Highlights
-
•
Protease producing Bacillus subtilis was isolated from animal slaughter house waste.
-
•
Protease was covalently immobilized on amino-functionalized magnetic nanoparticles (AMNPs).
-
•
To our knowledge, for the first time these magnetic nano biocatalyst was used for the synthesis of series of novel glycinamides.
-
•
Parameters for glycinamides synthesis such as, pH, temperature and time were optimized using Response Surface Methodology.
-
•
Reusability study showed that protease immobilized MNPs retain up to 70% of initial activity after 8th cycles of reuse for the synthesis.
Keywords: Protease, Magnetic nanoparticles, Immobilization, Glycinamides, Response surface methodology
Abstract
In the present investigation, Bacillus subtilis was isolated from slaughterhouse waste and screened for the production of protease enzyme. The purified protease was successfully immobilized on magnetic nanoparticles (MNPs) and used for the synthesis of series of glycinamides. The binding and thermal stability of protease on MNPs was confirmed by FTIR spectroscopy and TGA analysis. The surface morphology of MNPs before and after protease immobilization was carried out using SEM analysis. XRD pattern revealed no phase change in MNPs after enzyme immobilization. The processing parameters for glycinamides synthesis viz. temperature, pH, and time were optimized using Response Surface Methodology (RSM) by using Design Expert (9.0.6.2). The maximum yield of various amides 2 butyramidoacetic acid (AMD-1,83.4%), 2-benzamidoacetic acid (AMD-2,80.5%) and 2,2′((carboxymethyl) amino)-2-oxoethyl)-2-hydroxysuccinyl)bis(azanediyl))diacetic acid (AMD-3,80.8%) formed was observed at pH-8, 50 °C and 30 min. The synthesized immobilized protease retained 70% of the initial activity even after 8 cycles of reuse.
1. Introduction
The amide functionality is ubiquitous in life and as protein plays a crucial role in virtually all biological processes conducted for the sustenance of life. It is a common feature in small or complex synthetic or natural molecules and is a key component of various important chemicals such as medicinal chemicals, agrochemicals, hormones, pesticide, polymers, and other natural products [1], [2], [3]. Amide bonds are typically formed from amines and unactivated carboxylic acid using stoichiometric amounts of coupling reagents such as carbodiimides or pre-activated carboxylic acid derivatives such as acyl halides, acyl azides, anhydrides, or esters. All of these derivatization methods possess considerable drawbacks; harsh reaction conditions, use of toxic reactive reagents, significant by-product formation, production of stoichiometric amounts of hazardous chemical wastes during the reaction processes, product purification and show poor atom economy. The preponderance of the amide bond in natural products and its importance in industrial and pharmaceutical chemistry, there is an immense need to develop clean, catalytic, environmentally benign, ambient, waste-free and cost effective methods for amide bond formation [1], [3], [4], [5].
Enzymes are versatile biocatalysts, serve as a key enabling technology for chemical synthesis and are considered as “green chemicals” due to their eco-friendly nature [6]. Proteases are the group of proteolytic enzymes which are capable of peptide bond hydrolysis but there is evidence that it can efficiently catalyze the peptide synthesis. Protease represents one of the most important industrial enzymes and has wide application in food, pharmaceutical, detergent and leather industry. Proteolytic enzyme plays an essential role in cellular metabolic process and support the immune system such as to digest the unwanted debris in blood cell [7], [8]. Protease catalyzed synthesis of amid has a numerous advantages over chemical synthesis methods such as extremely mild reaction condition, stability over wide range of pH and temperature, industrial scale-up scope, high reaction rates, high reaction specificity with fewer side reactions, regio and stereospecificity, the absence of racemization, no requirement of side-chain protection, lower reprocessing and purification steps and less pollution [9], [10], [11]. Besides these applications considering the industrial interests recently, protease have been successfully used to have anti-biofilm properties by immobilizing on chitosan [12], [13].
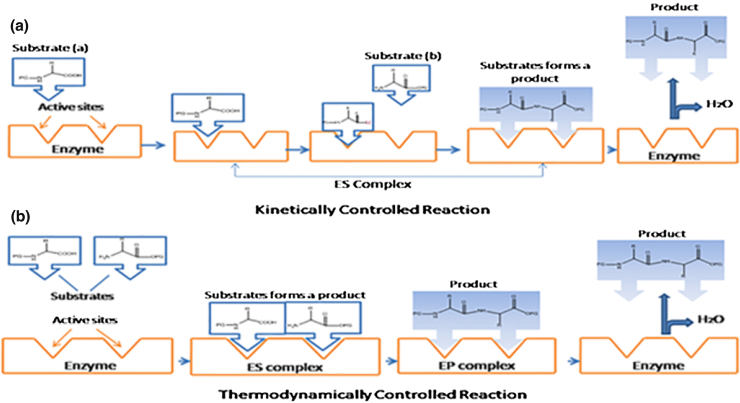
There are two basic strategies towards enzymatic amide synthesis, either the kinetically controlled or the thermodynamically controlled. In kinetically controlled amide synthesis, N-terminally protected acyl donor reacts with enzyme and forms an acyl-enzyme intermediate, which subsequently react with the C-terminally protected acyl acceptor as shown in Fig. 1a. In thermodynamically controlled amide synthesis, C-protected acyl acceptor directly reacts with the N-terminally protected acyl donor as shown in Fig. 1b. As compared to kinetically controlled reaction thermodynamically controlled reaction is rather slow and gives lower yield. Under thermodynamic control, manipulation of reaction conditions are required to shift the equilibrium in the direction of amide synthesis instead of their hydrolysis by, product precipitation, water removal or addition of organic solvents to suppress the ionization of the starting materials [6], [14], [15], [16], [17].
Fig 1.
Mechanism of enzymatic reaction (a) kinetically controlled (b) thermodynamically controlled.
The activity of the enzyme is mostly affected by temperature, pH, substrate concentrations, and type of solvents used to carry out the reaction. In aqueous medium, an enzyme-catalyzed amide synthesis require optimal and altered reaction conditions to overcome the preference to hydrolysis. In this paper, we have reported that in an aqueous medium the equilibrium can also be shifted toward amide synthesis by immobilizing the enzyme on a solid support [18], [19]. The aim of immobilization for the biocatalyst is to explore the economics of biocatalytic processes since the technique enables (1) operational stability, feasibility, and increased functional efficiency of enzyme (2) reuse of the enzyme (3) enhanced reproducibility of the results (4) minimum reaction time and (5) simpler catalyst separation from the product [20], [21], [22], [23], [24]. Depending upon the type of bond involved on solid supports or matrix, there are several strategies of immobilization including adsorption, covalent bonding, encapsulation, entrapment in inorganic and organic matrices and copolymerization on polysaccharides such as chitosan, anionic polysaccharides, oligosaccharide derivatives (polyglucuronic acid) etc. [12], [13], [25]. Among these, copolymerization of enzyme on solid support through glutaraldehyde has been extensively investigated [21]. Recently the use of combination of nanotechnology and biotechnology as nanobiocatalysts, have received a great deal of attention owing to their high surface area and have lead to excellent loading and high catalytic activity and smaller particle size [17], [20].
In this work efficiently protease producing bacteria was isolated from a slaughterhouse waste. The purified protease was then further immobilized on an amino-functionalized magnetic nanoparticle and used for the synthesis and optimization of novel glycinamides using response surface methodology.
