Abstract
Like many aquatic vertebrates, whales have blue-shifting spectral tuning substitutions in the dim-light visual pigment, rhodopsin, that are thought to increase photosensitivity in underwater environments. We have discovered that known spectral tuning substitutions also have surprising epistatic effects on another function of rhodopsin, the kinetic rates associated with light-activated intermediates. By using absorbance spectroscopy and fluorescence-based retinal release assays on heterologously expressed rhodopsin, we assessed both spectral and kinetic differences between cetaceans (killer whale) and terrestrial outgroups (hippo, bovine). Mutation experiments revealed that killer whale rhodopsin is unusually resilient to pleiotropic effects on retinal release from key blue-shifting substitutions (D83N and A292S), largely due to a surprisingly specific epistatic interaction between D83N and the background residue, S299. Ancestral sequence reconstruction indicated that S299 is an ancestral residue that predates the evolution of blue-shifting substitutions at the origins of Cetacea. Based on these results, we hypothesize that intramolecular epistasis helped to conserve rhodopsin's kinetic properties while enabling blue-shifting spectral tuning substitutions as cetaceans adapted to aquatic environments. Trade-offs between different aspects of molecular function are rarely considered in protein evolution, but in cetacean and other vertebrate rhodopsins, may underlie multiple evolutionary scenarios for the selection of specific amino acid substitutions.
Keywords: protein structure–function, epistasis, retinal release, meta II stability, spectral tuning
1. Introduction
Cetaceans (whales, dolphins, porpoises) are obligatorily aquatic mammals that descended from terrestrial ancestors, and so are regarded as one of the most stunning examples of macroevolutionary transitions. In recent years, several whole genome projects have provided motivation for computational investigations into the molecular variation underlying cetacean sensory and anatomical adaptations [1]. Though computationally detected selection patterns have generated many hypotheses concerning protein evolution, they are only just beginning to be subjected to experimental testing (e.g. [2,3]). Nevertheless, this approach has enormous potential for linking the genotypic foundations of the terrestrial–aquatic transition with adaptive phenotypic changes in protein function.
The visual pigment opsins remain one of the best model systems for comparative studies of the relationship between sequence variation and protein function in vertebrates [4–6]. The dim-light pigment, rhodopsin, in particular has served as a model system for the study of G protein-coupled receptors (GPCRs), and was the first for which a high resolution X-ray crystal structure could be determined [7]. The rhodopsin pigment itself comprises a heptahelical transmembrane protein moiety with a central pocket that houses a covalently bound chromophore, 11-cis retinal, derived from vitamin A [7]. Rhodopsin activation is triggered by light-induced isomerization of the chromophore to all-trans retinal, which results in a rapid series of conformational shifts through several intermediates. The biologically active intermediate, metarhodopsin II (meta II) [8], binds the G protein transducin, and initiates the phototransduction cascade within the rod photoreceptor cell [9]. Eventually, hydrolysis of the covalent Schiff base linkage releases the all-trans retinal, which must be replaced by a new 11-cis molecule for rhodopsin to regain photosensitivity [9].
Specializations in dim-light vision are thought to have played a major role in the evolution of many vertebrate lineages adapting to nocturnal [10], aquatic [11], and fossorial lifestyles [12], but by far the majority of visual pigment evolutionary studies have centred on spectral tuning of the peak absorbance wavelength (λmax). Blue-shifts in the visual pigments of aquatic vertebrates are thought to allow maximum photon capture in the more blue-shifted light environments found underwater [13,14]. A number of studies have suggested specific sites may be responsible for these shifts. In particular, the role of sites 83, 292 and 299 in spectral blue-shifts was first established in studies of the rhodopsin pigments from deep-dwelling cottoid fish in Lake Baikal [14], and the same substitutions have convergently evolved in several deep-sea teleost fish lineages [11]. These three sites were later identified as under positive and divergent selection in cetaceans [3,15], and site-directed mutagenesis experiments in both bovine and killer whale rhodopsin showed they are responsible for the majority of spectral tuning variation observed among experimentally characterized cetacean rhodopsins [3,16].
For two of the sites (83 and 299), however, the effect on spectral tuning in the killer whale rhodopsin background was found to be minimal (less than 2 nm) [3]. Moreover, recent studies that have mutated spectral tuning sites in other rhodopsins including the great bowerbird [17], echidna [18] and a variety of African cichlids [19,20], have demonstrated that functional shifts caused by the same substitution are not always consistent across species. This finding agrees with studies that show the functional effects of amino acid substitutions are often context-dependent due to intramolecular epistasis, which arises from among-site interactions within the protein that can varyingly suppress or exaggerate the functional effects of the same substitution in different backgrounds [21]. Such effects can lead to potentially misleading cases where parallel or convergent molecular changes do not necessarily correspond to convergent functional shifts. Cetacean rhodopsin may be one of these cases, where the evolutionary importance of 83 and 299 to spectral tuning in the lineage has been overestimated due to the more pronounced effects of those sites in other vertebrates.
Only in recent years have functional aspects of rhodopsin, aside from spectral tuning, been explored experimentally using comparative approaches [17,20,22]. Site 83 has experienced a parallel D83N substitution in several vertebrate groups (including cetaceans) where it has been associated with blue-shifts in absorption maxima [11,14,16]. However, a key study from Sugawara et al. [20] showed that the N83 residue also accelerates formation of the active meta II intermediate, a shift the authors hypothesized was an adaptation to improve dim-light photosensitivity. This acceleration may be the result of a more stable meta II intermediate, as evidenced by reduced rates of all-trans retinal release following light activation in vitro in association with N83 [17,18]. Site 83 may thus have an adaptive role in both the spectral tuning and activation kinetics of vertebrate rhodopsins. In cetaceans, because sites 83 and 299 seem to have minor spectral tuning roles [3], non-spectral selection pressures may have influenced their evolution. Unlike site 83, however, the impact of site 299 on metarhodopsin kinetics has not been experimentally investigated. The evolutionary importance of these functions in cetaceans is also unknown as, until now, there have been no experimental studies of light-activated kinetic rates in any cetacean visual pigment.
