Highlights
-
•
Use of culture filtrates of Termitomyces sp. OE147.
-
•
Decolourization and degradation of Reactive blue 21.
-
•
Cellobiose dehydrogenase and laccase as effective redox couple.
Keywords: Termitomyces sp. OE 147, Fungal culture filtrates, Complex phthalocyanine dyes, Cellobiose dehydrogenase/laccase, Textile effluents
Abstract
This study evaluates culture filtrate, rich in cellobiose dehydrogenase and laccases, of Termitomyces sp. OE 147, in decolouration and degradation of Reactive blue (RB) 21. About 35% decolouration was achieved at low volumes of the culture supernatant without addition of external redox mediators. An optimized dye to culture fluid ratio (75 ppm: 0.1 ml) at a pH of 4–5 resulted in removal of colour by 60%. The degradation products of RB21 were analysed by Electron Spray Ionization-Mass Spectrometry and several small molecules (of m/z 106–199) were detected. These were concluded to be o-Xylene, 2,3-Dihydro-1H-isoindole, Isoindole-1,3-dione, 2,Benzenesulfonyl-ethanol, (4-Hydroxy-phenyl)-sulfamic acid, 2,3-Dihydro-1H-isoindole-5-sulfonic acid and proposed to result from joint action of cellobiose dehydrogenase, laccase, peroxidases and unidentified oxidoreductases present in the culture fluids. Based on the products formed and the known reactions of these enzymes, a degradation pathway was proposed for RB21. The culture fluid was also effective in decolouration (by about 50%) and detoxification (by ∼25%) of the combined effluent collected from a local mill indicating a treatment process that bypasses use of H2O2 and toxic mediators.
1. Introduction
Wastewaters from textile dyeing industries are one of the most polluting sources of water bodies [17]. The synthetic dyes used in the textile industries have complex aromatic molecular structures making them recalcitrant to biological degradation [15]. In terms of chemical structures, out of nearly 30 dye classes, the most common ones belong to azo, anthraquinone, triarylmethane and pthalocyanine categories. The dyes are produced to the extent of 2.2 × 107 tons per year [46] and it is estimated that 10–15% of the dyes are discharged during the dyeing process [25] resulting in major contamination of water bodies around the world.
Many physical (adsorption, coagulation, precipitation, filtration) and chemical (oxidation, advanced oxidation) methods have been used for treatment of dye-containing effluents [19], [36], but these are either expensive to apply or result in formation of sludge. Biological treatment involving either fungal biomass or enzymes are good alternatives [19]. A variety of extracellular enzymes such as peroxidases (lignin peroxidase or LiP, manganese peroxidase or MnP, horseradish peroxidases or HRP) [20], [31], [35], oxidoreductases (cellobiose dehydrogenases or CDH, laccases) produced by fungi [50], [53] and other biological systems have been shown to be effective against a variety of synthetic dyes. Out of these, the oxidoreductases are gaining importance due to their requirement of simple co-factors and/or molecular oxygen. CDHs are heme containing flavo-proteins important in cellulose [41] and lignocellulose [21] degradation. The action of these enzymes is mediated by production of hydroxyl radicals by Fenton reaction [2]. The hydroxyl radicals are effective for action on wood, as they transform non-phenolics and facilitate their degradation by other oxidative enzymes such as MnP or laccases [22]. Laccases (p-diphenol:dioxygen oxidoreducatse; EC 1.10.3.2) use molecular oxygen for their activity on phenolic substrates and aromatic amines [6]. The substrate range of these enzymes can be extended to non-phenolic compounds by inclusion of small mediators such as 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) or ABTS, Hydroxybenzotriazole (HOBT) and Violuric acid. Laccase mediator combinations have been shown to be effective against azo and phthalocyanine (PC) dyes [7], [9], [12]. The major drawback of this method is the high cost and the toxicity of the mediators. Recently, culture fluids of several fungi, containing high amounts of laccase along with CDH, have been used successfully for decolouration of mono-, di-, tri-, poly-azo, anthroquinonic, mono-azo Cr-complexed dyes and dye mixtures [11], [14], [43], [48]. The PC dyes are used extensively in the industry due to their brilliant blue/green colour but are listed as highly polluting as very few enzymes are active on these. Peroxidases have been reported to be effective in decolouration of Reactive blue 15 and Reactive blue 38 [20]. RB21 has been reported to be decolourized by turnip peroxidase [45] and engineered laccases of Cyathus bulleri [24], the latter requiring participation of ABTS. Both peroxidases and laccases suffer from the disadvantage of being dependent on hydrogen peroxide and mediators respectively.
In our previous study [5], the basidiomycete Termitomyces sp. OE147 was shown to produce a wide array of oxidoreductases (CDH, laccases and some uncharacterized oxidoreductases) in the culture fluids when cultivated on cellulose. In this work, effective decolouration and degradation of RB21 is shown using this culture fluid. The effectiveness of the culture fluid was also demonstrated in removing color and phytotoxicity of combined effluent obtained from a local mill.
2. Materials and methods
2.1. Chemicals
All chemicals were of analytical grade. Cellulose (microcrystalline) powder, malt extract, agar, yeast extract, and mycological peptone were purchased from Hi-Media Laboratories Pvt. Ltd. (Mumbai, India). All substrates (ABTS, 2,6-Dichlorophenol-indophenol or DCIP, 2,6-Dimethoxyphenol, Veratryl alcohol) used for measurement of enzyme activities were procured from Sigma-Aldrich Corp. (St. Louis, USA). Reactive blue (RB) 21, Reactive orange (RO) 16, Reactive red (RR) 198, Reactive violet (RV) 5 and Reactive yellow (RY) 42 were obtained from Department of Textile Technology, Indian Institute of Technology Delhi, New Delhi India. The solvents were from J.T. Baker (U.S.A.).
2.2. Organism and culture conditions
Termitomyces sp. OE 147 was obtained from The Directorate of Mushroom Research, Solan, India and maintained on malt extract agar slants at 28 °C. It was sub-cultured every three months. Submerged cultivation was carried out in medium containing malt extract (1%, w/v), yeast extract (0.4%, w/v) and glucose (1%, w/v). The cultures were incubated at 25 °C at 150 rpm. After 5 days, the culture was used to inoculate modified Ludwig medium [18] containing yeast extract (1.75%, w/v), mycological peptone (1.75%, w/v), NH4H2PO4 (0.8%, w/v) and cellulose (3%, w/v) as sole carbon source. The cultures were incubated at 25 °C at 150 rpm. Culture broth was collected at maximum CDH activity (day 5) and clarified by centrifugation at 16000 × g for 30 min at 4 °C. The resulting supernatant was used to perform the decolouration experiments. Activities of CDH and laccase in the culture supernatant were 1.98 U/ml and 1.16 U/ml respectively.