2. Materials and methods
2.1. Reagents
Di-potassium hydrogen phosphate (K2HPO4) and potassium di-hydrogen phosphate (KH2PO4) were purchased from Thomas Bakers, Mumbai, India. Sodium hydroxide pellets (NaOH) was purchased from Merck Specialties (Mumbai, India). Ferrous chloride tetra hydrate (FeCl2·4H2O), Ferric chloride hexa hydrate (FeCl3·6H2O) and DNS (3,5-dinitrosalicylic acid) were the product of HiMedia Lab. Pvt. Ltd (Mumbai, India). APTES (3-aminopropyltriethoxysilane) was purchased from Sigma-Aldrich (Bangalore, India). Glutaraldehyde 25% (w/v), butyric acid, citric acid, glycine, and benzoic acid were obtained from SD Fine Chem Ltd. (SDFCL Mumbai, India). All the other chemicals used further were of analytical grade with the highest purity. The water used for all the experiments was of Milli-Q System (Millipore).
2.2. Enzyme production
The protease enzyme was produced and isolated from the Bacillus subtilis. The procedure for production and purification was carried out as per the reported literature, Badhe et al. [26].
2.3. Synthesis of amino-functionalized magnetic nanoparticles (AMNPs)
Iron oxide magnetic nanoparticles (MNPs) were synthesized by using simple chemical co-precipitation method as per the method previously described by Reza et al. [27] and Talekar et al. [28]. The synthesized MNPs were amino functionalized using APTES reagent. For this, 1 gm of MNPs were added to 100 mL of ethanol and water (1:1) mixture. The mixture was then sonicated for the complete dispersion of MNPs in the solution. APTES (3 mL) was added drop wise to the reaction mixture while shaking. The complete reaction mixture was then kept for stirring (8 h). The APTES coated MNPs were separated using an external magnet and washed several times with deionized water and then with ethanol once to remove unbound APTES to get amino functionalized MNPs.
2.4. Immobilization of protease on functionalized MNPs
Protease was immobilized on AMNPs using glutaraldehyde as a coupling agent. For this, the requisite amount of amino functionalized MNPs were taken in the boric acid buffer (pH 9.0) and enzyme (liquid) was added drop-wise to the reaction mixture and kept for stirring (30 min). After completion of stirring, glutaraldehyde was added to the reaction mixture and flask was kept for stirring for another 8 h. The enzyme immobilized MNPs were then magnetically separated and washed with buffer several times [29]. After each wash amount of supernatant were collected in order to estimate the amount of protein bound to the MNPs using Bradford Method [30]. The percentage loading was calculated using the mass balance equation:
Where, Ci = Initial protein content (mg/mL).
Vi = Initial volume of reaction mixture (mL).
Co = Final protein content (mg/mL).
Vo = Final volume of the reaction mixture (mL).
2.5. Assay of immobilized protease enzyme
The activity of immobilized proteases enzyme was determined by standard Folin-Lowery assay method described by Badhe et al., [26] with some modification. In 5 mL of phosphate buffer (pH = 8), 0.5 mg of an immobilized protease enzyme and 0.5 mL of casein solution were added and then incubated in a shaker incubator at 35 °C for 30 min at 180 rpm. After that, 5 mL of 0.11 M trichloroacetic acid was added to stop the reaction and the reaction mixture was then filtered. After the filtration 5 mL of sodium carbonate and 0.5 mL of Folin's reagent were added to the filtrate and kept for 30 min at 35 °C. The optical density of the solution was measured by using UV-Spectrophotometer (UV-1800, Shimadzu) at a wavelength of 420 nm. One unit of immobilized protease enzyme activity (U) was defined as the amount of immobilized enzyme required to produce 1 μg of tyrosine mL−1 min−1 under the optimal experimental conditions.
2.6. Physicochemical characterization of MNPs and immobilized enzyme
Immobilization of protease was confirmed by FTIR analysis using Shimadzu IR-Affinity 1-spectrometer (Japan). The crystal structure of the nanoparticles was measured by an X-ray diffractometer (Lab X, XRD 6100, SHIMADZU, Germany) with Cu K radiation. A continuous scan mode was used to collect 2Θ data over 10° to 80 °C, at a constant rate of 4 °C/min. The surface morphology of the naked magnetic nanoparticles and protease immobilized MNPs were studied by using field emission gun-scanning electron microscopy (FEG-SEM) analysis (TESCAN MIRA 3 model). All the surface of magnetic nanoparticles and immobilized enzyme was sputter coated in a vacuum evaporator, with platinum. A thermo gravimetric analysis (TGA) DTG-60H EME instrument was used to calculate the percentage weight loss in protease immobilized MNPs over 30 to 500 °C in nitrogen atmosphere with a heating rate of 10 °C/min.
2.7. Synthesis of glycinamides
Enzymatic reactions were performed at pH of 5, 8, and 11 in Na-phosphate buffer and at 30, 50, and 70 °C. In a stoppered flask, 1 mM (0.088 g) of butyric acid was dissolved in an appropriate volume of phosphate buffer and 1 mg of immobilized protease enzyme was added. The reaction mixtures were shaken at 30, 50 and 70 °C for 15, 30, and 45 min and the progress of the reaction was monitored by TLC using n-butanol–acetone–water–25% ammonium hydroxide solution (3:2.5:2:1) as the mobile phase. After complete reaction of butyric acid, 1.5 mM (0.113 g) glycine was added to the reaction mixture and the reaction progress was monitored by TLC, n-butanol-acetic acid-water (3:1:1) as a mobile phase. The product formation was checked by TLC with the solvents chloroform and methanol in a 10:1 ratio (v/v) as a mobile phase and visualization of spots was achieved by spraying ninhydrin solution, followed by heating the plates at 120 °C. After the completion of the reaction, whole mixture was filtered by means of magnetic separation to separate the magnetic nanoparticles and the filtrate was subjected to rota-evaporation to evaporate the water. The mixture was recrystallised with 95% ethanol. Same protocol were followed for the synthesis of 2-benzamidoacetic acid (AMD-2) and 2,2′((carboxymethyl) amino)-2-oxoethyl)-2hydroxysuccinyl)bis(azanediyl))diacetic acid (AMD-3).
2.8. Experimental design
RSM is a group of mathematical and statistical techniques which require a few experimental runs to optimize the reaction conditions and provide useful and precise information to show the implication of the observed trends. Design-Expert 9.0.6.2 was adopted for experimental design and statistical analysis for the synthesis of series of glycinamides using immobilized protease enzyme as a biocatalyst. A Box–Behnken statistical experimental design with three variables was carried out in order to obtain the optimal reaction conditions for the synthesis of various glycinamides, 2-butyramidoacetic acid (AMD-1), 2-benzamidoacetic acid (AMD-2) and 2,2′((carboxymethyl)amino)-2-oxoethyl)-2hydroxysuccinyl)bis(azanediyl))diacetic acid (AMD-3). Temperature, pH and time were used as the variables to maximize the response and Table 1 gives the range of variables employed in the work. The experimental design requires 17 experiments with three variables conducted with five replicates at the central point for the estimation of pure error sum of squares for each experiment [31], [32], [33]. The actual experimental data to establish the relationship between variables (temperature, pH, and time) and responses (% yield conversion) that are carried out for developing the model are shown in Table 2, for the immobilized protease catalyzed synthesis of AMD-1, AMD-2 and AMD-3.
Table 1.
Variables and range of variations.
| Variables | Range of variations |
|
|---|---|---|
| −1 | +1 | |
| Temperature (°C) | 30 | 70 |
| Time (min) | 15 | 45 |
| pH | 5 | 11 |
Table 2.
