domatium samples, which in turn contained more OTUs than the greenhouse domatium samples (figure 2a). At a rarefaction depth of 1000 sequences, these differences were highly significant (leaves versus Kenyan domatia: t5.3 = 6.0, p < 0.01; Kenyan versus greenhouse domatia: t66.8 = 12.8, p < 0.01). However, the Kenyan domatia contained similar numbers of OTUs irrespective of ant or location (ant: F2,53 = 2.1, p = 0.13; location: t48.5 = 0.34, p = 0.74).
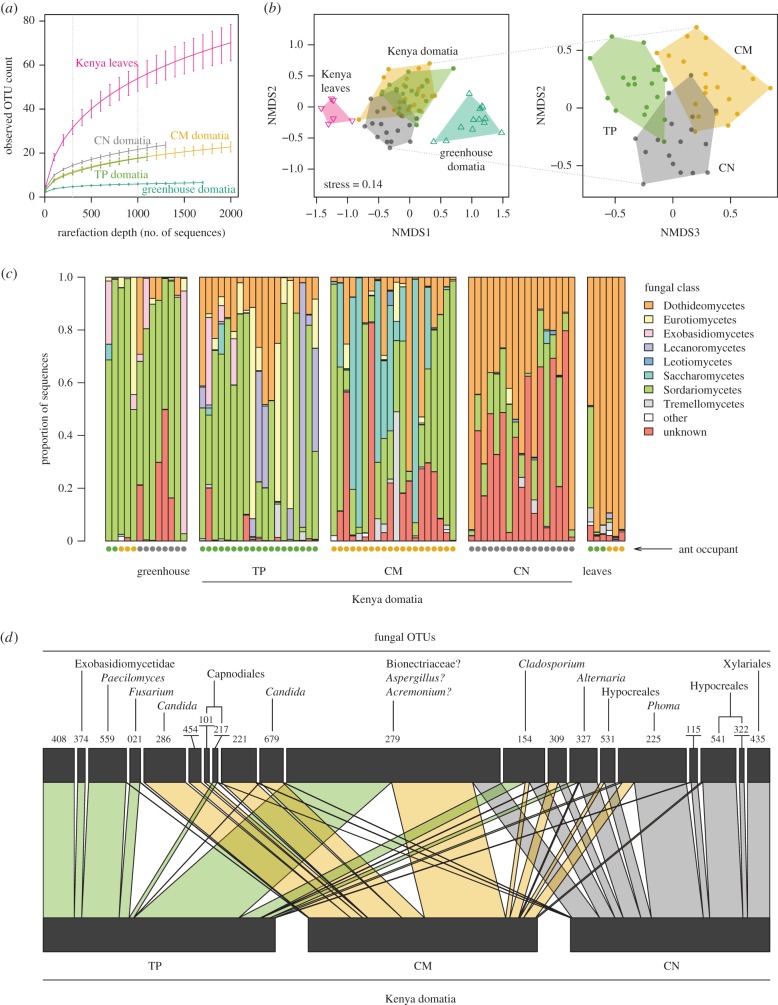
Figure 2.
(a) Kenyan domatium communities differ in taxonomic diversity compared with leaf sample communities and greenhouse domatium communities. Error bars show standard errors. (b) Non-metric multidimensional scaling (NMDS) based on Sørensen distances for rarefied dataset. Greenhouse domatia and leaf samples were distinct from Kenyan domatium samples. Kenyan domatium communities also differed among ant species. (c) Relative abundances of fungi aggregated to class differ between sample types and between ant occupants. Each column represents one sample. (d) Bipartite graph of ant species against most abundant fungal OTUs in rarefied dataset. Connections reflect sequence counts.
Fungal community composition varied significantly among sample types when evaluated using Sørensen distances between samples rarefied to 1000 sequences. The greenhouse domatia and the leaf samples were consistently distinct from the Kenyan domatium samples (figure 2b; adonis pseudo-F2,72 = 9.4, p < 0.01), reflecting in part the differences in alpha diversity among these sample types. But the Kenyan domatium communities also differed among the ants, as well as between sampling locations (adonis results for ant: pseudo-F2,50 = 5.4, p < 0.01; location: pseudo-F1,50 = 2.6, p < 0.01; ant × location: pseudo-F2,50 = 1.6, p < 0.02). The community differences among the ants were apparent even after aggregating the fungi by class (figure 2c). Although most abundant OTUs were present with more than one ant, some OTUs showed stronger associations with some ants than with others (figure 2d; 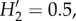 p < 0.01).
p < 0.01).
