Abstract
Phylogenetic and biogeographic analyses can enhance our understanding of multispecies interactions by placing the origin and evolution of such interactions in a temporal and geographical context. We use a phylogenomic approach—ultraconserved element sequence capture—to investigate the evolutionary history of an iconic multispecies mutualism: Neotropical acacia ants (Pseudomyrmex ferrugineus group) and their associated Vachellia hostplants. In this system, the ants receive shelter and food from the host plant, and they aggressively defend the plant against herbivores and competing plants. We confirm the existence of two separate lineages of obligate acacia ants that convergently occupied Vachellia and evolved plant-protecting behaviour, from timid ancestors inhabiting dead twigs in rainforest. The more diverse of the two clades is inferred to have arisen in the Late Miocene in northern Mesoamerica, and subsequently expanded its range throughout much of Central America. The other lineage is estimated to have originated in southern Mesoamerica about 3 Myr later, apparently piggy-backing on the pre-existing mutualism. Initiation of the Pseudomyrmex/Vachellia interaction involved a shift in the ants from closed to open habitats, into an environment with more intense plant herbivory. Comparative studies of the two lineages of mutualists should provide insight into the essential features binding this mutualism.
Keywords: multispecies interactions, phylogenomics, ultraconserved elements, convergence, Mesoamerica, Formicidae
1. Introduction
An outstanding question in the study of multispecies symbioses concerns the influence of phylogeny and biogeography on the origin, evolution and functional dynamics of such interactions. By considering multispecies interactions in a phylogenetic and geographical context, we gain insight into factors propelling, impeding or modifying the associations, and the extent to which selective pressures vary across different landscapes and phylogenetic lineages [1–6]. Phylogenies also allow instances of convergent evolution of symbiotic associations to be more rigorously identified and analysed [6,7].
Recent years have seen a proliferation of studies on ant/plant mutualisms, often employing phylogenetic approaches to make inferences about the extent of coevolution between partners, the biogeographic context in which evolution has occurred, the factors favouring elaboration or dissolution of the mutualism and the effects of the mutualism on diversification rates [7–15]. When we consider ant/plant mutualisms that involve domatia-bearing plants and their specialized, protective ants, two contrasting aspects are evident: on the one hand, there are unique features associated with particular taxa and geographical settings [16–18]; on the other hand, we see replication of pattern and process across different sets of interacting partners [6,18]. Multiple case studies, employing a phylogenetic/historical framework, are needed to provide a balanced perspective on these contrasting elements of contingency and convergence.
Here we examine the evolutionary history of a classic ant/plant mutualism: the association of Central American ants in the Pseudomyrmex ferrugineus group with swollen-thorn acacias (Vachellia species). In this system, the ants receive nesting space in the form of swollen stipular thorns, and food from extrafloral nectaries and specialized leaf-tip food bodies (Beltian bodies). The ants in return protect their host plant from herbivores and competing plants, by patrolling aggressively, removing or repelling intruders and clipping competing vegetation [16,19–23]. The system has also been invaded to a limited degree by parasitic (non-protective) ants [24,25], although certain features of the ants and plants appear to constrain this [23,26–28]. There are aggressive Pseudomyrmex associated with other domatia-bearing plants, such as Triplaris and Tachigali [11,13,29], but most species in this genus have very different habits: they are timid ants, nesting opportunistically in dead, insect-bored twigs of many species of woody plants [16,30]. Similarly, other species of Vachellia do not display the constellation of mutualism-associated traits—inhabitable swollen thorns, food bodies and enlarged extrafloral nectaries—shown by the Mesoamerican ant-acacias [20].
In this study, we employ a phylogenomic approach, ultraconserved element (UCE) sequence capture [31,32] and comprehensive taxon sampling to address the following questions: (i) what are the phylogenetic relationships of the obligate acacia ants? More specifically, do phylogenomic data support recent inferences [13,23], based on Sanger sequencing, that the acacia ants evolved more than once? (ii) what is the biogeographic history of the acacia ants? Where and when did they originate? By what sequence of events did they come to occupy much of Mesoamerica? and (iii) what has been the role of habitat and host plant use during the evolution of acacia ants? Although our focus is on the ants, we take into account the available information about the phylogeny and distribution of the plant partners. Our findings highlight a strong biogeographic component to the evolution of this system, and potent forces of convergent evolution in the ants. These results generate additional questions about the selective forces driving and restraining this iconic mutualism.
2. Material and methods
(a). Taxon sampling
We sampled all 10 species of obligate acacia ants in the P. ferrugineus group [33]; two other generalist species recently implicated as part of the P. ferrugineus group [13]; and five other closely related species of Pseudomyrmex. Based on current taxonomic knowledge [13,33–36], this represents a comprehensive sampling of the species of Pseudomyrmex that are related to the acacia ants. For eight of the 10 species of acacia ants, we sampled multiple populations, drawn from across the known range of the species [33]. Our taxon set comprises 29 samples belonging to 18 species. Details of the species, sample names and voucher specimens are given in the electronic supplementary material, table S1 (see also the electronic supplementary material, figure S1). Pseudomyrmex depressus, determined from previous work [13] to be the most distantly related of the six outgroup species, was used to root the tree.
(b). DNA extraction, library preparation and target enrichment
DNA was extracted from single ants, usually workers or worker pupae, using the DNeasy Blood and Tissue Kit (Qiagen Inc., Valencia, CA, USA), and quantified using a Qubit fluorometer (HS Assay Kit, Life Technologies Inc., Carlsbad, CA, USA). We sheared 10–138 ng input DNA to a target size of approximately 600 bp by sonication using a Diagenode BioRuptor (Diagenode Inc., Denville, NJ, USA), and this product served as input for a modified genomic DNA library preparation protocol (Kapa Hyper Prep Library Kit, Kapa Biosystems) that included SPRI bead cleanup using an AMPure substitute [37] and custom dual-indexing barcodes [38].
For UCE enrichment, we pooled 8–10 libraries together at equimolar concentrations and adjusted pool concentrations to 147 ng µl−1. For each enrichment, we used a total of 500 ng of DNA (3.4 µl each pool), and we performed enrichments using a custom RNA bait library developed for ants [39] and synthesized by MYcroarray (Ann Arbor, MI, USA). The probe set includes 9446 probes, targeting 2524 UCE loci. Although these loci are likely to be present across Hymenoptera, this set is ant-specific because the probes used were designed from ant genomes (Harpegnathos saltator and Atta cephalotes). For each enrichment, we hybridized the RNA bait libraries to sequencing libraries at 65°C for 24 h and we followed a standardized, in-solution enrichment protocol (v. 1.5; protocol available from http://ultraconserved.org). Following enrichment, we quantified the DNA concentration of enriched pools using qPCR and we used these values to make an equimolar pool-of-pools, containing up to 102 individual samples, which were submitted to the High Throughput Genomics Core Facility, Huntsman Cancer Institute, University of Utah, where they were quality checked, quantified with qPCR and sequenced on an Illumina HiSeq 2500 (125 cycle paired-end sequencing, v. 4 chemistry).