2.3. Dye decolouration
The molecular structures and characteristics of dyes used in the study are summarized in supplementary Fig. 1 and Table 1 respectively. Stock solutions of dyes (other than RB21) were prepared in 50 mM sodium acetate buffer (pH 4.0). For RB21, 0.1% stock solution was prepared in 50 mM of sodium citrate buffer (for pH 3.0), sodium acetate buffer (for pH 4.0 and 5.0) and sodium phosphate buffer (for pH 6.0 and 7.0) and stored in dark at 4 °C. The reactions (final volume 20 ml) were carried out in a 50 ml shake flask as described previously [24] except that the culture filtrate (containing 0.2 U/ml CDH and 0.1 U/ml of laccase) replaced purified laccase and the time of incubation was increased to 12 h. Samples were removed periodically to estimate residual dye concentration by measuring the change in absorbance at λmax of the dyes in PerkinElmer EnSpire® Multimode Plate Reader. Dye decolouration was also studied by inclusion of 200 μM ABTS in the reaction vials. Control experiments were also run wherein the dye was incubated with heat treated culture fluids. Percent decolouration was calculated as follows:
| % Decolouration = [(Initial Absorbance − Final Absorbance)/Initial Absorbance] × 100 |
Table 1.
Characteristics of the dyes used in the present study.
| Dye | Class | λmax (nm) | CI Name | CI Number | Structure |
|---|---|---|---|---|---|
| Remazol Turquoise Blue G-133 | Phthalocyanine | 620 | Reactive Blue 21 | – | Supplementary Fig. 1 (a) |
| Reactive Orange 3R | Monoazo | 490 | Reactive Orange 16 | 17757 | Supplementary Fig. 1 (b) |
| Remazol Red 198 | Monoazo | 520 | Reactive Red 198 | 18221 | Supplementary Fig. 1 (c) |
| Reactive Violet 5 | Monoazo | 550 | Reactive Violet 5 | 18097 | Supplementary Fig. 1 (d) |
| Reactive Yellow 42 | Monoazo | 430 | Reactive Yellow 42 | – | Supplementary Fig. 1 (e) |
Effect of common process parameters was investigated on decolouration of RB21 by varying the concentrations of dye (from 25 to 400 ppm), culture filtrate (from 0.1 to 1.0 U/ml), pH (from 3 to 7) and mixing (from 50 to 250 rpm). To investigate if peroxidase dependent activities were involved, concentration of H2O2 was varied (from 50 to 200 μM) in parallel experiments.
2.4. Characterization of metabolites by ESI–MS
Preliminary identification of the products obtained from degradation of RB21 was done by thin-layer chromatography (TLC). For this, the untreated and the treated dye samples were resolved using F254 silica plates and 80% methanol:20% water as the mobile phase as reported [24]. Plates were dried and observed under UV at 254 nm. The metabolites were extracted from the plates and dissolved in 500 μl of acidified methanol and vortexed for 1 h. It was centrifuged and filtered with a 0.22-μm membrane filter and concentrated to 100 μl using a Speed Vac. The extracted sample (20 μl) was diluted with methanol and injected into an ESI–MS equipped with a hybrid Q-TOF detector (AB Sciex, USA). The spectrum was monitored in positive ion mode with the following conditions: ion spray voltage, 5500 V; nebulizer gas, 20 lb/in2; curtain gas, 25 lb/in2; declustering potential, 60 V; focusing potential, 265 V; and flow rate, 5 μl/min. The spectra were acquired using a mass range (m/z) of 100–500 atomic mass units.
Tentative assignment of structures to various metabolites, liberated from degradation of RB21, was carried out by comparing the acquired mass spectra to spectra in the MS Database using the CambridgeSoft ChemOffice® 2002 program, as well as from the reaction chemistries reported in literature [24], [39].
2.5. Decolouration of textile effluent using culture fluids of Termitomyces sp. OE147
Efficiency of culture fluids was also evaluated in decolouration and detoxification of real effluent obtained from a local denim dying textile mill. The combined effluent (pH 4.8) was collected at the outlet of the neutralization tank and used without any prior treatment. It was suitably diluted to achieve absorbance of <1 O.D. in the visible spectrum. The reactions (final volume 20 ml) were carried out in a 50 ml shake flask containing appropriate volume of culture fluid (equivalent to 1.0 U/ml CDH, 0.5 U/ml Laccase) and incubated shaken at 200 rpm. Aliquots were removed at regular intervals and UV–vis absorption spectrum recorded between 350 and 750 nm with a PerkinElmer EnSpire® Multimode Plate Reader.
2.6. Phytotoxicity studies
The toxicity studies were carried out according to [26] with some modifications. Mung bean (Vigna radiata) was used for toxicity analysis. The seeds, purchased from local market, were sterilized in a 5% sodium hypochlorite solution for 5–10 min [47] followed by thorough rinsing with sterilized deionized water. Sterilized seeds were subsequently placed in moist cotton in a dark incubator at 25 ± 1 °C. After 24 h, the seeds were checked for germination and the sprouted seeds were used in the tests. The toxicity tests were conducted in a Petri dish (87 mm × 18 mm) in three replicate test units per treatment. Each test unit contained a filter paper dipped in either distilled water (control), untreated effluent or treated effluent. Ten (10) germinated seedlings were placed just above the surface of the paper in the test units. The units were placed in dark incubator at 25 ± 1 °C. After 24 h incubation period, the seedlings were separated and the length of the radicle measured to the nearest 10−3 m with a ruler [26].
2.7. Analytical methods
2.7.1. Enzyme activity assays
CDH activity was determined by monitoring decrease in absorbance of DCIP at 520 nm using ε520 nm as 6800/M.cm as described [3]. Lactose was used as an electron donor. Sodium fluoride (10 mM) was used to inhibit laccase activity. One enzyme unit (U) was defined as the amount of enzyme that reduced 1 μmol DCIP per min under the experimental conditions.
Laccase activity was determined by monitoring oxidation of 500 μM ABTS (ε420 nm = 36000/M.cm), buffered with 50 mM sodium tartrate buffer (pH 4.5). One enzyme unit (U) is defined as the amount of laccase transforming 1 μmol ABTS to its cation radical per min [13]. LiP activity was measured using veratryl alcohol as the substrate at 310 nm as described [42]. The reaction was started by addition of 0.4 mM H2O2. One unit (U) was defined as 1 μmol of veratryl alcohol oxidized per min. MnP activity was determined by measuring the oxidation of 2, 6-dimethoxyphenol (0.5 mM) in 50 mM sodium tartrate buffer at 468 nm as described [51]. As with LiP, reaction was initiated by adding 0.4 mM H2O2. One enzyme unit (U) was defined as 1 μmol reaction product formed per min.
