Abstract
Large networks are becoming a widely used abstraction for studying complex systems in a broad set of disciplines, ranging from social network analysis to molecular biology and neuroscience. Despite an increasing need to analyze and manipulate large networks, only a limited number of tools are available for this task.
Here, we describe Stanford Network Analysis Platform (SNAP), a general-purpose, high-performance system that provides easy to use, high-level operations for analysis and manipulation of large networks. We present SNAP functionality, describe its implementational details, and give performance benchmarks. SNAP has been developed for single big-memory machines and it balances the trade-off between maximum performance, compact in-memory graph representation, and the ability to handle dynamic graphs where nodes and edges are being added or removed over time. SNAP can process massive networks with hundreds of millions of nodes and billions of edges. SNAP offers over 140 different graph algorithms that can efficiently manipulate large graphs, calculate structural properties, generate regular and random graphs, and handle attributes and meta-data on nodes and edges. Besides being able to handle large graphs, an additional strength of SNAP is that networks and their attributes are fully dynamic, they can be modified during the computation at low cost. SNAP is provided as an open source library in C++ as well as a module in Python.
We also describe the Stanford Large Network Dataset, a set of social and information real-world networks and datasets, which we make publicly available. The collection is a complementary resource to our SNAP software and is widely used for development and benchmarking of graph analytics algorithms.
CCS Concepts: Information systems → Data management systems, Computing platforms, Data mining, Main memory engines
Additional Key Words and Phrases: Networks, Graphs, Graph Analytics, Open-Source Software, Data Mining
1. INTRODUCTION
The ability to analyze large networks is fundamental to study of complex systems in many scientific disciplines [Easley and Kleinberg 2010; Jackson 2008; Newman 2010]. With networks, we are able to capture relationships between entities, which allows us to gain deeper insights into the systems being analyzed [Newman 2003]. This increased importance of networks has sparked a growing interest in network analysis tools [Batagelj and Mrvar 1998; Hagberg et al. 2008; Kyrola et al. 2012; Malewicz et al. 2010].
Network analysis tools are expected to fulfill a set of requirements. They need to provide rich functionality, implementing a wide range of graph and network analysis algorithms. Implementations of graph algorithms must be able to process graphs with 100s of millions of nodes. Graphs need to be represented in a compact form with a small memory footprint, since many algorithms are bound by the memory throughput. Powerful operators are required for modifying graph structure, so that nodes and edges in a graph can be added or removed, or new graphs can be constructed from existing ones. Additionally for a wide system adoption, it is desirable that the source code is available under an open source license.
While there has been significant amount of work on systems for processing and analyzing large graphs, none of the existing systems fulfills the requirements outlined above. In particular, research on graph processing in large-scale distributed environments [Gonzalez et al. 2012; Malewicz et al. 2010; Kang et al. 2009; Salihoglu and Widom 2013; Xin et al. 2013] provides efficient frameworks, but these frameworks only implement a handful of most common graph algorithms, which in practice is not enough to make these tools useful for practitioners. Similarly, there are several user-friendly libraries that implement dozens of network analysis algorithms [Batagelj and Mrvar 1998; Csardi and Nepusz 2006; Gregor and Lumsdaine 2005; Hagberg et al. 2008; O’Madadhain et al. 2005]. However, the limitations of these systems are that they might not scale to large graphs, can be slow, hard to use, or do not include support for dynamic networks. Thus, there is a need for a system that addresses those limitations and provides reasonable scalability, is easy to use, implements numerous graph algorithms, and supports dynamic networks.
Here, we present Stanford Network Analysis Platform (SNAP), which was specifically built with the above requirements in mind. SNAP is a general-purpose, high-performance system that provides easy to use, high-level operations for analysis and manipulation of large networks. SNAP has been developed for single big-memory multiple-cores machines and as such it balances the trade-off between maximum performance, compact in-memory graph representation, and the ability to handle dynamic graphs where nodes and edges are being added or removed over time.
SNAP offers methods that can efficiently manipulate large graphs, calculate structural properties, generate regular and random graphs, and handle attributes on nodes and edges. Besides being able to handle large graphs, an additional strength of SNAP is that network structure and attributes are fully dynamic, they can be modified during the computation via low cost operations.
Overall, SNAP implements 8 graph and network types, 20 graph generation methods/models, 20 graph manipulation methods, and over 100 graph algorithms, which provides in total over 200 different functions. It has been used in a wide range of applications, such as network inference [Gomez-Rodriguez et al. 2014], network optimization [Hallac et al. 2015], information diffusion [Leskovec et al. 2009; Suen et al. 2013], community detection [Yang and Leskovec 2014], and geo-spatial network analysis [Leskovec and Horvitz 2014]. SNAP is provided for major operating systems as an open source library in C++ as well as a module in Python. It is released under the BSD open source license and can be downloaded from http://snap.stanford.edu/snap.
Complementary to the SNAP software, we also maintain public Stanford Large Network Dataset Collection, an extensive set of social and information networks with about 80 different network datasets. The collection includes online social networks with rich dynamics and node attributes, communication networks, scientific citation networks, collaboration networks, web graphs, Internet networks, online reviews, as well as social media data. The network datasets can be obtained at http://snap.Stanford.edu/data.
The remainder of the paper is organized as follows. We discuss related graph analysis systems in Section 2. The next two sections describe key principles behind SNAP. We give an overview of basic graph and network classes in SNAP in Section 3, while Section 4 focuses on graph methods. Implementational details are discussed in Section 5. An evaluation of SNAP and comparable systems with benchmarks on a range of graphs and graph algorithms is presented in Section 6. Next, in Section 7, we describe Stanford Large Network Dataset Collection and, in Section 8, SNAP documentation and its distribution license. Section 9 concludes the paper.
2. RELATED NETWORK ANALYSIS SYSTEMS
In this section we briefly survey related work on systems for processing, manipulating, and analyzing networks. We organize the section into two parts. First, we discuss single-machine systems and then proceed to discuss how SNAP relates to distributed systems for graph processing.
One of the first single-machine systems for network analysis is Pajek [Batagelj and Mrvar 1998], which is able to analyze networks with up to ten million nodes. Pajek is written in Pascal and is distributed as a self-contained system with its own GUI-based interface. It is only available as a monolithic Windows executable, and thus limited to the Windows operating system. It is hard to extend Pajek with additional functionality or use it as a library in another program. Originally, networks in Pajek are represented using doubly linked lists [Batagelj and Mrvar 1998] and while linked lists make it easy to insert and delete elements, they can be slow to traverse on modern CPUs, where sequential access to memory is much faster than random access.
Other widely used open source network analysis libraries that are similar in functionality to SNAP are NetworkX [Hagberg et al. 2008] and iGraph [Csardi and Nepusz 2006]. NetworkX is written in Python and implements a large number of network analysis methods. In terms of the speed vs. flexibility trade-off, NetworkX offers maximum flexibility at the expense of performance. Nodes, edges and attributes in NetworkX are represented by hash tables, called dictionaries in Python. Using hash tables for all graph elements allows for maximum flexibility, but imposes performance overhead in terms of a slower speed and a larger memory footprint than alternative representations. Additionally, since Python programs are interpreted, most operations in NetworkX take significantly longer time and require more memory than alternatives in compiled languages. Overall, we find SNAP to be one to two orders of magnitude faster than NetworkX, while also using around 50 times less memory. This means that, using the same hardware, SNAP can process networks that are 50 times larger or networks of the same size 100 times faster.
Similar to NetworkX in functionality but very different in implementation is the iGraph package [Csardi and Nepusz 2006]. iGraph is written in the C programming language and can be used as a library. In addition, iGraph also provides interfaces for Python and R programming languages. In contrast to NetworkX, iGraph emphasizes performance at the expense of the flexibility of the underling graph data structure. Nodes and edges are represented by vectors and indexed for fast access and iterations over nodes and edges. Thus graph algorithms in iGraph can be very fast. However, iGraph’s representation of graphs is heavily optimized for fast execution of algorithms that operate on a static network. As such, iGraph is prohibitively slow when making incremental changes to the graph structure, such as node and edge additions or deletions. Overall, we find SNAP uses about three times less memory than iGraph due to extensive use of indexes in iGraph, while being about three times slower executing a few algorithms that benefit from indexes and fast vector access. However, the big difference is in flexibility of the underlying graph data structure. For example, SNAP was five orders of magnitude faster than iGraph in our benchmarks of removal of individual nodes from a graph.
