Abstract
The dark data extraction or knowledge base construction (KBC) problem is to populate a SQL database with information from unstructured data sources including emails, webpages, and pdf reports. KBC is a long-standing problem in industry and research that encompasses problems of data extraction, cleaning, and integration. We describe DeepDive, a system that combines database and machine learning ideas to help develop KBC systems. The key idea in DeepDive is that statistical inference and machine learning are key tools to attack classical data problems in extraction, cleaning, and integration in a unified and more effective manner. DeepDive programs are declarative in that one cannot write probabilistic inference algorithms; instead, one interacts by defining features or rules about the domain. A key reason for this design choice is to enable domain experts to build their own KBC systems. We present the applications, abstractions, and techniques of DeepDive employed to accelerate construction of KBC systems.
1. INTRODUCTION
The process of populating a structured relational database from unstructured sources has received renewed interest in the database community through high-profile start-up companies (e.g., Tamr and Trifacta), established companies like IBM’s Watson [5, 13], and a variety of research efforts [9, 24, 29, 38, 43]. At the same time, communities such as those of natural language processing and machine learning are attacking similar problems under the name knowledge base construction (KBC) [3, 11, 20]. While different communities place differing emphasis on the extraction, cleaning, and integration phases, all seem to be converging toward a common set of techniques that include a mix of data processing, machine learning, and engineers-in-the-loop.
The ultimate goal of KBC is to obtain high-quality structured data from unstructured information. The output databases produced are richly structured with tens of different entity types in complex relationships. Typically, quality is assessed using two complementary measures: precision (how often a claimed tuple is correct) and recall (of the possible tuples to extract, how many are actually extracted). These systems can ingest massive numbers of documents–far outstripping the document counts of even well-funded human curation efforts. Industrially, KBC systems are constructed by skilled engineers in a months-long (or longer) process–not a one-shot algorithmic task. Arguably, the most important question in such systems is how to best use skilled engineers’ time to rapidly improve data quality. In its full generality, this question spans a number of areas in computer science, including programming languages, systems, and HCI. We focus on a narrower question, with the axiom that the more rapidly the programmer moves through the KBC construction loop, the more quickly she obtains high-quality data.
This paper presents DeepDive, our open-source engine for knowledge base construction. DeepDive’s language and execution model are similar to other KBC systems: DeepDive uses a high-level declarative language [9, 29, 31]. From a database perspective, DeepDive’s language is based on SQL. From a machine learning perspective, DeepDive’s language is based on Markov Logic [10, 31]: DeepDive’s language inherits Markov Logic Networks’ (MLN’s) formal semantics. Moreover, it uses a standard execution model for such systems [9, 29, 31] in which programs go through two main phases: grounding, in which one evaluates a sequence of SQL queries to produce a data structure called a factor graph that describes a set of random variables and how they are correlated. Essentially, every tuple in the database or result of a query is a random variable (node) in this factor graph. The inference phase takes the factor graph from the grounding phase and performs statistical inference using standard techniques, e.g., Gibbs sampling [44, 47]. The output of inference is the marginal probability of every tuple in the database. As with Google’s Knowledge Vault [11] and others [32], DeepDive also produces marginal probabilities that are calibrated: if one examined all facts with probability 0.9, we would expect approximately 90% of these facts to be correct. To calibrate these probabilities, DeepDive estimates (i.e., learns) parameters of the statistical model from data. Inference is a subroutine of the learning procedure and is the critical loop. Inference and learning are computationally intense (hours on 1TB RAM/48-core machines).
In our experience with DeepDive, we found that KBC is an iterative process. In the past few years, DeepDive has been used to build dozens of high-quality KBC systems by a handful of technology companies, a number law enforcement agencies via DARPA’s MEMEX, and scientists in fields such as paleobiology, drug re-purposing, and genomics. Recently, we compared the quality of a DeepDive system’s extractions to those provided by human volunteers over the last ten years for a paleobiology database, and we found that the DeepDive system had higher quality (both precision and recall) on many entities and relationships. Moreover, on all of the extracted entities and relationships, DeepDive had no worse quality [34]. Additionally, the winning entry of the 2014 TAC-KBC competition was built on DeepDive [1]. In all cases, we have seen the process of developing KBC systems is iterative: quality requirements change, new data sources arrive, and new concepts are needed in the application. This led us to develop a set of techniques to make not only the execution of statistical inference and learning efficient, but also the entire pipeline incremental in the face of changes both to the data and to the DeepDive program.
This paper aims at giving a broad overview of DeepDive. The rest of the paper is organized as follows. Section 2 describes the KBC process, its scientific applications, and technical challenges. Section 3 presents our language for modeling KBC systems inside DeepDive. We discuss the different techniques in Section 4 and give pointers for readers who are interested in each technique.
2. APPLICATIONS AND CHALLENGES
Knowledge base construction (KBC) is the process of populating a knowledge base with facts extracted from unstructured data sources such as text, tabular data expressed in text and in structured forms, and even maps and figures, In sample-based science [34], one typically assembles a large number of facts (typically from the literature) to understand macroscopic questions, e.g., about the amount of carbon in the Earth’s atmosphere throughout time, the rate of extinction of species, or all the drugs that interact with a particular gene. To answer such questions, a key step is to construct a high-quality knowledge base, and some sciences have undertaken decade-long sample collection efforts, e.g., PaleoDB.org and PharmaGKB.org.
In parallel, KBC has attracted interest from industry [13, 49] and academia [2, 3, 6, 12, 21, 23, 29, 32, 35, 38, 40, 45]. To understand the common patterns in KBC systems, we are actively collaborating with scientists from a diverse set of domains, including geology [46], paleontology [34], pharmacology for drug repurposing, and others. We first describe one KBC application we built, called PaleoDeepDive, then present a brief description of other applications built with similar purposes, and then finally discuss the challenges.
2.1 PaleoDB and PaleoDeepDive
Paleontology is based on the description and biological classification of fossils, an enterprise that has been recorded in and an untold number of scientific publications over the past four centuries. One central task for paleontology is to construct a knowledge base about fossils from scientific publications, and an existing knowledge base compiled by human volunteers has greatly expanded the intellectual reach of paleontology and led to many fundamental new insights into macroevolutionary processes and the nature of biotic responses to global environmental change. However, the current process of using human volunteers is usually expensive and time-consuming. For example, PaleoDB, one of the largest such knowledge bases, took more than 300 professional paleontologists and 11 human years to build over the last two decades, resulting in PaleoDB.org. To get a sense of the impact of this database on this field, at the time of writing, this dataset has contributed to 205 publications, of which 17 have appeared in Nature or Science.
