Abstract
The present study aims to evaluate and validate a statistical model for maximizing biosurfactant productivity by Bacillus brevis using response surface methodology. In this respect, twenty bacterial isolates were screened for biosurfactant production using hemolytic activity, oil spreading technique, and emulsification index (E24). The most potent biosurfactant-producing bacterium (B. brevis) was used for construction of the statistical response surface model. The optimum conditions for biosurfactant production by B. brevis were: 33 °C incubation temperature at pH 8 for 10 days incubation period and 8.5 g/L glucose concentration as a sole carbon source. The produced biosurfactant (BS) (73%) exhibited foaming activity, thermal stability in the range 30–80 °C for 30 min., pH stability, from 4 to 9 and antimicrobial activity against (Escherichia coli). The BS gave a good potential application as an emulsifier.
Keywords: Biosurfactants, Optimization, Emulsification, Response surface
1. Introduction
Surfactants are widely used for industrial, agricultural, food, cosmetics and pharmaceutical applications. Most of these compounds are chemically synthesized and potentially causing environmental and toxicological problems [19], [30]. Therefore, microbial-derived surface-active compounds attract attention essentially due to their low toxicity, biodegradable nature [32,34], better environmental compatibility “Green Technology” and easily operated [5]. Recently, biosurfactants received much attention in nano biotechnology criteria [33], [26]. Furthermore, biosurfactants have antibacterial (inhibition activity of cell wall synthesis) [14], [27], antifungal and antiviral properties. They inhibit tumor growth and toxic effects, they also are immune stimulant and cell lysis (haemolysis) [4], they are less allergic, can be used as adhesive agents also, in vaccines and gene therapy [11]. Biosurfactants can be found in detergents, laundry formulations, household cleaning products, herbicides or pesticides, bioremediation, agriculture, textile, paper, petroleum industries, pharmaceutical and food-processing industry [6], [24]. Also, in enzyme stimulation and bio-regulatory effects [25]. They are important in plant pathogenicity, effective on migration of human neutrophils, respiratory action (anti-asthma activity) and food digestion [20], paint, cement, beer, beverages hygiene and cosmetics [23]. Furthermore, biosurfactants are usually effective at extreme environmental conditions and can be produced from renewable resources [9].
In this investigation, the power of response surface method using central composite design (CCD) had been explored to optimize biosurfactant production by Bacillus brevis. Therefore, in this study, the effect of temperature (A), pH (B), incubation period (C) and glucose concentration (D) for maximizing biosurfactant production by B. brevis using central composite design had been evaluated and validated, experimentally.
2. Materials and methods
2.1. Microorganism and culture conditions
Different samples were collected from oil contaminated soil and sediment of mangrove trees. To isolate bacteria, these sample were cultured on the following medium [3] (g/L): NaNO3 (2.0), KCl (0.5), Na2HPO4H2O (1.0), KH2PO4 (1.0), CaCl2 (0.025); MgSO4 (0.1), FeSO4.7·H2O (0.001) and 2 ml/L trace element solution containing the following ingredients (mg/L): FeCl3·6H2O (60), ZnSO4·7H2O (600), MnSO4·H2O (200), CuSO4·5H2O (590), CoCl2·6H2O (60). The pH of the medium was adjusted to 7.0 and sterilized by autoclaving at 121 ∘C for 20 min. A potent biosurfactant-producing bacterium has been isolated from the sediment of mangrove trees (Makadi vallige, Hurghada region, Egypt), purified and characterized. This isolate has been identified based on 16S-rRNA. The pure culture was preserved at (4 °C) and subculturing was done every month.
3. Biosurfactant productivity tests
3.1. Hemolytic activity
A pure culture of each bacterial isolate was streaked on the freshly prepared blood agar and incubated at 37 °C for 48–72 h. Results were recorded based on the type of clear zone observed [21], [35].
3.2. Oil spreading method
Oil spreading technique was carried out according to the method described by Satpute et al. [31]. Briefly, 50 mL of distilled water was added to the Petri plate followed by addition of 100 μL of olive oil to the surface of the water. Then, 10 μL of cell-free culture broth was dropped on the crude oil surface. The diameter of the clear zone on the oil surface was measured and compared to 10 μL of distilled water as a negative control.
3.3. Emulsification activity (E24)
The emulsification activity was measured using the method described by Plaza et al. [22]. About 2 mL of olive (crude oil) and 2 mL of cell-free medium (supernatant) were inoculated to a test tube and homogenized by vortexing at high speed for 2 min. After 24 h, the emulsification activity was calculated using following formula:
E24 (%) = total height of the emulsified layer/total height of the liquid layer [15].
3.4. Identification of bacterial isolate
The most efficient biosurfactant producer bacterial isolate was then identified as B. brevis using 16S rRNA analysis Procedure, which has been performed at Macrogen company (Korea) and used for the current investigation.
3.5. Foam height analysis
Foaming ability was determined according to Abou seoud et al. [1]. B. brevis was grown in 250 mL Erlenmeyer flask, containing 50 mL of nutrient broth medium. The flask was incubated at 33 °C on a shaker incubator (200 rpm) for 96 h. Foam activity was detected as the duration of foam stability, foam height and foam shape in the graduated cylinder.