Our tentative taxonomic assignments (electronic supplementary material, S4) are dominated by ascomycetes and include many matches to plant pathogens and saprophytic fungi whose presence in a natural plant environment is quite plausible (e.g. Fusarium, Alternaria, Phoma, Capnodiales). Nonetheless, assigning taxonomic names to our fungal OTUs using the short ITS1 reads generally proved challenging. In some cases, the barcode region shows little variation among relatively distantly related taxa. In other cases, larger databases such as the NCBI's nucleotide database often returned matches to unidentified or apparently mislabelled sequences, while smaller curated databases such as UNITE often failed to yield a good match. For example, in the case of OTU 279, the most abundant OTU in our dataset, the top five BLAST matches against the nucleotide database all scored more than 99% identity with more than 86% query coverage; yet these matches included samples labelled as Bionectriaceae (JQ905678.1), Aspergillus (EU139858.1 and AM176687.1), Cephalosporium (i.e. Acremonium, AM176712.1), plus an uncultured ascomycete clone (AY273329.2). Taxonomic assignments often showed consistency across OTUs—e.g. Candida appeared against multiple OTUs such as OTUs 286 and 679 that tended to be strongly C. mimosae associated—suggesting a degree of reliability. But, for the most part, we regard our taxonomic assignments as tentative and subject to verification (e.g. through further culturing and/or metagenomic sequencing).
(c). Fungi carried by alates
DNA yields from alates were generally low, reflecting in part the small size of each specimen. Nonetheless, 13 of the C. nigriceps alates and three of the T. penzigi alates each yielded more than 300 fungal sequence reads, which we deemed sufficient for comparing the alate and domatium fungal communities, although the sample sizes especially for T. penzigi provide only a tentative interpretation.
Individual alates showed low alpha diversity, and as a result did not recapitulate the typical domatium community composition for either C. nigriceps or T. penzigi. In aggregate, however, the alates contained matches for the majority of sequences from domatia occupied by the same ant species; domatia occupied by different ant species were less well matched. The three T. penzigi alates contained matches for, on average, 72% of the sequences we obtained from T. penzigi-occupied domatia, compared with only 34% of sequences from those occupied by C. mimosae, and 20% of those occupied by C. nigriceps (figure 3a). The 13 C. nigriceps alates contained matches for, on average, 84% of the sequences we obtained from C. nigriceps-occupied domatia, compared with 81% of sequences from those occupied by T. penzigi, and 41% of those occupied by C. mimosae.
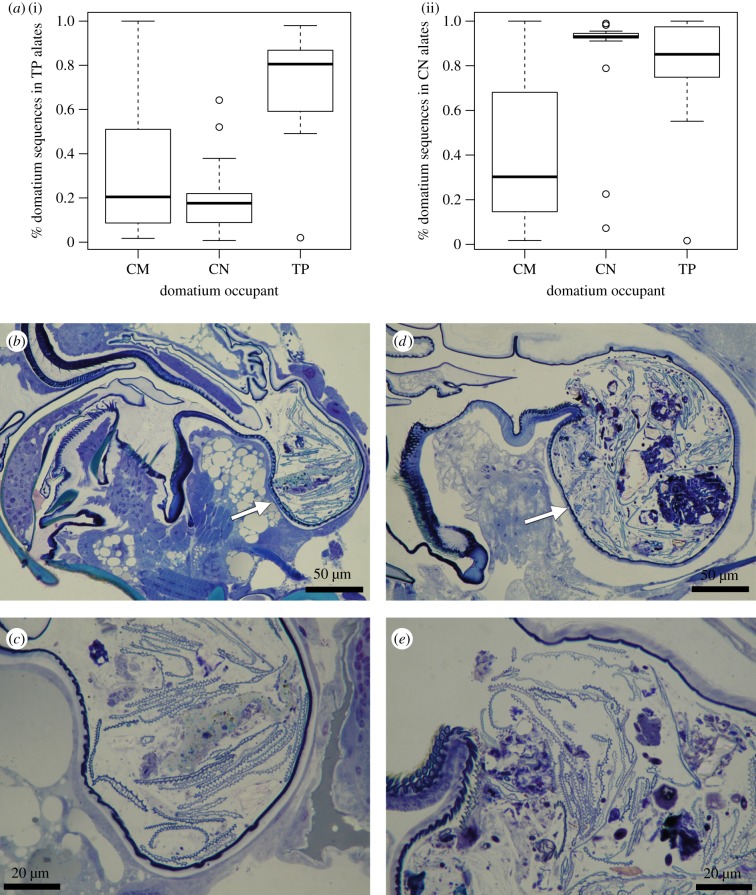
Figure 3.