The design point combinations and the corresponding experimental responses for AMD 1, AMD 2 and AMD 3.
| Run | Variable | Levels | Responses in | Yield (%) |
||
|---|---|---|---|---|---|---|
| Temperature (°C) |
pH | Time (Min.) | AMD1 | AMD2 | AMD3 | |
| 1 | 50 | 5 | 45 | 58.2 | 50.4 | 54 |
| 2 | 50 | 8 | 30 | 83 | 80.2 | 79.5 |
| 3 | 30 | 5 | 30 | 40 | 37 | 38.2 |
| 4 | 50 | 11 | 45 | 76.5 | 72.6 | 73.2 |
| 5 | 50 | 8 | 30 | 83.4 | 79.9 | 79.5 |
| 6 | 30 | 8 | 15 | 63.9 | 55 | 59 |
| 7 | 70 | 11 | 30 | 70.1 | 68 | 63 |
| 8 | 50 | 5 | 15 | 58 | 52.7 | 51.5 |
| 9 | 50 | 8 | 30 | 83.2 | 80.5 | 80.4 |
| 10 | 50 | 8 | 30 | 83 | 80.2 | 80.3 |
| 11 | 70 | 5 | 30 | 51.2 | 46.5 | 49 |
| 12 | 30 | 11 | 30 | 59.6 | 53.5 | 57 |
| 13 | 30 | 8 | 45 | 63 | 59.3 | 61.2 |
| 14 | 70 | 8 | 45 | 74.6 | 69.2 | 70 |
| 15 | 50 | 11 | 15 | 76.8 | 70 | 69 |
| 16 | 70 | 8 | 15 | 74 | 70 | 67.4 |
| 17 | 50 | 8 | 30 | 82.8 | 80.5 | 80.8 |
AMD 1: 2-butyramidoacetic acid.
AMD 2: 2-benzamidoacetic acid.
AMD3: 2,2′((carboxymethyl)amino)-2-oxoethyl)-2-hydroxysuccinyl)bis(azanediyl))diacetic acid.
The effects of variables on the response could be described as a second order polynomial quadratic equation:
| (1) |
where, Y is the predicted response used as a dependent variable, Xi and Xj are the levels of variables, βo the constant term, βi the coefficient of the linear terms, βij the coefficient of the quadratic terms, and βii the coefficient of the cross-product terms [33], [34].
2.9. Reusability study
The reusability study of the immobilized enzyme was carried out in the batch mode. For this, a known amount of protease immobilized MNPs were taken in the reaction mixture and reaction was carried out under optimal conditions for a given period of time [35]. The immobilized protease was then separated from the reaction mixture using external magnet and were washed with phosphate buffer (pH = 8) thrice. After washing, the protease immobilized MNPs were again subjected for repeated use, by adding fresh substrate for the synthesis of glycinamide as described above. The percentage yield and the conversion of glycinamide in first run was considered as 100% [36].
3. Results and discussion
3.1. Fourier transform infrared (FT-IR) analysis
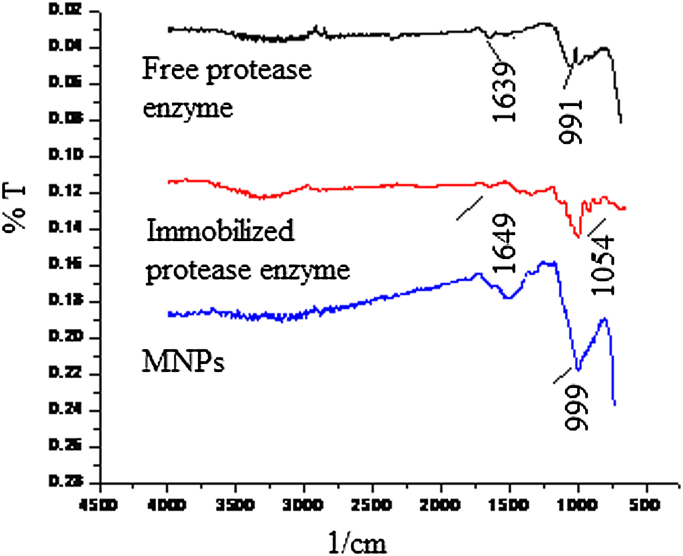
The binding of protease on MNPs is confirmed by FTIR spectroscopy. The IR of MNPs, free enzyme and protease immobilized MPNs are shown in Fig. 2. Characteristic band present at 1639 cm−1 confirms the presence of amide bond in the free enzyme. In spectra of enzyme immobilized MNPs the appearance of strong peak at 1054 and 1649 cm−1 confirms the presence of Fe—O—Si bond and the formation of bond between enzyme and MNPs respectively which revealed the immobilization of the enzyme on MNPs [36], [37].
Fig. 2.
FTIR analysis of magnetic nanoparticles, free protease enzyme, and immobilized protease on MNPs.
3.2. SEM analysis
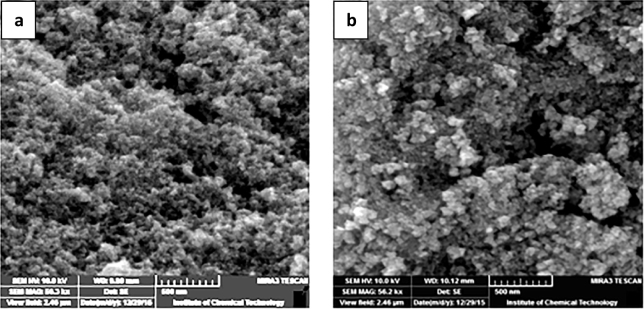
The typical size distribution and surface morphology of the magnetic nanoparticles and protease immobilized MNPs are shown in Fig. 3. From the figures it was clear that the naked MNPs were smooth and monodispersed and had of size of about 100 nm which was comparable to the particle size obtained by the particle size analyzer. After immobilization of protease on these MNPs the particles remained discrete and had a mean diameter of about 100 nm which is close to that of bare one. The results indicated that immobilization process did not significantly affect in the formation of agglomerates and did not alter the size of MNPs. This indicates to the fact that reaction occurred only on the surface of the MNPs [38].
Fig. 3.
SEM images of (a) Magnetic nanoparticles, (b) Immobilized protease enzyme on magnetic nanoparticles.