All enzyme assays were performed in a PerkinElmer Lambda 25 spectrophotometer. Enzyme activities were measured thrice in duplicate flasks set up over a period of three months. The reported values are an average of these. Standard deviation among the samples was between 5 and 8%.
2.7.2. Electrophoresis and zymogram
The zymogram of CDH and laccase was performed directly by loading small volumes of culture filtrates. Briefly, 20 μl (for CDH) and 5, 10 and 20 μl (for laccase) of crude filtrate was loaded on to a 10% Native-PAGE [37]. After electrophoretic run, gel was incubated in 3 μM DCIP (in 100-mM sodium acetate buffer, pH 5.0) for CDH and 200 μM ABTS solution (in 50 mM sodium-citrate buffer, pH 4.0) for laccase activity visualization. For CDH, after uniform staining, few drops of 300 mM lactose solution were added on the surface of gel to visualize the enzyme band [4]. For laccase, the gels were incubated for 5–10 min till green bands appeared.
3. Results and discussion
Both bacterial [32], [33], [38] and fungal [16], [19], [23] systems have been used for removal of textile dyes from the effluents. Although cell biomass has been suggested as a promising way to remove the colour through biosorption [33], it results in accumulation of biological sludge. Further, the toxic dyes also restrict the growth of the organisms [49]. Enzymes, primarily the lignin degrading enzymes, have been successfully used for decolouration of a variety of dyes [1], [10], [40]. Laccase, in combination with CDH, is reported to be effective against mono-, di-, tri-, poly-azo and some anthroquinone dyes [11], [14]. Termitomyces sp. OE147 secretes high amounts of CDH along with moderate amounts of laccase [5], [18] which have not been evaluated for dye decolouration. The data presented below shows effectiveness of this culture filtrate in decolouration and degradation of RB21 as well as real textile effluent.
3.1. Enzyme production by Termitomyces sp. OE 147 in liquid culture
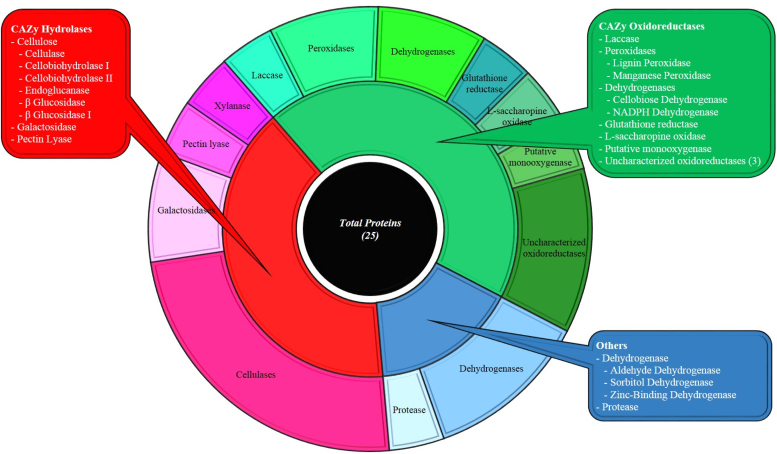
Fig. 1 shows the distribution of CAZy hydrolases, CAZy oxidoreductases and other enzymes in the secretome of Termitomyces sp. OE 147. On day 5, culture fluids exhibited ∼2000 U/l of CDH and 1160 U/l laccase activity. Low peroxidase (LiP and MnP <0.03 U/ml) activities were also detected in culture filtrates. Although high laccase and low CDH activities have been reported in Pycnoporus sanguiness [48], Trametes pubescens [14] and Trametes trogii [43] on cellulose, Termitomyces sp. OE147 was distinct in producing high amounts of CDH relative to laccase. The extracellular proteins in the culture filtrate were separated on Native-PAGE followed by zymogram analysis and the results are shown in supplementary Fig. 2. As seen, a single isoform of CDH and multiple forms of laccases were observed.
Fig. 1.
Pie chart showing the distribution of various enzyme activities present in the secretome of Termitomyces sp. OE147 cultivated on cellulose. Categories are (i) CAZy Hydrolases, (ii) CAZy oxidoreductases and (iii) Others.
3.2. Decolouration of different dyes by the culture filtrate of Termitomyces sp. OE147
The broad-spectrum decolouration efficiency of the culture filtrate (with no externally added mediators) was assessed using chemically different dyes and the rates of decolouration are shown in Table 2. A time course profile of decolouration is shown in Supplementary Fig. 3. Maximum decolouration and highest rate of decolouration of 25 ± 0.4 nmoles/min was obtained with RB21 followed by 8–11 nmoles/min for RY42 and RO16. Decolouration of RO16 and RY42, in absence of mediators, is important as both the dyes are highly recalcitrant. Moderate (40%) decolouration of RO16 has been reported with enzymes of culture filtrate of Ganoderma sp. En3 [27]. However, it is important to perform these experiments under carefully controlled conditions as contaminating bacteria often grow leading to adsorption of dyes by bacterial cells. Advanced oxidation processes such as ozonation [44] or treatment with UV/H2O2 [30] are currently the only effective methods for removal of color from RO16. Addition of ABTS accelerated rates of decolouration of all dyes and resulted in >80% removal of color. This process was, however, useless as coloured polymers formed at the end of the reactions. In place of ABTS, HOBT has also been reported to be very effective in decolouration of all synthetic dyes [14]. Since the culture fluids were effective on RB21, for which only peroxidases or laccase mediator systems have been found to be effective, further work was restricted to RB21.
Table 2.