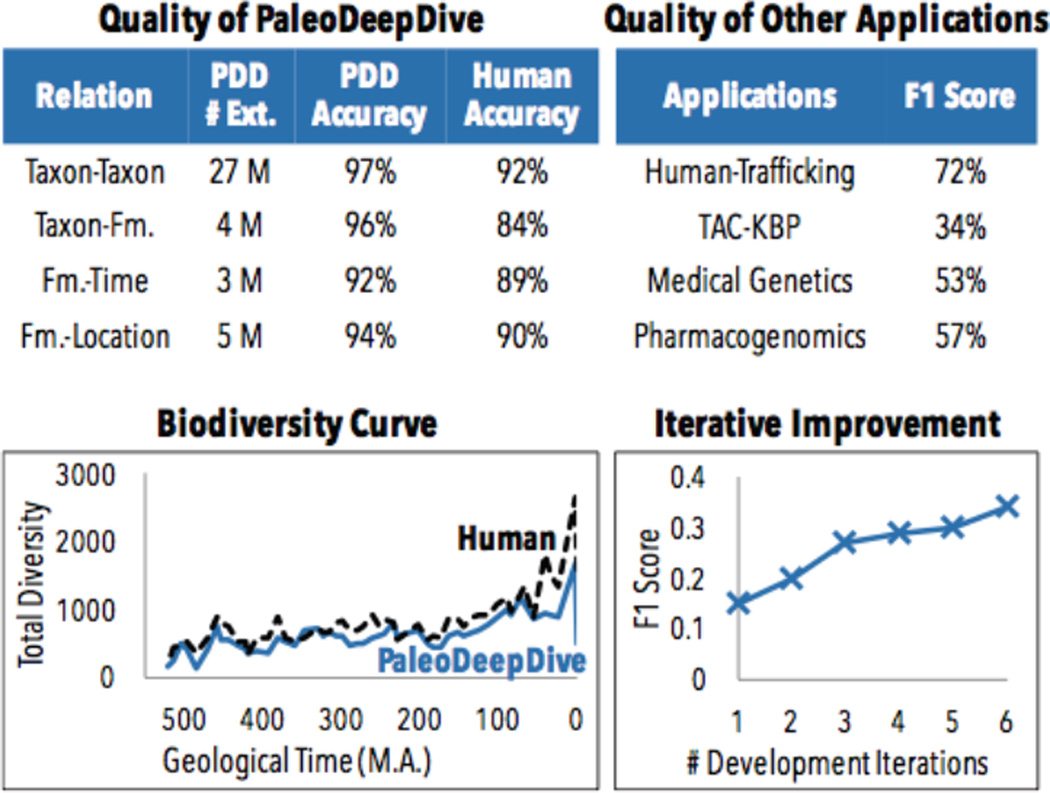
This provided an ideal test bed for our KBC research. In particular, we constructed a prototype called PaleoDeepDive [34] that takes in PDF documents. This prototype attacks challenges in optical character recognition, natural language processing, information extraction, and integration. Some statistics about the process are shown in Figure 3. As part of the validation of this system, we performed a double-blind experiment to assess the quality of the system versus the PaleoDB. We found that the KBC system built on DeepDive has achieved comparable—and sometimes better—quality than a knowledge base built by human volunteers over the last decade [34]. Figure 3 illustrates the accuracy of the results in PaleoDeepDive.
Figure 3.
Quality of KBC systems built with DeepDive. On many applications, KBC systems built with DeepDive achieves comparable (and sometimes better) quality than professional human volunteers, and leads to similar scientific insights on topics such as biodiversity. This quality is achieved by iteratively integrating diverse sources of data- often quality scales with the amount of information we enter into the system.
2.2 Beyond Paleontology
The success of PaleoDeepDive motivates a series of other KBC applications in a diverse set of domains including both natural and social sciences. Although these applications focus on very different types of KBs, they are usually built in a way similar to PaleoDeepDive. This similarity across applications motivate our study of building DeepDive as a unified framework to support these diverse applications.
Human-Trafficking
MEMEX is a DARPA program that explores how next generation search and extraction systems can help with real-world use cases. The initial application is the fight against human trafficking, hi this application, the input is a portion of the publicly-indexed and "dark" web in which human traffickers are likely to (surreptitiously) post supply and demand information about illegal labor, sex workers, and more. DeepDive processes such documents to extract evidential data such as names, addresses, phone numbers, job types, job requirements, information about rates of service, etc. Some of these data items are difficult for trained human annotators to accurately extract and have never been previously available, but DeepDive-based systems have high accuracy (Precision and Recall in the 90s, which may surpass that of non-experts). Together with provenance information, such structured, evidential data are then passed on to both other collaborators on the MEMEX program as well as law enforcement for analysis and consumption in operational applications. MEMEX has been featured extensively in the media and is supporting actual investigations. For example, every human trafficking investigation pursued by the Human Trafficking Response Unit in New York City now involves MEMEX, for which DeepDive is the main extracted data provider. In addition, future use cases such as applications in the war on terror are under active consideration.
Medical Genetics
The body of literature in life sciences has been growing at an accelerating speed, to the extent that it has been unrealistic for scientists to perform research solely based on reading and/or keyword search. Numerous manually-curated structured knowledge bases are likewise unable to keep pace with exponential increases in the number of publications available online. For example, OMIM is an authoritative database of human genes and mendelian genetic disorders which dates back to the 1960s, and so far contains about 6,000 hereditary diseases or phenotypes, growing at a rate of roughly 50 records / month for many years. Conversely, almost 10,000 publications were deposited into PubMed Central per month last year. In collaboration with Prof. Gill Bejerano at Stanford, we are developing DeepDive applications to create knowledge bases in the field of medical genetics. Specifically, we use DeepDive to extract mentions of genes, gene variants, and phenotypes from the literature, and statistically infer their relationships, presently being applied to clinical genetic diagnostics & reproductive counseling.
Pharmacogenomics
Understanding the interactions of chemicals in the body is key for drug discovery. However, the majority of this data resides in the biomedical literature and cannot be easily accessed. The Pharmacogenomics Knowledgebase is a high quality database that aims to annotate the relationships between drugs, genes, diseases, genetic variation, and pathways in the literature. With the exponential growth of the literature, manual curation requires prioritization of specific drugs or genes in order to stay up to date with current research. In collaboration with Emily Mallory and Prof. Russ Altman [27] at Stanford, we are developing DeepDive applications in the field of pharmacogenomics. Specifically, we use DeepDive to extract relations between genes, diseases, and drugs in order to predict novel pharmacological relationships.
TAC-KBP
TAC-KBP is a NIST-sponsored research competition where the task is to extract common properties of people and organizations (e.g., age, birthplace, spouses, and shareholders) from a 1.3 million newswire and web documents – this task is also termed Slot Filling. In the 2014 evaluation, 31 US and international teams participated in the competition, including a solution based on DeepDive from Stanford [1]. The DeepDive based solution achieved the highest precision, recall, and Fl among all submissions.
2.3 Challenges
On all the applications mentioned above, KBC systems built with DeepDive achieved high quality as illustrated in Figure 3. Achieving this high quality level requires that we solve a set of challenges.