(a) Proportion of each Kenyan domatium's sequences that were also recovered from the three pooled T. penzigi alate samples (i) and the 12 pooled C. nigriceps alate samples (ii). Sagittal view of (b,c) T. penzigi and (d,e) C. nigriceps alates showing debris in the infrabuccal pockets (arrowed). Higher magnification images in (c) and (e) show details of debris present in the pocket.
Sagittal sections through the heads of female T. penzigi and C. nigriceps alates showed a pellet of mixed debris in the infrabuccal pocket, supporting our assumption that fungi sequenced from the surface-sterilized alate heads are being carried in the infrabuccal pocket (figure 3b–e).
4. Discussion
(a). Ant associated differences in fungal communities
Our sequencing results show that domatium fungal community composition differed among V. drepanolobium trees occupied by different ant species. To the best of our knowledge, this is the first record of such ant-specific fungal community-level differences on the same myrmecophytic host species. Domatia of V. drepanolobium that have never been occupied by ants have no entry holes (as these are created by foundress queens), and we were unable to amplify fungal ITS from nine unopened domatia, indicating that the fungal community begins to assemble after ant occupation. Fungal community composition showed a large and significant main effect of ant species across our two Kenyan sampling sites.
In contrast to the recent work in other ant–plant–fungal systems [18], Chaetothyriales fungi are not prominent in our sequence libraries. Although PCR biases could also be responsible, we think it unlikely: we had good sequencing depth for most samples and have no reason to believe that Chaetothyriales would be especially prone to poor amplification. It is more likely that these fungi were not highly abundant in our samples. However, it is possible that sampling from a different part of the domatium (e.g. the domatium wall instead of the domatium contents), or using culture-based methods to target these fungi [19,20], would yet reveal Chaetothyriales in our domatia.
(b). Origins of fungal community differences
The differences in fungal community composition and richness between leaves and domatia indicate that domatium communities are shaped by processes specific to the domatia, and can be viewed as ‘extended phenotypes’ of the ants that inhabit them. Fungal dispersal may influence communities—including both passive dispersal (e.g. by air or water movement, or on ants' legs and bodies) and active vectoring by ants (discussed further below). Ant-specific differences in the domatia, such as in the size and number of domatium holes, may tend to favour different fungi. The ants also differ in the extent to which they forage in the tree canopy, or on the ground off the tree, which might lead to them accumulating different fungi from their environment. The different substrates contributed by the ants—i.e. loose fibrous particles, old dried leaflets and carton for T. penzigi, C. nigriceps and C. mimosae, respectively—may have been exposed to different fungi prior to being handled by ants, and are likely to select differently for fungal growth. The ants might also play an active role in manipulating community composition, e.g. weeding and grooming, or applying metapleural gland secretions, may help remove entomopathogens [21–23] or phytopathogens [24]. The fungal community could also be shaped by the untargeted trimming of fungal growth to keep the ants' living space free from obstructions and entanglements [25], or by ant-mediated changes to domatium characteristics such as chemistry, temperature or humidity.
Behavioural differences among the ant species, as suggested by our observations, provide a plausible route to differentiated fungal communities, though further investigation will be required to determine what role ant behaviours may play. It remains unknown, for example, to what extent T. penzigi's fungus-removing behaviour might vary across fungal taxa, or exactly how ant behaviour might shape domatium fungal communities. For example, the Phoma sp. fungus that we frequently cultured from domatia and which was so readily removed by T. penzigi workers in our behavioural trial was relatively uncommon in our 454 sequencing from T. penzigi domatia. This might reflect that T. penzigi workers are mainly working to suppress this fungus. Alternatively, workers of T. penzigi could lower the abundance of Phoma by consuming it, but since Phoma is relatively fast growing, it may be replaced rapidly by re-growth, such that our 454 data may under-represent the contribution of Phoma to biological activity in the domatia.