Rate and% decolouration of dyes treated with culture filtrate of Termitomyces sp. OE147.
| Dye | Molecular Mass (g/mol) | % Decolouration | Rate of Decolouration (nmoles/min) |
|---|---|---|---|
| Reactive Blue 21 | 987.4 | 36 | 25 ± 0.4 |
| Reactive Orange 16 | 617.5 | 5 | 11 ± 0.3 |
| Reactive Red 198 | 968.21 | 10 | 4 ± 0.2 |
| Reactive Violet 5 | 735.59 | 5 | 5 ± 0.6 |
| Reactive Yellow 42 | 662.6 | 8 | 8 ± 0.3 |
3.3. Factors affecting decolouration of RB21
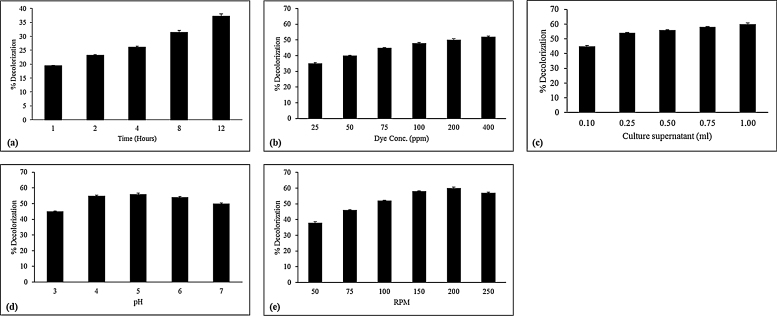
Effect of operating conditions (static vs shaking, time of treatment, dye: culture fluid ratio, pH) was evaluated on RB21 decolouration and the results are shown in Fig. 2(a). Decolouration was more effective under shaking than under static conditions. Maximum colour removal was 37.19 ± 0.87% in 12 h (for 25 ppm) after which there was no further increase. Higher concentrations of the dye (at fixed volume of culture fluid) did not lead to decrease in the rates of decolouration (Fig. 2b) and 50% of the dye was decolourized at 100 ppm levels indicating non toxicity of the reaction products towards enzyme activities. This was contrary to earlier reports [31], [39] where turnip peroxidase was effective at low concentration (30–40 ppm) of RB21. An optimum ratio was arrived at between the dye (75 ppm) and culture fluid whereby 56 ± 0.4% decolouration was obtained. As seen in Fig. 2(d), more than 55 ± 0.4% of the colour was removed between pH 4.0–5.0 suggesting involvement of laccase and CDH that are most effective at the acidic pH values. An increase in rpm to 200 resulted in an increase in colour removal to 60% and was attributed to better mixing (Fig. 2e).
Fig. 2.
Effect of process parameters (a) time of contact (b) dye concentration, (c) culture supernatant, (d) pH and (e) RPM on decolouration of RB21 when incubated with the culture filtrate of Termitomyces sp. OE 147.
H2O2 acts as a co-substrate to activate the enzymatic action of peroxidase radical. However, excess of this reagent can inhibit enzyme activity while at low concentrations it can limit the reaction rates [52]. Since low levels of peroxidases were detected in the culture supernatant and to ascertain if these played a major role in removal of colour, H2O2 concentration was varied. However, no significant increase in decolouration was observed (data not shown). This is important as in almost all studies involving degradation of RB21, peroxidases were indicated to play a major role and H2O2 was found to have a stimulatory effect.
3.4. Characterization of degradation products of RB21 by ESI–MS
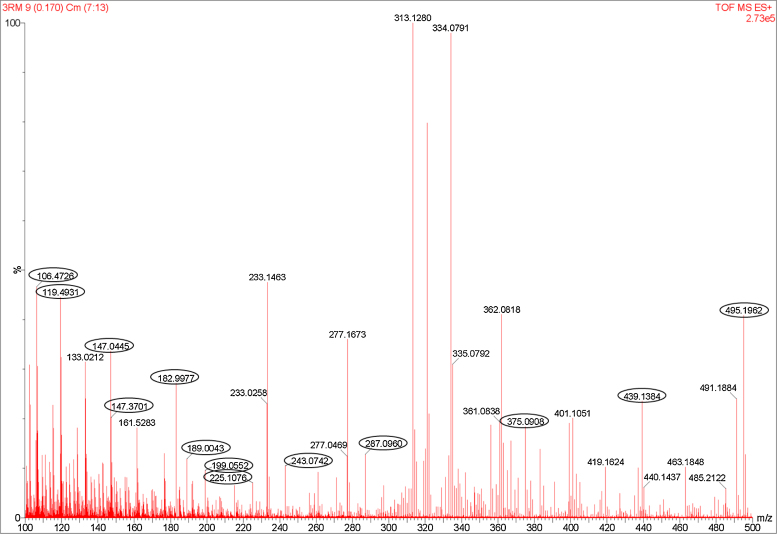
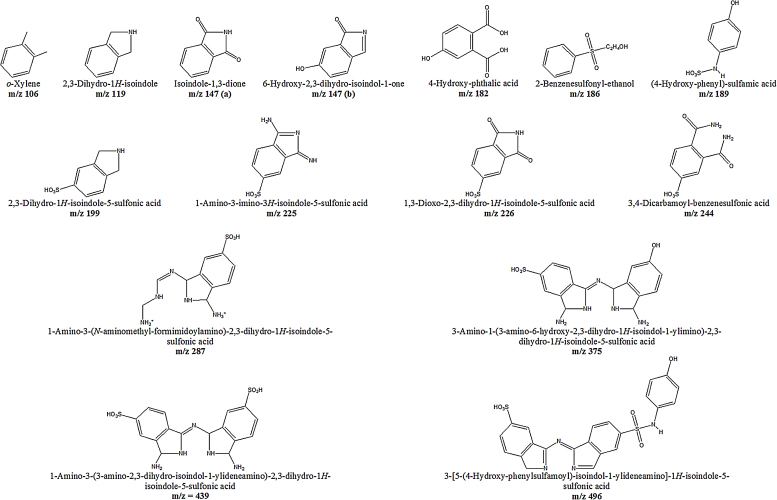
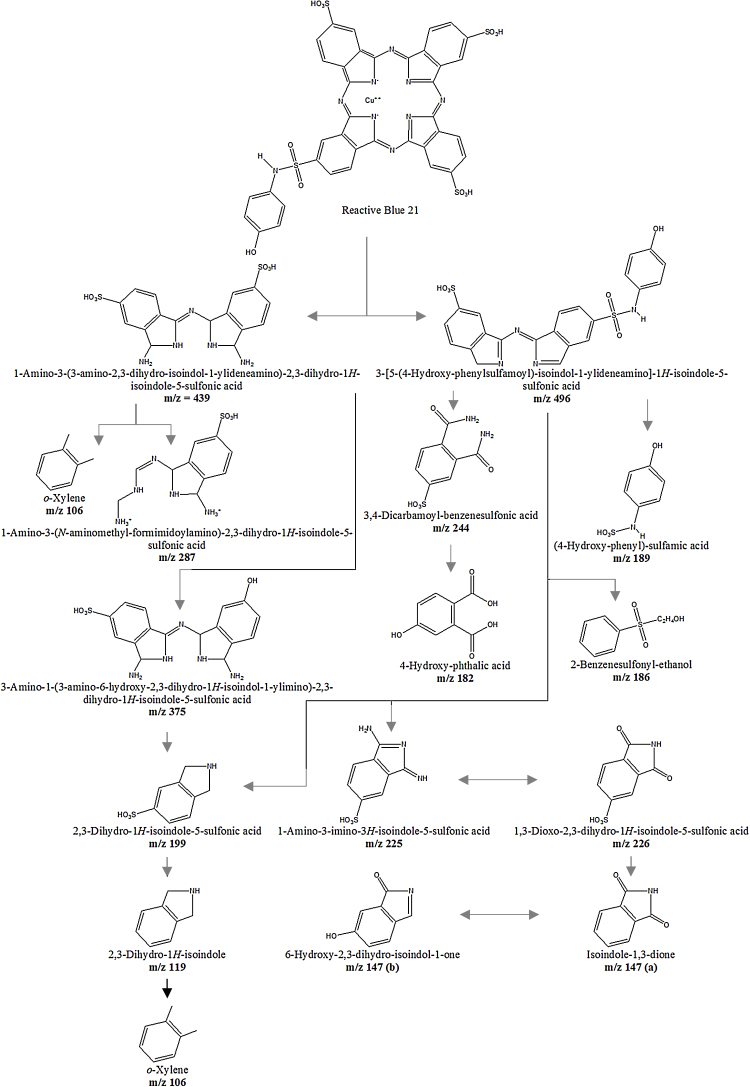
UV–vis spectrophotometry of the treated RB21 showed decrease in absorbance at 620 nm (Supplementary Fig. 3a). In order to identify the intermediates as well as the end products of degradation, aliquots were removed at regular intervals and different components separated by preparative TLC. The UV absorbing materials were visualized and extracted in methanol. The mass spectrum (Fig. 3) of the extracted metabolites indicated a number of prominent peaks in the lower mass range. The assigned structures of these are shown in Fig. 4. The products obtained were compared with the products generated by action of laccase + ABTS and peroxidase on RB21, as reported in literature [24], [39]. As seen (Table 3), only a few peaks m/z 147, m/z 244, m/z 439] were common with the products identified in earlier studies. Several additional peaks were observed indicating extensive breakdown of RB21 on account of action of various enzymes present in the culture fluid. Few moieties (m/z 182, m/z 244) were similar to the products obtained by the Palladium based oxidation of RB21 [29].