3.6. Experimental design
Central composite design (CCD) model, based on four factors and five levels was used to study the effect and interactions between temperature (A) in the range between 25 and 35 °C, pH (B) in the range between 6 and 8, incubation period (C) in the range between 3 and 7 days and glucose concentration (D) between 5 and 15 g/L for maximum production of biosurfactant by B. brevis (Table 1). Experimental designs were performed using Design-Expert software (Stat-Ease Inc., Minneapolis, MN, USA, ver 7.0.0). A total of 108 experiments were employed in CCD to estimate curvature and interaction effects of selected variables, and finally, significance of the obtained model was checked by t-test (calculated p-value) and goodness of fit by multiple correlations as well as determination 2 coefficients. Emulsification index (E24) was the measured experimental response.
Table 1.
Central Composite design runs showing factors and their levels (based on actual value).
| Run | Run type | (A) Incubation temperature (°C) | (B) pH |
(C) Incubation period (days) |
(D) Glucose concentration (g/L) |
|---|---|---|---|---|---|
| 1, 26, 59, 71 | Factorial | 25 | 6 | 3 | 5 |
| 2, 20, 94, 100 | Factorial | 35 | 6 | 7 | 5 |
| 3, 9, 34, 37, 42, 50, 57, 58, 65, 70, 84, 95 | Center | 30 | 7 | 5 | 10 |
| 4, 19, 55, 81 | Factorial | 35 | 6 | 3 | 5 |
| 5, 47, 88, 91 | Factorial | 25 | 8 | 7 | 5 |
| 6, 15, 68, 108 | Factorial | 25 | 8 | 7 | 15 |
| 7, 38, 83, 96 | Factorial | 25 | 6 | 7 | 5 |
| 8, 52, 63, 99 | Axial | 30 | 5 | 5 | 10 |
| 10, 54, 79, 90 | Axial | 30 | 7 | 1 | 10 |
| 11, 21, 62, 92 | Axial | 20 | 7 | 5 | 10 |
| 12, 13, 93, 78 | Factorial | 25 | 6 | 7 | 15 |
| 14, 29, 101, 102 | Factorial | 35 | 8 | 3 | 5 |
| 16, 46, 77, 98 | Factorial | 35 | 6 | 3 | 15 |
| 17, 22, 60, 64 | Factorial | 35 | 6 | 7 | 15 |
| 18, 43, 76, 103 | Factorial | 35 | 8 | 3 | 15 |
| 23, 32, 56, 61 | Axial | 30 | 7 | 5 | 0 |
| 24, 33, 87, 104 | Factorial | 35 | 8 | 7 | 5 |
| 25, 31, 67, 89 | Factorial | 25 | 8 | 3 | 5 |
| 27, 30, 74, 105 | Factorial | 25 | 6 | 3 | 15 |
| 28, 53, 82, 97 | Axial | 30 | 9 | 5 | 10 |
| 35, 40, 75, 107 | Axial | 30 | 7 | 9 | 10 |
| 36, 41, 73, 86 | Axial | 40 | 7 | 5 | 10 |
| 39, 51, 66, 80 | Factorial | 25 | 8 | 3 | 15 |
| 44, 45, 69, 85 | Factorial | 35 | 8 | 7 | 15 |
| 48, 49, 72, 106 | Axial | 30 | 7 | 5 | 20 |
3.7. Experimental validation of statistical model
The response surface model and the optimum conditions were tested and validated in four replicas and recorded as (mean ± standard deviation).
3.8. Statistical analysis
Analysis of variance (ANOVA) was used to estimate the statistical parameters for optimization of culture conditions. A probability value of <0.05 was used as the criterion for statistical significance.
3.9. Extraction and recovery of biosurfactant
According to optimized conditions, B. brevis was grown in 500 mL Erlenmeyer flask containing 100 ml mineral salt broth medium. To extract the biosurfactant, the bacterial cells were removed by centrifugation and the remaining supernatant was filtered through a 0.50 mm pore size filter. The cell free supernatant was acidified, using 1 M H2SO4. Then, equal volume of chloroform: methanol (2:1) was added, this mixture was shaken well. The solvent layer was separated from aqueous phase and lift overnight for evaporation to concentrate biosurfactant. For further purification the crude surfactant was dissolved in distilled water at pH 7.0 and dried at 60 °C. The dry product was extracted with Chloroform: Methanol (65:15), filtered and the solvent evaporated. Sediment was obtained as a result i.e., the biosurfactant [31]. The Grey white precipitate thus obtained was centrifuged for 20 min, dried, and was gravimetrically weighted expressed as g/L [13].
3.10. Temperature and pH stability profiles
For thermal stability, the cell-free broth of B. brevis was maintained at constant temperatures in the range 30–80 °C for 30 min [17]. and then cooled to room temperature, before measuring the emulsification activity. For pH stability, the cell-free supernatant was adjusted to various pH values from 4 to 9 with 1 N HCl or 1 N NaOH [16]. The emulsifying indexes were measured after fifteen minutes.