3.3. TGA analysis
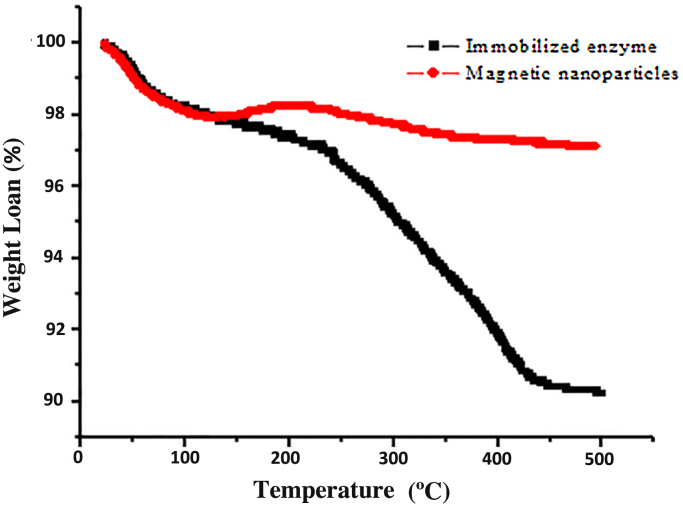
TGA is frequently used to confirm the immobilization of protease on MNPs by determining the percentage loss of weight of the naked MNPs and enzyme immobilized MNPs. For this, MNPs and enzyme immobilized MNPs were subjected to the temperature range of 30–500 °C. The weight loss curves of naked MNPs and protease immobilized MNPs are shown in Fig. 4. The thermogram profile of MNPs and protease immobilized MNPs show relatively same weight loss of about 0.3% at temperatures ranging from 65 to 120 °C, which is mainly due to the loss of physically adsorbed water. Further increase in the temperature above 150 to 500 °C, the total loss of about 2.9% was observed in MNPs which might be because of remaining organic residues used for coating (APTES and GA).The weight loss of enzyme immobilized MNPs was about 9.86% in a broad temperature range between 100 and 500 °C. Particularly, the weight loss of protease immobilized MNPs was observed to be around 7.86% between 150 and 450 °C which confirms the binding of protease on MNPs [39]. The mass balance was calculated on the basis of amount of enzyme added and amount of enzyme immobilized using Bradford assay which is carried out to check the efficiency of immobilization. From the mass balance equation the good conformity were observed between the Bradford Method and TGA data.
Fig. 4.
TGA curve of magnetic nanoparticles and immobilized protease on MNPs.
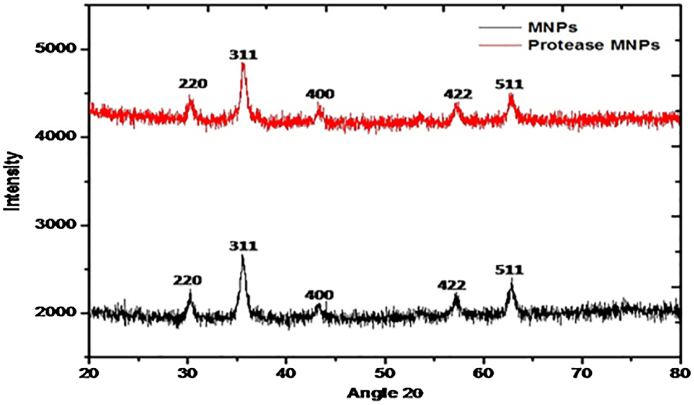
3.4. XRD analysis
In order to check the purity as well as the crystallinity of magnetic nanoparticles and MNPs after protease immobilization, XRD analysis was carried out. Fig. 5 shows the XRD patterns for the naked magnetic nanoparticles and protease immobilized MNPs. Five characteristic peaks for Fe3O4 (2θ = 30.1, 35.5, 43.1, 57.0 and 62.6°), marked by their indices (220), (311), (400), (422), and (511), were almost same for both samples. This implies that there was no phase change in MNPs after enzyme immobilization [36]. From both XRD patterns, it was observed that the sharp diffraction peaks clearly indicates the spinal magnetite product as well defined crystallites, without any impurity diffraction peaks, which showed synthesized magnetite nanoparticles in a pure phase.
Fig. 5.
XRD analysis of magnetic nanoparticles and immobilized protease on MNPs.
3.5. Model development
In the processes of model development, verifying the efficiency of the selected model and fitting an appropriate model are the two main steps. Table 2 represents the relationship between experimental variables (temperature, pH and time) and the responses (% yield) for immobilized protease catalyzed synthesis of AMD-1, AMD-2 and AMD-3 measured at each point. The regression analysis and the determination of correlation coefficient (R2) is the general approach to test the fit of the model. Results shown in the model fit summary (Table 3) confirms, that the quadratic model adequately described the relationship between variables and the % yield of AMD-1, AMD-2 and AMD-3. In this study, the value of the determination coefficient (R2) of the model was (0.99, 0.99 and 0.99 for AMD-1, AMD-2 and AMD-3 respectively) suggests a highly satisfactory representation of the real relationships among the selected reaction parameters studied in this work [31], [40], [41].
Table 3.
Model fit summary of determination coefficients of AMD 1, AMD 2 and AMD 3.
| Source | Sequential p-value | R-Squared | Predicted R-Squared | Sequential p-value | R-Squared | Predicted R-Squared | Sequential p-value | R- Squared | Predicted R-Squared | |
|---|---|---|---|---|---|---|---|---|---|---|
| AMD 1 | AMD 2 | AMD 3 | ||||||||
| Linear | 0.1208 | 0.3508 | −0.0300 | 0.1239 | 0.3480 | −0.0019 | 0.2067 | 0.2873 | −0.1000 | |
| 2FI | 0.9999 | 0.3511 | −0.9664 | 0.9918 | 0.3542 | −0.8094 | 0.9982 | 0.2898 | −1.0203 | |
| Quadratic | < 0.0001 | 0.9998 | 0.9984 | < 0.0001 | 0.9992 | 0.9908 | < 0.0001 | 0.9988 | 0.9852 | |
| Cubic | 0.3220 | 0.9999 | 0.1451 | 0.9998 | 0.1358 | 0.9996 | ||||
The coefficient of independent variables for the second order polynomial model for the% yield of AMD-1, AMD-2 and AMD-3 are explained in the form of regression equation (Eq. (2), (3) and (4)).
| (2) |
| (3) |
| (4) |
Where, Y is the percentage yield of the glycinamides, and A, B and C are the coded value of temperature, pH and time respectively. Synergetic and antagonistic effects of the mutual interacting parameters and independent variables are illustrated by positive and negative sign in front of the term respectively. Furthermore, the corresponding large F- value coupled with a very small P-value demonstrates the corresponding coefficient in the ANOVA more significant. Therefore the result obtained in this study clearly reveals that the reaction temperature and pH has the largest effect on the% yield of AMD-1, AMD-2 and AMD-3 respectively [42], [43].
An analysis of variance (ANOVA) was carried out to investigate the selection of appropriate models and evaluate the statistical parameters used for the optimization of the reaction parameters. ANOVA results are tabulated in Tables 4–6 respectively, for the synthesis of AMD-1, AMD-2 and AMD-3 respectively. The efficiency and significance of the model was investigated by performing a lack-of-fit test, P value and F value. F-test shows the relationship between the effect of independent variables (A, B and C) and dependent variables (AB, AC, BC, A2, B2, C2) on the percentage yield of all the products. In the above order A, B and C represent the terms temperature, pH and time respectively. In this study, ANOVA results show that the “Lack of Fit of F- value” of 1.6, 3.20 and 3.37 implies the lack of fit is insignificant for the quadratic model that was chosen for the percentage yield of AMD-1, AMD-2 and AMD-3 respectively. The quadratic model that was chosen for the percentage yield of the glycinamides, the model F-value of 4597.79, 997.34 and 624.22 indicate the model is significant. There is only a 0.01% chance that an F-value this large could occur due to noise. For the present model A, B, AC, A2, B2, C2 (AMD-1), A, B, AB, AC, BC, A2, B2, C2 (AMD-2) and A, B, C, AB, A2, B2, C2 are the order of model terms regarding the significance and the value of “Prob > F” less than 0.05 indicate model terms are significant. The value of P- value and F-value indicate the models are significant at 95% confidence interval [33], [40], [41].
Table 4.