We used the killer whale rhodopsin in vitro expression system established in our previous work [3] to determine whether retinal release rates differ between cetaceans and their nearest living outgroups, Hippopotamidae and Ruminantia (represented by the hippo, Hippopotamus amphibius, and the bovine, Bos taurus). If both spectral and kinetic functional shifts are important for aquatic visual adaptation, we would expect killer whale rhodopsin to be functionally distinguishable from terrestrial outgroups. To investigate the mechanisms of underlying functional differences between species, we used site-directed mutagenesis and retinal release fluorescence assays to compare the effects of sites 83, 292 and 299 on rhodopsin light-activated kinetics. Lastly, to investigate whether functional differences arising from these substitutions support ancestral increases in underwater photosensitivity, we performed ancestral sequence reconstruction for sites 83, 292 and 299 on a cetartiodactyl species tree. The results of our study support the idea that spectral blue-shifts are a derived cetacean trait, while slower retinal release kinetics are an ancestral cetartiodactyl trait. We show that shifts in both aspects of function can arise from substitutions that occurred on the ancestral cetacean branch (D83N, A292S), and that the kinetic effect of D83N in killer whale depends on a specific epistatic interaction with site 299. Our study provides new insight into how visual pigment evolution may reflect trade-offs between different aspects of rhodopsin function, and that these trade-offs may be mediated by specific structural interactions among amino acid residues in the protein.
2. Results
(a). Killer whale rhodopsin is blue-shifted, but both killer whale and hippo have greater metarhodopsin II stability than bovine rhodopsin
The wild-type rhodopsin coding sequences from killer whale, bovine and hippo were expressed and purified in vitro; the resulting killer whale pigment had a peak spectral absorbance (λmax) at 487 nm as reported previously [3], while both the bovine and hippo were 499 nm (figure 1a and table 1). This is the first in vitro measurement of λmax in hippo rhodopsin, though our bovine measurement is consistent with prior literature [10,11,17]. Because hippopotamids are the closest living relatives to cetaceans, this comparison establishes the killer whale blue-shift as a derived cetacean trait. To investigate meta II stability, we used a fluorescence assay to measure the rate of all-trans retinal release from rhodopsin following light activation [24]. Our half-life estimate for bovine rhodopsin (approx. 15 min, table 1) agreed with prior literature [17,22,24]. However, in contrast to spectral tuning, both the killer whale and hippo retinal release half-lives were significantly longer (approx. 23 min) than bovine (one-tail t-tests: t = 11.05, d.f. = 19, p < 0.001 for killer whale and t = 7.61, d.f. = 17, p < 0.001 for hippo), but were not significantly different from each other (one-tail t-test: t = 0.12, d.f. = 4, p = 0.91) (figure 1b and table 1).
Figure 1.
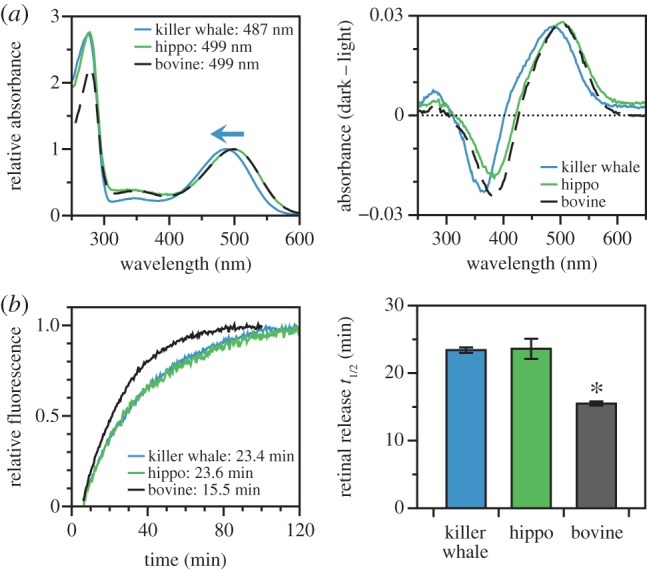
Functional characteristics of wild-type killer whale and hippo rhodopsin, as compared with bovine rhodopsin. (a) Spectral absorbance curves of dark state rhodopsin (left) and dark–light difference spectra (right). Indicated spectral peaks (λmax) were estimated according to the curve-fitting methodology of Govardovskii et al. [23] showing standard vertebrate peaks for the bovine and hippo, but a blue-shifted peak for the killer whale. (b) Fluorescence assays of retinal release rates following light activation of rhodopsin, with indicated half-lives estimated by fitting time courses to first-order exponential curves (left) where the killer whale has a half-life similar to the hippo, but significantly greater than the bovine (right).
Table 1.
Retinal release half-lives and spectral tuning of rhodopsins expressed in vitro.
| species | mutant | 83, 292, 299a | t1/2 (min) | λmax(nm) |
|---|---|---|---|---|
| B. taurus | WT | D A A | 15.5 ± 0.3 (17) | 498.9 |
| B. taurus | D83N | N A Ab | 27.3 ± 0.9 (3) | 494.7 |
| B. taurus | A292S | D S Ab | 11.4 ± 0.5 (3) | 489.3 |
| B. taurus | A299S | D A S | 18.1 ± 0.1 (2) | 500.4 |
| B. taurus | D83N/A292S | N S Ab | 20.5 ± 1.3 (3) | 485.2 |
| B. taurus | D83N/A299S | N A S | 33.0 ± 2.2 (2) | 496.1 |
| H. amphibius | WT | D A S | 23.6 ± 1.5 (2) | 498.7 |
| O. orca | WT | N S S | 23.4 ± 0.4 (4) | 486.8 |
| O. orca | N83D | D S S | 20.7 ± 1.5 (3) | 489.3 |
| O. orca | S292A | N A S | 24.4 ± 1.2 (3) | 497.6 |
| O. orca | S299A | N S A | 18.6 ± 0.4 (3) | 484.9 |
| O. orca | N83D/S292A | D A S | 22.6 ± 1.1 (2) | 500.7 |
| O. orca | N83D/S299A | D S A | 12.3 ± 0.3 (2) | 488.1 |
a83, 292 and 299 refer to the amino acid positions of the rhodopsin protein sequence. The amino acid identities are indicated with standard abbreviations (A, alanine; D, aspartic acid; N, asparagine; S, serine). λmax values are the mean of three dark spectrum measurements (at most ±0.5 nm), and retinal release half-lives are ±standard error with sample size in brackets.
bThese pigments include data points published in van Hazel et al. [19].