(c). Assembly and alignment
We performed initial bioinformatics steps, including read cleaning, assembly and alignment, using the software package Phyluce v. 1.5 [40]. Demultiplexed FASTQ output was cleaned and trimmed using Illumiprocessor [41], a wrapper program around Trimmomatic [42]. Cleaned reads were assembled de novo using Trinity (v. trinityrnaseq_r2013-02-25) [43]. After assembly, we used Phyluce to identify UCE loci from the assembled pool of contigs and to remove any potential paralogs. We performed this step using default settings (80% for min-coverage and min-identity). We then separated and aligned individual loci using a wrapper script around the alignment program MAFFT [44]. We performed this step using default Phyluce settings, except for the ‘incomplete-matrix’ and ‘no-trim’ flag, which we used to allow for missing data and to prevent default alignment trimming. Each alignment was trimmed using a wrapper around Gblocks v. 0.91b [45], run with reduced stringency settings of b1 = 0.5, b2 = 0.5, b3 = 12 and b4 = 7. To reduce missing data, we chose a subset of trimmed UCE alignments in which the loci were represented in at least 95% of taxa (more than 27 out of 29 taxa). This subset with 95% taxon completeness had 1672 loci, and 10.8% missing data across all cells in the concatenated matrix. The complete dataset included 1 321 987 bp of sequence data, of which 58 173 were informative. Assembly and matrix stats are provided in the electronic supplementary material, table S2.
(d). Phylogenetic analyses
Most phylogenetic analyses, including divergence dating, were performed using either the CIPRES Science Gateway [46] or the Smithsonian Institution's high-performance computer cluster (Hydra). For concatenated maximum-likelihood (ML) analysis, we used RAxML v. 8.2 [47] with the GTR + Γ model and we performed best tree plus rapid bootstrap searches using 100 bootstrap replicates. We ran analyses under three partitioning schemes: (i) unpartitioned, (ii) the best-fit scheme chosen by PartitionFinder v. 1.1.1 [48] under the hcluster algorithm [49], with each UCE locus corresponding to a separate data block, and (iii) the best-fit scheme chosen by PartitionFinder v. 2.0 [50] under the kmeans algorithm [51]. To reduce the possible influence of nucleotide frequency heterogeneity and saturation, we also ran an unpartitioned ML treatment with RY-coding.
We used ExaBayes v. 1.4.1 [52] for Bayesian analysis of the concatenated data matrix, with the GTR + Γ model and the same three partition schemes employed in RAxML. Each ExaBayes analysis included two independent runs, each having four coupled chains, and each run was performed for 500 000 generations, sampling every 100 generations. Burnin was set to 25% and Markov chain Monte Carlo (MCMC) convergence was confirmed with Tracer v. 1.6.0 [53].
For species tree analysis, we employed ASTRAL [54] using v. 4.8.0, which allows multiple individuals to be assigned to the same species. We ran one analysis with the 500 best genes (those with highest average bootstrap scores) and one analysis with all 1672 loci. We also ran a third analysis with a newer version of ASTRAL (v. 4.10.8) which generates branch lengths in coalescent units but does not allow for species assignment; this third analysis used the 500 best genes. All analyses were performed using 100 multi-locus bootstrap replicates [55].
(e). Divergence dating
As input for divergence dating with BEAST v. 1.8.3 [56], we used several different constraint topologies: (0) all 29 taxa, kmeans-partitioned ML topology; (1) 18 taxa, kmeans-partitioned ML topology and (2) 18 taxa, kmeans-partitioned Bayesian (ExaBayes) topology. Hereafter we refer to these as ‘topology 0’, ‘topology 1’ and ‘topology 2’, respectively. The 18-taxon topologies included only a single exemplar for each species, and we generated these trees by pruning taxa from the trees containing all taxa using the R package APE [57]. We pruned taxa arbitrarily except for the two ‘paraphyletic’ species, P. ferrugineus and Pseudomyrmex mixtecus, in which we retained the population that was closest to the divergent ingroup species (Pseudomyrmex janzeni and Pseudomyrmex veneficus, respectively).
For each constrained topology, we had two node age calibrations: the root node was assigned a normal prior, with mean 20.3 ± 10 Ma, based on results in Chomicki et al. [13]; and the Dominican amber fossil Pseudomyrmex baros was used to calibrate stem Pseudomyrmex haytianus, an internal node to which we assigned the same gamma prior as Chomicki et al. [13] (offset = 15 Ma; shape α = 3; scale β = 3.8, median = 25 Ma). For each analysis, we chose 50 randomly sampled loci from the complete (1672 locus) dataset (same locus set used for topology 1 and 2, with alignments pruned to 18 taxa; different locus set for topology 0); and we performed three independent BEAST runs, each for 100 million generations, sampling every 2000 generations, under a GTR + Γ substitution model and uncorrelated lognormal clock. We also performed one MCMC run in which only the prior was sampled. For all runs, we used a birth–death model for the tree prior and we turned off all tree search operators, thus constraining tree topology to the user-supplied input tree. Convergence and run performance was assessed with Tracer v. 1.6.0 [53] by examining post-burnin parameter values across all runs.
(f). Biogeographic analysis
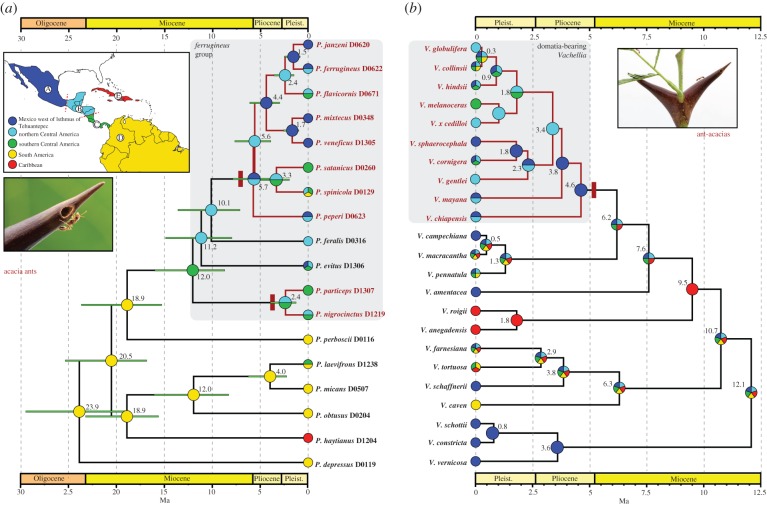
We used the R package BioGeoBEARS [58,59] for ancestral range estimation of the ants, evaluating six models: dispersal–extinction–cladogenesis (DEC) [60], DEC + J, where J is a parameter allowing for founder event speciation [58], DIVALIKE [61], DIVALIKE + J, BAYAREALIKE [62] and BAYAREALIKE + J. We recognized five areas, based on the distribution ranges of the ants [33] and historical biogeographic barriers in the region [63–65]: (i) Mexico north and west of the Isthmus of Tehuantepec, (ii) northern Central America: Isthmus of Tehuantepec to northern Nicaragua, (iii) southern Central America: central Nicaragua to eastern Panama, (iv) South America, and (v) the Caribbean (figure 2). We used the BEAST chronograms from the two 18-taxon analyses (topology 1 and topology 2), where each species is represented by a single exemplar. We coded each species for its entire range, except that two outgroup species (Pseudomyrmex perboscii, Pseudomyrmex obtusus), which are subtended by long branches and whose main distribution is in South America, were coded as occurring in area D only.