ANOVA test for the synthesis of AMD 1.
| Source | Sum of Squares | df | Mean Square | F Value | p-value Prob > F |
|
|---|---|---|---|---|---|---|
| Model | 2707.44 | 9 | 300.83 | 4597.79 | <0.0001 | significant |
| A-Temperature | 235.45 | 1 | 235.45 | 3598.50 | <0.0001 | |
| B-pH | 714.42 | 1 | 714.42 | 10919.08 | <0.0001 | |
| C-time | 0.020 | 1 | 0.020 | 0.31 | 0.5976 | |
| AB | 0.12 | 1 | 0.12 | 1.87 | 0.2135 | |
| AC | 0.56 | 1 | 0.56 | 8.60 | 0.0220 | |
| BC | 0.063 | 1 | 0.063 | 0.96 | 0.3609 | |
| A2 | 731.14 | 1 | 731.14 | 11174.68 | <0.0001 | |
| B2 | 907.07 | 1 | 907.07 | 13863.51 | <0.0001 | |
| C2 | 4.45 | 1 | 4.45 | 67.94 | <0.0001 | |
| Residual | 0.46 | 7 | 0.065 | |||
| Lack of Fit | 0.25 | 3 | 0.083 | 1.60 | 0.3220 | not significant |
| Pure Error | 0.21 | 4 | 0.052 | |||
| Total | 2707.90 | 16 |
Table 5.
ANOVA test for the synthesis of AMD 2.
| Source | Sum of Squares | df | Mean Square | F Value | p-value Prob > F |
|
|---|---|---|---|---|---|---|
| Model | 3019.16 | 9 | 335.46 | 997.34 | <0.0001 | significant |
| A-Temperature | 298.90 | 1 | 298.90 | 888.64 | <0.0001 | |
| B-pH | 750.78 | 1 | 750.78 | 2232.10 | <0.0001 | |
| C-Time | 1.81 | 1 | 1.81 | 5.37 | 0.0537 | |
| AB | 6.25 | 1 | 6.25 | 18.58 | 0.0035 | |
| AC | 6.50 | 1 | 6.50 | 19.33 | 0.0032 | |
| BC | 6.00 | 1 | 6.00 | 17.85 | 0.0039 | |
| A^2 | 758.30 | 1 | 758.30 | 2254.45 | <0.0001 | |
| B^2 | 994.68 | 1 | 994.68 | 2957.22 | <0.0001 | |
| C^2 | 44.34 | 1 | 44.34 | 131.82 | <0.0001 | |
| Residual | 2.35 | 7 | 0.34 | |||
| Lack of Fit | 1.66 | 3 | 0.55 | 3.20 | 0.1451 | not significant |
| Pure Error | 0.69 | 4 | 0.17 | |||
| Total | 3021.52 | 16 |
Table 6.
ANOVA test for the synthesis of AMD 3.
| Source | Sum of Squares | df | Mean Square | F Value | p-value Prob > F |
|
|---|---|---|---|---|---|---|
| Model | 2658.51 | 9 | 295.39 | 624.22 | <0.0001 | significant |
| A-Temperature | 144.50 | 1 | 144.50 | 305.36 | <0.0001 | |
| B-pH | 603.78 | 1 | 603.78 | 1275.92 | <0.0001 | |
| C-Time | 16.53 | 1 | 16.53 | 34.93 | 0.0006 | |
| AB | 5.76 | 1 | 5.76 | 12.17 | 0.0101 | |
| AC | 0.040 | 1 | 0.040 | 0.085 | 0.7797 | |
| BC | 0.72 | 1 | 0.72 | 1.53 | 0.2565 | |
| A2 | 707.48 | 1 | 707.48 | 1495.05 | < 0.0001 | |
| B2 | 1003.44 | 1 | 1003.44 | 2120.47 | < 0.0001 | |
| C2 | 33.90 | 1 | 33.90 | 71.64 | < 0.0001 | |
| Residual | 3.31 | 7 | 0.47 | |||
| Lack of Fit | 2.37 | 3 | 0.79 | 3.37 | 0.1358 | not significant |
| Pure Error | 0.94 | 4 | 0.23 | |||
| Total | 2661.82 | 16 |
3.6. Effect of parameters
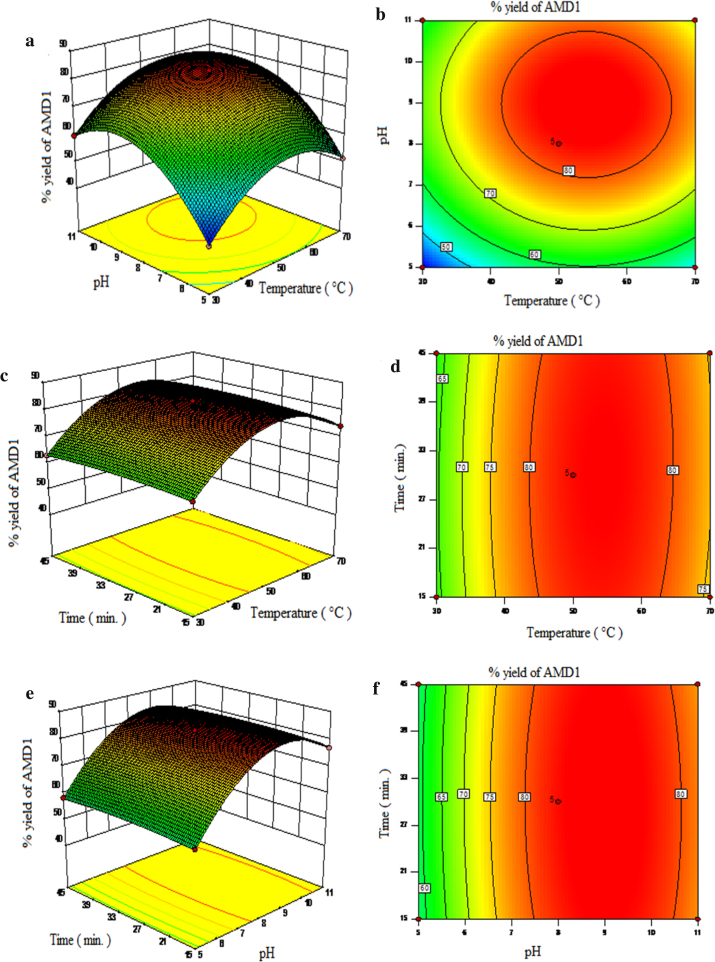
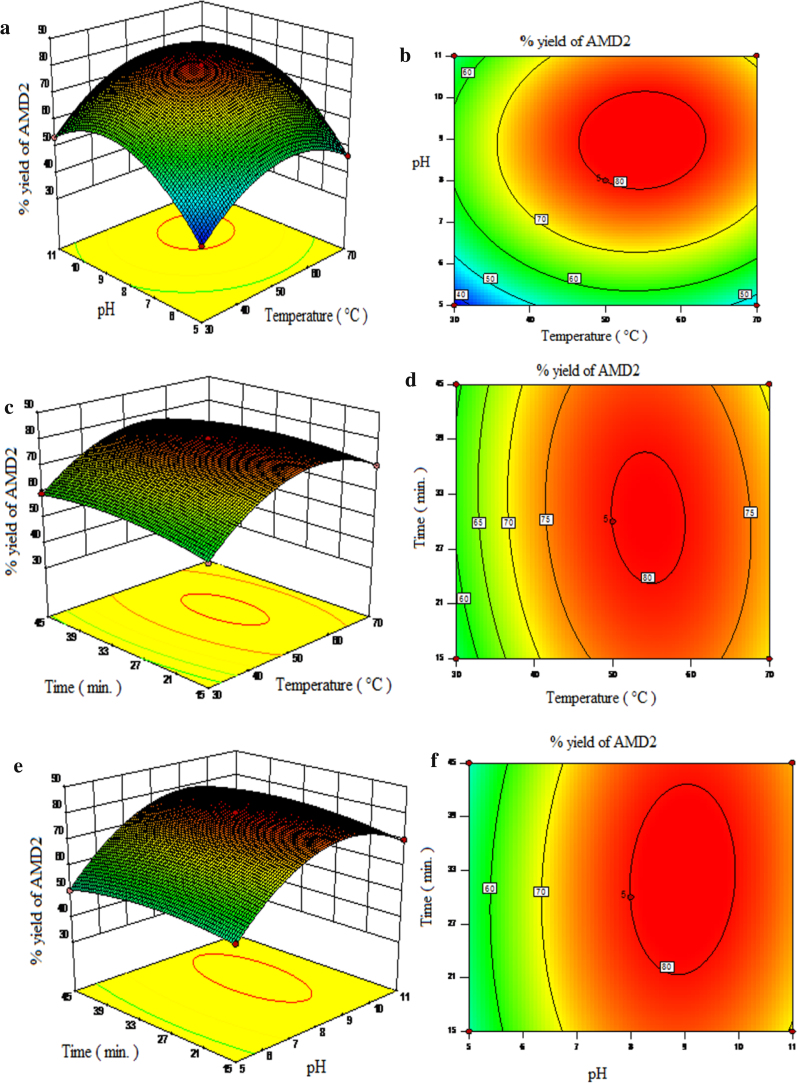
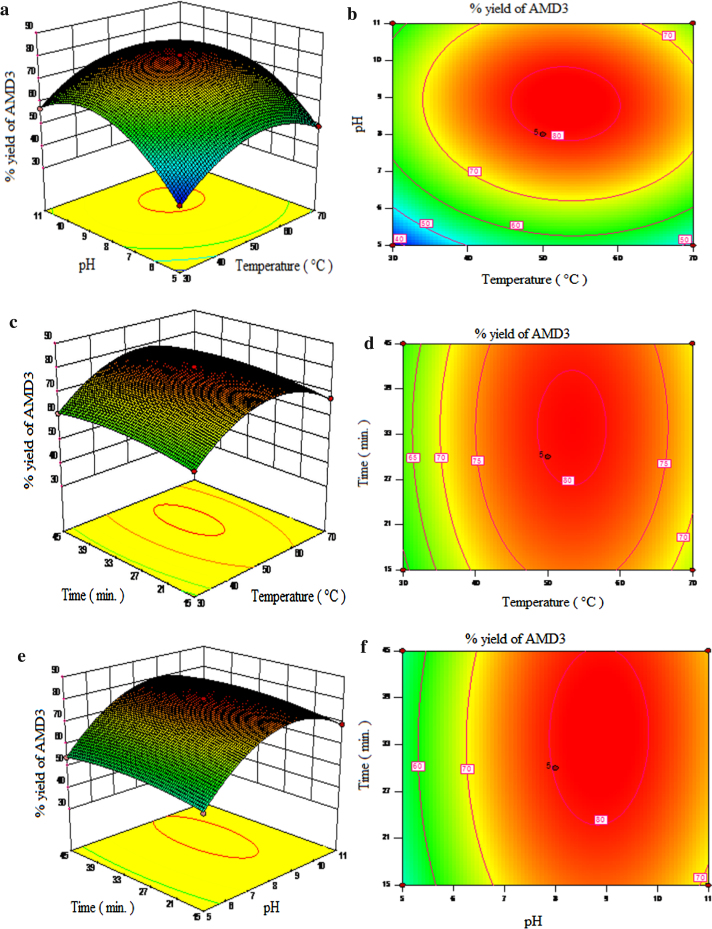
The parameters like pH and temperature plays a vital role in the activity and thermal stability of the immobilized enzymes. It is mandatory to find the optimum pH and temperature to get the maximum activity of the enzymes, which ultimately increases the overall yield of the product. Hence, a the simultaneous effect of reaction parameters, pH (5, 8 and 11), temperature (30 °C, 50 °C and 70 °C) and time (15 min, 30 min and 45 min) was studied. To visualize the effect of reaction variables and their responses on immobilized protease catalyzed synthesis of AMD-1, AMD-2 and AMD-3, 3-D surface plot and their corresponding 2-D counter plot were generated based on the predicted model. Fig. 6, Fig. 7, Fig. 8 depict the response surface plots and corresponding counter plot as a function of two variables at a time, keeping the third variable fixed at their centre point values for AMD-1, AMD-2 and AMD-3 respectively. From Fig. 6, Fig. 7, Fig. 8 it is seen that the reaction temperature and pH are the most significant interaction whereas the interaction of time with temperature and interaction of time with pH are the least significant interactions on the percentage yield of AMD-1, AMD-2 and AMD-3.