While SNAP was designed to work on a single large-memory machine, an alternative approach would be to use a distributed system to perform network analysis. Examples of such systems include Pregel [Malewicz et al. 2010], PowerGraph [Gonzalez et al. 2012], Pegasus [Kang et al. 2009], and GraphX [Xin et al. 2013]. Distributed graph processing systems can in principle process larger networks than a single machine, but are significantly harder to program, and more expensive to maintain. Moreover, none of the existing distributed systems comes with a large suite of graph processing functions and algorithms. Most often, graph algorithms, such as community detection or link prediction, have to be implemented from scratch.
We also note a recent trend where, due to decreasing RAM prices, the need for distributed graph processing systems has diminished in the last few years. Machines with large RAM of 1TB or more have become relatively inexpensive. Most real-world graphs comfortably fit in such machines, so multiple machines are not required to process them [Perez et al. 2015]. Multi-machine environments also impose considerable execution overhead in terms of communication and coordination costs, which further reduces the benefit of distributed systems. A single machine thus provides an attractive platform for graph analytics [Perez et al. 2015].
3. SNAP FOUNDATIONS
SNAP is a system for analyzing graphs and networks. In this section we shall provide an overview of SNAP, starting by introducing some basic concepts. In SNAP we define graphs to consist of a set of nodes and a set of edges, each edge connecting two nodes. Edges can be either directed or undirected. In multigraphs, more than one edge can exist between a pair of nodes. In SNAP terminology networks are defined as graphs, where attributes or features, like “age”, “color”, “location”, “time” can be associated with nodes as well as edges of the network.
SNAP is designed in such a way that graph/network methods are agnostic to the underling graph/network type/representation. As such most methods work on any type of a graph/network. So, for most of the paper we will be using terms graphs and networks interchangeably, meaning graph and I or network and the specific meaning will be evident from the context.
An alternative terminology to the one we use here is to use the term graph to denote mathematical objects and the term network for real-world instances of graphs, such as an online social network, a road network, or a network of protein interactions. However, inside the SNAP library we use the terminology where graphs represent the “wiring diagrams”, and networks are graphs with data associated with nodes and edges.
3.1. Graph and Network Containers
SNAP is centered around core foundational classes that store graphs and networks. We call these classes graph and network containers. The containers provide several types of graphs and networks, including directed and undirected graphs, multigraphs, and networks with node and edge attributes. In order to optimize for execution speed and memory usage, an application can chose the most appropriate container class so that critical operations are executed efficiently.
An important aspect of containers is that they all have a unified interface for accessing the graph/network structure as well as for traversing nodes and edges. This common interface is used by graph methods to implement more advanced graph algorithms. Since the interface is the same for all graph and network containers, these advanced methods in SNAP are generic in a sense that each method can work on a container of any type. Implementation of new algorithms is thus simplified as each method needs to be implemented only once and can then be executed on any type of a graph or a network. At the same time, the use of SNAP library is also streamlined. It is easy to substitute one type of graph container for another at the container creation time, and the rest of the code usually does not need to be changed.
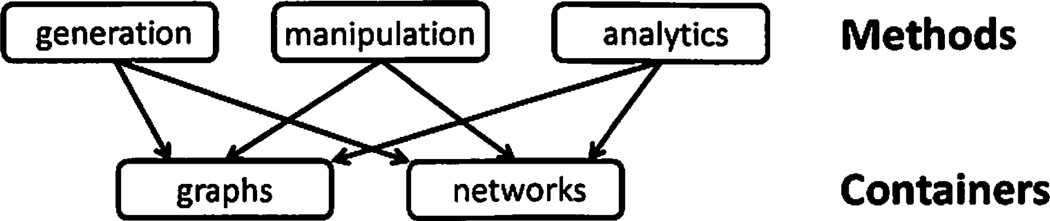
Methods that operate on graph/network containers can be split into several groups (Figure 1): graph generation methods which create new graphs as well as networks, graph manipulation methods which manipulate the graph structure, and graph analytic methods which do not change the underlying graph structure, but compute specific graph statistics. Graph methods are discussed further in Section 4.
Fig. 1.
SNAP components: graph and network containers and methods.
Table I describes the multiple graph and network containers provided by SNAP. Each container is optimized for a particular type of graph or network.
Table I.
SNAP Graph and Network Containers.
| Graph Containers | |
| TUNGraph | Undirected graphs |
| TNGraph | Directed graphs |
| TNEGraph | Directed multigraphs |
| TBPGraph | Bipartite graphs |
| Network Containers | |
| TNodeNet | Directed graphs with node attributes |
| TNodeEDatNet | Directed graphs with node and edge attributes |
| TNodeEdgeNet | Directed multigraphs with node and edge attributes |
| TNEANet | Directed multigraphs with dynamic node and edge attributes |
Graph containers are TUNGraph, TNGraph, TNEGraph, and TBPGraph, which correspond to undirected graphs where edges are bidirectional, directed graphs where edges have direction, directed multigraphs where multiple edges can exist between a pair of nodes, and bipartite graphs, respectively. Network containers are TNodeNet, TNodeEDatNet, TNodeEdgeNet, and TNEANet, which correspond to directed graphs with node attributes, directed graphs with node and edge attributes, directed multigraphs with node and edge attributes and directed multigraphs with dynamic node and edge attributes, respectively.
In all graph and network containers, nodes have unique identifiers (ids), which are non-negative integers. Node ids do not have to be sequentially ordered from one to the number of nodes, but can be arbitrary non-negative integers. The only requirement is that each node has a unique id. In simple graphs edges have no identifiers and can be accessed by providing an pair of node ids that the edge connects. However, in multigraphs each edge has a unique non-negative integer id and edges can be accessed either by providing an edge id or a pair of node ids.
The design decision to allow arbitrary node (and edge) ids is important as it allows us to preserve node identifiers as the graph structure is being manipulated. For example, when extracting a subgraph of a given graph, the node as well as edge ids get preserved.
Network containers, except TNEANet, require that types of node and edge attributes are specified at compile time. These attribute types are simply passed as template parameters in C++, which provides a very efficient and convenient way to implement networks with rich data on nodes and edges. Types of node and edge attributes in the TNEANet container can be provided dynamically, so new node and edge attributes can be added or removed at run time.
Graph and network containers vary in how they represent graphs and networks internally, so time and space trade-offs can be optimized for specific operations and algorithms. Further details on representations are provided in Section 5.
3.2. Functionality of Graph Containers
Container interface allows that the same commonly used primitives are used by containers of all types. This approach results in significant reduction of the effort needed to provide new graph algorithms in SNAP, since most algorithms need to be implemented only once and can then be used for all the graph and network container types.
Common container primitives are shown in Table II. These provide basic operations for graph manipulation. For example, they include primitives that add or delete nodes and edges, and primitives that save or load the graph.
Table II.