3.11. Antibacterial activity
Antibacterial activity of partially purified BS of B. brevis was evaluated using agar diffusion method [18]. Twenty ml nutrient agar medium were poured in Petri plate. An aliquot (0.05 ml) of (Escherichia coli) inoculum was introduced to the molten agar. After solidification, the appropriate well was made on agar plate using sterile cork-borer, 6.0 mm, in which, 50 μl of partially purified BS was added, and distilled water was added to another plate as control. The plates were incubated at 30 °C for 48 hours. The presence of clear zone marked the antibacterial activity of BS (all experiments were performed in duplicates).
4. Results and discussion
Biosurfactants are attracting a pronounced interest owing to their potential advantages over their chemical counterparts [8]. In accordance with Saimmai et al. [29], twenty bacterial isolates were screened with different screening tests (hemolytic activity, oil spreading technique and Emulsification activity) to find the most efficient biosurfactant producer (Table 2).
Table 2.
Screening for biosurfactant producing isolates by preliminary and complementary screening methods.
| Isolate No. |
Hemolytic activity | Oil displacement area (cm2) |
Emulsification index (%) |
|---|---|---|---|
| 1 | − | 2.2 | 0 |
| 2 | − | 1.8 | 0 |
| 3 | + | 4.4 | 0 |
| 4 | − | 1.4 | 6.4 |
| 5 | + | 4.6 | 0 |
| 6 | + | 4.2 | 0 |
| 7 | +++++ | 28.2 | 46.6 |
| 8 | + | 3.6 | 0 |
| 9 | ++ | 2.8 | 24.2 |
| 10 | + | 2.6 | 20.2 |
| 11 | +++ | 12.2 | 26.4 |
| 12 | ++ | 2.2 | 22.6 |
| 13 | + | 3.1 | 18.8 |
| 14 | +++ | 16.8 | 24.4 |
| 15 | ++ | 3.6 | 22.6 |
| 16 | ++ | 2.8 | 23.6 |
| 17 | ++ | 2.9 | 24.3 |
| 18 | + | 1.4 | 16.6 |
| 19 | ++ | 2.8 | 22.4 |
| 20 | ++ | 2.4 | 22.6 |
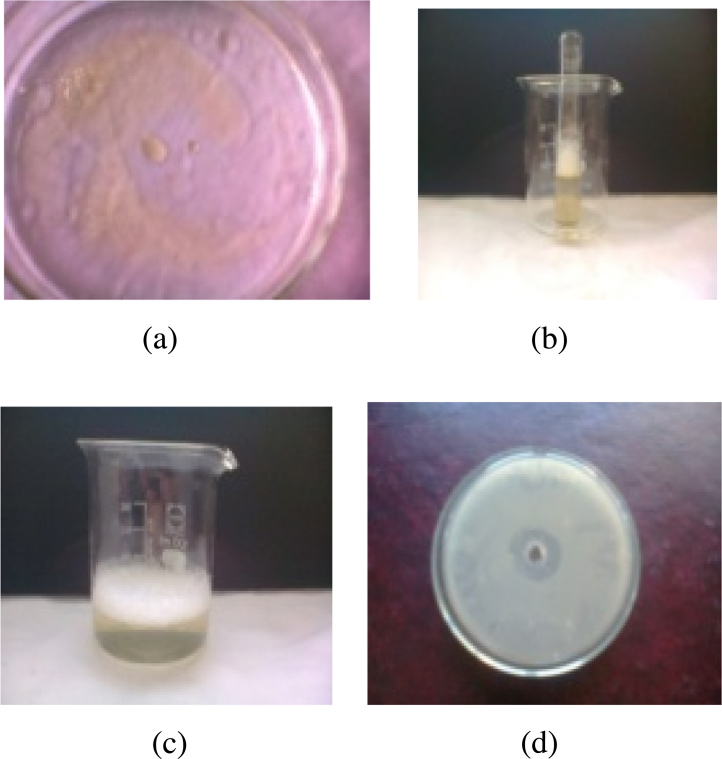
B. brevis (number 7) showed good ability to emulsify olive oil and to disperse the oil (Fig. 2).
Fig. 2.
(a) Biosurfactant positive Bacillus brevis shows oil spreading; (b): emulsification test for Bacillus brevis; (c) foam forming activity for Bacillus brevis; (d): zone of clearance showing antimicrobial activity of Bacillus brevis.
4.1. Identification of the efficient biosurfactant producer
Strain No.7 was identified morphologically and physiologically according to Holt Manual of Determinative Bacteriology [12]. Cells of B. brevis were gram-positive, aerobically rods, motile, spore-former, with positive catalase activity, amylase negative, casein negative, gelatinase positive, and indole negative, with the optimal growth of 35–55 °C.