Joint Statistical Inference
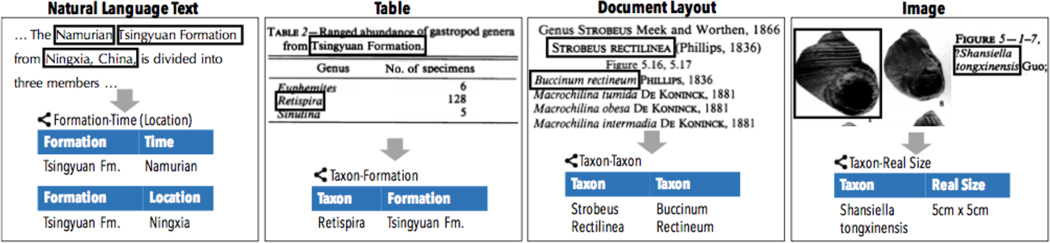
We have found that text is often not enough: often, the data that are interesting to scientists are located in the tables, figures, and images of articles. For example, in geology, more than 50% of the facts that we are interested in are buried in tables [14]. For paleontology, the relationship between taxa, as known as taxonomy, is almost exclusively expressed in section headers [34]. For pharmacology, it is not uncommon for a simple diagram to contain a large number of metabolic pathways. To build a KBC system with the quality that scientists will be satisfied with, we need to deal with these diverse sources of input. Additionally, external sources of information (other knowledge bases) typically contain high-quality signals (e.g., Freebase and Macrostrat). Leveraging these sources in information extraction is typically not studied in the classical information extraction context. To perform high-quality and high-coverage knowledge extraction, one needs a model that is able to ingest whatever sources present themselves, opportunistically—that is, a model which is not tied solely to text but can handle more general extraction and integration from multiple source types simultaneously.
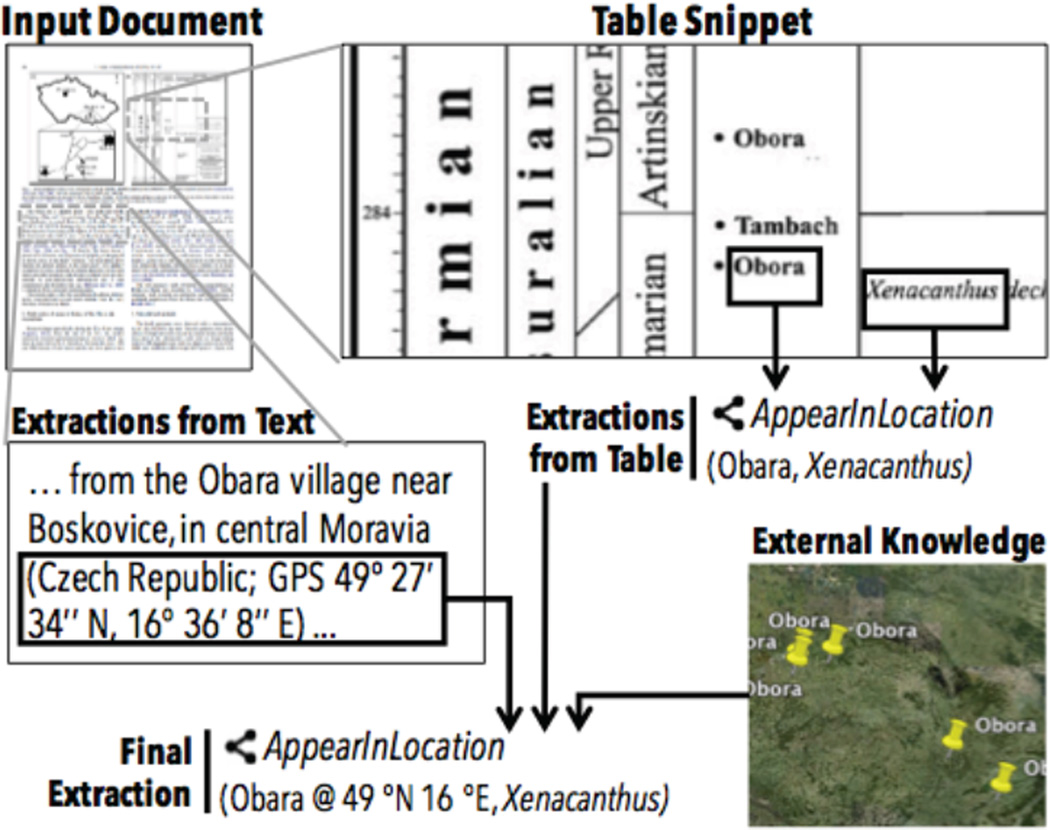
This challenge becomes more serious when the information from different sources are all noisy. Take Figure 4 for example, to reach the extraction that the genus Xenacanthus appears in the location of the name Obara, the extraction system needs to consult extractions from text, tables, and external structured sources. These extractions are often associated with a confidence score. To join these extractions with difference confidence level together, one needs a principled framework. The DeepDive approach to this challenge is based on a Bayesian probabilistic approach. DeepDive treats all these information sources as one joint probabilistic inference problem, with all predictions modeled as random variables within a factor graph model. This probabilistic framework ensures all facts that are produced by DeepDive are associated with a marginal probability. These marginal probabilities are meaningful in DeepDive, i.e., the empirical accuracy that one should expect for the extracted mentions, and provide a guideline to the developer to improve the KBC system built using DeepDive. In Section 3, we present a declarative language inside DeepDive to help developers specify a joint statistical inference problem easily.
Figure 4.
One challenge of building high-quality KBC systems is dealing with diverse sources jointly to make predictions. In this example page of a Paleontology journal article, information extracted from tables, text, and external structured knowledge bases are all required to reach the final extraction. This problem becomes even more challenging when many extractors are not 100% accurate, thus motivating the joint probabilistic inference engine inside DeepDive.