Fig. 6.
Response surfaces of % yield of AMD-1: (a) pH and temperature at 30 min, (c) time and temperature at pH 8, (e) time and pH at 50 °C, and their corresponding contour plots: (b) pH and temperature at 30 min, (d) time and temperature at pH 8, (f) time and pH at 50 °C.
Fig. 7.
Response surfaces of % yield of AMD-2: (a) pH and temperature at 30 min, (c) time and temperature at pH 8, (e) time and pH at 50 °C, and their corresponding contour plots: (b) pH and temperature at 30 min, (d) time and temperature at pH8, (f) time and pH at 50 °C.
Fig. 8.
Response surfaces of % yield of AMD-3: (a) pH and temperature at 30 min, (c) time and temperature at pH 8, (e) time and pH at 50 °C, and their corresponding contour plots: (b) pH and temperature at 30 min, (d) time and temperature at pH8, (f) time and pH at 50 °C.
3.6.1. Effect of temperature
Increase in the temperature normally affects the activity of the enzyme, solubility of the reactants and simultaneously that of products, rate of reaction and the direction of the equilibrium process involved in amidation reaction. Figs. 6 (a–d) and 8 (a–d) show the surface response plot and their corresponding counter plot elucidating the effect of the temperature with pH at fixed time and similarly that of time at fix pH respectively on the percentage yield of immobilized protease catalyzed synthesis of AMD-1, AMD-2 and AMD-3 respectively. Observing the surface plot and contour plot it can be concluded that the an increase the reaction temperature up to the optimum point (50 °C) shows an increase in the percentage yield for the conversion of AMD-1 (83.4%), AMD-2 (80.5%) and AMD-3 (80.8%), while further increase in temperature reverses the trend, highest yield of the product was obtained at 50 °C and pH of 8. This reason could be explained by; (1) higher temperature provides more heat energy making the enzyme molecules mobile increasing the number of collision due to higher kinetic energy and this will increase the rate of reaction, (2) “ the lock and key” theory attributed this to an increase in the collision between enzyme molecules resulting in the more enzyme substrate complex formation increasing the rate of the reaction, (3) due to possible changes of the enzyme structure after glutaraldehyde cross-linking, resulting in the increased resistance to temperature. While further increase in temperature the% yield of the products, AMD-1 (74.6%), AMD-2 (69.2%) and AMD-3 (70%) decreases at 70 °C and pH-8, probably due to the thermal deactivation of the enzyme. For chemical as well as enzymatic reactions, Arrhenius equation is used to correlate the effect of temperature on the reaction rate, admittedly the explored temperature range in this work is quite limited. If the temperature of the reaction is too high, the enzyme molecule posses higher energies and tendency to move faster, acquire the sufficient amount of the energy to break the bond of the enzyme molecules and thermal deactivation follows and thus halting the progress of the reaction [44], [45], [46].