4.2. Partial sequencing
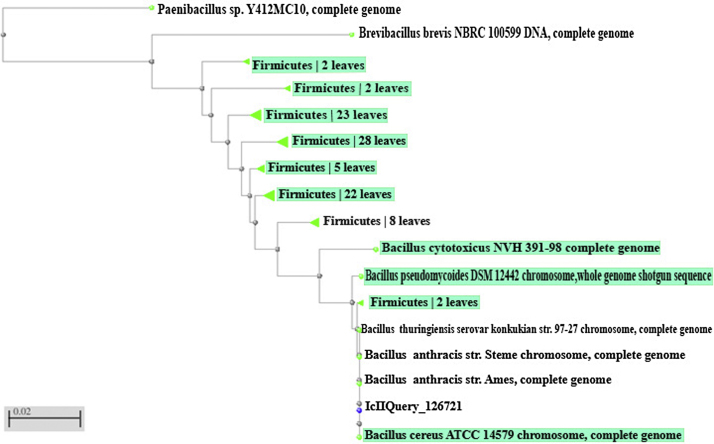
The strain was reclassified into genus Brevibacillus. Where identification was confirmed with 16S rDNA sequence analysis. 16S rDNA gene was amplified by polymerase chain reaction (PCR) using forward and reverse primers. Hence, the strain was identified as B. brevis as shown in the phylogenetic tree (Fig. 1).
Fig. 1.
Phylogenetic tree based on 16S rDNA gene sequencing, showing the phylogenetic relationship of Bacillus brevis within representative species of the genus Bacillus.
In this investigation, we have explored the power of response surface method using central composite design (CCD) to optimize biosurfactant production by B. brevis. Also we evaluated and validated, experimentally, the effect of Temperature (A), pH (B), Incubation period (C) and Glucose concentration (D) on maximization of biosurfactant production by B. brevis using central composite design. Based on the CCD, the experimental levels of Emulsification Index (E24) under each set of conditions were determined and compared with the corresponding predicted levels suggested by Design-Expert (Table 3).
Table 3.
Central composite design runs with actual and predicted response values.
| Run | Emulsification index (E24)% |
Run | Emulsification index (E24)% |
Run | Emulsification index (E24)% |
|||
|---|---|---|---|---|---|---|---|---|
| Actual value |
Predicted value |
Actual value |
Predicted value |
Actual value |
Predicted value |
|||
| 1 | 28.88 | 30.55 | 37 | 57.33 | 57.40 | 73 | 28.88 | 38.02 |
| 2 | 51.55 | 43.72 | 38 | 36 | 37.44 | 74 | 31.11 | 36.31 |
| 3 | 57.77 | 57.40 | 39 | 42.66 | 43.98 | 75 | 63.11 | 66.09 |
| 4 | 44.44 | 36.43 | 40 | 62.22 | 66.62 | 76 | 66.66 | 62.64 |
| 5 | 50.22 | 41.01 | 41 | 30.22 | 38.54 | 77 | 29.33 | 37.11 |
| 6 | 44 | 49.12 | 42 | 58.66 | 57.40 | 78 | 34.88 | 38.69 |
| 7 | 36.44 | 37.44 | 43 | 66.44 | 63.16 | 79 | 54.22 | 53.66 |
| 8 | 42.88 | 36.53 | 44 | 72.24 | 68.70 | 80 | 32 | 43.46 |
| 9 | 57.55 | 57.40 | 45 | 70.66 | 68.70 | 81 | 44 | 35.90 |
| 10 | 54.44 | 54.18 | 46 | 29.55 | 37.64 | 82 | 55.55 | 65.09 |
| 11 | 17.55 | 13.08 | 47 | 51.11 | 41.01 | 83 | 35.55 | 36.92 |
| 12 | 33.77 | 39.21 | 48 | 63.55 | 47.59 | 84 | 56.44 | 56.88 |
| 13 | 34.66 | 39.21 | 49 | 62.66 | 47.59 | 85 | 71.77 | 68.18 |
| 14 | 60 | 55.60 | 50 | 56.88 | 57.40 | 86 | 30.66 | 38.02 |
| 15 | 42.22 | 49.12 | 51 | 43.11 | 43.98 | 87 | 71.11 | 65.14 |
| 16 | 29.77 | 37.64 | 52 | 41.33 | 36.53 | 88 | 51.33 | 40.48 |
| 17 | 35.77 | 40.42 | 53 | 56.44 | 65.61 | 89 | 38.22 | 30.83 |
| 18 | 66.66 | 63.16 | 54 | 54 | 54.18 | 90 | 53.33 | 53.66 |
| 19 | 43.11 | 36.43 | 55 | 43.11 | 35.90 | 91 | 49.77 | 40.48 |
| 20 | 51.11 | 43.72 | 56 | 18.88 | 37.74 | 92 | 17.33 | 12.56 |
| 21 | 17.77 | 13.08 | 57 | 56.88 | 56.88 | 93 | 35.11 | 38.69 |
| 22 | 34.66 | 40.42 | 58 | 55.55 | 56.88 | 94 | 52.22 | 43.20 |
| 23 | 17.55 | 38.26 | 59 | 29.33 | 30.03 | 95 | 57.77 | 56.88 |
| 24 | 70.66 | 65.66 | 60 | 34.22 | 39.89 | 96 | 36.88 | 36.92 |
| 25 | 37.78 | 31.35 | 61 | 18.66 | 37.74 | 97 | 56 | 65.09 |
| 26 | 29.55 | 30.55 | 62 | 18.22 | 12.56 | 98 | 28.88 | 37.11 |
| 27 | 33.33 | 36.83 | 63 | 42.88 | 36.00 | 99 | 42.26 | 36.00 |
| 28 | 55.11 | 65.61 | 64 | 35.55 | 39.89 | 100 | 50.66 | 43.20 |
| 29 | 62.22 | 55.60 | 65 | 57.33 | 56.88 | 101 | 61.77 | 55.08 |