Scalability and Efficiency
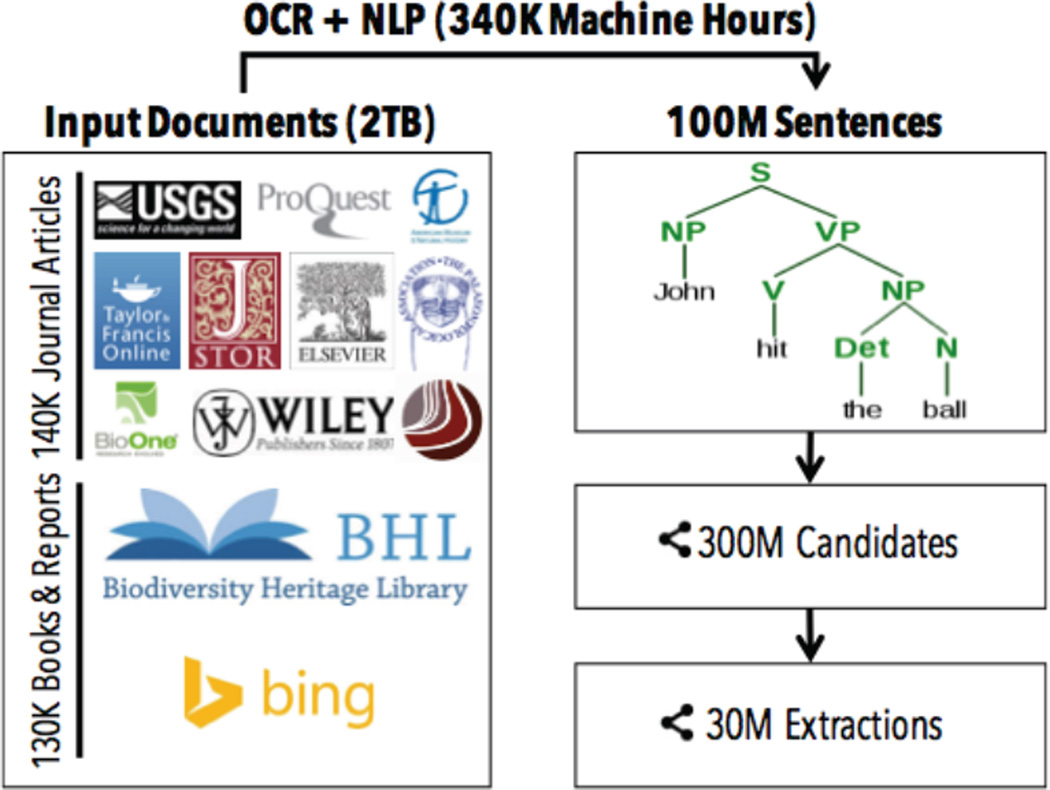
Performance is also a major challenge. In our KBC systems using DeepDive, we may need to perform inference and learning on billions of highly correlated random variables. For example, Figure 5 illustrates the data flow of PaleoDeepDive. The input to PaleoDeepDive contains nearly 300K journal articles and books, whose total size exceeds 2TB. These raw inputs are then processed with tools such as OCR and linguistic parsing, which are computationally expensive and may take hundreds of thousands of machine hours. The outputs of these tools are then used by DeepDive to construct factor graphs which contain more than 300 million variables as candidates for predictions (where over 30 million of these variables have probability ⩾ 0.9 and are thus output as final predictions). Therefore, one of our technical focus areas has been to speed up probabilistic inference [30, 31, 33, 47, 48]. In Section 4, we briefly describe these techniques and provide pointers to readers who are interested in further details.
Figure 5.
Another challenge of building high-quality KBC systems is that one usually needs to deal with data at the scale of tera-bytes. These data are not only processed with traditional relational operations, but also operations involving machine learning and statistical inference. Thus, DeepDive consists of a set of techniques to speed up and scale up inference tasks involving billions of correlated random variables.
3. KBC USING DEEPDIVE
We describe DeepDive, an end-to-end framework for building KBC systems with a declarative language.
3.1 Definitions for KBC Systems
The input to a KBC system is a heterogeneous collection of unstructured, semi-structured, and structured data, ranging from text documents to existing but incomplete KBs. The output of the system is a relational database containing relations extracted from the input and put into an appropriate schema. Creating the knowledge base may involve extraction, cleaning, and integration.
Example 3.1
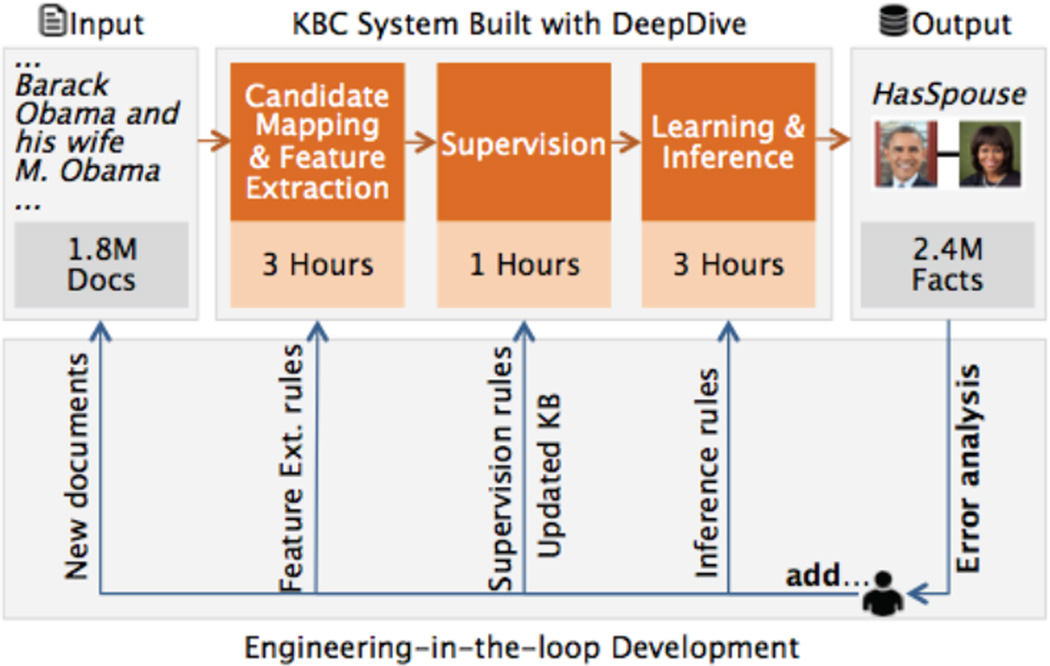
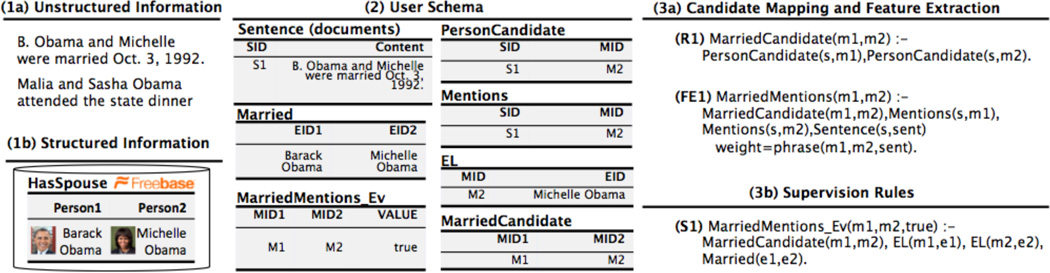
Figure 6 illustrates a new running example: a knowledge base with pairs of individuals that are married to each other. The input to the system is a collection of news articles and an incomplete set of married persons; the output is a KB containing pairs of person that are asserted to be married by the input sources. A KBC system extracts linguistic patterns, e.g., “… and his wife…” between a pair of mentions of individuals (e.g., “Barack Obama” and ”M. Obama“). Roughly, these patterns are then used as features in a classifier deciding whether this pair of mentions indicates that they are married (in the HasSpouse) relation.
Figure 6.