Fig. 3.
Mass spectrum of reaction products produced by degradation of RB21.
Fig. 4.
Predicted structures of products produced by degradation of RB21.
Table 3.
Comparison of metabolites generated from Reactive blue 21 by action of laccase, peroxidases and culture filtrate of Termitomyces sp. OE147.
| Product m/z | Product Name | Laccase [24] | Peroxidase [39] | Palladium catalyst [29] |
CDH, Laccase, Oxidoreductases Present Study |
|---|---|---|---|---|---|
| 106 | o-Xylene | + | |||
| 119 | 2,3-Dihydro-1H-isoindole | + | |||
| 147 (a) | Isoindole-1,3-dione | + | |||
| 147 (b) | 6-Hydroxy-2,3-dihydro-isoindol-1-one | + | + | ||
| 182 | 4-Hydroxy-phthalic acid | + | + | ||
| 186 | 2-Benzenesulfonyl-ethanol | + | |||
| 189 | (4-Hydroxy-phenyl)-sulfamic acid | + | |||
| 199 | 2,3-Dihydro-1H-isoindole-5-sulfonic acid | + | |||
| 225 | 1-Amino-3-imino-3H-isoindole-5-sulphonic acid | + | |||
| 226 | 1,3-Dioxo-2,3-dihydro-1H-isoindole-5-sulphonic acid | + | |||
| 244 | 3,4-Dicarbamoyl-benzenesulfonic acid | + | + | + | |
| 287 | 1-Amino-3-(N-aminomethyl-formimidoylamino)-2,3-dihydro-1H-isoindole-5-sulfonic acid | + | |||
| 375 | 3-Amino-1-(3-amino-6-hydroxy-2,3-dihydro-1H-isoindole-1-ylimino)-2,3-dihydro-1H-isoindole-5-sulfonic acid | + | |||
| 439 | 1-Amino-3-(3-amino-2,3-dihydro-isoindole-1-ylideneamino)-2,3-dihydro-1H-isoindole-5-sulfonic acid | + | + | ||
| 496 | 3-[5-(4-Hydroxy-phenylsufamoyl)-isoindole-1-ylideneamino]-1H-isoindole-5-sulfonic acid | + |
+ Indicates the presence of the named metabolite (see column 2) in the indicated study shown in the column heading. The names of the enzyme responsible for generating the product are also listed.
Based on the reaction chemistries of laccase, peroxidase and CDH, a pathway of degradation of RB21 is proposed (Fig. 5). The high molecular weight products formed (m/z of 496, 439) resulted from cleavage of the nitrogen bonds in the inner ring of the phthalocyanine molecule by mediator assisted action of laccase, as reported earlier [24]. One of these (m/z 496) was concluded to be 3-[5-(4-Hydroxy-phenylsulfamoyl)-isoindol-1-ylideneamino]-1H-isoindole-5-sulfonic acid, and the other at m/z of 439 was the same as reported by action of turnip peroxidase on RB21 [39] indicating low levels of peroxidases present in the culture filtrate to be effective agents for degradation. The smaller fragment could also result from a fragmentation of the larger molecule of m/z 496. Two other metabolites m/z = 243 (3,4-Dicarbamoyl-benzenesulfonic acid) and m/z = 147 (6-Hydroxy-2,3-dihydro-isoindol-1-one) have been reported earlier [24] and result from oxidative action of laccase and ABTS. In the present study, CDH could be involved in direct oxidation of dyes through generation of OH• radicals through Fenton chemistry or by participation of the Flavin domain, as proposed for cellulose [34]. All known CDH’s contain an N-terminus heme domain and a C-terminus flavin domain. The flavin domain is considered as a part of the widespread glucose-methanol-choline oxidoreductase superfamily [8] whereas the likes of heme domains are only found in fungi [53]. In the event of the oxidation mediated by Flavin domain, the electrons are transferred to heme domain. Laccases can accept electrons from the heme domain of reduced CDH by virtue of presence of copper and therefore the two can serve as an effective redox couple. Participation of molecular oxygen for laccase oxidation and recycling allows the system to work effectively in the absence of any external mediators.
Fig. 5.
Suggested degradation pathway of RB21 by enzyme mixtures in culture filtrate of Termitomyces sp. OE 147.