(b). Killer whale metarhodopsin II stability is affected by magnitude epistasis involving sites 83 and 299
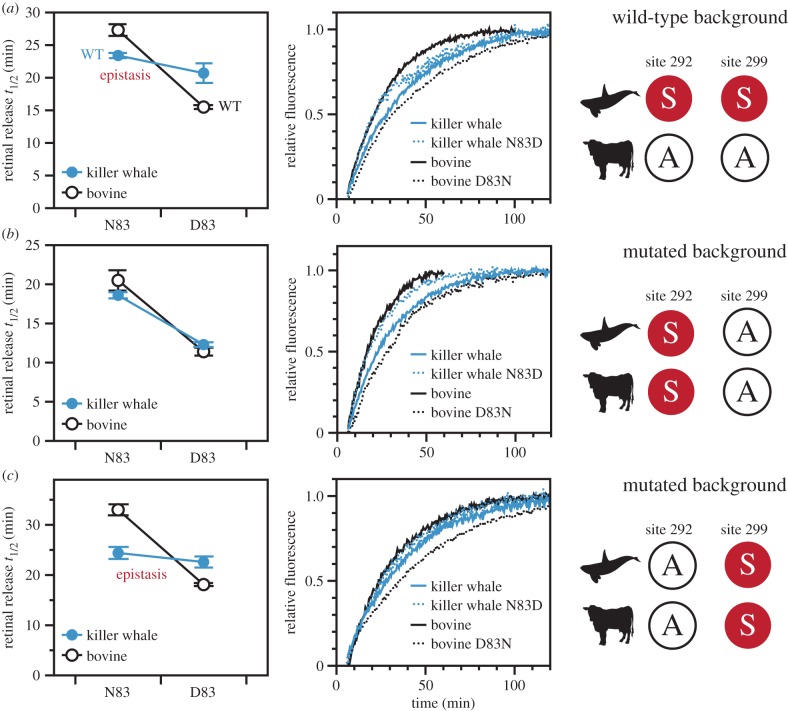
When we mutated site 83 in the wild-type killer whale and bovine rhodopsin backgrounds, the resulting shifts to the retinal release rate were directionally consistent, but differed greatly in magnitude (figure 2a and table 1). The D83N substitution in the bovine background increased the retinal release half-life by approximately 12 min, but N83D in killer whale reduced the half-life only by approximately 3 min. This result is especially surprising given that in other mammals and birds N83D tends to reduce half-lives to a greater extent, between 6 and 15 min [17,18]. To statistically confirm the magnitude epistasis between the species, we used a two-way factorial ANOVA to show that a significant amount of the variance in our retinal release half-life estimates could be explained by an interaction effect between site 83 and species background (F1,8 = 24.2, p = 0.00116) (electronic supplementary material, table S1). The main effect from the site 83 residue itself was also significant (F1,8 = 68.5, p = 0.00003). Spectral shifts showed a similar pattern to the retinal release half-lives, where N83D caused a smaller change to killer whale λmax relative to D83N in bovine (a 2 nm red-shift versus a 4 nm blue-shift, table 1).
Figure 2.
The effect of site 83 on retinal release in different mutant backgrounds of killer whale rhodopsin, as compared with bovine rhodopsin. (a) Mutations at site 83 conferred a significant shift in retinal release half-life under a bovine background, but not in a killer whale background; an example of magnitude epistasis. (b) When amino acid identities at sites 292 and 299 were mutated to be identical between species, there was no longer any epistasis under an S292/A299 background. Both killer whale and bovine had similar half-lives with N83 being significantly greater than D83. (c) However, there was still an epistatic pattern under an A292/S299 background. For ANOVA statistics see electronic supplementary material, tables S1–S5.
Killer whale and bovine rhodopsin have different residues at 292 and 299, so these sites are potential candidates for explaining the epistasis associated with killer whale site 83. To experimentally investigate this hypothesis, we used site-directed mutagenesis to engineer identical residues at sites 292/299 in both killer whale and bovine rhodopsins, and then re-evaluated how site 83 shifted retinal release rates under these conditions. First, we tested the S292/A299 combination (achieved by mutating killer whale S299A and bovine A292S) and found that, surprisingly, there was no longer any evidence for epistatic effects at site 83 (interaction effect: F1,4 = 2.28, p = 0.20539, electronic supplementary material, table S2). In these mutated backgrounds, N83D decreased the killer whale retinal release half-life by about 7 min, while D83N caused a similar increase in bovine by about 9 min (site 83 main effect: F1,4 = 385, p = 0.00004) (figure 2b and table 1). N83D shifted the retinal release rate by a greater amount in this killer whale S299A mutant than in the killer whale wild-type. Therefore, S299 is a likely culprit for explaining the unusual resilience of the wild-type killer whale retinal release rate to N83D.
These patterns were in stark contrast to our investigations in an A292/S299 background (achieved by mutating killer whale S292A and bovine A299S), which produced results comparable to the unmutated wild-type backgrounds (figure 2c and table 1; electronic supplementary material, table S3). Site 292 thus seems not to contribute significantly to the epistasis associated with site 83 in killer whale rhodopsin retinal release, an unexpected result given the recognized importance of 292 in spectral tuning. For both 292 and 299, spectral tuning shifts were fairly consistent between species; bovine A292S resulted in a 10 nm blue-shift and killer whale S292A resulted in a 10 nm red-shift, while site 299 caused minimal shifts in both backgrounds (1–2 nm) (table 1). Unlike site 83, neither of these sites showed a prominent epistatic effect on retinal release (electronic supplementary material, tables S4 and S5).