A KBC system takes as input unstructured documents and outputs a structured knowledge base. The runtimes are for the TAC-KBP competition system. To improve quality, the developer adds new rules and new data with error analysis conducted on the result of the current snapshot of the system. DeepDive provides a declarative language to specify each type of different rules and data, and techniques to incrementally execute this iterative process.
We adopt standard terminology from KBC, e.g., ACE. There are four types of objects that a KBC system seeks to extract from input documents, namely entities, relations, mentions, and relation mentions. An entity is a real-world person, place, or thing. For example, “Michelle_Obama_1” represents the actual entity for a person whose name is “Michelle Obama”; another individual with the same name would have another number. A relation associates two (or more) entities, and represents the fact that there exists a relationship between the participating entities. For example, “Barack_Obama_1” and “Michelle_Obama_1” participate in the HasSpouse relation, which indicates that they are married. These real-world entities and relationships are described in text; a mention is a span of text in an input document that refers to an entity or relationship: “Michelle” may be a mention of the entity “Michelle_Obama_1.” A relation mention is a phrase that connects two mentions that participate in a relation such as “(Barack Obama, M. Obama)”. The process of mapping mentions to entities is called entity linking.
3.2 The DeepDive Framework
DeepDive is an end-to-end framework for building KBC systems, as shown in Figure 6. We walk through each phase. DeepDive supports both SQL and datalog, but we use datalog syntax for this exposition. The rules we describe in this section are manually created by the user of DeepDive and the process of creating these rules is application specific.
Candidate Mapping and Feature Extraction
All data in DeepDive is stored in a relational database. The first phase populates the database using a set of SQL queries and user-defined functions (UDFs) that we call feature extractors. By default, DeepDive stores all documents in the database in one sentence per row with markup produced by standard NLP pre-processing tools, including HTML stripping, part-of-speech tagging, and linguistic parsing. After this loading step, DeepDive executes two types of queries: (1) candidate mappings, which are SQL queries that produce possible mentions, entities, and relations, and (2) feature extractors that associate features to candidates, e.g., “… and his wife …” in Example 3.1.
Example 3.2
Candidate mappings are usually simple. Here, we create a relation mention for every pair of candidate persons in the same sentence (s):
(R1) MarriedCandidate(m1, m2):-
PersonCandidate(s, m1),PersonCandidate(s, m2).
Candidate mappings are simply SQL queries with UDFs that look like low-precision but high-recall ETL scripts. Such rules must be high recall: if the union of candidate mappings misses a fact, DeepDive has no chance to extract it.
We also need to extract features, and we extend classical Markov Logic in two ways: (1) user-defined functions (UDFs) and (2) weight tying, which we illustrate by example.
Example 3.3
Suppose that phrase(m1, m2, sent) returns the phrase between two mentions in the sentence, e.g., “and his wife” in the above example. The phrase between two mentions may indicate whether two people are married. We would write this as:
(FE1) MarriedMentions(m1, m2):-
MarriedCandidate(m1, m2),Mention(s, m1),
Mention(s, m2),Sentence(s, sent)
weight = phrase(m1, m2, sent).
One can think about this like a classifier: This rule says that whether the text indicates that the mentions m1 and m2 are married is influenced by the phrase between those mention pairs. The system will infer based on training data its confidence (by estimating the weight) that two mentions are indeed indicated to be married.
Technically, phrase returns an identifier that determines which weights should be used for a given relation mention in a sentence. If phrase returns the same result for two relation mentions, they receive the same weight. We explain weight tying in more detail in Section 3.3. In general, phrase could be an arbitrary UDF that operates in a per-tuple fashion. This allows DeepDive to support common examples of features such as “bag-of-words” to context-aware NLP features to highly domain-specific dictionaries and ontologies. In addition to specifying sets of classifiers, DeepDive inherits Markov Logic’s ability to specify rich correlations between entities via weighted rules. Such rules are particularly helpful for data cleaning and data integration.
Supervision
Just as in Markov Logic, DeepDive can use training data or evidence about any relation; in particular, each user relation is associated with an evidence relation with the same schema and an additional field that indicates whether the entry is true or false. Continuing our example, the evidence relation MarriedMentions_Ev could contain mention pairs with positive and negative labels. Operationally, two standard techniques generate training data: (1) hand-labeling, and (2) distant supervision, which we illustrate below.
Example 3.4
Distant supervision [17, 28] is a popular technique to create evidence in KBC systems. The idea is to use an incomplete KB of married entity pairs to heuristically label (as True evidence) all relation mentions that link to a pair of married entities:
(S1) MarriedMentions_Ev(m1, m2, true):-
MarriedCandidates(m1, m2),EL(m1, e1),
EL(m2, e2),Married(e1, e2).
Here, Married is an (incomplete) list of married real-world persons that we wish to extend. The relation EL is for “entity linking” that maps mentions to their candidate entities. At first blush, this rule seems incorrect. However, it generates noisy, imperfect examples of sentences that indicate two people are married. Machine learning techniques are able to exploit redundancy to cope with the noise and learn the relevant phrases (e.g., “and his wife”). Negative examples are generated by relations that are largely disjoint (e.g., siblings). Similar to DIPRE [4] and Hearst patterns [16], distant supervision exploits the “duality” [4] between patterns and relation instances; furthermore, it allows us to integrate this idea into DeepDive’s unified probabilistic framework.
Learning and Inference
In the learning and inference phase, DeepDive generates a factor graph, similar to Markov Logic, and uses techniques from Tuffy [31]. The inference and learning are done using standard techniques (Gibbs Sampling) that we describe below after introducing the formal semantics.
Error Analysis
DeepDive runs the above three phases in sequence, and at the end of the learning and inference, it obtains a marginal probability p for each candidate fact. To produce the final KB, the user often selects facts in which DeepDive is highly confident, e.g., p > 0.95. Typically, the user needs to inspect errors and repeat the previous steps, a process that we call error analysis. Error analysis is the process of understanding the most common mistakes (incorrect extractions, too-specific features, candidate mistakes, etc.) and deciding how to correct them [36]. To facilitate error analysis, users write standard SQL queries.
3.3 Discussion of Design Choices
We have found the following key aspects of the DeepDive approach that we believe enable non-computer scientists to build sophisticated KBC systems: (1) there is no reference in a DeepDive program to the underlying machine learning algorithms. Thus, DeepDive programs are declarative in a strong sense. Probabilistic semantics provide a way to debug the system independent of the algorithm it uses. (2) DeepDive allows users to write feature extraction code (UDFs) in familiar languages (Python, SQL, and Scala). (3) DeepDive fits into the familiar SQL stack, which allows standard tools to inspect and visualize the data. (4) The user constructs an end-to-end system and then refines the quality of the system in a pay-as-you-go way [26]. In contrast, traditional pipeline-based ETL scripts may lead to user’s time and effort over-spent on a specific extraction or integration step–without the ability to evaluate how important each step is for the quality of the end result. Anecdotally, pay-as-you-go leads to more informed decisions about how to improve quality.