Common Graph and Network Methods.
| Nodes | |
| AddNode | Adds a node |
| DelNode | Deletes a node |
| IsNode | Tests, if a node exists |
| GetNodes | Returns the number of nodes |
| Edges | |
| AddEdge | Adds an edge |
| DelEdge | Deletes an edge |
| IsEdge | Tests, if an edge exists |
| GetEdges | Returns the number of edges |
| Graph Methods | |
| Clr | Removes all nodes and edges |
| Empty | Tests, if the graph is empty |
| Dump | Prints the graph in a human readable form |
| Save | Saves a graph in a binary format to disk |
| Load | Loads a graph in a binary format from disk |
| Node and Edge Iterators | |
| BegNI | Returns the start of a node iterator |
| EndNI | Returns the end of a node iterator |
| GetNI | Returns a node (iterator) |
| NI++ | Moves the iterator to the next node |
| BegEI | Returns the start of an edge iterator |
| EndEI | Returns the end of an edge iterator |
| GetEI | Returns an edge (iterator) |
| EI++ | Moves the iterator to the next edge |
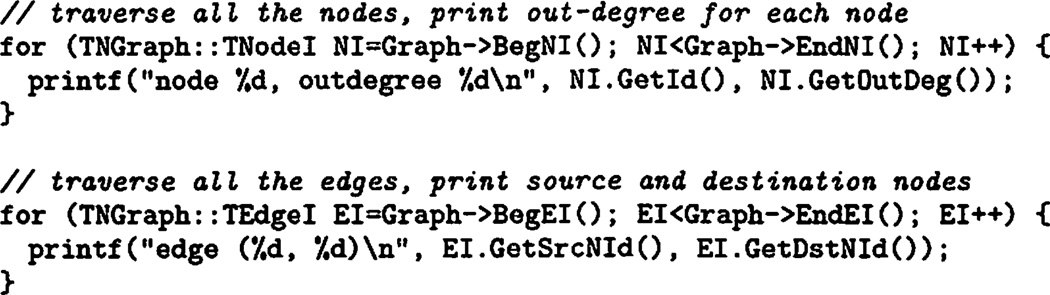
Expressive power of SNAP comes from iterators that allow for a container independent traversal of nodes and edges. Listing 1 illustrates the use of iterators by providing examples of how all the nodes and edges in the graph can be traversed.
The iterators are used consistently and extensively throughout the SNAP code base. As a result, existing graph algorithms in SNAP do not require any changes in order to be applied to new graph and network container types.
Special attention has been paid in SNAP to performance of graph load and save operations. Since large graphs with billions of edges can take a long time to load or save, it is important that these operations are as efficient as possible. To support fast graph saving and loading operations, SNAP can save graphs directly in a binary format, which avoids a computationally expensive step of data serializing and deserializing.
4. GRAPH METHODS
SNAP provides efficient implementations of commonly used traditional algorithms for graph and network analysis, as well as recent algorithms that employ machine learning techniques on graph problems, such as community detection [Yang and Leskovec 2013; Yang and Leskovec 2014; McAuley and Leskovec 2014], statistical modeling of networks [Kim and Leskovec 2012b; Kim and Leskovec 2013], network link and missing node prediction [Kim and Leskovec 2011b], random walks [Lofgren et al. 2016], network structure inference [Gomez-Rodriguez et al. 2010; Gomez-Rodriguez et al. 2013]. These algorithms have been developed within our research group or in collaboration with other groups. They use SNAP primitives extensively and their code is made available as part of SNAP distributions.
Graph methods can be split into the following groups: graph creation, graph manipulation, and graph analytics. Graph creation methods, called generators, are shown in Table III. They implement a wide range of models for generation of regular and random graphs, as well as graphs that model complex real-world networks. Table IV shows major families of graph manipulation and analytics methods. Next, we describe advanced graph methods in more detail.
Table III.
Graph generators in SNAP.
| Category | Graph Generators |
|---|---|
| Regular graphs | Complete graphs, circles, grids, stars, and trees; |
| Basic random graphs | Erdõs-Rényi graphs, Bipartite graphs, |
| Graphs where each node has a constant degree, | |
| Graphs with exact degree sequence; | |
| Advanced graph models | Configuration model [Bollobás 1980], |
| Ravasz-Barabasi model [Ravasz and Barabási 2003], | |
| Copying model [Kumar et al. 2000], | |
| Forest Fire model [Leskovec et al. 2005], | |
| Geometric preferential model [Flaxman et al. 2006], | |
| Barabasi-Albert model [Barabási and Albert 1999], | |
| Rewiring model [Milo et al. 2003], | |
| R-MAT [Chakrabarti et al. 2004], | |
| Graphs with power-law degree distribution, | |
| Watts-Strogatz model [Watts and Strogatz 1998], | |
| Kronecker graphs [Leskovec et al. 2010], | |
| Multiplicative Attribute Graphs [Kim and Leskovec 2012b]. |
Table IV.
Graph manipulation and analytics methods in SNAP.
| Category | Graph Manipulation and Analytics |
|---|---|
| Graph manipulation | Graph rewiring, decomposition to connected components, subgraph extraction, graph type conversions; |
| Connected components | Analyze weakly, strongly, bi- and 1-connected components; |
| Node connectivity | Node degrees, degree distribution, in-degree, out-degree, combined degree, Hop plot, Scree plot; |
| Node centrality algorithms | PageRank, Hits, degree-, betweenness-, closeness-, farness-, and eigen-centrality, personalized PageRank; |
| Triadic closure algorithms | Node clustering coefficient, triangle counting, clique detection; |
| Graph traversal | Breadth first search, depth first search, shortest paths, graph diameter; |
| Community detection | Fast modularity, clique percolation, link clustering, Community-Affiliation Graph Model, BigClam, CoDA, CESNA, Circles; |
| Spectral graph properties | Eigenvectors and eigenvalues of the adjacency matrix, spectral clustering; |
| K-core analysis | Identification and decomposition of a given graph to k-cores; |
| Graph motif detection | Counting of small subgraphs; |
| Information diffusion | Infopath, Netinf; |
| Network link and node prediction | Predicting missing nodes, edges and attributes. |
4.1. Community Detection
Novel SNAP methods for community detection are based on the observation that overlaps between communities in the graph are more densely connected than the non-overlapping parts of the communities [Yang and Leskovec 2014]. This observation matches empirical observations in many real-world networks, however, it has been ignored by most traditional community detection methods.
The base method for community detection is the Community-Affiliation Graph Model (AGM) [Yang and Leskovec 2012]. This method has been extended in several directions to cover networks with millions of nodes and edges [Yang and Leskovec 2013], networks with node attributes [Yang et al. 2013], and 2-mode communities [Yang et al. 2014].
Community-Affiliation Graph Model identifies communities in the entire network. SNAP also provides a complementary approach to network wide community detection. The Circles method [McAuley and Leskovec 2012] uses the friendship network connections as well as user profile information to categorize friends from a person’s ego network into social circles [McAuley and Leskovec 2014].
4.2. Predicting Missing Links, Nodes, and Attributes in Networks
The information we have about a network might often be partial and incomplete, where some nodes, edges or attributes are missing from the available data. Only a subset of nodes or edges in the network is known, the rest of the network elements are unknown. In such cases, we want to predict the unknown, missing network elements.
SNAP methods for these prediction tasks are based on the multiplicative attribute graph (MAG) model [Kim and Leskovec 2012b]. The MAG model can be used to predict missing nodes and edges [Kim and Leskovec 2011a], missing node features [Kim and Leskovec 2012a], or network evolution over time [Kim and Leskovec 2013].
4.3. Fast Random Walk Algorithms
Random walks can be used to determine the importance or authority of nodes in a graph. In personalized PageRank, we want to identify important nodes from the point of view of a given node [Benczur et al. 2005; Lofgren et al. 2014; Page et al. 1999].
SNAP provides a fast implementation of the problem of computing personalized PageRank scores for a distribution of source nodes to a given target node [Lofgren et al. 2016]. In the context of social networks, this problem can be interpreted as finding a source node that is interested in the target node. The fast personalized PageRank algorithm is birectional. First, it works backwards from the target node to find a set of intermediate nodes ’near’ it and then generates random walks forwards from source nodes in order to detect this set of intermediate nodes and compute a provably accurate approximation of the personalized PageRank score.
4.4. Information Diffusion
Information diffusion and virus propagation are fundamental network processes. Nodes adopt pieces of information or become infected and then transmit the information or infection to some of their neighbors. A fundamental problem of diffusion over networks is the problem of network inference [Gomez-Rodriguez et al. 2010]. The network inference task is to use node infection times in order to reconstruct the transmissions as well as the network that underlies them. For example, in an epidemic, we can usually observe just a small subset of nodes being infected, and we want to infer the underlying network structure over which the epidemic spread.