Figure 2.
Chronogram and ancestral range estimation of acacia ants and relatives (a) and New World Vachellia (b). The ant chronogram is based on a BEAST analysis of topology 1 (figure 1b); the plant chronogram is from Chomicki et al. [13]. Ancestral range estimates are from the model with the highest likelihood in BioGeoBEARS analyses (DEC + J for both ants and plants), with one area constraint for the ants (ant analysis no. 2) and disjunct continental areas disallowed for plants (plant analysis no. 2) (see text). The ancestral range(s) of highest probability is depicted for each node. Results are similar using other models and with alternate topologies and area constraints (see the electronic supplementary material, figures S4 and S5). Photographic images courtesy of Alex Wild (www.alexanderwild.com).
For all models, we created two time intervals, one from 0 to 5 Ma (approximating the time of close proximity of Central and South America, culminating in final closure of the Isthmus of Panama about 3 Ma [65]) and 5–25 Ma (reduced proximity). For dispersal multipliers, we used a value of 1.0 for all adjacent, connected areas, 0.5 for southern Central America and South America prior to 5 Ma, and 0.01 for distant unconnected areas. We ran two sets of analyses: (i) in analysis 1 we allowed all area possibilities; (ii) in analysis 2 we excluded range combinations containing DE, because the outcome of analysis 1 was an ancestral range of DE at the base of the tree—a result that is implausible and seemed to be driven by the occurrence among the outgroup species of a single Caribbean endemic. Use of 0–5 Ma as the period of close proximity of Central and South America was based on current thinking about the formation of the Isthmus of Panama [65], but we ran additional sensitivity analyses in which we employed two other sets of time intervals: 0–8 Ma/8–25 Ma and 0–3 Ma/3–25 Ma. Results (not shown) yielded very similar inferences about biogeographic history, with the few differences having no effect on our overall conclusions.
We also estimated ancestral ranges in the domatia-bearing Vachellia and their relatives, using the same six models in BioGeoBEARS and the same five biogeographic areas, time intervals (0–5 Ma/5–25 Ma) and dispersal multipliers. Information on plant distributions was taken from GBIF [66], and filtered for dubious records, i.e. those not based on verified specimen records. We carried out three sets of analyses: (i) all areas allowed; (ii) all disjunct continental areas disallowed, but all connected continental areas containing E allowed; and (iii) all areas that included E disallowed except for widespread ranges: ABCDE, BCDE, ABCE and CDE. Our input tree was the Vachellia chronogram generated by Chomicki et al. [13].
(g). Ancestral trait reconstruction
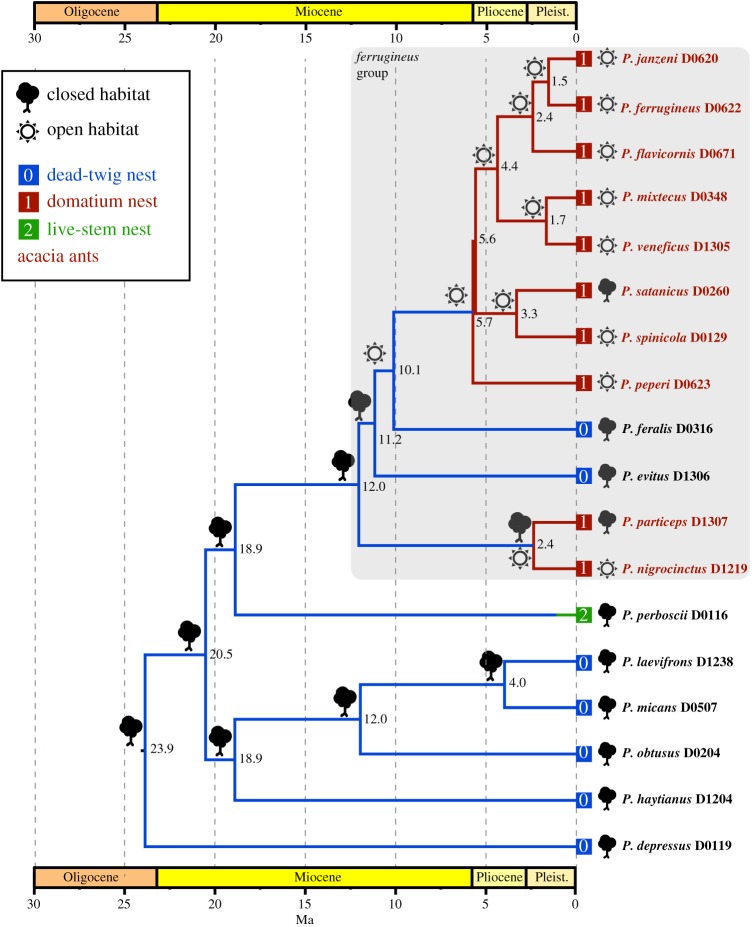
We used an ML approach as implemented in the Ace function of APE [57] to estimate the history of changes in habitat use among ant species, comparing equal rates (ER), symmetrical rates (SYM) and all rates different (ARD) models. Ant taxa were coded according to whether they occur predominantly in (0) closed habitats (rainforest and tropical moist forest) or (1) open habitats (tropical dry forest, pastures, roadsides). As the input tree we used the BEAST topology 1 chronogram. We evaluated the best trait model by performing likelihood ratio tests among competing models. We used the same approach for estimating ancestral nesting habits in the ants, using the following three discrete states: (0) nesting in dead twigs of various plants, non-aggressive; (1) nesting in Vachellia domatia, aggressive, and (2) nesting in live branches of various trees, non-aggressive. Trait data on the ants were taken from published sources [13,20,33–36].