Our alate sequencing results suggest that fungi may also be vectored in ants' infrabuccal pockets when the ants disperse between host trees. These structures in adult ants are typically filled with material that the ants ingest while foraging, feeding or cleaning [25,26]. Workers may periodically expel pellets of such material onto the colony's waste piles, potentially aiding microbial dispersal [25,27]. Infrabuccal pockets may also allow microbes to be carried by dispersing alates—leafcutter ant foundresses, for example, use the infrabuccal pocket to carry a fungal inoculum when starting a new colony [21,26], and our dissections revealed infrabuccal pellets in many alates.
A substantial fraction of the fungal sequences from our T. penzigi domatia was recovered from among the three surface-sterilized T. penzigi alates; likewise, most of the sequences in C. nigriceps domatia were recovered from among the 13 surface-sterilized C. nigriceps alates. The small number of alates from which we obtained sequence and the difference in sample sizes between the two ant species mean that our results require some caution, because individual alates showed substantial variation; and indeed the fact that some of our dissected alates failed to yield sequence data may also reflect variation in the amount of fungal material being carried. But this notwithstanding, the shared sequences show the ant-species specificity that we would expect if fungi are carried from domatia by dispersing alates: the majority of sequences from our C. mimosae and C. nigriceps domatia were not recovered from the T. penzigi alates, and the majority of sequences from C. mimosae domatia were not recovered from the C. nigriceps alates. The C. nigriceps alates contained a high proportion of the T. penzigi domatium sequences; these sequences generally belonged to OTUs that were found at low abundance in C. nigriceps domatia and at high abundance in T. penzigi domatia, as would be expected given the distinction in domatium communities among the ant species. These shared OTUs were different from those responsible for the high overlap between C. nigriceps domatia and C. nigriceps alates—those OTUs were present at high abundance in C. nigriceps domatia and lower abundance in T. penzigi domatia. The C. nigriceps and T. penzigi alate samples also contained fungi that were not recovered from our domatium samples. This might reflect alates acquiring fungi from sources other than their domatia, but might also reflect variation among host plants not captured in our sampling—the alates were collected from different trees than the domatium samples, as the alates came from previous, opportunistic collections.
The variation among individual alates deserves further investigation. Individual alates showed much lower alpha diversity than the domatium samples. As a result, the fungal sequences recovered from individual alates did not closely resemble entire domatium communities. It is likely that the small size of the alate samples—and especially the small amount of material contained in the infrabuccal pocket—contributed to the limited DNA yield from these samples. Together with the finite sensitivity of 454 sequencing, this could make the alate samples appear to contain fungal communities that are less diverse and less representative of the domatium communities than they really are. Our surface sterilization procedure may also have lowered DNA recovery from the infrabuccal pocket—lengthier immersions in bleach typically lower total yield, indicating that the action of the bleach is not completely restricted to the outside surface. But our results might also represent true heterogeneity in the fungi carried by alates. For example, if fungi are heterogeneously distributed inside the domatium, then alates sampling from the domatium might acquire a non-representative selection. If alates contribute to fungal communities when founding a nest, however, then we might expect this individual-level variation in the alates to destroy the consistency observed in the domatium communities; this in turn suggests that inoculation by alates would operate alongside other species-specific forces shaping the growing domatium fungal communities, such as differences in growth conditions in the domatium. Alternatively, the colonization of trees by multiple foundresses, as has been documented in this system [28] might also facilitate the reconstruction of a more diverse community in the domatium.
(c). Potential roles for fungi
Fungi may comprise part of the T. penzigi diet, though further observations and experiments remain necessary to confirm whether the ants gain nutritional benefits. Our recruitment experiment showed T. penzigi workers readily removing mycelium, but adult ants are not generally thought to be capable of processing solid food. Instead, it is more likely that workers accumulate hyphae in their infrabuccal pockets and, in a natural setting, later transfer the hyphal pellet to larvae for digestion. The larvae of pseudomyrmecine ants, which include Tetraponera, possess a characteristic ventral pouch known as a trophothylax [29,30]. Despite wide variation in nesting habits and other aspects of lifestyle in the Pseudomyrmecinae (e.g. [31]), the trophothylax is often thought to be involved in larval feeding, with workers storing pellets of foraged material in their infrabuccal pockets and transferring it to larvae for digestion.