4. TECHNIQUES
A DeepDive program is a set of rules with weights specified using the language we described above. During inference, the values of all weights are assumed to be known, while, in learning, one finds the set of weights that maximizes the probability of the evidence. The execution of a DeepDive program consists of two phases, namely grounding and statistical inference and learning. In this section, we briefly describe the techniques we developed in each phase to make DeepDive performant and scalable.
4.1 Grounding
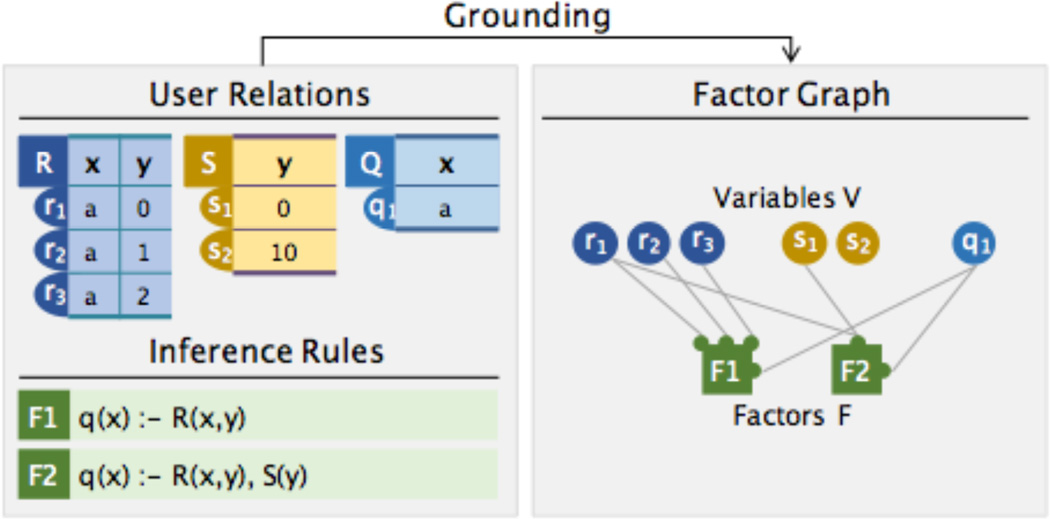
As in Figure 8, DeepDive explicitly constructs a factor graph for inference and learning using a set of SQL queries. A factor graph is a triple (𝖵, F, ŵ) in which 𝖵 is a set of nodes that correspond to Boolean random variables, 𝖥 is a set of hyperedges (for 𝖿 ∈ 𝖥, 𝖿 ⊆ 𝖵), and ŵ : 𝖥 × {0, 1}𝖵 → ℝ is a weight function. In DeepDive, each hyperedge 𝖿 corresponds to the set of groundings for a rule. In DeepDive, 𝖵 and 𝖥 are explicitly created using a set of SQL queries, and this process is called grounding.
Figure 8.
Schematic illustration of grounding. Each tuple corresponds to a Boolean random variable and node in the factor graph. We create one factor for every set of groundings.
Example 4.1
Take the database instances and rules in Figure 8 as an example: each tuple in relation 𝖱, 𝖲, and 𝖰 is a random variable, and 𝖵 contains all random variables. The inference rules 𝖥 and 𝖥2 ground factors with the same name in the factor graph as illustrated in Figure 8. Both 𝖥1 and 𝖥2 are implemented as SQL statements in DeepDive.
Incremental Grounding
Because DeepDive is based on SQL, we are able to take advantage of decades of work on incremental view maintenance. The input to this phase is the same as the input to the grounding phase, a set of SQL queries and the user schema. The output of this phase is how the output of grounding changes, i.e., a set of modified variables Δ𝖵 and their factors Δ𝖥. Since 𝖵 and 𝖥 are simply views over the database, any view maintenance techniques can be applied to incremental grounding. DeepDive uses the DRed algorithm [15] which handles both additions and deletions. Recall that in DRed, for each relation 𝖱𝗂 in the user’s schema, we create a delta relation, , with the same schema as 𝖱𝗂 and an additional column count. For each tuple t, t.count represents the number of derivations of 𝗍 in 𝖱𝗂. On an update, DeepDive updates delta relations in two steps. First, for tuples in , DeepDive directly updates the corresponding counts. Second, a SQL query called a “delta rule” is executed which processes these counts to generate modified variables Δ𝖵 and factors Δ𝖥. We found that the overhead of DRed is modest and the gains may be substantial, so DeepDive always runs DRED–except on initial load.
4.2 Statistical Inference and Learning
The main task that DeepDive conducts on factor graphs is statistical inference, i.e. determining for a given node what the marginal probability is that this node takes the value 1. Since a node takes value 1 when a tuple is in the output, this process computes the marginal probability values returned to users. In general, computing these marginal probabilities is ♯𝖯-hard [42]. Like many other systems, DeepDive uses Gibbs sampling [37] to estimate the marginal probability of every tuple in the database.
Efficiency and Scalability
There are two components to scaling statistical algorithms: statistical efficiency, roughly how many steps an algorithm takes to converge, and hardware efficiency, how efficient each of those step is. We introduced this terminology and studied this extensively in a recent paper [48].
DimmWitted, the statistical inference and learning engine in DeepDive [48] is built upon our research of how to design a high-performance statistical inference and learning engine on a single machine [25, 30, 47, 48]. DimmWitted models Gibbs sampling as a “column-to-row access” operation: each row corresponds to one factor, each column to one variable, and the non-zero elements in the matrix correspond to edges in the factor graph. To process one variable, DimmWitted fetches one column of the matrix to get the set of factors, and other columns to get the set of variables that connect to the same factor. In standard benchmarks, DimmWitted was 3.7× faster than GraphLab’s implementation without any application-specific optimization. Compared with traditional work, the main novelty of DimmWitted is that it considers both hardware efficiency and statistical efficiency for executing an inference and learning task.
Hardware Efficiency DeepDive takes into consideration the architecture of modern Non-uniform memory access (NUMA) machines. A NUMA machine usually contains multiple nodes (sockets), where each sockets contains multiple CPU cores. To achieve high hardware efficiency, one wants to decrease the communication across different NUMA nodes.
Statistical Efficiency Pushing hardware efficiency to the extreme might decrease statistical efficiency because the lack of communication between nodes might decrease the rate of convergence of a statistical inference and learning algorithm. DeepDive takes advantage of the theoretical results of model averaging [50] and our own results about lock-free execution [25, 30].
On the whole corpus of Paleobiology, the factor graph contains more than 0.2 billion random variables and 0.3 billion factors. On this factor graph, DeepDive is able to run Gibbs sampling on a machine with 4 sockets (10 cores per socket), and we find that we can generate 1,000 samples for all 0.2 billion random variables in 28 minutes. This is more than 4× faster than a non-NUMA-aware implementation.