3. Results
(a). Phylogenetic relationships
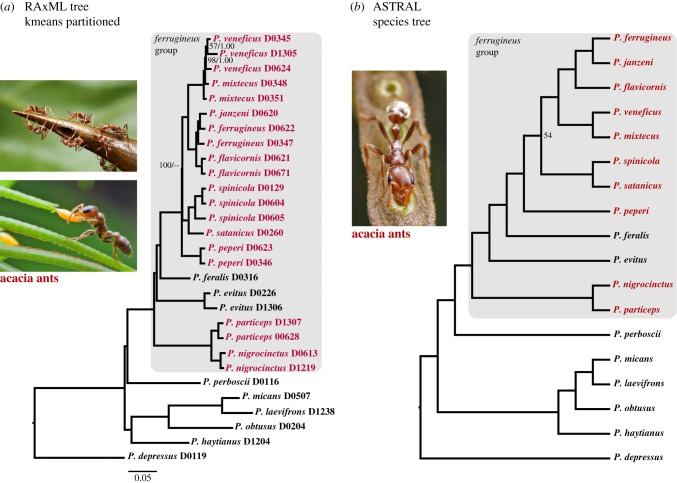
Except for the position of one species, Pseudomyrmex peperi, the same tree topology was obtained in nearly all analytical treatments, with maximum support at most nodes (figure 1; electronic supplementary material, figure S2). The P. ferrugineus group is shown to be a strongly supported clade (ML bootstrap percentages 100, Bayesian posterior probabilities 1.00, ASTRAL multi-locus bootstraps 100) that includes the 10 species of obligate acacia ants and also contains two species of generalist (dead twig-inhabiting) ants, Pseudomyrmex evitus and Pseudomyrmex feralis. This effectively splits the acacia ants into two different subgroups, here termed the Pseudomyrmex nigrocinctus subgroup and the P. ferrugineus subgroup, separated by two maximally supported nodes (figure 1). This result was also obtained by Chomicki et al. [13] with a 10-gene dataset, although the two critical nodes upholding this arrangement had less than full support.
Figure 1.
Phylogeny of Pseudomyrmex acacia ants and related species, derived from genomic data. (a) ML tree from kmeans-partitioned RAxML analysis of 1672 UCE loci. All nodes have 100% bootstrap support and 1.00 posterior probability (from a separate kmeans-partitioned Bayesian analysis—see the electronic supplementary material, figure S2f), except where indicated otherwise and then given as bootstrap percentage/posterior probability. (b) Species tree from ASTRAL (v.4.8.0) analysis of 500 best loci (those with highest average bootstrap support). All nodes have 100% bootstrap support except where indicated otherwise. Taxon names in dark red refer to obligate acacia ants. Photographic images courtesy of Alex Wild (www.alexanderwild.com).
The P. nigrocinctus subgroup comprises only two species, with the remaining eight acacia ants falling in the P. ferrugineus subgroup. At the base of this subgroup is a poorly resolved three-way split among P. peperi, and two other lineages, here referred to as the Pseudomyrmex spinicola complex (two species) and the P. ferrugineus complex (five species). Most treatments recover P. peperi as sister to all other members of the P. ferrugineus subgroup (kmeans-partitioned ML bootstrap 100, RY-coding ML bootstrap 86, ASTRAL bootstraps 45, 54, 54), but in some analyses P. peperi is situated one node shallower, as sister to the P. ferrugineus complex (electronic supplementary material, table S3). Within the P. ferrugineus complex, there are two sister groups, with two and three species, respectively, and each sister group contains an example where one species is embedded within another, rendering the latter paraphyletic (figure 1a).
The sister group of the P. ferrugineus group is P. perboscii, a species occurring from Costa Rica to Brazil, which nests in the live branches of a wide variety of plants including Albizia, Cordia, Bombax, Pseudobombax and Macrolobium. This species is neither a dead twig inhabitant nor a specialized ant–plant mutualist; rather it is a timid, non-specific live-stem nester, that apparently does not provide protection to the plants it inhabits [30,34]. Sister to this more inclusive group of (P. perboscii + P. ferrugineus group) is a clade comprising (i) P. haytianus, an isolated species endemic to Hispaniola, and (ii) the Pseudomyrmex goeldii group, which is centred in South America (figure 1a). These more distantly related species are all generalist inhabitants of dead twigs [36].
(b). Divergence dating
Divergence dates from the BEAST analyses are similar across three alternate topologies (table 1 and figure 2; electronic supplementary material, figure S3), with slightly older ages being estimated with topology 0 (29 taxa) than topologies 1 and 2 (18 taxa). We estimate the crown age of the P. ferrugineus subgroup to be 6.7 Ma (95% highest posterior density (HPD) 9.0–4.8 Ma) with topology 0, 5.7 Ma (95% HPD: 7.9–4.0 Ma) with topology 1, and 5.7 Ma (95% HPD: 7.8–4.1 Ma) with topology 2. Equivalent estimates for the other clade of obligate acacia ants, the P. nigrocinctus subgroup, are 2.5 (3.7–1.5) Ma, 2.4 (3.9–1.3) Ma and 2.4 (3.9–1.3) Ma, respectively.
Table 1.
Estimated crown ages and 95% highest probability density (HPD) values for selected taxa in the P. ferrugineus group. (Results are taken from BEAST analyses across three alternate topologies: (0) all 29 taxa, kmeans-partitioned ML topology; (1) 18 taxa, kmeans-partitioned ML topology and (2) 18 taxa, kmeans-partitioned Bayesian (ExaBayes) topology. Topology 1 was also obtained with ASTRAL species tree analyses (figure 1b).)
| clade | topology 0 |
topology 1 |
topology 2 |
|||
|---|---|---|---|---|---|---|
| age (Ma) | 95% HPD | age (Ma) | 95% HPD | age (Ma) | 95% HPD | |
| P. ferrugineus group | 13.5 | 17.5–10.1 | 12.0 | 16.0–8.7 | 12.0 | 15.9–8.8 |
| P. ferrugineus subgroup | 6.7 | 9.0–4.8 | 5.7 | 7.9–4.0 | 5.7 | 7.8–4.1 |
| P. nigrocinctus subgroup | 2.5 | 3.7–1.5 | 2.4 | 3.9–1.3 | 2.4 | 3.9–1.3 |
| P. spinicola complex | 4.5 | 6.3–3.0 | 3.3 | 5.0–2.0 | 3.4 | 5.1–2.0 |
| P. ferrugineus complex | 4.8 | 6.5–3.3 | 4.4 | 6.1–3.0 | 4.3 | 6.0–3.0 |
Within the P. ferrugineus subgroup, the P. ferrugineus complex has estimated crown ages of 4.3–4.8 Ma, and for the P. spinicola complex these estimates are 3.3–4.5 Ma (table 1). For the two paraphyletic–monophyletic species pairs within the P. ferrugineus complex, the divergence dates are quite recent, in the order of 1.5 Ma (figure 2; electronic supplementary material, figure S3). The P. ferrugineus group as a whole appears to have a crown age of about 12–14 Ma.
(c). Biogeographic inference and trait evolution
Among the six models evaluated with BioGeoBEARS, DEC + J consistently received the highest likelihood, and provided a significantly better fit than its null model, DEC, across two alternate ant topologies and analyses (electronic supplementary material, table S4). The addition of a jump dispersal parameter always improved the models DIVALIKE and BAYAREALIKE too. Here we focus on the ancestral range estimates obtained with topology 1/analysis 2 and the DEC + J model (figure 2a), but other analytical treatments yielded broadly similar results (electronic supplementary material, figure S4).