Another metabolite with m/z of 226 (1,3-Dioxo-2,3-dihydro-1H-isoindole-5-sulfonic acid) has been reported from the degradation of Remazol turquoise blue G 133 by soybean peroxidase [28] and in the present system, could result from action of peroxidases in the culture fluids. Many of the small products were formed by reduction and cleavage of the nitrogen bonds in the inner ring of the PC dye due to oxidative action of hereto uncharacterized enzymes followed by hydrolytic reactions. It has also been stated [29] that some of these smaller products could be related to precursor molecules used for the synthesis of these dyes. The discrepancy of one unit mass observed in some molecules was attributed to gain of proton in the side chain.
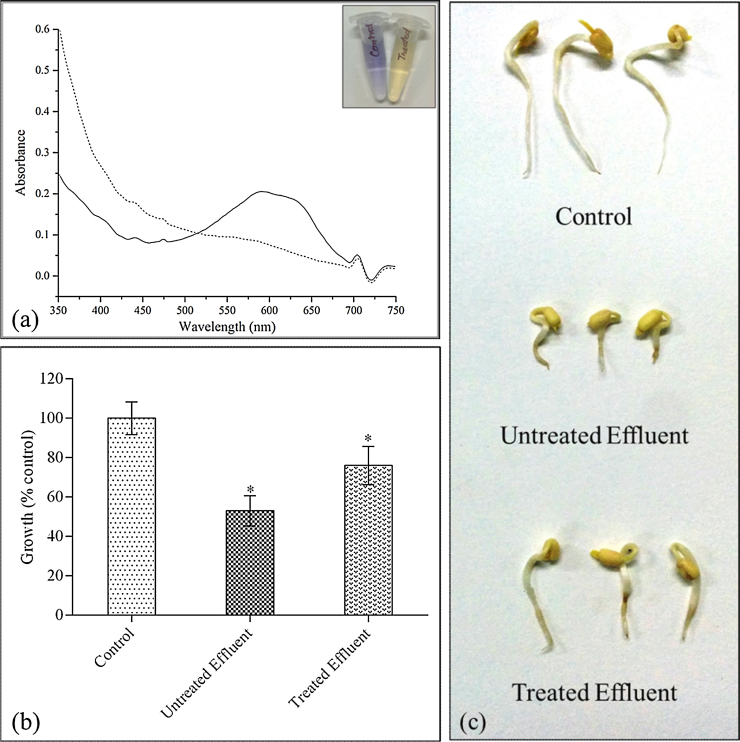
3.5. Decolouration of textile effluent using culture fluids of Termitomyces sp. OE147 and analysis of phytotoxicity
The experiments were carried out following a procedure similar to that used for the pure dyes. Fig. 6a shows the absorption spectrum of the textile effluent before and after treatment with the culture fluid. A decrease in O.D. was observed at all wavelengths (corresponding to absorption from the dyes) indicating the effectiveness of the culture filtrate on effluent with about 52% reduction in colour. An increase in O.D. values was observed in the far-UV range indicating formation of intermediates with UV absorbing properties. The phytotoxicity analysis of the untreated and the culture filtrate treated effluent was carried out with the purpose of evaluating whether the reaction products would be more toxic than the raw effluent. Fig. 6b shows the seedling growth of mung bean exposed to effluent before and after treatment, expressed as a percentage of growth in the control treatment group. It was observed that the growth of the seedlings was adversely affected (residual growth of 53%) by untreated textile effluent. Less toxic affect was noticed for the treated effluent and better growth (76%) of radicles was noticed indicating lowering of toxicity by treatment with the culture fluids. Additionally, browning of the root tips was detected in the exposed roots (Fig. 6c). Further studies need to be carried out to see the impact at cellular level.
Fig. 6.
(a) UV–vis absorption spectrum of treated (.....) and untreated (___) textile effluent. The inset shows the effluent before and after treatment. (b) Seedling growth of Vignus radiata exposed to textile effluent before and after treatment with culture fluids of Termitomyces sp. OE147. Sterile deionized water was used as control. The values are mean and standard error of means (Mean ± S.E.) of 10 samples with three replicates (n = 10, P = 0.05). *Significant at 5% level. (c) Actual pictures of the seedlings after exposure to untreated and treated effluent. Control (exposed to deionized water) seeds are shown for comparison.
4. Conclusions
The results showed that culture fluids of Termitomyces sp. OE417, obtained on cellulose, were highly effective for decolouration and degradation of the reactive phthalocyanine dye RB21. Optimization of dye: culture fluid ratio, pH and mixing lead to 60% removal of colour. Based on the products of degradation, many of which have not been previously reported, it was concluded that oxidoreductases including CDH and laccase, were effective without addition of mediators. The culture filtrate was also effective in decolouration of real effluent by 52% without addition of external mediators. Evaluation of toxicity confirmed reduction of phytotoxicity of the effluent after treatment. Given that this fungus can be cultivated in a bioreactor with cellulose as a sole carbon source, the study presents an effective strategy wherein the culture filtrates can serve as a source of oxidoreductases for application in treatment of textile wastewaters.
Conflict of interest
Dr. Mishra has nothing to disclose.