Incremental Inference
Due to our choice of incremental grounding, the input to DeepDive’s inference phase is a factor graph along with a set of changed variables and factors. The goal is to compute the output probabilities computed by the system. Our approach is to frame the incremental maintenance problem as approximate inference. Previous work in the database community has looked at how machine learning data products change in response to both to new labels [22] and to new data [7, 8]. In KBC, both the program and data change on each iteration. Our proposed approach can cope with both types of change simultaneously.
The technical question is which approximate inference algorithms to use in KBC applications. We choose to study two popular classes of approximate inference techniques: sampling-based materialization (inspired by sampling-based probabilistic databases such as MCDB [18]) and variational-based materialization (inspired by techniques for approximating graphical models [41]). Applying these techniques to incremental maintenance for KBC is novel, and it is not theoretically clear how the techniques compare. Thus, we conducted an experimental evaluation of these two approaches on a diverse set of DeepDive programs. We found these two approaches are sensitive to changes along three largely orthogonal axes: the size of the factor graph, the sparsity of correlations, and the anticipated number of future changes. The performance varies by up to two orders of magnitude in different points of the space. Our study of the tradeoff space highlights that neither materialization strategy dominates the other. To automatically choose the materialization strategy, we developed a simple rule-based optimizer [39].
5. RELATED WORK
Knowledge Base Construction (KBC) has been an area of intense study over the last decade [2, 3, 6, 12, 21, 23, 29, 35, 38, 40, 45, 49]. Within this space, there are a number of approaches.
Rule-Based Systems
The earliest KBC systems used pattern matching to extract relationships from text. The most well-known example is the “Hearst Pattern” proposed by Hearst [16] in 1992. In her seminal work, Hearst observed that a large number of hyponyms can be discovered by simple patterns, e.g., “X such as Y.” Hearst’s technique has formed the basis of many further techniques that attempt to extract high-quality patterns from text. Rule-based (pattern matching-based) KBC systems, such as IBM’s SystemT [23, 24], have been built to aid developers in constructing high-quality patterns. These systems provide the user with a (declarative) interface to specify a set of rules and patterns to derive relationships. These systems have achieved state-of-the-art quality on tasks such as parsing [24].
Statistical Approaches
One limitation of rule-based systems is that the developer needs to ensure that all rules provided to the system are high-precision rules. For the last decade, probabilistic (or machine learning) approaches have been proposed to allow the system to select from a range of a priori features automatically. In these approaches, the extracted tuple is associated with a marginal probability that it is true. DeepDive, Google’s knowledge graph, and IBM’s Watson are built on this approach. Within this space, there are three styles of systems that based on classification-based frameworks [2, 3, 6, 12, 45], maximum a posteriori (MAP) [21, 29, 40], and probabilistic graphical models [10, 35, 49]. Our work on DeepDive is based on graphical models.
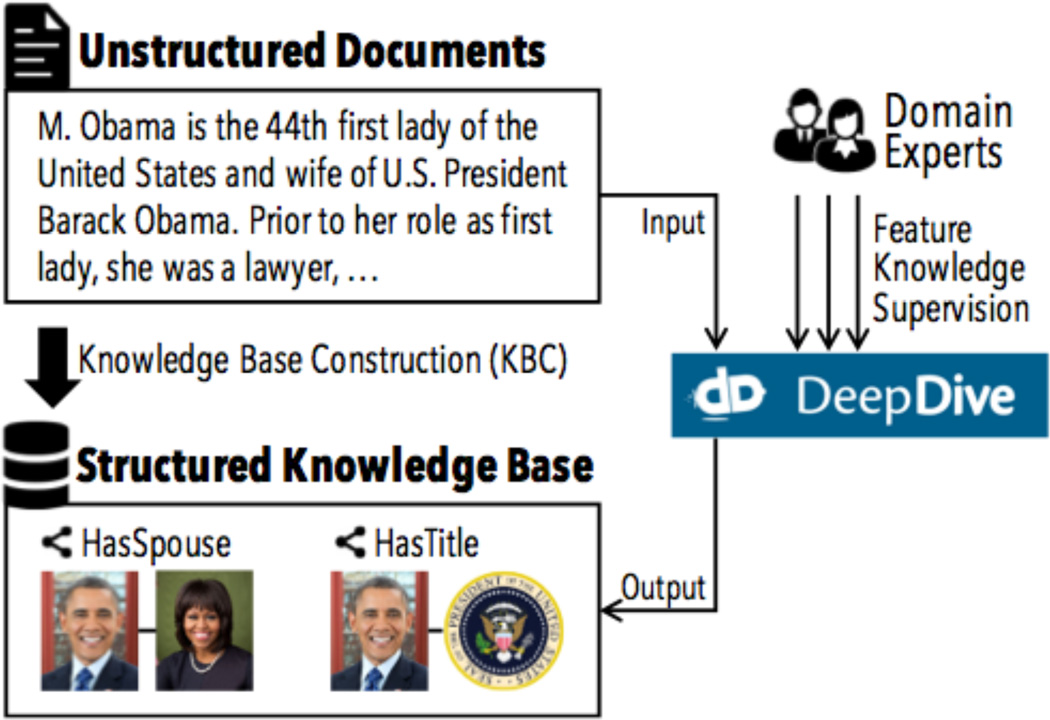
Figure 1.
Knowledge Base Construction (KBC) is the process of populating a structured relational knowledge base from unstructured sources. DeepDive is a system aimed at facilitating the KBC process by allowing domain experts to integrate their domain knowledge without worrying about algorithms.
Figure 2.
Example KBC Application Built with DeepDive.
Figure 7.
An example KBC system. See Section 3.2 for details.
Acknowledgments
We gratefully acknowledge the support of the Defense Advanced Research Projects Agency (DARPA) XDATA program under No. FA8750-12-2-0335 and DEFT program under No. FA8750-13-2-0039, DARPA’s MEMEX program and SIMPLEX program, the National Science Foundation (NSF) CAREER Award under No. IIS-1353606, the Office of Naval Research (ONR) under awards No. N000141210041 and No. N000141310129, the National Institutes of Health Grant U54EB020405 awarded by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) through funds provided by the trans-NIH Big Data to Knowledge (BD2K) initiative, the Sloan Research Fellowship, the Moore Foundation, American Family Insurance, Google, and Toshiba. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of DARPA, AFRL, NSF, ONR, NIH, or the U.S. government.
Footnotes
There is a justification for probabilistic reasoning as Cox’s theorem asserts (roughly) that if one uses numbers as degrees of belief, then one must either use probabilistic reasoning or risk contradictions in one’s reasoning system, i.e., a probabilistic framework is the only sound system for reasoning in this manner. We refer the reader to Jaynes [19].
For more information, including examples, please see http://deepdive.Stanford.edu. Note that our engine is built on Postgres and Greenplum for all SQL processing and UDFs. There is also a port to MySQL.
For example, for the grounding procedure illustrated in Figure 8, the delta rule for 𝖥1 is qδ (x) : −𝖱δ (x, y).