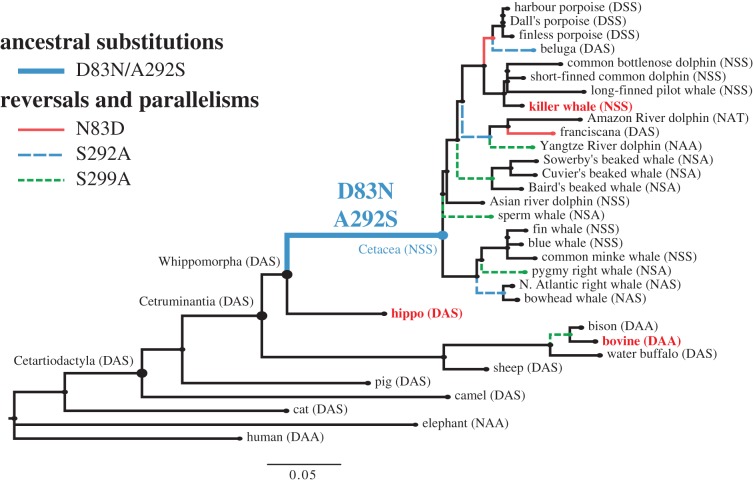
(c). N83 and S292 are derived, while S299 is ancestral in cetaceans
To reconstruct the history of sites 83, 292, and 299 on the cetartiodactyl species tree, we implemented codon-based random sites models of evolution in the codeml program of the PAML package [25]. The amino acid residues at these three sites in the last common cetacean ancestor (Cetacea) and the last common ancestor of cetaceans and hippos (Whippomorpha) were extracted from the best-fitting model according to Akaike's information criterion, model M7 (electronic supplementary material, table S6). Among mammals outside of Cetacea, these three sites are overwhelmingly conserved as either a D83/A292/S299 (DAS) or D83/A292/A299 (DAA) residue combination. Other than marine mammals, the D83N substitution occurs rarely (e.g. African elephant, guinea pig, some bats), and A292S does not occur in any terrestrial mammals [5]. The terrestrial–aquatic transition between Whippomorpha and Cetacea was characterized by a double D83N/A292S substitution, resulting in the NSS combination as the basal cetacean state that has persisted in the killer whale (figure 3). Interestingly, these three sites occur in a wide variety of combinations within Cetacea, including several parallelisms and reversals. For example, there are at least three instances of S292A reversions, and the bovine substitution S299A has also occurred separately in four cetacean lineages (figure 3).
Figure 3.
Cetartiodactyl species tree showing substitutions at rhodopsin sites 83, 292 and 299, as derived from ancestral sequence reconstruction using codon-based random sites models. The ancestral cetacean branch representing the transition from terrestrial to aquatic ancestors is characterized by a double D83N/A292S mutation, resulting in the N83/S292/S299 identity that persists in living delphinids such as the killer whale. Relative to other mammals, cetaceans have evolved a surprisingly diverse number of amino acid combinations at these three sites, including reversals to terrestrial residues.
3. Discussion
Here, we provide the first experimental results with respect to rhodopsin kinetics in a cetacean visual pigment. Even though we found slower retinal release rates in the killer whale relative to the well-studied bovine rhodopsin model, our outgroup comparison with the hippo and our ancestral sequence reconstructions support this phenotype as ancestral, not derived, in cetaceans. Furthermore, our site-directed mutagenesis experiments offer new insight into the structural mechanisms of the D83N substitution, which we found interacts epistatically with S299 in the killer whale to affect meta II stability. Because sites 83, 292 and 299 have been highlighted as potentially giving rise to convergent functional shifts through parallel substitutions in cetacean rhodopsins [3], our study also has implications for how patterns of convergence in coding sequences can be interpreted in light of epistasis.
Our comparative approach highlights the intramolecular epistatic effects on protein function that can emerge from detailed site-directed mutagenesis investigations of specific amino acid substitutions in different genetic backgrounds. Such effects can be obscured by over-reliance on model organisms for in vitro structure–function experiments, but are increasingly being recognized as having critical roles in shaping fitness landscapes, protein evolutionary trajectories, and evolvability (or, capacity for adaptive evolution) in many protein systems [21,26,27]. Intramolecular epistasis is generally defined as a non-additive effect on protein function arising from multiple substitutions, which can then lead to inconsistent phenotypic effects from the same substitution in different genotypic backgrounds (e.g. [28]). This phenomenon has been experimentally demonstrated to constrain several protein evolutionary pathways including increased oxygen affinity in high-altitude haemoglobins [29], bacterial antibiotic resistance [27] and glucocorticoid receptor specificity [30]. In the opsins, however, spectral tuning effects have generally been treated as additive, despite mounting evidence to the contrary [31].
A variety of prior studies have highlighted the multiple occurrences of the D83N substitution in vertebrate rhodopsins, particularly for its contribution to spectral tuning [11,16] and metarhodopsin kinetics [17,18,20]. In fact, D83 is a highly conserved residue not only in visual pigments, but also in family A GPCRs where it is thought to be a key component of a complex network of stabilizing interhelical hydrogen bonds [7,32,33]. In rhodopsin, a central role of D83 appears to be stabilization of the dark state via H-bonds to N55 and a water molecule that is bonded to other residues [7,33,34]. Site N55 forms a H-bond with the carbonyl backbone of A299 in the dark state, which creates an indirect H-bond link between site 83 and site 299 (D83-N55-A299) [33,35]. During light activation, breakage and reformation of H-bonds in this network are key mechanisms by which the transmembrane helices rotate into the meta II conformation [34]. In particular, recent work using solid-state NMR spectroscopic techniques have confirmed that the N55-A299 bond is broken during the transition to meta II [35]. This breakage may be due in part to reorientation of the N55 amine group away from A299 and towards D83 as seen in the bovine meta II crystal structure (figure 4a), which strengthens the D83–N55 bond [35,36]. Though the D83N substitution likely decouples it from N55 in the dark [32], our killer whale meta II homology model suggests the N83 amine group may be near enough to the free 299 carbonyl oxygen to form a H-bond, perhaps replacing the broken N55–299 bond (figure 4b). As such, our working hypothesis is that the D83N substitution facilitates the helical movements associated with light activation by changing the H-bond dynamics in the vicinity of the retinal chromophore. The more stable meta II conformation that results could potentially explain how D83N both accelerates meta II formation [20] and decreases retinal release rates, as indicated by this study and others [17,18].
Figure 4.