Acknowledgements
This research was funded by Department of Biotechnology (Govt. of India) through a grant awarded to SM (RP02875). The authors would like to express sincere thanks to Dr. Tenzin Kenzom for interpretation of the mass data and structural assignments to various compounds.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2016.10.002.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Abadulla E., Robra K.-H., Gübitz G.M., Silva L.M., Cavaco-Paulo A. Enzymatic decolorization of textile dyeing effluents. Text. Res. J. 2000;70:409–414. [Google Scholar]
- 2.Baminger U., Ludwig R., Galhaup C., Leitner C., Kulbe K.D., Haltrich D. Continuous enzymatic regeneration of redox mediators used in biotransformation reactions employing flavoproteins. J. Mol. Catal. B: Enzym. 2001;11:541–550. [Google Scholar]
- 3.Baminger U., Nidetzky B., Kulbe K.D., Haltrich D. A simple assay for measuring cellobiose dehydrogenase activity in the presence of laccase. J. Microbiol. Methods. 1999;35:253–259. doi: 10.1016/s0167-7012(99)00022-6. [DOI] [PubMed] [Google Scholar]
- 4.Bao W.J., Lymar E., Renganathan V. Optimization of cellobiose dehydrogenase and β-glucosidase production by cellulose-degrading cultures of Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1994;42:642–646. [Google Scholar]
- 5.Bashir H., Gangwar R., Mishra S. Differential production of lignocellulolytic enzymes by a white rot fungus Termitomyces sp. OE147 on cellulose and lactose. BBA − Proteins Proteomics. 2015;1854:1290–1299. doi: 10.1016/j.bbapap.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Basosi R., Della Lunga G., Pogni R. Copper biomolecules in solution. In: Eaton S.R., Eaton G.R., Berliner L.J., editors. Biomedical EPR, Part A: Free Radicals, Metals, Medicine, and Physiology. Springer; US, Boston, MA: 2005. pp. 385–416. [Google Scholar]
- 7.Camarero S., Ibarra D., Martínez M.J., Martínez Á.T. Lignin-derived compounds as efficient laccase mediators for decolorization of different types of recalcitrant dyes. Appl. Environ. Microbiol. 2005;71:1775–1784. doi: 10.1128/AEM.71.4.1775-1784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavener D.R. GMC oxidoreductases: a newly defined family of homologous proteins with diverse catalytic activities. J. Mol. Biol. 1992;223:811–814. doi: 10.1016/0022-2836(92)90992-s. [DOI] [PubMed] [Google Scholar]
- 9.Chhabra M., Mishra S., Sreekrishnan T.R. Mediator-assisted decolorization and detoxification of textile dyes/dye mixture by Cyathus bullerilaccase. Appl. Biochem. Biotechnol. 2008;151:587–598. doi: 10.1007/s12010-008-8234-z. [DOI] [PubMed] [Google Scholar]
- 10.Ciullini I., Gullotto A., Tilli S., Sannia G., Basosi R., Scozzafava A., Briganti F. Enzymatic decolorization of spent textile dyeing baths composed by mixtures of synthetic dyes and additives. Appl. Microbiol. Biotechnol. 2012:1–11. doi: 10.1007/s00253-011-3809-y. [DOI] [PubMed] [Google Scholar]
- 11.Ciullini I., Tilli S., Scozzafava A., Briganti F. Fungal laccase, cellobiose dehydrogenase, and chemical mediators: combined actions for the decolorization of different classes of textile dyes. Bioresour. Technol. 2008;99:7003–7010. doi: 10.1016/j.biortech.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Couto S.R., Sanromán M.Á. The effect of violuric acid on the decolourization of recalcitrant dyes by laccase from Trametes hirsuta. Dyes Pigm. 2007;74:123–126. [Google Scholar]
- 13.Eggert C., Temp U., Eriksson K.E. The ligninolytic system of the white rot fungus Pycnoporus cinnabarinus: purification and characterization of the laccase. Appl. Environ. Microbiol. 1996;62:1151–1158. doi: 10.1128/aem.62.4.1151-1158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enayatzamir K., Tabandeh F., Yakhchali B., Alikhani H.A., Rodríguez Couto S. Assessment of the joint effect of laccase and cellobiose dehydrogenase on the decolouration of different synthetic dyes. J. Hazard. Mater. 2009;169:176–181. doi: 10.1016/j.jhazmat.2009.03.088. [DOI] [PubMed] [Google Scholar]
- 15.Fewson C.A. Biodegradation of xenobiotic and other persistent compounds: the causes of recalcitrance. Trends Biotechnol. 1988;6:148–153. [Google Scholar]
- 16.Fu Y., Viraraghavan T. Fungal decolorization of dye wastewaters: a review. Bioresour. Technol. 2001;79:251–262. doi: 10.1016/s0960-8524(01)00028-1. [DOI] [PubMed] [Google Scholar]
- 17.Groff K.A., Kim B.R. Textile wastes. J. Water Pollut. Control Fed. 1989;61:872–876. [Google Scholar]
- 18.Gupta G., Gangwar R., Gautam A., Kumar L., Dhariwal A., Sahai V., Mishra S. Production of cellobiose dehydrogenase from a newly isolated white rot fungus Termitomyces sp. OE147. Appl. Biochem. Biotechnol. 2014;173:1–17. doi: 10.1007/s12010-014-1010-3. [DOI] [PubMed] [Google Scholar]
- 19.Gupta V.K., Khamparia S., Tyagi I., Jaspal D., Malviya A. Decolorization of mixture of dyes: a critical review. Glob. J. Environ. Sci. Manag. 2015;1:71–94. [Google Scholar]
- 20.Heinfling A., Bergbauer M., Szewzyk U. Biodegradation of azo and phthalocyanine dyes by Trametes versicolor and Bjerkandera adusta. Appl. Microbiol. Biotechnol. 1997;48:261–266. [Google Scholar]
- 21.Henriksson G., Johansson G., Pettersson G. A critical review of cellobiose dehydrogenases. J. Biotechnol. 2000;78:93–113. doi: 10.1016/s0168-1656(00)00206-6. [DOI] [PubMed] [Google Scholar]
- 22.Hildén L., Johansson G., Pettersson G., Li J., Ljungquist P., Henriksson G. Do the extracellular enzymes cellobiose dehydrogenase and manganese peroxidase form a pathway in lignin biodegradation? FEBS Lett. 2000;477:79–83. doi: 10.1016/s0014-5793(00)01757-9. [DOI] [PubMed] [Google Scholar]
- 23.Kaushik P., Malik A. Fungal dye decolourization: recent advances and future potential. Environ. Int. 2009;35:127–141. doi: 10.1016/j.envint.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Kenzom T., Srivastava P., Mishra S. Structural insights into 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS)-mediated degradation of Reactive Blue 21 by engineered Cyathus bulleri laccase and characterization of degradation products. Appl. Environ. Microbiol. 2014;80:7484–7495. doi: 10.1128/AEM.02665-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langhals H. 3rd ed. WILEY-VCH Verlag; 2004. Color Chemistry. Synthesis, Properties and Applications of Organic Dyes and Pigments. [Google Scholar]