SNAP implements an efficient algorithm for network inference, where the problem is to find the optimal network that best explains a set of observed information propagation cascades [Gomez-Rodriguez et al. 2012]. The algorithm scales to large datasets and in practice gives provably near-optimal performance. For the case of dynamic networks, where edges are added or removed over time and we want to infer these dynamic network changes, SNAP provides an alternative algorithm [Gomez-Rodriguez et al. 2013].
5. SNAP IMPLEMENTATION DETAILS
SNAP is written in the C++ programming language and optimized for compact graph representation while preserving maximum performance. In the following subsections we shall discuss implementational details of SNAP.
5.1. Representation of Graphs and Networks
Our key requirement when designing SNAP was that data structures are flexible in allowing for efficient manipulation of the underlying graph structure, which means that adding or deleting nodes and edges must be reasonably fast and not prohibitively expensive. This requirement is needed, for example, for the processing of dynamic graphs, where graph structure is not known in advance, and nodes and edges get added and deleted over time. A related use scenario is motivated by on-line graph algorithms, where an algorithm incrementally modifies existing graphs as new input becomes available.
Furthermore, we also want our algorithms to offer high performance and be as fast as possible given the flexibility requirement. These opposing needs of flexibility and high performance pose a trade-off between graph representations that allow for efficient structure manipulation and graph representations that are optimized for speed. In general, flexibility is achieved by using hash table based representations, while speed is achieved by using vector based representations. An example of the former is NetworkX [Hagberg et al. 2008], an example of the latter is iGraph [Csardi and Nepusz 2006].
SNAP graph and network representation
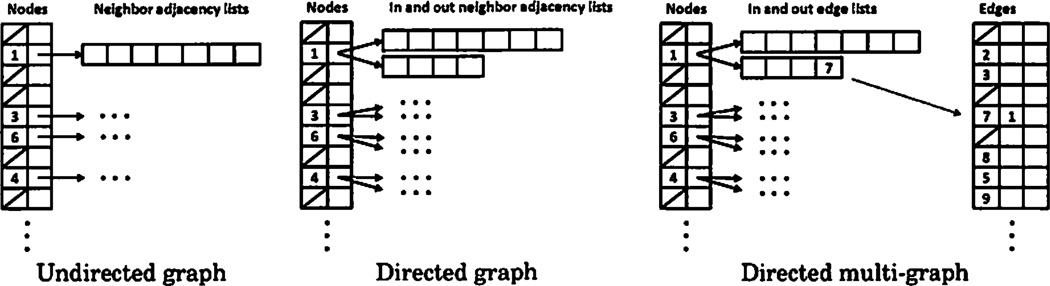
For SNAP, we have chosen a middle ground between all-hash table and all-vector graph representations. A graph in SNAP is represented by a hash table of nodes in the graph. Each node consists of a unique identifier and one or two vectors of adjacent nodes, listing nodes that are connected to it. Only one vector is used in undirected graphs, while two vectors, one for outgoing and another one for incoming nodes/edges, are used in directed graphs. In simple graphs, there are no explicit edge identifiers, edges are treated as pairs of a source and a destination node instead. In multigraphs, edges have explicit identifiers, so that two edges between the same pair of nodes can be distinguished. An additional hash table is required in this case for the edges, mapping edge ids to the source and destination nodes. Figure 2 summarizes graph representations in SNAP.
Fig. 2.
A diagram of graph data structures in SNAP. Node ids are stored in a hash table, and each node has one or two associated vectors of neighboring node or edge ids.
The values in adjacency vectors are sorted for faster access. Since most of the real-world networks are sparse with node degrees significantly smaller than the number of nodes in the network, while at the same time exhibiting a power law distribution of node degrees, the benefits of maintaining the vectors in a sorted order significantly outweigh the overhead of sorting. Sorted vectors also allow for fast and ordered traversal and selection of node’s neighbors, which are common operations in graph algorithms.
As we show in experiments (Section 6), SNAP graph representation also optimizes memory usage for large graphs. Although it uses more memory for storing nodes than some alternative representations, it requires less memory for storing edges. Since a vast majority of relevant networks have more edges than nodes, the overall memory usage in SNAP is smaller than representations that use less memory per node but more per edge. A compact graph representation is important for handling very large networks, since it determines the sizes of networks that can be analyzed on a computer with a given amount of RAM. With a more compact graph representation and smaller RAM requirements, larger networks can fit in the RAM available and can thus be analyzed. Since many graph algorithms are bound by memory throughput, an additional benefit of using less RAM to represent graphs is that the algorithms execute faster, since less memory needs to be accessed.
Time complexity of key graph operations
Table V summarizes time complexity of key graph operations in SNAP. It can be seen that most of the operations complete in constant time of O(1), and that the most time consuming are edge operations, which depend on the node degree. However, since most of the nodes in real-life networks have low degree, edge operations overall still perform faster than alternative approaches. One such alternative approach is to maintain neighbors in a hash table rather than in a sorted vector. This alternative approach does not work well in practice, because hash tables are faster than vectors only when the number of elements stored is large. But most nodes in real-time networks have a very small degree, and hash tables will be slower than vectors for these nodes. We find that a small number of large degree nodes does not compensate for the time lost with a large number of small degree nodes. Additionally, an adjacency hash table would need to be maintained for each node, leading to significantly increased complexity with hundreds of millions of hash tables for graphs with hundreds of millions of nodes.
Table V.
Time complexity of key graph operations in SNAP. degmax denotes the maximum node degree in the graph.
| Operation | Time Complexity |
|---|---|
| Get node, get next node | O(1) |
| Get edge, get next edge | O(1) |
| Add, delete, test an existence of a node | O(1) |
| Add, delete an edge | O(degmax) |
| Test an existence an edge | O(log(degmax)) |
As we show in the experimental section (Section 6), the representation of graphs in SNAP is able to provide high performance and compact memory footprint, while allowing for efficient additions or deletions of nodes and edges.
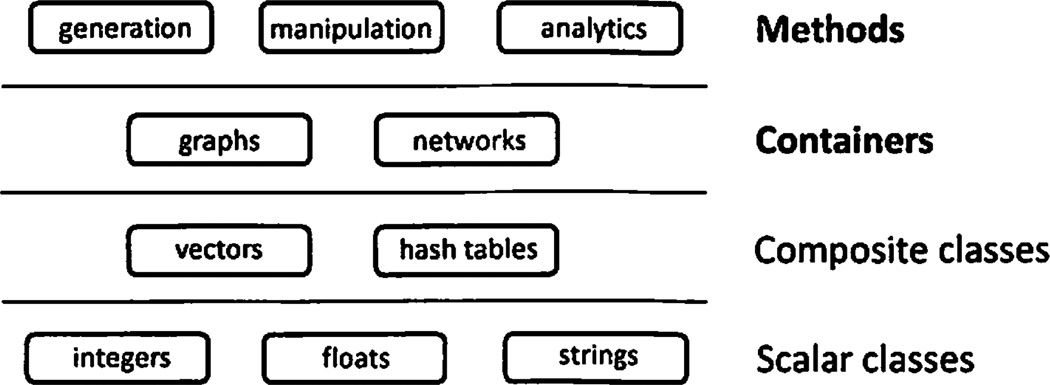
5.2. Implementation Layers
SNAP is designed to operate in conceptual layers (see Figure 3). Layers are designed in such a way that every level abstracts out the complexity of the lower level. The bottom layer comprises of basic scalar classes, like integers, floats, and strings. Next layer implements composite data structures, like vectors and hash tables. A layer above them are graph and network containers. And the last layer contains graph generation, manipulation, and analytics methods. SNAP implementation takes advantage of GLib, a general purpose C++ STL-like library (Standard Template Library), developed at Jozef Stefan Institute in Ljubljana, Slovenia. GLib is being actively developed and used in numerous academic and industrial projects.
Fig. 3.
Different layers of SNAP design.