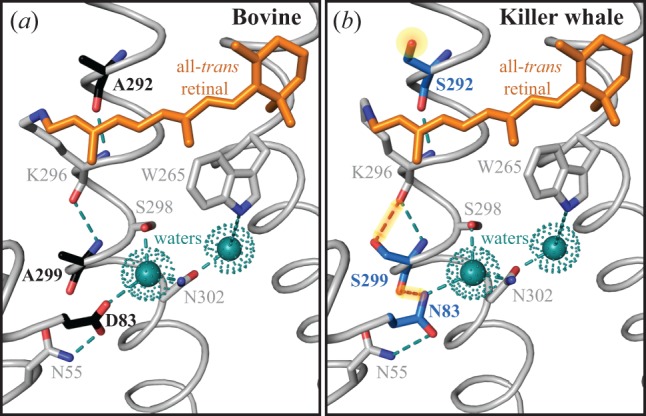
The crystal structure of wild-type bovine meta II (a) as compared with a homology model of wild-type killer whale meta II (b). Key differences highlighted between the species include the alternate residues at 83, 292 and 299 (black: bovine, blue: killer whale), and associated hydrogen bonds (dashed lines). The polar S292 side-chain near the killer whale retinal chromophore, and the hypothesized additional H-bonds in killer whale meta II that may account for slower retinal release rates and epistasis (N83–S299 and S299–K296) are highlighted.
The location of site 299 between site 83 and the retinal chromophore (figure 4) is also consistent with its ability to mediate the effect of site 83 on retinal release. Because our results suggest A299S has a stabilizing influence on meta II, the presence of both S299 and N83 in killer whale rhodopsin may result in some structural redundancy contributing to meta II stability. In the bovine meta II crystal structure, the side-chain of 299 is close to the main-chain of K296, which is covalently bound to the chromophore by a protonated Schiff base (figure 4a). The stabilizing effect of A299S could be explained by the additional hydroxyl group from the serine residue, which may allow additional H-bonds to form, or alternatively may affect chromophore isomerization (figure 4b). In our killer whale rhodopsin experiments, the otherwise destabilizing effect of N83D was thus compensated for by S299, giving rise to functional epistasis in retinal release rates. Interestingly, in the bovine rhodopsin A299S mutant, D83N shifted the retinal release half-life by a larger amount than in the killer whale wild-type, so there also seems to be other unknown site(s) epistatically impacting these mechanisms in bovine rhodopsin.
Relative to site 83, the effects of sites 292 and 299 on meta II stability were minor. This is an intriguing result considering both are closer to the chromophore than site 83, but is consistent with the more prominent role site 83 plays in the H-bond dynamics of light activation. Curiously, while A299S seemed to slightly stabilize meta II, the same substitution at site 292 had the opposite effect in bovine rhodopsin. The structural context of substitutions is thus highly relevant to predicting their functional consequences. The large spectral blue-shift from A292S is thought to be due to the close proximity of its hydroxyl group to the protonated Schiff base, where it can stabilize the ground state of the retinal chromophore and thereby increase the energy required for photon absorption [37]. The smaller impact of 83 and 299 on spectral tuning is thus consistent with their greater distance from the Schiff base.
Generally, epistasis is thought to decrease the likelihood of amino acid convergence and parallelism as a function of sequence divergence because few substitutions will have consistent fitness benefits in divergent protein backgrounds [26,38]. However, our experimental results suggest that parallel substitutions could evolve if they have alternative functional benefits under different background sequence contexts and environmental pressures. Under varying evolutionary scenarios, D83N may be important for blue-shifting λmax [19], for the boost it gives to meta II stability on its own [17], or for balancing functional shifts caused by other substitutions like A292S. The D83N substitution is rare and scattered in its distribution over the vertebrate rhodopsin phylogeny, with no obvious correlations to life history and ecology. Perhaps such correlations are obscured not only because D83N can affect multiple functions, but also because the functional shifts it causes can be mediated by intramolecular epistasis.
In cetacean rhodopsin, we propose that selective pressures were primarily acting on the large A292S blue-shift during the terrestrial–aquatic transition. The combined effect that A292S can have on both spectral tuning and kinetics means evolutionary scenarios that favour both a spectral blue-shift and high meta II stability may require mechanisms to compensate for the destabilizing effect of S292 on meta II. Such a scenario has been proposed for aquatic vertebrates, which are thought to rely both on blue-shifted spectral tuning and greater meta II stability to optimize photosensitivity in underwater environments [20]. The adaptive relevance of D83N in early cetaceans could have been to compensate for the effect of A292S on meta II kinetics. Our killer whale–hippo comparison suggests slower meta II kinetics are an ancestral cetartiodactyl trait, and assuming this trait was conserved in ancestral cetartiodactyls due to functional constraints, the mitigating effect from D83N may have been advantageous. Alternatively, D83N may have been important in early cetaceans for the slight synergistic effect we observed from D83N/A292S on spectral tuning. In either case, shifts to metarhodopsin kinetic rates were likely minimized by the presence of the ancestral S299 residue. This concept that pre-stabilizing, permissive substitutions create tolerance for later function-changing mutations is well-explored in the literature (e.g. [30,39]), and in some cases even allows human disease substitutions to persist in other species [40]. Decreased meta II stability in animals such as bovine ruminants (e.g. from the S299A substitution) may then reflect a relaxed constraint due to increased reliance on daylight vision. In cetaceans, D83N may also be important for other related functions such as G protein activation, thermal stability, or dark adaptation.
There is evidence that dark adaptation (recovery of rods from photo-bleaching) may be limited by meta II decay and retinal release under partial retinal bleaches, and by chromophore synthesis in the retinal pigment epithelium under complete bleaches [41], conditions that may approximate dim-light versus bright-light environments. Thus, in addition to their association with lower meta II stability, faster retinal release rates could also indicate faster rod dark adaptation in dim-light. For deep-diving cetacean species (e.g. sperm whales and beaked whales), faster rod recovery may be an advantage for resolving the bioluminescent signals of deep dwelling prey species, or for accommodating the rapidly changing light levels experienced during a dive from the surface down into the aphotic zones. Consequently, a link between rhodopsin kinetics and dark adaptation may help to explain the occurrence of A299 in these deepest diving cetacean lineages (figure 3). Site 299 is under strong positive selection in cetacean rhodopsin, and falls into a divergent site-class partition that includes the deep divers [3]. Furthermore, the S299A substitution, which occurred in parallel on the deep-diving lineages, accelerated retinal release in our killer whale rhodopsin experiments. Dark adaptation could distinguish the deep diving cetaceans from other cetacean species and deep dwelling teleost fish, for which meta II stability may be more prioritized [20]. However, the relationship between retinal release and dark adaptation, both in a mechanistic sense and in relevance to photosensitivity at the organismal level, is currently uncertain. Because the killer whale tends to forage closer to the surface, exploring these relationships would require kinetic assays on the rhodopsins from deep diving marine mammal species.