REFERENCES
- 1.Angeli G, et al. Stanford’s 2014 slot filling systems. TAC KBP. 2014 [Google Scholar]
- 2.Banko M, et al. Open information extraction from the Web. IJCAI. 2007 [Google Scholar]
- 3.Betteridge J, Carlson A, Hong SA, Hruschka ER, Jr, Law EL, Mitchell TM, Wang SH. Toward never ending language learning. AAAI Spring Symposium. 2009 [Google Scholar]
- 4.Brin S. Extracting patterns and relations from the world wide web. WebDB. 1999 [Google Scholar]
- 5.Brown E, et al. Tools and methods for building watson. IBM Research Report. 2013 [Google Scholar]
- 6.Carlson A, et al. Toward an architecture for never-ending language learning. AAAI. 2010 [Google Scholar]
- 7.Chen F, Doan A, Yang J, Ramakrishnan R. Efficient information extraction over evolving text data. ICDE. 2008 [Google Scholar]
- 8.Chen F, et al. Optimizing statistical information extraction programs over evolving text. ICDE. 2012 [Google Scholar]
- 9.Chen Y, Wang DZ. Knowledge expansion over probabilistic knowledge bases. SIGMOD. 2014 [Google Scholar]
- 10.Domingos P, Lowd D. Markov Logic: An Interface Layer for Artificial Intelligence. Morgan & Claypool. 2009 [Google Scholar]
- 11.Dong XL, et al. From data fusion to knowledge fusion. VLDB. 2014 [Google Scholar]
- 12.Etzioni O, et al. Web-scale information extraction in KnowItAll: (preliminary results) WWW. 2004 [Google Scholar]
- 13.Ferrucci D, et al. Building Watson: An overview of the DeepQA project. AI Magazine. 2010 [Google Scholar]
- 14.Govindaraju V, et al. Understanding tables in context using standard NLP toolkits. ACL. 2013 [Google Scholar]
- 15.Gupta A, Mumick IS, Subrahmanian VS. Maintaining views incrementally. SIGMOD Rec. 1993 [Google Scholar]
- 16.Hearst MA. Automatic acquisition of hyponyms from large text corpora. COLING. 1992 [Google Scholar]
- 17.Hoffmann R, et al. Knowledge-based weak supervision for information extraction of overlapping relations. ACL. 2011 [Google Scholar]
- 18.Jampani R, et al. MCDB: A Monte Carlo approach to managing uncertain data. SIGMOD. 2008 [Google Scholar]
- 19.Jaynes ET. Probability Theory: The Logic of Science. Cambridge University Press; 2003. [Google Scholar]
- 20.Jiang S, et al. Learning to refine an automatically extracted knowledge base using Markov logic. ICDM. 2012 [Google Scholar]
- 21.Kasneci G, et al. The YAGO-NAGA approach to knowledge discovery. SIGMOD Rec. 2009 [Google Scholar]
- 22.Koc ML, Ré C. Incrementally maintaining classification using an RDBMS. PVLDB. 2011 [Google Scholar]
- 23.Krishnamurthy R, et al. SystemT: A system for declarative information extraction. SIGMOD Rec. 2009 [Google Scholar]
- 24.Li Y, Reiss FR, Chiticariu L. SystemT: A declarative information extraction system. HLT. 2011 [Google Scholar]
- 25.Liu J, et al. An asynchronous parallel stochastic coordinate descent algorithm. ICML. 2014 [Google Scholar]
- 26.Madhavan J, et al. Web-scale data integration: You can only afford to pay as you go. CIDR. 2007 [Google Scholar]
- 27.Mallory EK, et al. Large-scale extraction of gene interactions from full text literature using deepdive. Bioinformatics. 2015 doi: 10.1093/bioinformatics/btv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mintz M, et al. Distant supervision for relation extraction without labeled data. ACL. 2009 [Google Scholar]
- 29.Nakashole N, et al. Scalable knowledge harvesting with high precision and high recall. WSDM. 2011 [Google Scholar]
- 30.Niu F, et al. Hogwild!: A lock-free approach to parallelizing stochastic gradient descent. NIPS. 2011 [Google Scholar]
- 31.Niu F, et al. Tuffy: Scaling up statistical inference in Markov logic networks using an RDBMS. PVLDB. 2011 [Google Scholar]
- 32.Niu F, et al. Elementary: Large-scale knowledge-base construction via machine learning and statistical inference. Int. J. Semantic Web Inf. Syst. 2012 [Google Scholar]
- 33.Niu F, et al. Scaling inference for Markov logic via dual decomposition. ICDM. 2012 [Google Scholar]
- 34.Peters SE, et al. A machine reading system for assembling synthetic Paleontological databases. PloS ONE. 2014 doi: 10.1371/journal.pone.0113523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon H, Domingos P. Joint inference in information extraction. AAAI. 2007 [Google Scholar]
- 36.Ré C, et al. Feature engineering for knowledge base construction. IEEE Data Eng. Bull. 2014 [Google Scholar]
- 37.Robert CP, Casella G. Monte Carlo Statistical Methods. Secaucus, NJ, USA: Springer-Verlag New York, Inc.; 2005. [Google Scholar]
- 38.Shen W, et al. Declarative information extraction using datalog with embedded extraction predicates. VLDB. 2007 [Google Scholar]
- 39.Shin J, et al. Incremental knowledge base construction using deepdive. PVLDB. 2015 doi: 10.14778/2809974.2809991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suchanek FM, et al. SOFIE: A self-organizing framework for information extraction. WWW. 2009 [Google Scholar]
- 41.Wainwright M, Jordan M. Log-determinant relaxation for approximate inference in discrete Markov random fields. Trans. Sig. Proc. 2006 [Google Scholar]
- 42.Wainwright MJ, Jordan MI. Graphical models, exponential families, and variational inference. FTML. 2008 [Google Scholar]
- 43.Weikum G, Theobald M. From information to knowledge: Harvesting entities and relationships from web sources. PODS. 2010 [Google Scholar]
- 44.Wick M, et al. Scalable probabilistic databases with factor graphs and MCMC. PVLDB. 2010 [Google Scholar]
- 45.Yates A, et al. TextRunner: Open information extraction on the Web. NAACL. 2007 [Google Scholar]
- 46.Zhang C, et al. GeoDeepDive: statistical inference using familiar data-processing languages. SIGMOD. 2013 [Google Scholar]
- 47.Zhang C, Ré C. Towards high-throughput Gibbs sampling at scale: A study across storage managers. SIGMOD. 2013 [Google Scholar]
- 48.Zhang C, Ré C. DimmWitted: A study of main-memory statistical analytics. PVLDB. 2014 [Google Scholar]
- 49.Zhu J, et al. StatSnowball: A statistical approach to extracting entity relationships. WWW. 2009 [Google Scholar]
- 50.Zinkevich M, et al. Parallelized stochastic gradient descent. NIPS. 2010:2595–2603. [Google Scholar]