Scalar classes
This foundational layer implements basic classes, such as integers, floating point numbers, and strings. A notable aspect of this layer is its ability to efficiently load and save object instances to a secondary storage device. SNAP saves objects in a binary format, which allows loading and storing of objects without any complex parsing and thus can be done at close to disk speeds.
Composite classes
The next layer implements composite classes on top of scalar classes. Two key composite classes are vectors, where elements are accessed by an integer index, and hash tables, where elements are accessed via a key. The elements and keys in hash tables can have an arbitrary type. SNAP expands fast load and save operations from scalar classes to vectors and hashes, so that these composite classes can be manipulated efficiently as well.
Graph and network containers
The layer above vectors and hash tables are graph and network containers. These were discussed in detail in Sections 3.1 and 5.1.
Graph and network methods
The top layer of SNAP implements graph and network algorithms. These rely heavily on node and edge iterators, which provide a unified interface to all graph and network classes in SNAP (Section 3.2). By using iterators, only one implementation of each algorithm is needed to provide the algorithm for all the graph/network containers. Without a unified iterator interface, a separate algorithm implementation would be needed for each container type, which would result in significantly larger development effort and increased maintenance costs.
For example, to implement a k-core decomposition algorithm [Batagelj and Zaveršnik 2002], one would in principle need to keep a separate implementation for each graph/network type (i.e., graph/network container). However, in SNAP all graph/network containers expose the same set of functions and interfaces to access the graph/network structure. In case of the k-core algorithm, we need functionality to traverse all of the nodes of the network (we use node iterators to do that), determine the degree of a current node, and then delete it. All graph/network containers in SNAP expose such functions and thus a single implementation of the k-core algorithm is able to operate on any kind of graph/network container (directed and undirected graphs, multigraphs as well as networks).
Memory management
In large software systems, memory management is an important aspect. All complex SNAP objects, from composite to network classes, employ reference counting, so memory for an object is automatically released, when no references are left that point to the object. Thus, memory management is completely transparent to the SNAP user and has minimal impact on performance, since the cost of reclaiming unused memory is spread in small chunks over many operations.
6. BENCHMARKS
In this section, we compare SNAP with existing network analytics systems. In particular, we contrast the performance of SNAP with two systems that are most similar in functionality, NetworkX [Hagberg et al. 2008] and iGraph [Csardi and Nepusz 2006].
NetworkX and iGraph are single machine, single thread graph analytics libraries that occupy two opposite points in the performance vs. flexibility spectrum. iGraph is optimized for performance, but not flexible in a sense that it supports primarily only static graph structure (dynamically adding/deleting nodes/edges is prohibitively expensive). On the other hand, NetworkX is optimized for flexibility at the expense of lower performance. SNAP lies in-between, providing flexibility while maximizing performance.
Furthermore, we also give a summary of our experiments with parallel versions of several SNAP algorithms [Perez et al. 2015]. These experiments demonstrate that a single large-memory multi-core machine provides an attractive platform for the analysis of all-but-the-largest graphs. In particular, we show that performance of SNAP on a single machine measures favorably when compared to distributed graph processing frameworks.
All the benchmarks were performed on a computer with 2.40GHz Intel Xeon E7-4870 processors and sufficient memory to hold the graphs in RAM. Since all the systems are non-parallel, benchmarks utilized only one core of the system. All benchmarks were repeated 5 times and the average times are shown.
6.1. Memory Consumption
A memory requirement to represent graphs is an important measure of a graph analytics library. Many graph operations are limited by available memory access bandwidth, and a smaller memory footprint allows for faster algorithm execution.
To determine memory consumption, we use undirected Erdõs-Rényi random graphs, G(n, m), where n represents the number of nodes, and m the number of edges in the graph. We measure memory requirements for G(n, m) graphs at three different sizes G(1M, 10M), G(1M, 100M), and G(10M, 100M), where 1M denotes 106. We have chosen those graph sizes to illustrate system scaling as the number of nodes or the average node degree increases.
Table VI shows the results. Notice, that SNAP can store a graph of 10M nodes, and 100M edges in mere 1.3GB of memory, while iGraph needs over 3.3GB and NetworkX requires nearly 55GB of memory to store the same graph. It is somewhat surprising that iGraph requires about 3 times more memory than SNAP, despite using vectors to represent nodes rather than a hash table. NetworkX uses hash tables extensively and it is thus not surprising that it requires over 40 times more memory than SNAP.
Table VI.
Memory requirements of undirected Erdõs-Rényi random graphs, the G(n, m) model. Memory usages are in MB. Overall, SNAP uses three times less memory than iGraph and over 40 times less memory than NetworkX.
| Graph size | Memory usage [MB] | |||
|---|---|---|---|---|
| Nodes | Edges | SNAP | iGraph | NetworkX |
| 1M | 10M | 137 | 344 | 5,423 |
| 1M | 100M | 880 | 3,224 | 43,806 |
| 10M | 100M | 1,366 | 3,360 | 54,171 |
We used the memory consumption measurements in Table VI to calculate the number of bytes required by each library to represent a node or an edge. As can be seen in Table VII, SNAP requires four times less memory per edge than iGraph and 50 times less memory per edge than NetworkX. Since graphs have usually significantly more edges than nodes, memory requirements to store the edges are the main indicator of the size of graphs that will fit in a given amount of RAM.
Table VII.
Memory requirements to represent a node or an edge, based on the measurements of the G(n, m) model. Memory usages are in bytes. SNAP uses four times less memory per edge than iGraph and over 50 times less memory per edge than NetworkX.
| Memory usage [bytes] | |||
|---|---|---|---|
| Item | SNAP | iGraph | NetworkX |
| Node | 54.4 | 24.0 | 1158.2 |
| Edge | 8.3 | 32.0 | 426.5 |
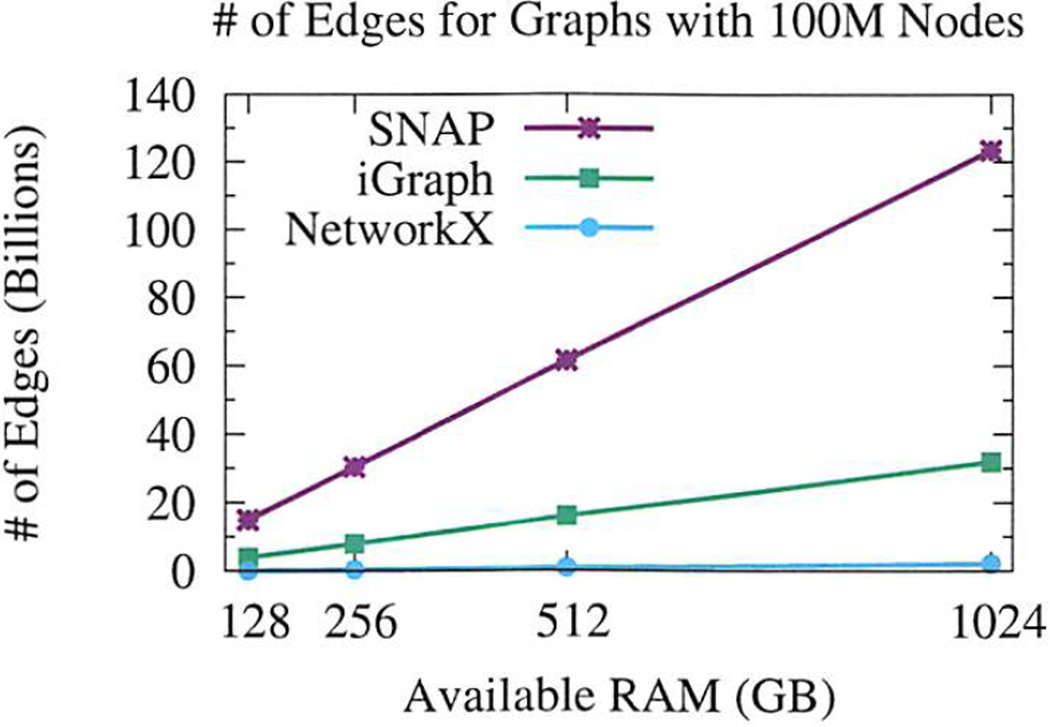
We illustrate the size of a graph that can be represented by each system in a given amount of RAM by fixing the number of nodes at 100 million and then calculating the maximum number of edges that fit in the remaining RAM, using numbers from Table VII. The results are shown in Figure 4. For 1024GB of RAM, SNAP can represent graphs with 123.5 billion edges, iGraph 31.9 billion edges, and NetworkX 2.1 billion edges.