In combination with BEAST divergence time estimates, our biogeographic analyses generate a number of well-supported inferences about the evolution of the P. ferrugineus group (figure 2a). This clade appears to have originated after dispersal of an ancestral species from South America to Mesoamerica between 12 Ma and 19 Ma, well before formation of the Isthmus of Panama [65]. This was presumably a generalist species, nesting in dead twigs and inhabiting wet tropical forest, as exemplified by the two extant species P. evitus and P. feralis. This supposition is supported by ancestral state reconstruction of habitat use and nest sites, which shows that use of open habitats and Vachellia domatia is derived (figure 3). We also uphold previous inferences [13,23] that inhabitation of domatia arose twice in this group of ants.
Figure 3.
Ancestral trait estimation for habitats and nesting sites of acacia ants and relatives. Analyses were conducted with the Ace function in APE [57]. Depicted here are the states with the highest probability under the ER (equal rates) model, the model favoured under a likelihood ratio test (electronic supplementary material, table S6). Outcomes from all three models, ER, SYM (symmetric rate) and ARD (all rates different), are shown in more detail in the electronic supplementary material, figure S6. (Online version in colour.)
We estimate a crown-group origin of the P. ferrugineus subgroup—the principal clade of obligate acacia ants—at about 6 Ma, most likely in northern Mesoamerica, and coincident with a shift to more open habitats (figures 2a and 3). This corresponds approximately to the estimated time and place of origin of domatia-bearing Vachellia (figure 2b). This was followed by dispersal and diversification of the ants and plants throughout Central America and Mexico. About 2.5 Ma the most recent common ancestor of the P. nigrocinctus subgroup appeared, most likely in southern Mesoamerica, apparently becoming a mutualist by exploiting an already existing association. The species that facilitated this was likely P. spinicola, a member of the P. ferrugineus subgroup, and by then a widespread mutualist species in southern Mesoamerica (figure 2a).
Within the P. ferrugineus complex, the two examples of species paraphyly can be explained by the times of origin and geographical distributions of the taxa. Both cases involve ‘daughter’ species (P. janzeni, P. veneficus) isolated in western Mexico, with the paraphyletic ‘parent’ species (P. ferrugineus, P. mixtecus) being more widespread in southern Mexico and northern Central America (range details in [33]). These are estimated to be recent divergences (approx. 1.5 Ma) for which there has presumably been insufficient time for complete lineage sorting in the parent species.
4. Discussion
Our findings confirm earlier reports [13,23] that the mutualistic acacia ants belong to two separate lineages, the P. ferrugineus subgroup and the P. nigrocinctus subgroup (figures 1 and 2). The initial association between ants and plants probably involved the P. ferrugineus subgroup, now comprised of eight species, which is inferred to have arisen in northern Mesoamerica at the end of the Miocene, in conjunction with a shift from closed to open habitats (figures 2 and 3). Such a shift in habitat use would have placed the ants in an environment where browsing mammals were a significant selective force on plants [20,67], arguably to a greater degree than in the ancestral closed forest habitat of these ants. The Late Miocene also corresponds to a time when drier habitats supporting more open vegetation became more widespread in the region [63,64].
The estimated crown age of the P. ferrugineus subgroup approximately matches that of the domatia-bearing Vachellia, as also observed by Chomicki et al. [13]. The P. nigrocinctus subgroup, by contrast, has an estimated crown age of only 2.5 Ma, is largely restricted to southern Mesoamerica, and has only two species. Of course, we cannot be certain when the mutualism with Vachellia arose in each lineage—the estimated stem age of the P. nigrocinctus subgroup is actually older than that of the P. ferrugineus subgroup—but the above lines of evidence from biogeography and observed species richness point to the P. ferrugineus subgroup as the initiating partner. This was also supported by stochastic character mapping of nesting traits across a comprehensive phylogeny of the entire genus Pseudomyrmex, in which domatia inhabitation was inferred to have arisen later on the branch subtending the P. nigrocinctus subgroup than on the stem of the P. ferrugineus subgroup [13].
An origin of the mutualism in northern Mesoamerica is also consistent with what is known about the phylogeny and distribution of Vachellia [13,68]. Based on current understanding, Vachellia chiapensis is sister to all other domatia-bearing congeners and is confined to northern Mesoamerica, as is the next-branching species, Vachellia mayana. Only in shallower parts of the tree do we find species that occur in southern Mesoamerica, and most of these are also found in northern regions. As expected, ancestral range inference with BioGeoBEARS supports northern Mesoamerica as the origin for domatia-bearing Vachellia, both with the DEC + J model (analysis 2) (figure 2b) and under alternate models (electronic supplementary material, figure S5 and table S5). It is worth emphasizing, however, that there is little evidence for strict co-cladogenesis between Vachellia and Pseudomyrmex [13,33]. There has been extensive host-plant switching and expansion by the ants, to the point where most acacia ants now occupy any species of domatia-bearing Vachellia occurring within their distribution range.
There is strong convergence between the P. ferrugineus subgroup and the P. nigrocinctus subgroup in several ‘classic’ traits associated with the ant/acacia mutualism [20]: (i) the association is obligate, i.e. the acacia ants nest only in Vachellia domatia; (ii) the workers are aggressive and sting much more readily than generalist twig-inhabiting species of Pseudomyrmex; (iii) the workers patrol the plants constantly; (iv) the ant colonies subsist on harvested Beltian bodies and extrafloral nectar, i.e. the workers are not generalist scavengers like most species of Pseudomyrmex; and (v) the workers have smaller eyes and more slender profemora than related non-mutualistic species [33,36].
This convergence is all the more striking because none of these traits are exhibited by the two generalist species, P. evitus and P. feralis, which are interpolated phylogenetically between the two acacia ant lineages. These generalists are timid, large-eyed, diurnal species that nest opportunistically in dead twigs and have no association with domatia-bearing Vachellia [36]. Although worker and queen morphology is different from that of the acacia ants, the male genitalia suggest an affinity to other members of the P. ferrugineus group [33,36], and this relationship is confirmed by the sequence data.
Despite these remarkable convergences there are some differences between the two groups of acacia ants. Workers of P. nigrocinctus and Pseudomyrmex particeps are smaller in size and more slender in body form than most species in the P. ferrugineus subgroup [33], and there are hints of differences in behaviour. The gaster is held straight by workers in the P. nigrocinctus subgroup, for example, whereas it is often curled forward by workers in the P. ferrugineus subgroup [69] (figure 1a), although the significance of this behaviour is unclear. At least three species in the P. ferrugineus subgroup possess derived physiological traits (reduced invertase and protease activity in adult workers) that adapt them to the nutritional rewards of their Vachellia hosts, an arrangement that evidently buffers the mutualism against cheaters [23,26,28,70]. We do not know the extent to which these traits are manifested in the P. nigrocinctus subgroup. Thus there is considerable scope for probing in greater detail the similarities and differences between these two groups of ant–plant specialists, and this could provide insight into the most essential elements of the symbiosis.