| 30 | 32.44 | 36.83 | 66 | 32.88 | 43.46 | 102 | 62.24 | 55.08 |
| 31 | 37.55 | 31.35 | 67 | 37.77 | 30.83 | 103 | 62.22 | 62.64 |
| 32 | 19.33 | 38.26 | 68 | 43.55 | 48.60 | 104 | 72.44 | 65.14 |
| 33 | 72 | 65.66 | 69 | 71.11 | 68.18 | 105 | 31.33 | 36.31 |
| 34 | 56.88 | 57.40 | 70 | 56.66 | 56.88 | 106 | 64.44 | 47.06 |
| 35 | 62.44 | 66.62 | 71 | 28.84 | 30.03 | 107 | 63.33 | 66.09 |
| 36 | 29.77 | 38.54 | 72 | 62.22 | 47.06 | 108 | 42.22 | 48.60 |
Quadratic model was found to be the “best fit model” for the Emulsification Index (E24) response with the highest -value in case of sequential model sum of squares and the lowest F-value in case of lack of fit test when compared to other models. The quadratic model has the standard deviation of 7.97, R-squared of 0.7643, adjusted R-squared of 0.6699 and PRESS of 8180.44. These results show that the model can be used for the navigation of biosurfactant model space.
The Model F-value of 21.31 implies the model is significant. There is only a 0.01% chance that a “model F-value” this large could occur due to noise. Values of “Prob > F” less than 0.05 indicate model terms are significant. In this case, A, B, C, D, AB, A2, D2 are significant model terms. The “lack of fit F-value” of 238.73 implies the lack of fit is significant. There is only a 0.01% chance that a “lack of fit -value” this large could occur due to noise.
The regression equation obtained after ANOVA (Table 4) indicated that the “Pred R-Squared” of 0.6699 is in reasonable agreement with the “Adj R-Squared” of 0.7284. “Adeq Precision” of 18.306-which measures the signal to noise ratio-indicates an adequate signal and this model can be used to navigate the design space.
Table 4.
Results of ANOVA for the produced Emulsification index (E24) quadratic model.
| Source | Sum of squares | Df | Mean square | F-value |
p-value prob > F |
|---|---|---|---|---|---|
| Model | 18939.63 | 14 | 1352.83 | 21.31 | <0.0001 |
| A—incubation temperature | 3888.251 | 1 | 3888.25 | 61.25 | <0.0001 |
| B—pH | 5075.914 | 1 | 5075.91 | 79.96 | <0.0001 |
| C—incubation period | 928.0241 | 1 | 928.02 | 14.62 | 0.0002 |
| D—glucose concentration | 521.7338 | 1 | 521.73 | 8.22 | 0.0051 |
| AB | 1350.379 | 1 | 1350.38 | 21.27 | <0.0001 |
| AC | 0.64 | 1 | 0.64 | 0.01 | 0.9202 |
| AD | 103.0225 | 1 | 103.02 | 1.62 | 0.2059 |
| BC | 30.52563 | 1 | 30.53 | 0.48 | 0.4898 |
| BD | 161.0361 | 1 | 161.04 | 2.54 | 0.1147 |
| CD | 81.49576 | 1 | 81.50 | 1.28 | 0.2601 |
| A2 | 5322.283 | 1 | 5322.28 | 83.84 | <0.0001 |
| B2 | 213.9541 | 1 | 213.95 | 3.37 | 0.0696 |
| C2 | 47.88008 | 1 | 47.88 | 0.75 | 0.3874 |
| D2 | 1118.049 | 1 | 1118.05 | 17.61 | <0.0001 |
| Residual | 5840.466 | 92 | 63.48 | ||
| Lack of fit | 5799.028 | 34 | 170.56 | 238.73 | <0.0001 |
| Pure error | 41.43782 | 58 | 0.71 | ||
| Cor total | 24787.48 | 107 |
* Values of “prob > F” less than 0.05 indicates model terms are significant.
4.3. Final equation in terms of coded factors
Emulsification Index (E24) = (57.14) + (6.36 × A) + (7.27 × B) + (3.11 × C) + (2.33 × D) + (4.59 × A × B) + (0.10 × A × C) − (1.27 × A × D) + (0.69 × B × C) + (1.59 × B × D) − (1.13 × C × D) − (7.90 × A2) − (1.58 × B2) + (0.75 × C2) − (3.62 × D2)
where, A: Incubation temperature (C); B: pH; C: Incubation period (days); D: Glucose concentration (g/L).