Fig. 4.
Maximum graph sizes for varying RAM availability. Number of nodes is fixed at 100 million, estimated maximum number of edges is shown. Using 1TB RAM, SNAP can fit over 120 billion edges, iGraph 30 billion, and NetworkX 2 billion.
6.2. Basic Graph Operations
Next, we measure execution times of basic graph operations for an Erdõs-Rényi random graph G(1M, 100M).
First, we examine the times for generating a graph, saving the graph to a file, and loading the graph from the file. Results are shown in Table VIII. We used a built-in function in each system to generate the graphs. For graph generation, SNAP is about two times slower than iGraph, and more than 5 times faster than NetworkX (Table VIII). However, graph generation in SNAP is inserting one edge at a time, while iGraph has an optimized implementation that inserts edges in bulk.
Table VIII.
Execution times for basic graph operations on Erdős-Rényi random graph G(1M, 100M). Times are in seconds. Overall, SNAP is about two times slower than iGraph at generating the graph but it is 15 times faster at loading and saving it to the disk. NetworkX is 5 to 200 times slower than SNAP.
| Execution time [seconds] | |||
|---|---|---|---|
| Operation | SNAP | iGraph | NetworkX |
| Generate | 139.3 | 74.2 | 748.7 |
| Save | 3.3 | 47.0 | 757.2 |
| Load | 4.6 | 87.8 | 522.0 |
The performance of graph loading and saving operations is often a bottleneck in graph analysis. For these operations, SNAP is over 15 times faster than iGraph and 100 times faster than NetworkX (Table VIII). The benchmark utilized an internal binary representation of graphs for SNAP, while a text representation was used for iGraph and NetworkX. SNAP and iGraph have similar performance when saving/loading graphs from/to a textual format. So, the advantage of SNAP over iGraph can be attributed to the SNAP support for the binary graph representation on the disk.
Second, we also benchmark the fundamental operations when working with graphs. We focus on the time it takes to test for the existence of a given edge (i, j). We performed an experiment where we generated larger and larger instances of Erdõs-Rényi random graphs and measured execution times for testing the presence of edges in a given graph. For each test, we generated a random source and destination node and tested for its existence in the graph. The number of test iterations is equal to the number of edges in the graph. Table IX gives the results and we notice that SNAP is about 10–20% faster than or comparable to iGraph and 3–5 times faster than NetworkX.
Table IX.
Testing edge existence. Edges are random, the number of tests is equal to the number of total edges in the graph. Times are in seconds. SNAP is about 10–20% faster than or comparable to iGraph, while being 3–5 times faster than NetworkX.
| Graph size | Execution time [seconds] | |||
|---|---|---|---|---|
| Nodes | Edges | SNAP | iGraph | NetworkX |
| 1M | 10M | 3.8 | 5.2 | 23.5 |
| 1M | 100M | 75.4 | 113.8 | 218.3 |
| 10M | 100M | 67.9 | 63.3 | 255.8 |
Last, we also estimate system flexibility, which tells us how computationally expensive it is to modify graph structure, by measuring the execution times of deleting 10% of nodes and their corresponding edges from G(1M, 10M). SNAP is much faster than iGraph and NetworkX when deleting nodes from the graph (Table X). Furthermore, the nodes in SNAP and NetworkX were deleted incrementally, one node at the time, while the nodes in iGraph were deleted in a single batch with one function call. When nodes were deleted one by one in iGraph as well, it took 334,720 seconds to delete 10% of nodes in the graph. The fact that SNAP is more than 5 orders of magnitude faster than iGraph indicates that iGraph’s graph data structures are optimized for speed on static graphs while also being less memory efficient. However, the iGraph data structure seems to completely fail in case of dynamic graphs where nodes/edges appear/disappear over time.
Table X.
Execution times for deleting 10% of nodes and their corresponding edges from Erdős-Rényi random graph G(1M, 10M). Times are in seconds. SNAP is four to five times faster than iGraph and NetworkX. However, if one deletes nodes from the graph one-by-one in iGraph, its performance slows down for five orders of magnitude.
| Execution time [seconds] | |||
|---|---|---|---|
| Operation | SNAP | iGraph | NetworkX |
| Deleting nodes | 0.7 | 3.0 | 4.1 |
6.3. Graph Algorithms
To evaluate system performance on a real-world graph, we used a friendship graph of the LiveJournal online social network [Leskovec and Krevl 2014]. The LiveJournal network has about 4.8M nodes and 69M edges. We measured execution times for common graph analytics operations: PageRank, clustering coefficient, weakly connected components, extracting 3-core of a network, and testing edge existence. For the PageRank algorithm, we show the time it takes to perform 10 iterations of the algorithm.
Table XI gives the results. We can observe that SNAP is only about 3 times slower than iGraph in some operations and about equal in others, while it is between 4 to 60 times faster than NetworkX (Table XI). As expected, NetworkX performs the best when the algorithms require mostly a large number of random accesses for which hash tables work well, while it performs poorly when the algorithm execution is dominated by sequential data accesses where vectors dominate.
Table XI.
Execution times for graph algorithms on the LiveJournal network with 4.8M nodes and 69M edges. Times are in seconds. Generally we observe that due to a hash based graph representation that allows efficient changes in the structure, SNAP is equal to iGraph in some graph operations while about 3 times slower in algorithms that benefit from fast vector access in iGraph. NetworkX is much slower than either SNAP or iGraph in most operations.
| Execution time [seconds] | |||
|---|---|---|---|
| Operation | SNAP | iGraph | NetworkX |
| PageRank | 40.9 | 10.6 | 2,720.8 |
| Clustering Coefficient | 143.3 | 58.5 | 4,265.4 |
| Connected Components | 13.3 | 5.8 | 60.3 |
| 3-core | 37.9 | 41.7 | 2,276.1 |
| Test Edge Existence | 45.7 | 35.2 | 158.6 |
In summary, we find that the SNAP graph data structure is by far the most memory efficient and also most flexible as it is able to add/delete nodes and edges the fastest. In terms of input/output operations SNAP also performs the best. And last, we find that SNAP offers competitive performance in executing static graph algorithms.
6.4. Comparison to Distributed Graph Processing Frameworks
So far we focused our experiments on SNAP performance on a sequential execution of a single thread on a single machine. However, we have also been studying how to extend SNAP to single machine multi-threaded architectures.
We have implemented parallel versions of several SNAP algorithms. Our experiments have shown that a parallel SNAP on a single machine can offer comparable performance to specialized algorithms and even frameworks utilizing distributed systems for network analysis and mining [Perez et al. 2015]. Results are summarized in Table XII. For example, triangle counting on the Twitter2010 graph [Kwak et al. 2010], which has about 42 million nodes and 1.5 billion edges, required 469s on a 6 core machine [Kim et al. 2014], 564s on a 200 processor cluster [Arifuzzaman et al. 2013], while the parallel SNAP engine on a single machine with 40 cores required 263s.
Table XII.
Execution times for graph algorithms on the Twitter2010 network with 42M nodes and 1.5B edges. Times are in seconds.
| Benchmark | System | Execution time [seconds] |
|---|---|---|
| Triangles | OPT, 1 machine, 6 cores [Kim et al. 2014] | 469 |
| PATRIC, 200 processor cluster [Arifuzzaman et al. 2013] | 564 | |
| SNAP, 1 machine, 40 cores | 263 | |
| PageRank | PowerGraph, 64 machines, 512 cores [Gonzalez et al. 2012] | 3.6 |
| SNAP, 1 machine, 40 cores | 6.0 | |
We obtained similar results by measuring execution time of the PageRank algorithm [Page et al. 1999] on the same graph. PowerGraph [Gonzalez et al. 2012], a state-of-the-art distributed system for network analysis running on 64 machines with 512 cores, took 3.6s per PageRank iteration, while our system needed 6s for the same operation using only one machine and 40 cores, a significantly simpler configuration and more than 12 times fewer cores.