A recent analysis of genome evolution in mutualist and non-mutualist species of Pseudomyrmex demonstrated convergent increases in rates of gene evolution in the mutualists [15]. This included a comparison between a species in the P. ferrugineus subgroup, Pseudomyrmex flavicornis, and the non-mutualist species, P. feralis (formerly called P. psw054). In our trees, we also find that P. feralis manifests consistently shorter branch lengths than its sister group, the P. ferrugineus subgroup (figure 1; electronic supplementary material, figure S2), but this appears not to be the case when the comparison is extended to the other closely related non-mutualist, P. evitus. Both species, P. feralis and P. evitus, deserve greater scrutiny: although they are timid species nesting in dead twigs, we know little else about their biology and they are infrequently encountered, suggesting that they may have other unusual or specialized characteristics.
Finally, the identification of P. perboscii, a generalized live-stem nesting ant, as the sister group of the P. ferrugineus group highlights the possible significance of this habit as a precursor to the development of more specialized ant/plant relationships [30]. It suggests the possibility that acacia ants evolved in a clade with a predisposition towards nesting in live plant cavities. This hypothesis could be explored by genetic comparisons of this larger clade (P. ferrugineus group + P. perboscii) with other related Pseudomyrmex clades that are strictly dead twig inhabitants, such as the P. goeldii group.
5. Conclusion
We investigate the evolutionary history behind the iconic ant/plant mutualism involving the P. ferrugineus group and swollen-thorn acacias in the genus Vachellia. Our results indicate that the mutualism is relatively young, having developed in the Late Miocene in northern Mesoamerica in a clade of ants, the P. ferrugineus subgroup, that shifted from closed to open environments, and from timid to aggressive behaviour. Sometime after this—approximately 3 Myr later—a second clade of ants, the P. nigrocinctus subgroup, independently evolved a mutualistic relationship with domatia-bearing Vachellia, apparently taking advantage of an already existing association. The arena for this second event was most likely southern Mesoamerica, to which members of the now-diversifying P. ferrugineus subgroup had dispersed. Convergence among these two groups of ants can be contrasted with what appears to be a single origin of domatia in the plants [13]. Despite being separated phylogenetically by non-mutualistic (dead twig-inhabiting) species the two clades of ant mutualists are nevertheless relatively closely related, sharing a common ancestor about 13 Ma, in contrast with several more distantly related congeners that have become obligate non-protective parasites of Vachellia, each independently of the other [13,33]. This suggests that there might be features of the P. ferrugineus group as a whole that favoured the development of mutualistic interactions. Additional comparisons at multiple phylogenetic depths can help to illuminate the historical context of these interactions, and the factors predisposing the development of either mutualistic or antagonistic relationships with the plants. The Pseudomyrmex/Vachellia system and other ant/plant mutualisms [7–18] add to a growing body of evidence for convergent evolution of complex multispecies interactions [6], tempered by particular ecological, phylogenetic and geographical conditions.
Supplementary Material
Supplementary Material
Acknowledgements
We are grateful to the following individuals for provision of ant specimens: Paulo Bettella, Lloyd Davis, Alain Dejean, Fernando Fernández, Stefanie Kautz, Jack Longino and William Mackay. We thank Marek Borowiec, Brendon Boudinot and Matthew Prebus for laboratory assistance and troubleshooting; the Brazelton Laboratory (University of Utah) for use of their Linux server; Guillaume Chomicki for sharing the Vachellia chronogram; and Jack Longino for stimulating discussion and support. We also thank Brian Dalley and Brett Milash from the University of Utah Genomics and Bioinformatics Core for assistance with sequencing. Phylogenetic analyses were performed using either the CIPRES Science Gateway or the Smithsonian Institution's high-performance computer cluster (Hydra). Guillaume Chomicki and two anonymous reviewers provided helpful comments that improved the manuscript.
Data accessibility
Raw sequence reads and Trinity contig assemblies representing UCE loci are available from the NCBI Sequence Read Archive (SRA) and GenBank, respectively (NCBI BioProject PRJNA357470). The concatenated matrix of 1672 aligned and trimmed loci and accompanying tree files are available from TreeBASE (accession S20346). Additional data, including alignments, alignment supermatrices, tree files, matrix partitioning files, BEAST xml files, trait and biogeography data files, and tables are available on Dryad: http://dx.doi.org/10.5061/dryad.3d31q [71].
Authors' contributions
P.S.W. coordinated the study, collected field samples and participated in the generation and analysis of data. M.G.B. participated in data generation and carried out phylogenetic and bioinformatic analyses. P.S.W. and M.G.B. co-wrote the paper.
Competing interests
We declare that we have no competing interests.
Funding
This research is part of the ADMAC (Ant Diversity of the Mesoamerican Corridor) Project supported by NSF grant no. DEB-1354996.
References
- 1.Yoder JB, Smith CI, Pellmyr O. 2010. How to become a yucca moth: minimal trait evolution needed to establish the obligate pollination mutualism. Biol. J. Linn. Soc. 100, 847–855. ( 10.1111/j.1095-8312.2010.01478.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cruaud A, et al. 2012. An extreme case of plant-insect codiversification: figs and fig-pollinating wasps. Syst. Biol. 61, 1029–1047. ( 10.1093/sysbio/sys068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray EA, Carmichael AE, Heraty JM. 2013. Ancient host shifts followed by host conservatism in a group of ant parasitoids. Proc. R. Soc. B 208, 20130495 ( 10.1098/rspb.2013.0495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segar ST, Pereira RAS, Compton SG, Cook JM. 2013. Convergent structure of multitrophic communities over three continents. Ecol. Lett. 16, 1436–1445. ( 10.1111/ele.12183) [DOI] [PubMed] [Google Scholar]
- 5.Weiblen GD, Treiber EL. 2015. Evolutionary origins and diversification of mutualism. In Mutualism (ed. Bronstein JL.), pp. 37–56. Oxford, UK: Oxford University Press. [Google Scholar]
- 6.Bittleston LS, Pierce NE, Ellison AM, Pringle A. 2016. Convergence in multispecies interactions. Trends Ecol. Evol. 31, 269–280. ( 10.1016/j.tree.2016.01.006) [DOI] [PubMed] [Google Scholar]
- 7.Dunn RR, Gove AD, Barraclough TG, Givnish TJ, Majer JD. 2007. Convergent evolution of an ant-plant mutualism across plant families, continents, and time. Evol. Ecol. Res. 9, 1349–1362. [Google Scholar]
- 8.Quek S-P, Davies SJ, Itino T, Pierce NE. 2004. Codiversification in an ant-plant mutualism: stem texture and the evolution of host use in Crematogaster (Formicidae: Myrmicinae) inhabitants of Macaranga (Euphorbiaceae). Evolution 58, 554–570. ( 10.1554/03-361) [DOI] [PubMed] [Google Scholar]