4.4. Point prediction and verification
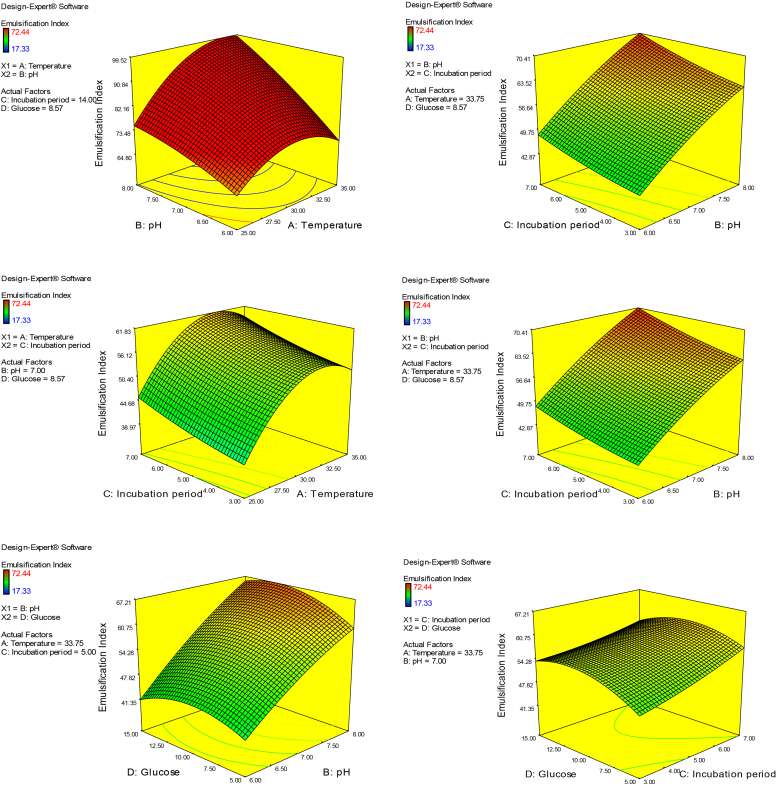
The optimum conditions for maximum biosurfactant productivity by B. brevis (79.96%) were predicted from the produced model as follows: 33 °C for incubation temperature, 8 for pH, 10 for incubation period and 8.5 for glucose concentration. This predicted point was experimentally verified and the emulsification index was 71.89 ± 0.56%. These results reveal a good correlation between the predicted and actual experimental values and this model is well-representing biosurfactant production by B. brevis (Fig. 3)
Fig. 3.
Response surface plot for the interactions between different selected factors.
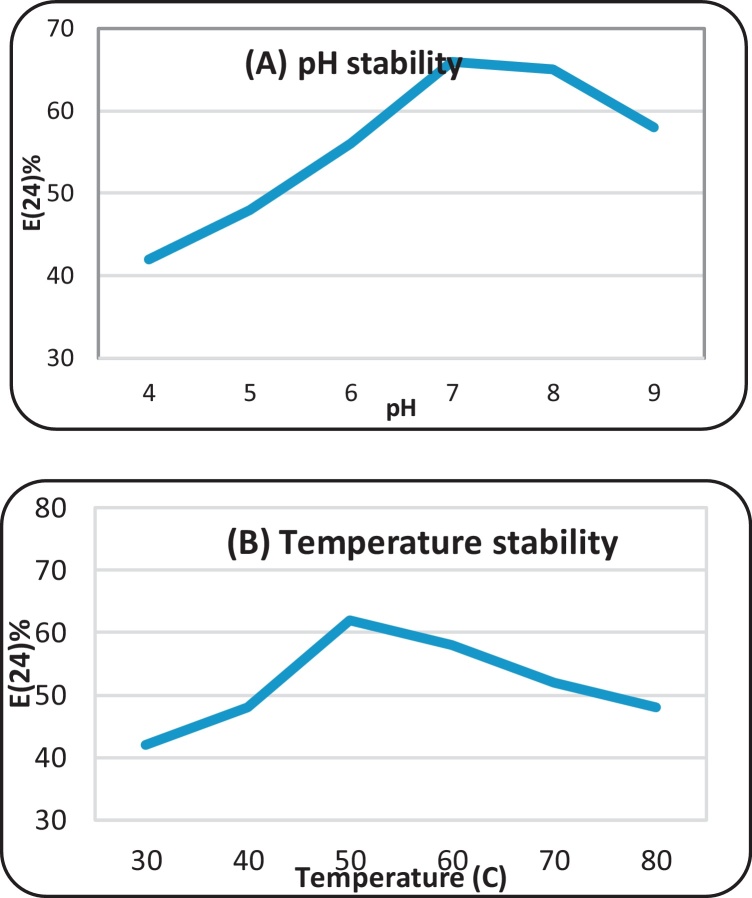
The semi-purified biosurfactant produced by B. brevis showed higher stability at alkaline conditions than acidic conditions. These results are in agreement with [10], [2]. In addition, B. brevis biosurfactant was thermally stable in a range of 30–80 °C. Similar behavior was observed with other strains [10], [28]. The aqueous solution of the partially recovered biosurfactant showed good foaming ability more than 50% and stability for more than 6 h. This result, is in accordance with El-Shahawy and Hussien [7] (Fig. 4).