Note also that SNAP uses only about 13GB of RAM to process the Twitter2010 graph, so the graph fits easily in the RAM of most modern laptops.
These results, together with the sizes of networks being analyzed, demonstrate that a single multi-core big-memory machine provides an attractive platform for network analysis of a large majority of networks [Perez et al. 2015].
7. STANFORD LARGE NETWORK DATASET COLLECTION
As part of SNAP, we are also maintaining and making publicly available the Stanford Large Network Dataset Collection [Leskovec and Krevl 2014], a set of around 80 different social and information real-world networks and datasets from a wide range of domains, including social networks, citation and collaboration networks, Internet and Web based networks, and media networks. Table XIII gives the types of datasets in the collection.
Table XIII.
Datasets in the Stanford Large Network Dataset Collection.
| Dataset type | Count | Sample datasets |
|---|---|---|
| Social networks | 10 | Facebook, Google+, Slashdot, Twitter, Epinions |
| Ground-truth communities | 6 | LiveJournal, Friendster, Amazon products |
| Communication networks | 3 | Email, Wikipedia talk |
| Citation networks | 3 | Arxiv, US patents |
| Collaboration networks | 5 | Arxiv |
| Web graphs | 4 | Berkeley, Stanford, Notre Dame |
| Product co-purchasing networks | 5 | Amazon product |
| Internet peer-to-peer networks | 9 | Gnutella |
| Road networks | 3 | California, Pennsylvania, Texas |
| Autonomous systems graphs | 5 | AS peering, CAIDA, Internet topology |
| Signed networks | 6 | Epinions, Wikipedia, Slashdot Zoo |
| Location-based social networks | 2 | Gowalla, Brightkite |
| Wikipedia networks | 6 | Navigation, voting, talk, elections, edit history |
| Memetracker and Twitter | 4 | Post hyperlinks, popular phrases, tweets |
| Online communities | 2 | Reddit, Flickr |
| Online reviews | 6 | BeerAdvocate, RateBeer, Amazon, Fine Foods |
The datasets were collected as part of our research in the past and in that sense represent typical graphs being analyzed. Table XIV gives the distribution of graph sizes in the collection. It can be observed that a vast majority of graphs are relatively small with less than 100 million edges and thus can easily be analyzed in SNAP. The performance benchmarks in Table XI are thus indicative of the execution times of graph algorithms being applied to real-world networks.
Table XIV.
Distribution of graph sizes in the Stanford Large Network Dataset Collection.
| Graph size (number of edges) |
Number of graphs |
|---|---|
| <0.1M | 18 |
| 0.1M – 1M | 24 |
| 1M – 10M | 17 |
| 10M – 100M | 7 |
| 100M – 1B | 4 |
| >1B | 1 |
8. RESOURCES
SNAP resources are available from our Web site at: http://snap.Stanford.edu.
The site contains extensive user documentation, tutorials, regular SNAP stable releases, links to the relevant GitHub repositories, a programming guide, and the datasets from the Stanford Large Network Dataset Collection.
Complete SNAP source code has been released under a permissive BSD type open source license. SNAP is being actively developed. We welcome community contributions to the SNAP code base and the SNAP dataset collection.
9. CONCLUSION
We have presented SNAP, a system for analysis of large graphs. We demonstrate that graph representation employed by SNAP is unique in the sense that it provides an attractive balance between the ability to efficiently modify graph structure and the need for fast execution of graph algorithms. While SNAP implements efficient operations to add or delete nodes and edges in a graph, it imposes only limited overhead on graph algorithms. An additional benefit of SNAP graph representation is that it is compact and requires lower amount of RAM than alternative representations, which is useful in analysis of large graphs.
We are currently extending SNAP in several directions. One direction is speeding up algorithms via parallel execution. Modern CPUs provide a large number of cores, which provide a natural platform for parallel algorithms. Another direction is exploring ways of how the graphs are constructed from data and then identify powerful primitives that cover a broad range of graph construction scenarios.
Listing 1.
Iterating over Nodes and Edges. Top example prints out the ids and out-degrees of all the nodes. Bottom example prints out all the edges as pairs of edge source node id and edge destination node id. These traversals can be executed on any type of a graph/network container.
Acknowledgments
Many developers contributed to SNAP. Top 5 contributors to the repository, excluding the authors, are Nicholas Shelly, Sheila Ramaswamy, Jaewon Yang, Jason Jong, and Nikhil Khadke. We also thank Jožef Stefan Institute for making available their GLib library.
This work has been supported in part by DARPA XDATA, DARPA SIMPLEX, NIH U54EB020405, IIS-1016909, CNS-1010921, IIS-1149837, Boeing, and Stanford Data Science Initiative.
Contributor Information
Jure Leskovec, Stanford University.
Rok Sosič, Stanford University.
REFERENCES
- Arifuzzaman S, Khan M, Marathe M. PATRIC: A parallel algorithm for counting triangles in massive networks; ACM International Conference on Information and Knowledge Management (CIKM); 2013. pp. 529–538. [Google Scholar]
- Barabási A-L, Albert R. Emergence of scaling in random networks. Science. 1999;286(5439):509–512. doi: 10.1126/science.286.5439.509. (1999) [DOI] [PubMed] [Google Scholar]
- Batagelj V, Mrvar A. Pajek-program for large network analysis. Connections. 1998;21(2):47–57. (1998) [Google Scholar]
- Batagelj V, Zaveršnik M. Generalized cores. ArXiv cs.DS/0202039. 2002 Feb; 2002. [Google Scholar]
- Benczur AA, Csalogany K, Sarlos T, Uher M. Spamrank–fully automatic link spam detection. International Workshop on Adversarial Information Retrieval on the Web. 2005 [Google Scholar]
- Bollobás B. A probabilistic proof of an asymptotic formula for the number of labelled regular graphs. European Journal of Combinatorics. 1980;1(4):311–316. (1980) [Google Scholar]
- Chakrabarti D, Zhan Y, Faloutsos C. SIAM International Conference on Data Mining (SDM) Vol. 4. SIAM; 2004. R-MAT: A recursive model for graph mining; pp. 442–446. [Google Scholar]
- Csardi G, Nepusz T. The igraph software package for complex network research. Inter Journal, Complex Systems. 2006;1695(5) (2006) [Google Scholar]
- Easley D, Kleinberg J. Networks, crowds, and markets: Reasoning about a highly connected world. Cambridge University Press; 2010. [Google Scholar]
- Flaxman AD, Frieze AM, Vera J. A geometric preferential attachment model of networks. Internet Mathematics. 2006;3(2):187–205. (2006) [Google Scholar]
- Gomez-Rodriguez M, Leskovec J, Balduzzi D, Schölkopf B. Uncovering the structure and temporal dynamics of information propagation. Network Science. 2014;2(01):26–65. (2014) [Google Scholar]
- Gomez-Rodriguez M, Leskovec J, Krause A. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD) ACM; 2010. Inferring networks of diffusion and influence; pp. 1019–1028. [Google Scholar]
- Gomez-Rodriguez M, Leskovec J, Krause A. Inferring networks of diffusion and influence. ACM Transactions on Knowledge Discovery from Data. 2012 Feb.5(4):37. Article 21 2012. [Google Scholar]
- Gomez-Rodriguez M, Leskovec J, Schölkopf B. ACM International Conference on Web Search and Data Mining (WSDM) ACM; 2013. Structure and dynamics of information pathways in online media; pp. 23–32. [Google Scholar]