- 9.Pringle EG, Ramírez SR, Bonebrake TC, Gordon DM, Dirzo R. 2012. Diversification and phylogeographic structure in widespread Azteca plant-ants from the northern Neotropics. Mol. Ecol. 21, 3576–3592. ( 10.1111/j.1365-294X.2012.05618.x) [DOI] [PubMed] [Google Scholar]
- 10.Weber MG, Agrawal AA. 2014. Defense mutualisms enhance plant diversification. Proc. Natl Acad. Sci. USA 111, 16 442–16 447. ( 10.1073/pnas.1413253111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez A. 2015. Fidelity and promiscuity in an ant-plant mutualism: a case study of Triplaris and Pseudomyrmex. PLoS ONE 10, e0143535 ( 10.1371/journal.pone.0143535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomicki G, Renner SS. 2015. Phylogenetics and molecular clocks reveal the repeated evolution of ant-plants after the late Miocene in Africa and the early Miocene in Australasia and the Neotropics. New Phytol. 207, 411–424. ( 10.1111/nph.13271) [DOI] [PubMed] [Google Scholar]
- 13.Chomicki G, Ward PS, Renner SS. 2015. Macroevolutionary assembly of ant/plant symbioses: Pseudomyrmex ants and their ant-housing plants in the Neotropics. Proc. R. Soc. B 282, 20152200 ( 10.1098/rspb.2015.2200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Treiber EL, Gaglioti AL, Romaniuc-Neto S, Madriñán S, Weiblen GD. 2016. Phylogeny of the Cecropieae (Urticaceae) and the evolution of an ant-plant mutualism. Syst. Bot. 41, 56–66. ( 10.1600/036364416X690633) [DOI] [Google Scholar]
- 15.Rubin BER, Moreau CS. 2016. Comparative genomics reveals convergent rates of evolution in ant–plant mutualisms. Nat. Commun. 7, 12679 ( 10.1038/ncomms12679) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler WM. 1942. Studies of Neotropical ant-plants and their ants. Bull. Mus. Comp. Zool. 90, 1–262. [Google Scholar]
- 17.Jolivet P. 1996. Ants and plants. An example of coevolution. Leiden, The Netherlands: Backhuys Publishers. [Google Scholar]
- 18.Davidson DW, McKey D. 1993. The evolutionary ecology of symbiotic ant-plant relationships. J. Hymenopt. Res. 2, 13–83. [Google Scholar]
- 19.Safford WE. 1922. Ant acacias and acacia ants of Mexico and Central America. Annu. Rep. Smithson. Inst. 1921, 381–394. [Google Scholar]
- 20.Janzen DH. 1966. Coevolution of mutualism between ants and acacias in Central America. Evolution 20, 249–275. ( 10.2307/2406628) [DOI] [PubMed] [Google Scholar]
- 21.Janzen DH. 1967. Interaction of the bull's-horn acacia (Acacia cornigera L.) with an ant inhabitant (Pseudomyrmex ferruginea F. Smith) in eastern Mexico. Univ. Kansas Sci. Bull. 47, 315–558. [Google Scholar]
- 22.Janzen DH. 1973. Evolution of polygynous obligate acacia-ants in western Mexico. J. Anim. Ecol. 42, 727–750. ( 10.2307/3134) [DOI] [Google Scholar]
- 23.Kautz S, Lumbsch HT, Ward PS, Heil M. 2009. How to prevent cheating: a digestive specialization ties mutualistic plant-ants to their ant-plant partners. Evolution 63, 839–853. ( 10.1111/j.1558-5646.2008.00594.x) [DOI] [PubMed] [Google Scholar]
- 24.Janzen DH. 1975. Pseudomyrmex nigropilosa: a parasite of a mutualism. Science 188, 936–937. ( 10.1126/science.188.4191.936) [DOI] [PubMed] [Google Scholar]
- 25.Clement LW, Köppen SCW, Brand WA, Heil M. 2008. Strategies of a parasite of the ant-Acacia mutualism. Behav. Ecol. Sociobiol. 62, 953–962. ( 10.1007/s00265-007-0520-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heil M, Orona-Tamayo D, Eilmus S, Kautz S, González-Tauber M. 2010. Chemical communication and coevolution in an ant–plant mutualism. Chemoecology 20, 63–74. ( 10.1007/s00049-009-0036-4) [DOI] [Google Scholar]
- 27.Orona-Tamayo D, Wielsch N, Blanco-Labra A, Svatos A, Farías-Rodríguez R, Heil M. 2013. Exclusive rewards in mutualisms: ant proteases and plant protease inhibitors create a lock–key system to protect Acacia food bodies from exploitation. Mol. Ecol. 22, 4087–4100. ( 10.1111/mec.12320) [DOI] [PubMed] [Google Scholar]
- 28.Heil M, Barajas-Barron A, Orona-Tamayo D, Wielsch N, Svatos A. 2014. Partner manipulation stabilises a horizontally transmitted mutualism. Ecol. Lett. 17, 185–192. ( 10.1111/ele.12215) [DOI] [PubMed] [Google Scholar]
- 29.Ward PS. 1999. Systematics, biogeography and host plant associations of the Pseudomyrmex viduus group (Hymenoptera: Formicidae), Triplaris- and Tachigali-inhabiting ants. Zool. J. Linn. Soc. 126, 451–540. ( 10.1111/j.1096-3642.1999.tb00157.x) [DOI] [Google Scholar]
- 30.Ward PS. 1991. Phylogenetic analysis of pseudomyrmecine ants associated with domatia-bearing plants. In Ant-plant interactions (eds Huxley CR, Cutler DF), pp. 335–352. Oxford, UK: Oxford University Press. [Google Scholar]
- 31.Faircloth BC, McCormack JE, Crawford NG, Harvey MG, Brumfield RT, Glenn TC. 2012. Ultraconserved elements anchor thousands of genetic markers spanning multiple evolutionary timescales. Syst. Biol. 61, 717–726. ( 10.1093/sysbio/sys004) [DOI] [PubMed] [Google Scholar]
- 32.Faircloth BC, Branstetter MG, White ND, Brady SG. 2015. Target enrichment of ultraconserved elements from arthropods provides a genomic perspective on relationships among Hymenoptera. Mol. Ecol. Resour. 15, 489–501. ( 10.1111/1755-0998.12328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward PS. 1993. Systematic studies on Pseudomyrmex acacia-ants (Hymenoptera: Formicidae: Pseudomyrmecinae). J. Hymenopt. Res. 2, 117–168. [Google Scholar]
- 34.Ward PS. 1989. Systematic studies on pseudomyrmecine ants: revision of the Pseudomyrmex oculatus and P. subtilissimus species groups, with taxonomic comments on other species. Quaest. Entomol. 24, 393–468. [Google Scholar]
- 35.Ward PS, Downie DA. 2005. The ant subfamily Pseudomyrmecinae (Hymenoptera: Formicidae): phylogeny and evolution of big-eyed arboreal ants. Syst. Entomol. 30, 310–335. ( 10.1111/j.1365-3113.2004.00281.x) [DOI] [Google Scholar]
- 36.Ward PS. In press A review of the Pseudomyrmex ferrugineus and Pseudomyrmex goeldii species groups: acacia-ants and their relatives (Hymenoptera: Formicidae). Zootaxa 4227, 524–542. ( 10.11646/zootaxa.4227.4.3) [DOI] [PubMed] [Google Scholar]
- 37.Rohland N, Reich D. 2012. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 22, 939–946. ( 10.1101/gr.128124.111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glenn TC, et al. 2016. Adapterama I: universal stubs and primers for thousands of dual-indexed Illumina libraries (iTru & iNext). bioRxiv. ( 10.1101/049114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Branstetter MG, Longino JT, Ward PS, Faircloth BC. In press. Enriching the ant tree of life: enhanced UCE bait set for genome-scale phylogenetics of ants and other Hymenoptera. Methods Ecol. Evol. ( 10.1111/2041-210X.12742) [DOI] [Google Scholar]
- 40.Faircloth BC. 2016. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32, 786–788. ( 10.1093/bioinformatics/btv646) [DOI] [PubMed] [Google Scholar]
- 41.Faircloth BC. 2013. Illumiprocessor: a trimmomatic wrapper for parallel adapter and quality trimming. See 10.6079/J9ILL (accessed 4 November 2016). [DOI]
- 42.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. ( 10.1093/nar/gkf436) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552. ( 10.1093/oxfordjournals.molbev.a026334) [DOI] [PubMed] [Google Scholar]
- 46.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proc. of the Gateway Computing Environments Workshop (GCE), 14 November 2010, pp. 1–8. New Orleans, LA, USA: Association for Computing Machinery.