If T. penzigi does derive nutrition from fungi in its domatia, the degree of specialization between ant and fungus would contrast with that observed in other insect agriculture systems such as attine ants, Chaetothyriales-focused ant–plant systems or stingless bees [19,20,32,33]. While the accumulation of plant material in domatia superficially resembles the collection of fungal garden substrate by lower attine ants, no obvious fungal cultivars are growing on the material in V. drepanolobium domatia. Moreover, our sequencing indicates that T. penzigi domatium communities are not consistently dominated by any single fungal crop species, and no reproductive or other specialized fungal structures were observed in domatia. Our working hypothesis is that Tetraponera workers constantly gather suitable material to feed their larvae, including by trimming fungal hyphae from their environment relatively indiscriminately. We propose that the combination of this scavenging and the unique domatium environment that they generate, for example, through the accumulation of loose fibrous particles, generates a distinctive domatium fungal community, without workers necessarily selecting or cultivating specific fungi, or restricting their foraging to particular taxa. However, we stress that nutritional benefits to the ants remain to be established, and may yet turn out to be limited, or secondary to other functions of fungus removal, such as the repression of pathogenic fungi.
Nonetheless, even small quantities of fungi may help relieve nitrogen or other nutrient constraints for phytoecious ants on V. drepanolobium [34]. Fungi might allow ants to access nutrients from plant-derived substrates in their domatia, or provide a way to recycle nutrients from ant waste products [35]. This is especially likely to be important for T. penzigi, which is rarely observed foraging for insect prey [11]. Nitrogen stable isotope values suggest that T. penzigi is functionally herbivorous [13], but this species' workers do not associate with scale insects, and they destroy their host plant's extrafloral nectaries [2,7,10,13]. Furthermore, while some ants overcome nitrogen constraints through gut bacterial associations [36], T. penzigi does not possess the bacterial gut pouch observed in other Tetraponera species (i.e. the ‘diverticulum’ in [37]), and fluorescence microscopy and sequencing indicate relatively few bacteria [36]. By easing T. penzigi's probable nutritional constraints, domatium fungi may play a role in facilitating the coexistence of T. penzigi alongside C. mimosae and C. nigriceps on V. drepanolobium host plants. However, additional work will be needed to determine whether this is the case.
Domatium fungi might also play a role in host plant growth and nutrition. Nutrients released by digesting plant material or ant waste may become available to the host via the domatium wall. Such nutrient uptake is known in other systems (e.g. [38]) and, in at least some cases, appears to be facilitated by fungi [35,39]. Alternatively, the repression of fungal pathogens may represent a benefit to the plant of hosting ants. We might expect partner fidelity feedback to select for such fungi-mediated contributions by ants to their hosts. In this respect, it is also possible that individual V. drepanolobium trees benefit synergistically from hosting all three ant species over time [9] in part because of the effects of turnover in fungal communities associated with turnover in ant inhabitants, and this, too, deserves further investigation.
5. Conclusion
The roles played by domatium fungi and other ‘third parties’ should be considered alongside the direct interactions among ants, herbivores and host plants in constructing a more complete picture of the ecology of the V. drepanolobium system. Our metabarcoding data show that the different ant species which occupy V. drepanolobium host trees generate distinctive domatium fungal communities that may be regarded as extended phenotypes of the ants. Our behavioural observations and additional metabarcode results suggest that ant behaviours, including dispersal by alates, may contribute to those community differences, though further work on these potential fungal community drivers is required. The fungal community differences are broad, rather than reflecting just one or two important taxa, highlighting the value of a community sequencing approach, and suggesting that these ant–fungal associations are less specialized and more diffuse than classic fungus farming mutualisms. But the functional roles of these fungi are still unknown. Our observations of the foraging behaviour of T. penzigi suggest that this species may be feeding on fungi in the domatia, and further work is needed to investigate this hypothesis. It also seems possible that the fungal communities in the domatia contribute to host plant growth, perhaps influencing the phenotypic differences exhibited by plants occupied by different species, and this, too, represents a promising area for future work.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful for help in the field from Paul Oyieng Angienda, Nkaunya ole Ngotik, Bernard Rono, Lesere ole Shimbale and Lomwogo ole Sian. The staff at the Mpala Research Centre, Kitengela and Suyian Ranch ensured that all our work in Kenya went smoothly. Special thanks to Anne Powys and Gilfrid Powys at Suyian, and Nani Croze and Eric Krystall at Kitengela for welcoming us into their homes in Kenya and letting us work on their land. Many thanks also to Christian Rabeling for helping to dissect the infrabuccal pockets of the alates; to Jack Boyle, Jignasha Rana and Katie Berry for helping collect the colonies used in the behavioural assay; to Bonnie Lei for assistance with the preliminary behavioural trials; to Stephanie Hays for her preliminary investigations of ant diet; and to An Vandoren for making the wonderful histological sections shown here. The computations in this paper were run on the Odyssey cluster supported by the FAS Research Computing Group at Harvard University. And, finally, thanks to Susanne Renner and Guillaume Chomicki for their efforts in coordinating such a fascinating collection of works for this special issue of Proceedings B.