Fig. 4.
Biosurfactant stability at different pH (A) and different temperatures (B).
5. Conclusion
The response surface method allowed the development of a polynomial model for the production of biosurfactant by B. brevis. The model was able to foresee accurately the BS production by changing pH, temperature, incubation period and glucose concentration. Application of such models is of great importance for making the process industrially viable.
References
- 1.Abou seoud M., Maachi R., Amranec A., Boudergua S., Nabi A. Evaluation of different carbon and nitrogen sources in the production of biosurfactant by Pseudomonas fluorescens. Desalination. 2008;223:143–151. [Google Scholar]
- 2.Al-Bahry S.N., Al-Wahaibi Y.M., Elshafie A.E., Al-Bemani A.S., Joshi S.J., Al-Makhmari H.S., Al-Sulaimani H.S. Biosurfactant production by Bacillus subtilis B20 using date molasses and its possible application in enhanced oil recovery. Int. Biodeter. Biodegrad. 2013;81:141–146. [Google Scholar]
- 3.Chandankere R., Jun Y., Choi M.M.F., Masakorala K., Chan Y. An efficient biosurfactant-producing and crude-oil emulsifying bacterium Bacillus methylotrophicus USTBa isolated from petroleum reservoir. Biochem. Eng. J. 2013;74:46–53. [Google Scholar]
- 4.Dehghan-Noudeh G., Housaindokht M., Bazzaz B.S. Isolation, characterization and investigation of surface and haemolytic activities of a lipopeptide biosurfactant produced by Bacillus subtilis ATCC 6633. J. Microbiol. 2005;43:272–276. [PubMed] [Google Scholar]
- 5.El-Shahawy M.R., Hussien H.A. Applications of crude biosurfactant produced by Egyptian local bacteria in enhanced oil recovery. J. Nucl. Tech. Appl. Sci. 2014;2(4):431–436. [Google Scholar]
- 6.Geys R., Soetaert W., Van Bogaert I. Biotechnological opportunities in biosurfactant production. Curr. Opin. Biotechnol. 2014;30:66–72. doi: 10.1016/j.copbio.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Ghayyomi Jazeh M., Forghani F., Deog-Hwan Oh Biosurfactant production by Bacillus sp. isolated from petroleum contaminated soils of Sirri island. American Journal of Applied Sciences. 2012;9(1):1–6. [Google Scholar]
- 8.Gudiña Eduardo J., Elisabete Rodrigues, Teixeira A., Rodrigues L.R. Biosurfactant production by Bacillus subtilis using corn steep liquor as culture medium. Fornt. Microbiol. 2015 doi: 10.3389/fmicb.2015.00059. Published : 06 February 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gudiña E.J., Rodrigues Ana I., Alves Eliana, Domingues M., Rosário, Teixeira J.A., Rodrigues, Lígia R. Bioconversion of agro-industrial by-products in rhamnolipids towards applications in enhanced oil recovery and bioremediation. Biores. Technol. 2015;177:87–93. doi: 10.1016/j.biortech.2014.11.069. [DOI] [PubMed] [Google Scholar]
- 10.Gudina E.J., Pereira J.F.B., Rodrigues L.R., Coutinho J.A.P., Teixeira J.A. Isolation and study of microorganisms from oil samples for application in microbial enhanced oil recovery. Int. Biodeterior. Biodegrad. 2012;68:56–64. [Google Scholar]
- 11.Harshada Kasar. Biosurfactant: a potent antimicrobial agent. J. Microbiol. Exp. 2014;1(5):1–5. [Google Scholar]
- 12.Holt J.G., Krieg N.R., Sneath P.H.A., Staley J.T., Williams T. In: Bergey’s Manual of Determinative Bacteriology. 9th edition. Hensyl W.R., editor. Williams & Wilkins; United States, Baltimore: 1994. pp. 40–169. [Google Scholar]
- 13.Ismail W., Al-Rowaihi I.S., Al-Humam A.A., Hamza R.Y., El Nayal A.M., Bououdina M. Characterization of a lipopeptide biosurfactant produced by a crude-oil- emulsifying Bacillus sp. I-15. Int Biodeter Biodegrad. 2013;84:168–178. [Google Scholar]
- 14.Joshi S., Bharucha C., Desai A.J. Production of biosurfactant and antifungal compound by fermented food isolate Bacillus subtilis 20B. Bioresour. Technol. 2008;99:4603–4608. doi: 10.1016/j.biortech.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 15.Lai C.C., Huang Y.C., Wei Y.H., Chang J.S. Biosurfactant-enhanced removal of total petroleum hydrocarbons from contaminated soil. J Hazard Mater. 2009;167:609–614. doi: 10.1016/j.jhazmat.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Lotfabad T.B., Shourian M., Roostaazad R., Najafabadi A.R., Adelzadeh M.R., Noghabi K.A. An efficient biosurfactant-producing bacterium Pseudomonas aeruginosa MR01, isolated from oil excavation areas in south of Iran. Colloids Surf B Biointerfaces. 2009;69:183–193. doi: 10.1016/j.colsurfb.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 17.Mabrouk M.E.M., Youssif E.M., Sabry S.A. Biosurfactant production by a newly isolated soft coral-associated marine Bacillus sp. E34: statistical optimization and characterization. Life Sci. J. 2014;11(10):756–768. [Google Scholar]