This study is the first to explicitly examine the role of among-site interactions on both spectral tuning and light-activated kinetic rates in rhodopsin, and our surprising discovery of epistasis in the kinetics of a cetacean rhodopsin highlights previously underappreciated ways by which substitutions can lead to functional evolution in visual pigments. The effects of intramolecular epistasis are being increasingly recognized in protein evolution [21,26], yet have been largely absent from the rhodopsin literature. We have shown that the highly conserved GPCR site 83, and the major rhodopsin spectral tuning site 292, evolved functionally relevant substitutions over the terrestrial–aquatic cetacean transition. In an extant cetacean rhodopsin model, these substitutions have a synergistic effect on spectral tuning, but an antagonistic effect on activation kinetics, the latter resulting in a negligible net-shift due to epistasis between site 83 and the ancestral cetartiodactyl S299 residue. While there is much literature on the role of substitutions that are alternately beneficial or deleterious to the evolution of amino acid convergence [26], the role of substitutions that affect multiple functions in different molecular contexts is rarely considered. Our study hints at intriguing scenarios where trade-offs among different protein functions could create multiple evolutionary contexts for the selection of particular amino acid combinations.
4. Material and methods
(a). Synthesis of rhodopsin coding sequences and site-directed mutagenesis
Protocols for rhodopsin sequence preparation and mutagenesis were as described in Dungan et al. [3] and van Hazel et al. [17]. In brief, wild-type rhodopsin coding sequences for each species (electronic supplementary material, table S7) were synthesized using the services of GeneArt (Invitrogen) with 5′ and 3′ restriction sites for insertion into the p1D4-hrGFP II expression vector [42]. For the killer whale and bovine sequences, site-directed mutagenesis (QuickChange II protocol) was used to introduce mutations at sites 83, 292 and 299, each species being substituted with the other's residue. All mutants were confirmed by double-stranded sequencing.
(b). Rhodopsin expression and functional assays
As previously described [3], expression vectors containing each wild-type and mutant rhodopsin coding sequence were transiently transfected into HEK293T cells and harvested after 48 h. Expressed rhodopsins were then regenerated with 11-cis-retinal, solubilized in 1% N-dodecyl-d-maltoside, and purified using the 1D4 monoclonal antibody in the dark. We used a Cary 4000 double-beam spectrophotometer (Varian) to measure the UV–visible absorption spectra of eluted rhodopsin samples (20°C in the dark). Spectral peak (λmax) values were then estimated by fitting a standardized template [23] to the dark absorbance spectra. To determine release rates of all-trans-retinal from light-activated rhodopsins, we measured intrinsic increases in tryptophan fluorescence that occur as residues are unquenched during chromophore migration from the binding pocket [24]. Fluorescence signals were measured with a Cary Eclipse fluorescence spectrophotometer (Varian) at 20°C following a 30 s light-bleach [17,22]. Retinal release half-life (t1/2) values were estimated by fitting the resulting time courses to first-order exponential curves (y = y0 + a(1 − e−kx), where t1/2 = ln(2)/k). Statistical procedures for retinal release half-life comparisons are elaborated in electronic supplementary material.
(c). Ancestral reconstruction of sites 83, 292 and 299
To reconstruct the evolutionary history of sites 83, 292 and 299 both within and at the origin of Cetacea, we downloaded the rhodopsin coding sequences of 32 mammals from GenBank: 22 cetaceans, seven other cetartiodactyls (including the hippopotamus and bovine), and three outgroups from Laurasiatheria (cat), Euarchontoglires (human), and Afrotheria (elephant) (electronic supplementary material, table S7). The sequences were aligned by codons using MUSCLE in MEGA6 [43], and we constructed a tree topology based on known mammal relationships [44–46]. The alignment and tree were then used to implement codon-based marginal ancestral sequence reconstructions using the codeml program of the PAML 4.8 software package [25]. Ancestral sequences were chosen from the best-fitting random-sites model, M7 (electronic supplementary material, table S6).
(d). Homology modelling of killer whale metarhodopsin II
We inferred the three-dimensional structure of killer whale meta II with homology modelling, using the bovine meta II crystal structure (PDB code: 3PQR [8]) as a template. One-hundred models were generated in the Modeller package [47], each ranked according to DOPE score [48]. The model with the lowest DOPE score was chosen for visualization in MacPyMOL (The PyMOL Molecular Graphics System, v. 1.7.4.4). The generated killer whale meta II structure had a total energy that was comparable to the bovine homolog (comparable z-scores in ProSA-web, [49]), and had amino acid bond conformations with high probabilities (assessed with ProCheck [50]).
Supplementary Material
Acknowledgements
The 11-cis retinal chromophore was generously provided by Rosalie Crouch (Medical University of South Carolina). We thank two anonymous reviewers of this manuscript for helpful suggestions and comments.
Data accessibility
Data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.5k0s6 [51].
Authors' contributions
B.S.W.C. and S.Z.D. conceived and designed the study. S.Z.D. acquired experimental data, conducted computational and statistical analyses, and drafted the manuscript. B.S.W.C. and S.Z.D. interpreted the results and revised the manuscript.
Competing interests
We have no competing interests.
Funding
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (to B.S.W.C.), an NSERC post-graduate scholarship (to S.Z.D.), and a Vision Science Research Program Fellowship (to S.Z.D.).