Data accessibility
Data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.q82kn [40].
Authors' contributions
D.J.M. has been observing ant acacia interactions for most of his life and introduced all other authors to their fascinating natural history. He made the original observations of Tetraponera penzigi foraging on fungus, collected alates for analysis and sectioning, and carried out fieldwork. N.E.P., D.J.M. and C.C.M.B. designed the project. C.C.M.B. performed behavioural experiments, sampling and molecular work, and conducted data analysis. J.N.P. dissected alates and carried out laboratory work. J.P.J.B. prepared sections and performed microscopy. All authors helped interpret data and write the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by FQEB grant no. RFP-12-06 from the National Philanthropic Trust to N.E.P., M.E.F. and A.P., and from NSF SES-0750480 to N.E.P. In addition, C.C.M.B. and D.J.M. were supported by fellowships from the Department of Organismic and Evolutionary Biology at Harvard University. C.C.M.B. was further supported by a Putnam Expedition grant from Harvard's Museum of Comparative Zoology. D.J.M.'s work in Kenya is supported by the National Geographic Society, the Turkana Basin Institute, Suyian Trust, the National Museums of Kenya and Nature Kenya. M.E.F. acknowledges funding from an NSERC Discovery Grant and the University of Toronto.
References
- 1.Madden D, Young TP. 1992. Symbiotic ants as an alternative defense against giraffe herbivory in spinescent Acacia drepanolobium. Oecologia 91, 235–238. ( 10.1007/BF00317789) [DOI] [PubMed] [Google Scholar]
- 2.Young TP, Stubblefield CH, Isbell LA. 1997. Ants on swollen-thorn acacias: species coexistence in a simple system. Oecologia 109, 98–107. ( 10.1007/s004420050063) [DOI] [PubMed] [Google Scholar]
- 3.Stanton ML, Palmer TM, Young TP, Evans A, Turner ML. 1999. Sterilization and canopy modification of a swollen thorn acacia tree by a plant-ant. Nature 401, 578–581. ( 10.1038/44119) [DOI] [Google Scholar]
- 4.Stanton ML, Palmer TM, Young TP. 2002. Competition-colonization trade-offs in a guild of African acacia-ants. Ecol. Monogr. 72, 347–363. ( 10.1890/0012-9615(2002)072%5B0347:CCTOIA%5D2.0.CO;2) [DOI] [Google Scholar]
- 5.Martins DJ. 2010. Not all ants are equal: obligate acacia ants provide different levels of protection against mega-herbivores. Afr. J. Ecol. 48, 1115–1122. ( 10.1111/j.1365-2028.2010.01226.x) [DOI] [Google Scholar]
- 6.Baker CCM, Bittleston LS, Sanders JG, Pierce NE. 2016. Dissecting host-associated communities with DNA barcodes. Phil. Trans. R. Soc. B 371, 20150328 ( 10.1098/rstb.2015.0328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hocking B. 1970. Insect associations with the swollen thorn acacias. Trans. R. Entomol. Soc. Lond. 122, 211–255. ( 10.1111/j.1365-2311.1970.tb00532.x) [DOI] [Google Scholar]
- 8.Palmer TM, Young TP, Stanton ML, Wenk E. 2000. Short-term dynamics of an acacia ant community in Laikipia, Kenya. Oecologia 123, 425–435. ( 10.1007/s004420051030) [DOI] [PubMed] [Google Scholar]
- 9.Palmer TM, Doak DF, Stanton ML, Bronstein JL, Kiers ET, Young TP, Goheen JR, Pringle RM. 2010. Synergy of multiple partners, including freeloaders, increases host fitness in a multispecies mutualism. Proc. Natl Acad. Sci. USA 107, 17 234–17 239. ( 10.1073/pnas.1006872107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palmer TM. 2003. Spatial habitat heterogeneity influences competition and coexistence in an African acacia ant guild. Ecology 84, 2843–2855. ( 10.1890/02-0528) [DOI] [Google Scholar]
- 11.Palmer TM, Stanton ML, Young TP, Goheen JR, Pringle RM, Karban R. 2008. Breakdown of an ant-plant mutualism follows the loss of large herbivores from an African savanna. Science 319, 192–195. ( 10.1126/science.1151579) [DOI] [PubMed] [Google Scholar]
- 12.Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. 2012. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. ( 10.1111/j.1365-294X.2012.05470.x) [DOI] [PubMed] [Google Scholar]
- 13.Martins DJ. 2011. Multi-species interactions in African ant-acacias. PhD dissertation, Harvard University, Cambridge, MA, USA.