- 47.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. ( 10.1093/bioinformatics/btu033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. ( 10.1093/molbev/mss020) [DOI] [PubMed] [Google Scholar]
- 49.Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 14, 82 ( 10.1186/1471-2148-14-82) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Calcott B. 2016. PartitionFinder 2. See https://github.com/brettc/partitionfinder (accessed 4 November 2016).
- 51.Frandsen PB, Calcott B, Mayer C, Lanfear R. 2015. Automatic selection of partitioning schemes for phylogenetic analyses using iterative k-means clustering of site rates. BMC Evol. Biol. 15, 13 ( 10.1186/s12862-015-0283-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aberer AJ, Kobert K, Stamatakis A. 2014. ExaBayes: massively parallel Bayesian tree inference for the whole-genome era. Mol. Biol. Evol. 31, 2553–2556. ( 10.1093/molbev/msu236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v 1.6. See http://beast.bio.ed.ac.uk/Tracer (accessed 4 November 2016).
- 54.Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T. 2014. ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30, i541–i548. ( 10.1093/bioinformatics/btu462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo TK. 2008. Calculating bootstrap probabilities of phylogeny using multilocus sequence data. Mol. Biol. Evol. 25, 960–971. ( 10.1093/molbev/msn043) [DOI] [PubMed] [Google Scholar]
- 56.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973. ( 10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. ( 10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 58.Matzke NJ. 2014. Model selection in historical biogeography reveals that founder-event speciation is a crucial process in island clades. Syst. Biol. 63, 951–970. ( 10.1093/sysbio/syu056) [DOI] [PubMed] [Google Scholar]
- 59.Matzke NJ. 2013. BioGeoBEARS: biogeography with Bayesian (and likelihood) evolutionary analysis in R scripts. Berkeley, CA: University of California. [Google Scholar]
- 60.Ree RH, Smith SA. 2008. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst. Biol. 57, 4–14. ( 10.1080/10635150701883881) [DOI] [PubMed] [Google Scholar]
- 61.Ronquist F. 1997. Dispersal-vicariance analysis: a new approach to the quantification of historical biogeography. Syst. Biol. 46, 195 ( 10.2307/2413643) [DOI] [Google Scholar]
- 62.Landis MJ, Matzke NJ, Moore BR, Huelsenbeck JP. 2013. Bayesian analysis of biogeography when the number of areas is large. Syst. Biol. 62, 789–804. ( 10.1093/sysbio/syt040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burnham RJ, Graham A, Burnham J. 1999. The history of Neotropical vegetation: new developments and status. Ann. Mo. Bot. Gard. 86, 546–589. ( 10.2307/2666185) [DOI] [Google Scholar]
- 64.Graham A. 2010. Late Cretaceous and Cenozoic history of Latin American vegetation and terrestrial environments. St. Louis, MO: Missouri Botanical Garden Press. [Google Scholar]
- 65.O'Dea A, et al. 2016. Formation of the Isthmus of Panama. Sci. Adv. 2, 1–12. ( 10.1126/sciadv.1600883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Global Biodiversity Information Facility. 2016. GBIF. Global biodiversity information facility. See www.gbif.org (accessed 25 August 2016).
- 67.Brown WL., Jr 1960. Ants, acacias and browsing mammals. Ecology 41, 587–592. ( 10.2307/1933346) [DOI] [Google Scholar]
- 68.Gómez-Acevedo S, Rico-Arce L, Delgado-Salinas A, Magallón S, Eguiarte LE. 2010. Neotropical mutualism between Acacia and Pseudomyrmex: phylogeny and divergence times. Mol. Phylogenet. Evol. 56, 393–408. ( 10.1016/j.ympev.2010.03.018) [DOI] [PubMed] [Google Scholar]
- 69.Janzen DH. 1983. Pseudomyrmex ferruginea (hormiga del cornizuelo, acacia-ant). In Costa Rican natural history (ed. Janzen DH.), pp. 762–764. Chicago, IL: University of Chicago Press. [Google Scholar]
- 70.Heil M, Rattke J, Boland W. 2005. Postsecretory hydrolysis of nectar sucrose and specialization in ant/plant mutualism. Science 308, 560–563. ( 10.1126/science.1107536) [DOI] [PubMed] [Google Scholar]
- 71.Ward PS, Branstetter MG. 2017. Data from: The acacia ants revisited: convergent evolution and biogeographic context in an iconic ant/plant mutualism. Dryad Digital Repository. ( 10.5061/dryad.3d31q) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Ward PS, Branstetter MG. 2017. Data from: The acacia ants revisited: convergent evolution and biogeographic context in an iconic ant/plant mutualism. Dryad Digital Repository. ( 10.5061/dryad.3d31q) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Raw sequence reads and Trinity contig assemblies representing UCE loci are available from the NCBI Sequence Read Archive (SRA) and GenBank, respectively (NCBI BioProject PRJNA357470). The concatenated matrix of 1672 aligned and trimmed loci and accompanying tree files are available from TreeBASE (accession S20346). Additional data, including alignments, alignment supermatrices, tree files, matrix partitioning files, BEAST xml files, trait and biogeography data files, and tables are available on Dryad: http://dx.doi.org/10.5061/dryad.3d31q [71].