References
- 1.McGowen MR, Gatesy J, Wildman DE. 2014. Molecular evolution tracks macroevolutionary transitions in Cetacea. Trends Ecol. Evol. 29, 336–346. ( 10.1016/j.tree.2014.04.001) [DOI] [PubMed] [Google Scholar]
- 2.Liu Z, Qi F-Y, Zhou X, Ren H-Q, Shi P. 2014. Parallel sites implicate functional convergence of the hearing gene prestin among echolocating mammals. Mol. Biol. Evol. 31, 2415–2424. ( 10.1093/molbev/msu194) [DOI] [PubMed] [Google Scholar]
- 3.Dungan SZ, Kosyakov A, Chang BSW. 2016. Spectral tuning of killer whale (Orcinus orca) rhodopsin: evidence for positive selection and functional adaptation in a cetacean visual pigment. Mol. Biol. Evol. 33, 323–336. ( 10.1093/molbev/msv217) [DOI] [PubMed] [Google Scholar]
- 4.Hauser FE, van Hazel I, Chang BSW. 2014. Spectral tuning in vertebrate short wavelength-sensitive 1 (SWS1) visual pigments: can wavelength sensitivity be inferred from sequence data? J. Exp. Zool. B Mol. Dev. Evol. 322, 529–539. ( 10.1002/jez.b.22576) [DOI] [PubMed] [Google Scholar]
- 5.Hunt DM, Carvalho LS, Cowing JA, Davies WL. 2009. Evolution and spectral tuning of visual pigments in birds and mammals. Phil. Trans. R. Soc. B 364, 2941–2955. ( 10.1098/rstb.2009.0044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokoyama S, Yang H, Starmer WT. 2008. Molecular basis of spectral tuning in the red- and green-sensitive (M/LWS) pigments in vertebrates. Genetics 179, 2037–2043. ( 10.1534/genetics.108.090449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palczewski K, et al. 2000. Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289, 739–745. ( 10.1126/science.289.5480.739) [DOI] [PubMed] [Google Scholar]
- 8.Choe H-W, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP. 2011. Crystal structure of metarhodopsin II. Nature 471, 651–655. ( 10.1038/nature09789) [DOI] [PubMed] [Google Scholar]
- 9.Lamb TD, Pugh EN. 2004. Dark adaptation and the retinoid cycle of vision. Prog. Retin. Eye Res. 23, 307–380. ( 10.1016/j.preteyeres.2004.03.001) [DOI] [PubMed] [Google Scholar]
- 10.Bickelmann C, Morrow JM, Du J, Schott RK, van Hazel I, Lim S, Muller J, Chang BS. 2015. The molecular origin and evolution of dim-light vision in mammals. Evolution 69, 2995–3003. ( 10.1111/evo.12794) [DOI] [PubMed] [Google Scholar]
- 11.Hunt DM, Dulai KS, Partridge JC, Cottrill P, Bowmaker JK. 2001. The molecular basis for spectral tuning of rod visual pigments in deep-sea fish. J. Exp. Biol. 204, 3333–3344. [DOI] [PubMed] [Google Scholar]
- 12.Schott RK, et al. 2016. Evolutionary transformation of rod photoreceptors in the all-cone retina of a diurnal garter snake. Proc. Natl Acad. Sci. USA 113, 356–361. ( 10.1073/pnas.1513284113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warrant EJ, Locket NA. 2004. Vision in the deep sea. Biol. Rev. 79, 671–712. ( 10.1017/S1464793103006420) [DOI] [PubMed] [Google Scholar]
- 14.Hunt DM, Fitzgibbon J, Slobodyanyuk SJ, Bowmakers JK. 1996. Spectral tuning and molecular evolution of rod visual pigments in the species flock of cottoid fish in Lake Baikal. Vision Res. 36, 1217–1224. ( 10.1016/0042-6989(95)00228-6) [DOI] [PubMed] [Google Scholar]
- 15.Meredith RW, Gatesy J, Emerling CA, York VM, Springer MS. 2013. Rod monochromacy and the coevolution of cetacean retinal opsins. PLoS Genet. 9, e1003432 ( 10.1371/journal.pgen.1003432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fasick JI, Robinson PR. 1998. Mechanism of spectral tuning in the dolphin visual pigments. Biochemistry 37, 433–438. ( 10.1021/bi972500j) [DOI] [PubMed] [Google Scholar]
- 17.van Hazel I, Dungan SZ, Hauser FE, Chang BSW, Endler JA. 2016. A comparative study of rhodopsin function in the great bowerbird (Ptilonorhynchus nuchalis): spectral tuning and light-activated kinetics. Protein Sci. 25, 1308–1318. ( 10.1002/pro.2902) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bickelmann C, Morrow JM, Müller J, Chang BSW. 2012. Functional characterization of the rod visual pigment of the echidna (Tachyglossus aculeatus), a basal mammal. Vis. Neurosci. 29, 1–7. ( 10.1017/S0952523812000223) [DOI] [PubMed] [Google Scholar]
- 19.Sugawara T, Terai Y, Imai H, Turner GF, Koblmüller S, Sturmbauer C, Shichida Y, Okada N. 2005. Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep-water cichlids from Lakes Tanganyika and Malawi. Proc. Natl Acad. Sci. USA 102, 5448–5453. ( 10.1073/pnas.0405302102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugawara T, Imai H, Nikaido M, Imamoto Y, Okada N. 2010. Vertebrate rhodopsin adaptation to dim light via rapid meta-II intermediate formation. Mol. Biol. Evol. 27, 506–519. ( 10.1093/molbev/msp252) [DOI] [PubMed] [Google Scholar]
- 21.Starr TN, Thornton JW. 2016. Epistasis in protein evolution. Protein Sci. 25, 1204–1218. ( 10.1002/pro.2897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrow JM, Chang BSW. 2015. Comparative mutagenesis studies of retinal release in light-activated zebrafish rhodopsin using fluorescence spectroscopy. Biochemistry 54, 4507–4518. ( 10.1021/bi501377b) [DOI] [PubMed] [Google Scholar]
- 23.Govardovskii VI, Fyhrquist N, Reuter T, Kuzmin DG, Donner K. 2000. In search of the visual pigment template. Vis. Neurosci. 17, 509–528. ( 10.1017/S0952523800174036) [DOI] [PubMed] [Google Scholar]
- 24.Farrens DL, Khorana HG. 1995. Structure and function in rhodopsin. Measurement of the rate of metarhodopsin II decay by fluorescence spectroscopy. J. Biol. Chem. 270, 5073–5076. ( 10.1074/jbc.270.10.5073) [DOI] [PubMed] [Google Scholar]
- 25.Yang Z. 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 26.Storz JF. 2016. Causes of molecular convergence and parallelism in protein evolution. Nat. Rev. Genet. 17, 239–250. ( 10.1038/nrg.2016.11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. 2006. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science 312, 111–114. ( 10.1126/science.1123539) [DOI] [PubMed] [Google Scholar]