- 14.Caporaso JG, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. ( 10.1038/nmeth.f.303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson RH, et al. 2010. An open source software package for automated extraction of ITS1 and ITS2 from fungal ITS sequences for use in high-throughput community assays and molecular ecology. Fungal Ecol. 3, 284–287. ( 10.1016/j.funeco.2010.05.002) [DOI] [Google Scholar]
- 16.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. ( 10.1093/bioinformatics/btq461) [DOI] [PubMed] [Google Scholar]
- 17.R Core Team. 2015. R: a language and environment for statistical computing. Version 3.1.0 ed. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 18.Mayer VE, Frederickson ME, McKey D, Blatrix R. 2014. Current issues in the evolutionary ecology of ant-plant symbioses. New Phytol. 202, 749–764. ( 10.1111/nph.12690) [DOI] [PubMed] [Google Scholar]
- 19.Voglmayr H, Mayer V, Maschwitz U, Moog J, Djieto-Lordon C, Blatrix R. 2011. The diversity of ant-associated black yeasts: insights into a newly discovered world of symbiotic interactions. Fungal Biol. 115, 1077–1091. ( 10.1016/j.funbio.2010.11.006) [DOI] [PubMed] [Google Scholar]
- 20.Nepel M, Voglmayr H, Schonenberger J, Mayer VE. 2014. High diversity and low specificity of chaetothyrialean fungi in carton galleries in a neotropical ant-plant association. PLoS ONE 9, e112756 ( 10.1371/journal.pone.0112756) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Currie CR, Stuart AE. 2001. Weeding and grooming of pathogens in agriculture by ants. Proc. R. Soc. Lond. B 268, 1033–1039. ( 10.1098/rspb.2001.1605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-Marín H, Zimmerman JK, Rehner SA, Wcislo WT. 2006. Active use of the metapleural glands by ants in controlling fungal infection. Proc. R. Soc. B 273, 1689–1695. ( 10.1098/rspb.2006.3492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oi DH, Pereira RM. 1993. Ant behavior and microbial pathogens (Hymenoptera, Formicidae). Fla. Entomol. 76, 63–74. ( 10.2307/3496014) [DOI] [Google Scholar]
- 24.Letourneau DK. 1998. Ants, stem-borers, and fungal pathogens: experimental tests of a fitness advantage in Piper ant-plants. Ecology 79, 593–603. ( 10.1890/0012-9658(1998)079%5B0593:ASBAFP%5D2.0.CO;2) [DOI] [Google Scholar]
- 25.Bailey IW. 1920. Some relations between ants and fungi. Ecology 1, 174–189. ( 10.2307/1929134) [DOI] [Google Scholar]
- 26.Quinlan RJ, Cherrett JM. 1978. Studies on the role of the infrabuccal pocket of the leaf-cutting ant Acromyrmex octospinosus (Reich) (Hym., Formicidae). Insectes Soc. 25, 237–245. ( 10.1007/Bf02224744) [DOI] [Google Scholar]
- 27.Eisner T, Happ GM. 1962. The infrabuccal pocket of a formicine ant: a social filtration device. Psyche 69, 107–116. ( 10.1155/1962/25068) [DOI] [Google Scholar]
- 28.Rubin BER, Anderson RM, Kennedy D, Palmer TM, Stanton ML, Lovette IJ. 2013. Polygyny in the nest-site limited acacia-ant Crematogaster mimosae. Insectes Soc. 60, 231–241. ( 10.1007/s00040-013-0287-5) [DOI] [Google Scholar]