3.6.2. Effect of pH
The effects of pH on the activity and the stability of the enzymes resemble in some respect the effect of the temperatures in the synthesis of AMD-1, AMD-2 and AMD-3. Figs. 6 (a,b,e and f) and 8 (a,b,e and f), shows the result of surface plot and their corresponding counter plot of the effect of pH with temperature at constant time and with time at constant temperature for the synthesis of AMD-1, AMD-2 and AMD-3 using immobilized protease enzyme respectively. When pH increases from pH 5–8, the% yield of the AMD-1 (83.4%), AMD-2 (80.5%) and AMD-3 (80.8%) were found to increase, while further increase in the pH from pH 8–11 the percentage yield of the AMD-1 (76.5%), AMD-2 (72.6%) and AMD-3 (73.2%) starts decreasing at 50 °C. Several factors govern this behavior which is: (1) the protonation of the functional group of the amino acids mainly situated on the surface of the enzyme and it is a reversible process, (2) denaturation of three dimensional structure of the enzyme molecule which is an irreversible process. Enzymes are amphoteric molecules consisting of large number of acidic and basic groups on their surface that affects the net charge on the enzyme surface and thus affect the catalytic behavior of enzyme. The pH of the reaction directly influences the binding of the substrate to the enzyme, ionization of amino acids situated on the enzyme surface and the ionization of substrate. Ionic bonds are present in tertiary structure of enzyme which is sensitive to H+ ion concentration in the solution. The reduction in the activity might be due to the breakdown of ionic bonds in presence of higher concentration of H+ ions which affects functional shape of active site resulting in the lower yield of product. At higher pH the yield of amide was decreases; as amide synthesis was carried out in a phosphate buffer; the amount of monovalent ion (K+) increases and compete with the substrate for active binding site on enzyme and thus caused the breaking of ionic bond to denature the enzyme [45].
3.6.3. Effect of time
In the synthesis of AMD-1, AMD-2 and AMD-3, the interaction of time with temperature and pH has no significant impact on the yield. In Fig. 6 (c, d e, and f)-8(c, d e, and f) it can be shown that maximum yield of for AMD-1 (83.4%), AMD-2 (80.5%) and AMD-3 (80.8%) were obtain in 30 min at 50 °C and pH 8, as time increases the yield of the product decreases. The possible reason for this is that as the amidation reaction was carried out in aqueous medium, as increase in the reaction time the amount of reactants decrease. Thus the concentration of free nucleophile decreases and competes with water to react the acyl-enzyme intermediate that leads to the hydrolysis of acyl-enzyme intermediate by water.
3.7. Optimization of reaction parameters
Design expert software based on RSM was employed to maximize the yield of AMD-1, AMD-2 and AMD-3 for given operating range listed in Table 2. Fig. S1 (Supplementary information) a–e shows the perturbation graph, showing the effect of independent variables on the yield of the product. The highest yield of the product (80–84%) was obtained at 50 °C, the reason is that the immobilized enzyme achieved its highest activity when the reaction temperature was 50 °C, lower than this is not able to open all the active sites present in the protease, hence less affinity of enzymes to substrate was observed giving lower yield. Temperature above the optimum also showed lower product yield. The reason could be that at higher temperature, enzymes active sites reduce due to the denaturation of the enzymes which ultimately decreases the overall yield of the product. From Fig. S1 a–e, it is seen that the maximum activity of the immobilized enzyme occurred at pH 8 and the maximum yield of the product was obtained. The reason for all this could be that the protease shows maximum activity at pH 8 hence it has a greater affinity for the substrate and that of the product at this pH which ultimately increases the overall yield. From the perturbation graph it is clear that when the temperature and pH increase the yield of the product was also increases and there was no significant impact of time on the yield of the product. The straight lines of the residuals suggest that the errors are normally distributed and insignificant with the operating parameters (Fig. S1 Supplementary information) [31], [33], [47].
3.8. Fourier transform infrared (FT-IR) analysis of AMD-1, AMD-2 and AMD-3
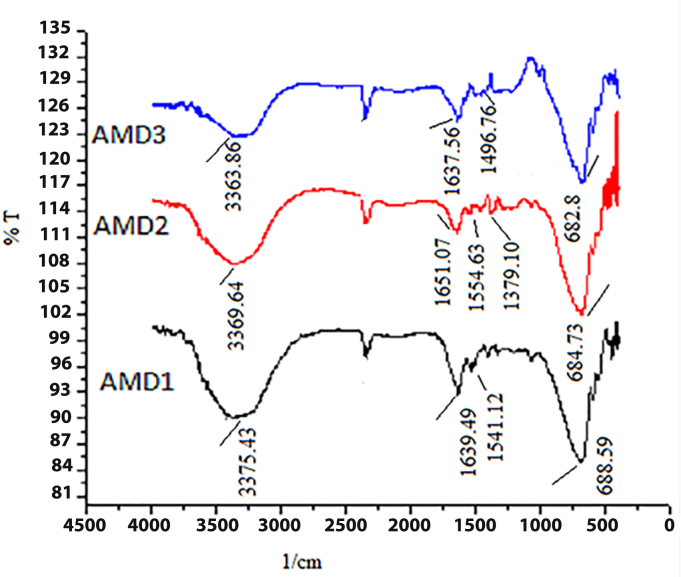
Fig. 9 shows the FTIR spectra of AMD-1, AMD-2 and AMD-3. In the FTIR spectrum characteristic peak at 1639.49, 1651.07 and 1637.56 cm−1 show the presence of C O stretching vibration of amide bond of AMD-1, AMD-2 and AMD-3 respectively. In the IR spectrum single peak appears at 3375.43, 3369.64 and 3363.86 cm−1 show the N—H stretching vibration and IR absorption bands at 1541.12, 1554.63 and 1496.76 cm−1 are due to the N—H bending vibration of amide bond. The characteristic peaks at 688.59, 684.73 and 682.8 cm−1 are mainly due to the O C—N bands of the amide. In AMD-2 absorption occurs at 1379.10 cm−1 show the presence of benzene ring [48], [49].
Fig. 9.
FTIR spectra of AMD-1, AMD-2, and AMD-3.
3.9. Reusability of immobilized protease enzyme
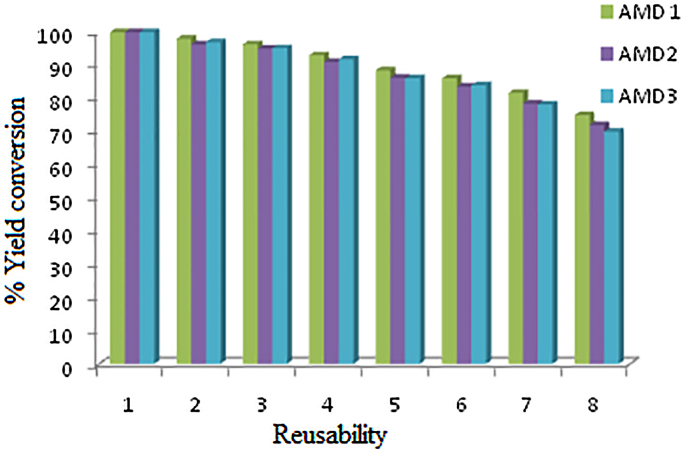
From an economic point of view, for an industrial application it is very necessary to check the reusability of the biocatalyst used to carry out the reaction. Use of magnetic nanoparticles as a carrier for enzyme augments the easy recoverability and reusability. Fig. 10 shows the reusability of the immobilized protease up to 8 successive cycles. From the results it was seen that, the immobilized protease gives up to 70% of yield for all the three amides even after 8 cycles of reuse (Fig. 10). The decrease in activity after each successive use might be because of enzyme denaturation and destruction of active sites because of repeated reuse. Distortion of the active site also results from the recurrent encountering of substrate to the active site of the immobilized enzyme [28], [50].