- 18.Mahalingam R., Ambikapathy V., Panneerselvam A. Studies on antifungal activities of some medicinal plants against Ceratocystis paradoxa causing pineapple disease. World J. Sci. Technol. 2011;1:10–13. [Google Scholar]
- 19.Makkar R.S., Rockne K.J. Comparison of synthetic surfactants and biosurfactants in enhancing biodegradation of polycyclic aromatic hydrocarbons. Environ. Toxicol. Chem. 2003;22:2280–2292. doi: 10.1897/02-472. [DOI] [PubMed] [Google Scholar]
- 20.Nitschke M., Costa S.G. Biosurfactants in food industry. Trends Food Sci. Technol. 2007;18:252–259. [Google Scholar]
- 21.Plaza G.A., Zjawiony I., Banat I.M. Use of different methods for detection of thermophilic biosurfactant-producing bacteria from hydrocarbon-contaminated and bioremediated soils. J. Petrol. Sci. Eng. 2006;50:71–77. [Google Scholar]
- 22.Rahman K.S.M., Rahman T.J., Lakshmanaperumalsamy P., Marchant R., Banat I.M. The potential of bacterial isolates for emulsification with a range of hydrocarbons. Acta Biotechnol. 2003;23:335–345. [Google Scholar]
- 23.Rai S.K., Mukherjee A.K. Statistical optimization of producing, purification and industrial application of a laundry detergent and organic solvent-stable subtilisn-like serine protease (Alzwiprase) from Bacillus subtilis DM-04. Biochem. Eng. J. 2010;48:173–180. [Google Scholar]
- 24.Rebello S., Asok A.K., Mundayoor S., Jisha M.S. Surfactants: toxicity, remediation, and green surfactants. Environ. Chem. Lett. 2014;12:275–287. [Google Scholar]
- 25.Rodrigues L.R., Teixeira J.A., Meib H.C., Oliveira R. Physicochemical and functional characterization of a biosurfactant produced by lactococcuslactis 53. Coll. Surf. B: Biointerfaces. 2006;49:79–86. doi: 10.1016/j.colsurfb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez A.P., Delgado D., Solinis M.A., Pedraz J.L., Echevarria E., Rodriguez J.M., Gascon A.R. Solid lipid nanoparticles as potential tools for gene therapy: in vitro portion expression after intravenous administration. Int. J. Pharm. 2010;385:157–162. doi: 10.1016/j.ijpharm.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 27.Sabate D.C., Carrillo L., Audisio M.C. Inhibition of paenibacillus larvae and ascosphaera apis by Bacillus subtilis isolated from honey bee gut and honey samples. Res. Microbiol. 2009;160:193–199. doi: 10.1016/j.resmic.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Saimmai A., Sobhon V., Maneerat S. Molasses as a whole medium for biosurfactants production by Bacillus strains and their application. Appl. Biochem. Biotechnol. 2011;165:315–335. doi: 10.1007/s12010-011-9253-8. [DOI] [PubMed] [Google Scholar]
- 29.Satpute S.K., Bhawsar B.D., Dhakephalkar P.K., Chopade B.A. Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Int. J. Mar. Sci. 2008;37:243–250. [Google Scholar]
- 30.Schramm L.L., Stasiuk E.N., Marangoni D.G. Surfactants and their applications. Ann. Rep. Program. Chem Sec. 2003;99:3–48. [Google Scholar]
- 31.Sharma Anayata, Soni Jitesh, Kaur Gurveer, Kaur Jaspreet. A Study on biosurfactant production in Lactobacillus and Bacillus sp. Int. J. Curr. Microbiol. Appl. Sci. 2014;3(11):723–733. [Google Scholar]
- 32.Singh S.P., Bharali P., Konwar B.K. Optimization of nutrient requirements and culture conditions for the production of rhamnolipid from Pseudomonas aeruginosa (MTCC 7815) using Mesua ferrea seed Oil. Indian. J. Micrbiol. 2013;53:467–476. doi: 10.1007/s12088-013-0403-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solanki J.N., Murthy Z.V.P. Higly monodisperse and sub-nano silver particles synthesis via microemulsion technique. Colloids Surf. A Physiocochem. Eng. Aspects. 2010;359:31–38. [Google Scholar]
- 34.Waghmode Samadhan, Kulkarni Chandrashekhar, Shukla Sneha, Sursawant Priyanka, Velhal Chaitanya. Low-cost production of biosurfactant from different substrates and their comparative study with commercially available chemical surfactant. Int. J. Sci. Technol. Res. 2014;3(3):146–149. [Google Scholar]
- 35.Youssef N.H., Duncan K.E., Nagle K.N., Savage Knapp R.M. Comparison of methods to detect biosurfactant production by diverse microorganisms. J. Microbiol. Methods. 2004;56:339–347. doi: 10.1016/j.mimet.2003.11.001. [DOI] [PubMed] [Google Scholar]