- 29.Wheeler WM, Bailey IW. 1920. The feeding habits of pseudomyrmine and other ants. Trans. Am. Philos. Soc. 22, 235–279. ( 10.2307/1005485) [DOI] [Google Scholar]
- 30.Wheeler GC, Wheeler J. 1956. The ant larvae of the subfamily Pseudomyrmecinae (Hymenoptera: Formicidae). Ann. Entomol. Soc. Am. 49, 374–398. ( 10.1093/aesa/49.4.374) [DOI] [Google Scholar]
- 31.Chomicki G, Ward PS, Renner SS. 2015. Macroevolutionary assembly of ant/plant symbioses: Pseudomyrmex ants and their ant-housing plants in the Neotropics. Proc. R. Soc. B 282, 20152200 ( 10.1098/rspb.2015.2200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blatrix R, Djiéto-Lordon C, Mondolot L, La Fisca P, Voglmayr H, McKey D. 2012. Plant-ants use symbiotic fungi as a food source: new insight into the nutritional ecology of ant–plant interactions. Proc. R. Soc. B 279, 3940–3947. ( 10.1098/rspb.2012.1403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menezes C, Vollet-Neto A, Marsaioli AJ, Zampieri D, Fontoura IC, Luchessi AD, Imperatriz-Fonseca VL. 2015. A Brazilian social bee must cultivate fungus to survive. Curr. Biol. 25, 2851–2855. ( 10.1016/j.cub.2015.09.028) [DOI] [PubMed] [Google Scholar]
- 34.Davidson DW, Cook SC, Snelling RR, Chua TH. 2003. Explaining the abundance of ants in lowland tropical rainforest canopies. Science 300, 969–972. ( 10.1126/science.1082074) [DOI] [PubMed] [Google Scholar]
- 35.Defossez E, Djiéto-Lordon C, McKey D, Selosse M-A, Blatrix R. 2010. Plant-ants feed their host plant, but above all a fungal symbiont to recycle nitrogen. Proc. R. Soc. B 278, 1419–1426. ( 10.1098/rspb.2010.1884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE. 2009. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc. Natl Acad. Sci. USA 106, 21 236–21 241. ( 10.1073/pnas.0907926106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Billen J, Buschinger A. 2000. Morphology and ultrastructure of a specialized bacterial pouch in the digestive tract of Tetraponera ants (Formicidae, Pseudomyrmecinae). Arthropod. Struct. Dev. 29, 259–266. ( 10.1016/s1467-8039(00)00029-3) [DOI] [PubMed] [Google Scholar]
- 38.Fischer RC, Wanek W, Richter A, Mayer V. 2003. Do ants feed plants? A 15N labelling study of nitrogen fluxes from ants to plants in the mutualism of Pheidole and Piper. J. Ecol. 91, 126–134. ( 10.1046/j.1365-2745.2003.00747.x) [DOI] [Google Scholar]
- 39.Leroy C, Séjalon-Delmas N, Jauneau A, Ruiz-González M-X, Gryta H, Jargeat P, Corbara B, Dejean A, Orivel J. 2011. Trophic mediation by a fungus in an ant–plant mutualism. J. Ecol. 99, 583–590. ( 10.1111/j.1365-2745.2010.01763.x) [DOI] [Google Scholar]
- 40.Baker CCM, Martins DJ, Pelaez JN, Billen JPJ, Pringle A, Frederickson ME, Pierce NE. 2017. Data from: Distinctive fungal communities in an obligate African ant-plant mutualism. Dryad Digital Repository. ( 10.5061/dryad.q82kn) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Baker CCM, Martins DJ, Pelaez JN, Billen JPJ, Pringle A, Frederickson ME, Pierce NE. 2017. Data from: Distinctive fungal communities in an obligate African ant-plant mutualism. Dryad Digital Repository. ( 10.5061/dryad.q82kn) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.q82kn [40].