- 26.Lee W.-M., Kwak J.I., An Y.-J. Effect of silver nanoparticles in crop plants Phaseolus radiatus and Sorghum bicolor: media effect on phytotoxicity. Chemosphere. 2012;86:491–499. doi: 10.1016/j.chemosphere.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 27.Ma L., Zhuo R., Liu H., Yu D., Jiang M., Zhang X., Yang Y. Efficient decolorization and detoxification of the sulfonated azo dye Reactive Orange 16 and simulated textile wastewater containing Reactive Orange 16 by the white-rot fungus Ganoderma sp. En3 isolated from the forest of Tzu-chin mountain in China. Biochem. Eng. J. 2014;82:1–9. [Google Scholar]
- 28.Marchis T., Avetta P., Bianco-Prevot A., Fabbri D., Viscardi G., Laurenti E. Oxidative degradation of remazol turquoise blue G 133 by soybean peroxidase. J. Inorg. Biochem. 2011;105:321–327. doi: 10.1016/j.jinorgbio.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 29.Matthews R.D., Bottomley L.A., Pavlostathis S.G. Palladium-catalyzed hydrogen reduction and decolorization of reactive phthalocyanine dyes. Desalination. 2009;248:816–825. [Google Scholar]
- 30.Mitrović J., Radović M., Bojić D., Andjelković T., Purenović M., Bojić A. Decolorization of textile azo dye reactive orange 16 with UV/H2O2 process. J. Serb. Chem. Soc. 2012;77:465–481. [Google Scholar]
- 31.Mohan S.V., Prasad K.K., Rao N.C., Sarma P.N. Acid azo dye degradation by free and immobilized horseradish peroxidase (HRP) catalyzed process. Chemosphere. 2005;58:1097–1105. doi: 10.1016/j.chemosphere.2004.09.070. [DOI] [PubMed] [Google Scholar]
- 32.Pandey A., Singh P., Iyengar L. Bacterial decolorization and degradation of azo dyes. Int. Biodeterior. Biodegrad. 2007;59:73–84. [Google Scholar]
- 33.Pearce C.I., Lloyd J.R., Guthrie J.T. The removal of colour from textile wastewater using whole bacterial cells: a review. Dyes Pigm. 2003;58:179–196. [Google Scholar]
- 34.Phillips C.M., Beeson W.T., Cate J.H., Marletta M.A. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 2011;6:1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 35.Preethi S., Anumary A., Ashokkumar M., Thanikaivelan P. Probing horseradish peroxidase catalyzed degradation of azo dye from tannery wastewater. SpringerPlus. 2013;2:1–8. doi: 10.1186/2193-1801-2-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson T., McMullan G., Marchant R., Nigam P. Remediation of dyes in textile effluent: a critical review on current treatment technologies with a proposed alternative. Bioresour. Technol. 2001;77:247–255. doi: 10.1016/s0960-8524(00)00080-8. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J., Fritsch E.F., T.M . Cold Spring Harbor Laboratory Press; NY, USA: 1989. Molecular Cloning: A Laboratory Manual. [Google Scholar]
- 38.Saratale R.G., Saratale G.D., Chang J.S., Govindwar S.P. Bacterial decolorization and degradation of azo dyes: a review. J. Taiwan Inst. Chem. E. 2011;42:138–157. [Google Scholar]
- 39.Silva M.C., Corrêa A.D., Amorim M.T.S.P., Parpot P., Torres J.A., Chagas P.M.B. Decolorization of the phthalocyanine dye reactive blue 21 by turnip peroxidase and assessment of its oxidation products. J. Mol. Catal. B: Enzym. 2012;77:9–14. [Google Scholar]
- 40.Singh R.L., Singh P.K., Singh R.P. Enzymatic decolorization and degradation of azo dyes −a review. Int. Biodeterior. Biodegrad. 2015;104:21–31. [Google Scholar]
- 41.Tan T.-C., Kracher D., Gandini R., Sygmund C., Kittl R., Haltrich D., Hallberg B.M., Ludwig R., Divne C. Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation. Nat. Commun. 2015;6:7542. doi: 10.1038/ncomms8542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tien M., Kirk T.K. Lignin-degrading enzyme from the hymenomycete Phanerochaete chrysosporium burds. Science. 1983;221:661–663. doi: 10.1126/science.221.4611.661. [DOI] [PubMed] [Google Scholar]
- 43.Tilli S., Ciullini I., Scozzafava A., Briganti F. Differential decolorization of textile dyes in mixtures and the joint effect of laccase and cellobiose dehydrogenase activities present in extracellular extracts from Funalia trogii. Enzyme Microb. Technol. 2011;49:465–471. doi: 10.1016/j.enzmictec.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Tizaoui C., Grima N. Kinetics of the ozone oxidation of Reactive Orange 16 azo-dye in aqueous solution. Chem. Eng. J. 2011;173:463–473. [Google Scholar]
- 45.Ulson de Souza S.M.A.G., Forgiarini E., Ulson de Souza A.A. Toxicity of textile dyes and their degradation by the enzyme horseradish peroxidase (HRP) J. Hazard. Mater. 2007;147:1073–1078. doi: 10.1016/j.jhazmat.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 46.UNSD, United Nations Statistics Division of the Department of Economics and Social Affairs (DESA). New York USA, http://data.un.org (2015).
- 47.The United States Environmental Protection Agency (USEPA), 1996. Ecological Effects Test Guidelines, OPPTS 850.4200, Seed Germination/Root Elongation Toxicity Test. EPA 712-C-96-154, Prevention, Pesticides and Toxic Substances, p. 7170.
- 48.Vanhulle S., Enaud E., Trovaslet M., Nouaimeh N., Bols C.-M., Keshavarz T., Tron T., Sannia G., Corbisier A.-M. Overlap of laccases/cellobiose dehydrogenase activities during the decolourisation of anthraquinonic dyes with close chemical structures by Pycnoporus strains. Enzyme Microb. Technol. 2007;40:1723–1731. [Google Scholar]
- 49.Vanhulle S., Trovaslet M., Enaud E., Lucas M., Taghavi S., van der Lelie D., van Aken B., Foret M., Onderwater R.C.A., Wesenberg D., Agathos S.N., Schneider Y.-J., Corbisier A.-M. Decolorization, cytotoxicity and genotoxicity reduction during a combined ozonation/fungal treatment of dye-contaminated wastewater. Environ. Sci. Technol. 2003;42:584–589. doi: 10.1021/es071300k. [DOI] [PubMed] [Google Scholar]
- 50.Viswanath B., Rajesh B., Janardhan A., Kumar A.P., Narasimha G. Fungal laccases and their applications in bioremediation. Enzyme Res. 2014;2014:21. doi: 10.1155/2014/163242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wariishi H., Valli K., Gold M.H. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J. Biol. Chem. 1992;267:23688–23695. [PubMed] [Google Scholar]
- 52.Wu J., Bewtra J.K., Biswas N., Taylor K.E. Effect of H2O2 addition mode on enzymatic removal of phenol from wastewater in the presence of polyethylene glycol. Can. J. Chem. Eng. 1994;72:881–886. [Google Scholar]
- 53.Zámocký M., Ludwig R., Peterbauer C., Hallberg M.B., Divne C., Nicholls P., Haltrich D. Cellobiose dehydrogenase − a flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr. Protein Pept. Sci. 2006;7:255–280. doi: 10.2174/138920306777452367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.