- 28.Odokonyero D, et al. 2013. Divergent evolution of ligand binding in the o-succinylbenzoate synthase family. Biochemistry 52, 7512–7521. ( 10.1021/bi401176d) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tufts DM, Natarajan C, Revsbech IG, Projecto-Garcia J, Hoffmann FG, Weber RE, Fago A, Moriyama H, Storz JF. 2015. Epistasis constrains mutational pathways of hemoglobin adaptation in high-altitude pikas. Mol. Biol. Evol. 32, 287–298. ( 10.1093/molbev/msu311) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. 2007. Crystal structure of an ancient protein: evolution by conformational epistasis. Science 317, 1544–1548. ( 10.1126/science.1142819) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yokoyama S, Xing J, Liu Y, Faggionato D, Altun A, Starmer WT. 2014. Epistatic adaptive evolution of human color vision. PLoS Genet. 10, e1004884 ( 10.1371/journal.pgen.1004884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Breikers G, Bovee-Geurts PH, DeCaluwé GL, DeGrip WJ. 2001. A structural role for Asp83 in the photoactivation of rhodopsin. Biol. Chem. 382, 1263–1270. ( 10.1515/BC.2001.157) [DOI] [PubMed] [Google Scholar]
- 33.Okada T, Fujiyoshi Y, Silow M, Navarro J, Landau EM, Shichida Y. 2002. Functional role of internal water molecules in rhodopsin revealed by X-ray crystallography. Proc. Natl Acad. Sci. USA 99, 5982–5987. ( 10.1073/pnas.082666399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pope A, Eilers M, Reeves PJ, Smith SO. 2014. Amino acid conservation and interactions in rhodopsin: probing receptor activation by NMR spectroscopy. Biochim. Biophys. Acta 1837, 683–693. ( 10.1016/j.bbabio.2013.10.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimata N, Pope A, Sanchez-Reyes OB, Eilers M, Opefi CA, Ziliox M, Reeves PJ, Smith SO. 2016. Free backbone carbonyls mediate rhodopsin activation. Nat. Struct. Mol. Biol. 23, 738–743. ( 10.1038/nsmb.3257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rath P, DeCaluwé L, Bovee-Geurts P, DeGrip WJ, Rothschild KJ. 1993. Fourier-transform infrared difference spectroscopy of rhodopsin mutants—light activation of rhodopsin causes hydrogen-bonding change in residue aspartic acid-83 during meta-II formation. Biochemistry 32, 10 277–10 282. ( 10.1021/bi00090a001) [DOI] [PubMed] [Google Scholar]
- 37.Lin SW, Kochendoerfer GG, Carroll KS, Wang D, Mathies RA, Sakmar TP. 1998. Mechanisms of spectral tuning in blue cone visual pigments. Visible and Raman spectroscopy of blue-shifted rhodopsin mutants. J. Biol. Chem. 273, 24 583–24 591. [DOI] [PubMed] [Google Scholar]
- 38.Goldstein RA, Pollard ST, Shah SD, Pollock DD. 2015. Nonadaptive amino acid convergence rates decrease over time. Mol. Biol. Evol. 32, 1373–1381. ( 10.1093/molbev/msv041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordes MHJ, Sauer RT. 1999. Tolerance of a protein to multiple polar-to-hydrophobic surface substitutions. Protein Sci. 8, 318–325. ( 10.1110/ps.8.2.318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Azevedo L, Suriano G, Vanasch B, Harding R, Amorim A. 2006. Epistatic interactions: how strong in disease and evolution? Trends Genet. 22, 581–585. ( 10.1016/j.tig.2006.08.001) [DOI] [PubMed] [Google Scholar]
- 41.Ala-Laurila P, et al. 2006. Visual cycle: dependence of retinol production and removal on photoproduct decay and cell morphology. J. Gen. Physiol. 128, 153–169. ( 10.1085/jgp.200609557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morrow JM, Chang BSW. 2010. The p1D4-hrGFP II expression vector: a tool for expressing and purifying visual pigments and other G protein-coupled receptors. Plasmid 64, 162–169. ( 10.1016/j.plasmid.2010.07.002) [DOI] [PubMed] [Google Scholar]
- 43.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. ( 10.1093/molbev/mst197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Asher RJ, Helgen KM. 2010. Nomenclature and placental mammal phylogeny. BMC Evol. Biol. 10, 102 ( 10.1186/1471-2148-10-102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gatesy J, Geisler JH, Chang J, Buell C, Berta A, Meredith RW, Srpinger MS, McGowen MR. 2013. A phylogenetic blueprint for a modern whale. Mol. Phylogenet. Evol. 66, 479–506. ( 10.1016/j.ympev.2012.10.012) [DOI] [PubMed] [Google Scholar]
- 46.McGowen MR, Spaulding M, Gatesy J. 2009. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phylogenet. Evol. 53, 891–906. ( 10.1016/j.ympev.2009.08.018) [DOI] [PubMed] [Google Scholar]
- 47.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 5.6.1–5.6.30, Chapter 5: Unit 5.6 ( 10.1002/0471250953.bi0506s15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen M-Y, Sali A. 2006. Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524. ( 10.1110/ps.062416606) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wiederstein M, Sippl MJ. 2007. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35(Web Server issue), W407–W410. ( 10.1093/nar/gkm290) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. Procheck—a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291. ( 10.1107/S0021889892009944) [DOI] [Google Scholar]
- 51.Dungan SZ, Chang BSW. 2017. Data from: Epistatic interactions influence terrestrial–marine functional shifts in cetacean rhodopsin. Dryad Digital Repository. ( 10.5061/dryad.5k0s6) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Dungan SZ, Chang BSW. 2017. Data from: Epistatic interactions influence terrestrial–marine functional shifts in cetacean rhodopsin. Dryad Digital Repository. ( 10.5061/dryad.5k0s6) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data are available from the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.5k0s6 [51].