- Gonzalez JE, Low Y, Gu H, Bickson D, Guestrin C. PowerGraph: Distributed graph-parallel computation on natural graphs. USENIX Symposium on Operating Systems Design and Implementation (OSDI) 2012;12:2. [Google Scholar]
- Gregor D, Lumsdaine A. The parallel BGL: A generic library for distributed graph computations. Parallel Object-Oriented Scientific Computing (POOSC) 2005;2:1–18. (2005) [Google Scholar]
- Hagberg A, Swart P, Chult DS. Technical Report. Los Alamos National Laboratory (LANL); 2008. Exploring network structure, dynamics, and function using NetworkX. [Google Scholar]
- Hallac D, Leskovec J, Boyd S. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD) ACM; 2015. Network lasso: Clustering and optimization in large graphs; pp. 387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MO. Social and economic networks. Vol. 3. Princeton: Princeton university press; 2008. [Google Scholar]
- Kang U, Tsourakakis CE, Faloutsos C. IEEE International Conference on Data Mining (ICDM) IEEE; 2009. Pegasus: A peta-scale graph mining system implementation and observations; pp. 229–238. [Google Scholar]
- Kim J, Han W-S, Lee S, Park K, Yu H. ACM SIGMOD International Conference on Management of Data (SIGMOD) ACM; 2014. OPT: a new framework for overlapped and parallel triangulation in large-scale graphs; pp. 637–648. [Google Scholar]
- Kim M, Leskovec J. Modeling social networks with node attributes using the multiplicative attribute graph model; Conference on Uncertainty in Artificial Intelligence (UAI); 2011a. [Google Scholar]
- Kim M, Leskovec J. The network completion problem: inferring missing nodes and edges in networks; SIAM International Conference on Data Mining (SDM); 2011b. pp. 47–58. [Google Scholar]
- Kim M, Leskovec J. Latent multi-group membership graph model; International Conference on Machine Learning (ICML); 2012a. [Google Scholar]
- Kim M, Leskovec J. Multiplicative attribute graph model of real-world networks. Internet Mathematics. 2012b;8(1–2):113–160. (2012) [Google Scholar]
- Kim M, Leskovec J. Nonparametric multi-group membership model for dynamic networks. Advances in Neural Information Processing Systems (NIPS) 2013:1385–1393. [Google Scholar]
- Kumar R, Raghavan P, Rajagopalan S, Sivakumar D, Tomkins A, Upfal E. Annual Symposium on Foundations of Computer Science. IEEE; 2000. Stochastic models for the web graph; pp. 57–65. [Google Scholar]
- Kwak H, Lee C, Park H, Moon S. What is Twitter, a social network or a news media? WWW ’10. 2010 [Google Scholar]
- Kyrola A, Blelloch G, Guestrin C. GraphChi: Large-scale graph computation on just a PC; USENIX Symposium on Operating Systems Design and Implementation (OSDI); 2012. pp. 31–46. [Google Scholar]
- Leskovec J, Backstrom L, Kleinberg J. Meme-tracking and the dynamics of the news cycle; ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD); 2009. pp. 497–506. [Google Scholar]
- Leskovec J, Chakrabarti D, Kleinberg J, Faloutsos C, Ghahramani Z. Kronecker graphs: An approach to modeling networks. Journal of Machine Learning Research. 2010;11:985–1042. (2010) [Google Scholar]
- Leskovec J, Horvitz E. Geospatial structure of a planetary-scale social network. IEEE Transactions on Computational Social Systems. 2014;1(3):156–163. (2014) [Google Scholar]
- Leskovec J, Kleinberg J, Faloutsos C. ACM SIGKDD International Conference on Knowledge Discovery in Data Mining (KDD) ACM; 2005. Graphs over time: densification laws, shrinking diameters and possible explanations; pp. 177–187. [Google Scholar]
- Leskovec J, Krevl A. SNAP Datasets: Stanford Large Network Dataset Collection. 2014 Jun; http://snap.stanford.edu/data 2014. [Google Scholar]
- Lofgren P, Banerjee S, Goel A. ACM International Conference on Web Search and Data Mining (WSDM) ACM; 2016. Personalized PageRank estimation and search: a bidirectional approach. [Google Scholar]
- Lofgren PA, Banerjee S, Goel A, Seshadhri C. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD) ACM; 2014. Fast-ppr: Scaling personalized pagerank estimation for large graphs; pp. 1436–1445. [Google Scholar]
- Malewicz G, Austern MH, Bik AJC, Dehnert JC, Horn I, Leiser N, Czajkowski G. ACM SIGMOD International Conference on Management of data (SIGMOD) ACM; 2010. Pregel: a system for large-scale graph processing; pp. 135–146. [Google Scholar]
- McAuley J, Leskovec J. Learning to discover social circles in ego networks. Advances in Neural Information Processing Systems (NIPS) 2012 [Google Scholar]
- McAuley J, Leskovec J. Discovering social circles in ego networks. ACM Transactions on Knowledge Discovery from Data. 2014 Feb.8(1):28. Article 4 2014. [Google Scholar]
- Milo R, Kashtan N, Itzkovitz S, Newman MEJ, Alon U. On the uniform generation of random graphs with prescribed degree sequences. arXiv preprint cond-mat/0312028. 2003 (2003) [Google Scholar]
- Newman M. The structure and function of complex networks. SLAM Rev. 2003;45(2):167–256. (2003) [Google Scholar]
- Newman M. Networks: An introduction. Oxford: OUP; 2010. [Google Scholar]
- O’Madadhain J, Fisher D, Smyth P, White S, Boey Y. Analysis and visualization of network data using JUNG. Journal of Statistical Software. 2005;10(2):1–35. (2005) [Google Scholar]
- Page L, Brin S, Motwani R, Winograd T. Technical Report. Stanford InfoLab; 1999. Nov, The pagerank citation ranking: Bringing order to the web. [Google Scholar]
- Perez Y, Sosič R, Banerjee A, Puttagunta R, Raison M, Shah P, Leskovec J. Ringo: Interactive graph analytics on big-memory machines; ACM SIGMOD International Conference on Management of Data (SIGMOD); 2015. pp. 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasz E, Barabasi A-L. Hierarchical organization in complex networks. Physical Review E. 2003;67(2):026112. doi: 10.1103/PhysRevE.67.026112. (2003) [DOI] [PubMed] [Google Scholar]
- Salihoglu S, Widom J. International Conference on Scientific and Statistical Database Management. ACM; 2013. GPS: A graph processing system; p. 22. [Google Scholar]
- Suen C, Huang S, Eksombatchai C, Sosič R, Leskovec J. International conference on World Wide Web (WWW) International World Wide Web Conferences Steering Committee; 2013. NIFTY: A system for large scale information flow tracking and clustering; pp. 1237–1248. [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of small-world networks. Nature. 1998;393(6684):440–442. doi: 10.1038/30918. (1998) [DOI] [PubMed] [Google Scholar]
- Xin RS, Gonzalez JE, Franklin MJ, Stoica I. ACM International Workshop on Graph Data Management Experiences and Systems. ACM; 2013. GraphX: A resilient distributed graph system on Spark; p. 2. [Google Scholar]
- Yang J, Leskovec J. IEEE International Conference on Data Mining (ICDM) IEEE; 2012. Community-affiliation graph model for overlapping network community detection; pp. 1170–1175. [Google Scholar]
- Yang J, Leskovec J. ACM International Conference on Web Search and Data Mining (WSDM) ACM; 2013. Overlapping community detection at scale: A nonnegative matrix factorization approach; pp. 587–596. [Google Scholar]
- Yang J, Leskovec J. Overlapping communities explain core-periphery organization of networks. Proc IEEE. 2014 Dec;102(12):1892–1902. 2014. [Google Scholar]
- Yang J, McAuley J, Leskovec J. Community detection in networks with node attributes; IEEE International Conference on Data Mining (ICDM); 2013. [Google Scholar]
- Yang J, McAuley J, Leskovec J. Detecting cohesive and 2-mode communities in directed and undirected networks; ACM International Conference on Web Search and Data Mining (WSDM); 2014. [Google Scholar]