Fig. 10.
Reusability study of immobilized protease on MNPs for the synthesis of AMD-1, AMD-2 and AMD-3.
4. Conclusions
The objective of this work was to evaluate the performance of the protease enzyme extracted from slaughter house waste in the synthesis of series of glycinamides and optimize the operational parameters using RSM. Bio-catalyzed amide synthesis is an alternative method over chemical synthesis with an advantage of novelty of the biological reagent, eco-friendly due to its nontoxic nature and formation of less of by products. The maximum yield was obtained at a temperature 50 °C, pH-8 and 30 min of operation for all the three products AMD-1, AMD-2 and AMD-3 respectively. The reusability of the immobilized biocatalyst indicates 70% retention in initial activity up to 8 consecutive cycles. The statistical model used predicted performance values with a 95% confidence level. The trend in the experimental data, could be evaluated through surface response analysis and contour diagrams. It showed that the process was well modelled and that the range of parametric variation studied for the process variables was adequate. Furthermore, the immobilized protease can also be used for several applications including pharmaceutical, biological, dairy industries etc.
Acknowledgement
We would like to acknowledge University Grand Commission (F.5-64/2007 BSR), New Delhi, India for financially supporting the research work.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.btre.2016.07.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Montalbetti C.A.G.N., Falque V., Park M., Ox A. Amide bond formation and peptide coupling. Tetrahedron Lett. 2005;61:10827–10852. [Google Scholar]
- 2.Sergeeva M.V., Paradkar V.M., Dordick S. Peptide synthesis using proteases dissolved in organic solvents. Enzyme Microb. Technol. 1997;20:623–628. [Google Scholar]
- 3.Liu H., Liu J., Zhang Y., Shao C., Yu J. Copper-catalyzed amide bond formation from formamides and carboxylic acids. Chin. Chem. Lett. 2015;26:11–14. [Google Scholar]
- 4.Kang S., Yim H., Won J., Kim M., Kim J., Kim H., Lee S., Yoon Y. Effective amidation of carboxylic acids using (4,5-Dichloro-6-oxo-6H-pyridazin-1-yl) phosphoric acid diethyl ester. Bull. Korean Chem. Soc. 2008;29:1025–1032. [Google Scholar]
- 5.Hayley C. Durham Theses. Durham University; 2012. Direct amide formation between carboxylic acids and amines: mechanism and development of novel catalytic solutions; pp. 1–204. [Google Scholar]
- 6.Bordusa F. Nonconventional amide bond formation catalysis: programming enzyme specificity with substrate mimetics. Braz. J. Med. Biol. Res. 2000;33:469–485. doi: 10.1590/s0100-879x2000000500001. [DOI] [PubMed] [Google Scholar]
- 7.Naidu K.S. Characterization and purification of protease enzyme. J. Appl. Pharm. Sci. 2011;01(03):107–112. [Google Scholar]
- 8.Gaertner H., Puigserver A. Kinetics and specificity of serine proteases in peptide synthesis catalyzed in organic solvents. Eur. J. Biochem. 1989;181:207–213. doi: 10.1111/j.1432-1033.1989.tb14712.x. [DOI] [PubMed] [Google Scholar]
- 9.Gotor V., Gotor-Fernandez V., Busto E. Hydrolysis and reverse hydrolysis: hydrolysis and formation of amides. Compr. Chirality. 2012;7:101–121. [Google Scholar]
- 10.Guzmán F., Barberis S., Illanes A. Peptide synthesis: chemical or enzymatic. Electron. J. Biotechnol. 2007;10–2:279–314. [Google Scholar]
- 11.Ghaffari-moghaddam M., Eslahi H., Aydin Y.A., Saloglu D. Enzymatic processes in alternative reaction media: a mini review. J. Biol. Methods. 2015;2:1–9. [Google Scholar]
- 12.Mati-Baouche N., Elchinger P.H., DeBaynast H., Pierre G., Delattre C., Michaud P. Chitosan as an adhesive. Eur. Polym. J. 2014;2014:198–213. [Google Scholar]
- 13.Elchinger P.H., Delattre C., Faure S., Roy O., Badel S., Bernardi T., Taillefumier C., Michaud P. Immobilization of proteases on chitosan for the development of films with anti-biofilm properties. Int. J. Biol. Macromol. 2015;72:1063–1068. doi: 10.1016/j.ijbiomac.2014.09.061. [DOI] [PubMed] [Google Scholar]
- 14.Nuijens T., Cusan C., Schepers A.C.H.M., Kruijtzer J.A.W., Rijkers D.T.S., Liskamp R.M.J., Quaedflieg P.J.L.M. Enzymatic synthesis of activated esters and their subsequent use in enzyme-based peptide synthesis. J. Mol. Catal. B. 2011;71:79–84. [Google Scholar]
- 15.Toplak A., Nuijens T., Quaedflieg P.J.L.M., Wu B., Janssen D.B. Peptide synthesis in neat organic solvents with novel thermostable proteases. Enzyme Microb. Technol. 2015;73–74:20–28. doi: 10.1016/j.enzmictec.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Yazawa K., Numata K. Recent advances in chemoenzymatic peptide syntheses. Molecules. 2014;19:13755–13774. doi: 10.3390/molecules190913755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen F., Zhang F., Wang A., Li H., Wang Q., Zeng Z., Wang S., Xie T. Recent progress in the chemo-enzymatic peptide synthesis. Afr. J. Pharm. Pharmacol. 2010;4:721–730. [Google Scholar]
- 18.Yasui M., Shiroya T., Fujimoto K., Kawaguchi H. Activity of enzymes immobilized on microspheres with thermosensitive hairs. Colloids Surf. B. 1997;8:311–319. [Google Scholar]
- 19.Ulijn R.V., Baragan B., Halling P.J., Flitsch S.L. Protease-catalyzed peptide synthesis on solid support. J. Am. Chem. Soc. 2002;124:10988–10989. doi: 10.1021/ja026912d. [DOI] [PubMed] [Google Scholar]
- 20.Yang D., Wang X., Shi J., Wang X., Zhang S., Han P., Jiang Z. In situ synthesized rGO-Fe3O4 nanocomposites as enzyme immobilization support for achieving high activity recovery and easy recycling. Biochem. Eng. J. 2016;105:273–280. [Google Scholar]
- 21.Zhang Z., He F., Zhuo R. Immobilized lipase on porous silica particles: preparation and application for biodegradable polymer syntheses in ionic liquid at higher temperature. J. Mol. Catal. B. 2013;94:129–135. [Google Scholar]
- 22.Mahmod S.S., Yusof F., Jami M.S., Khanahmadi S., Shah H. Development of an immobilized biocatalyst with lipase and protease activities as a multipurpose cross-linked enzyme aggregate (multi-CELA) Process Biochem. 2015;50:2144–2157. [Google Scholar]
- 23.Rezakhani N., Molaei A., Parivar K., Khayati M., Etemadzade S. Immobilization of protease in biopolymers (mixture of alginate-chitosan) J. Paramedical Sci. 2014;5:108–113. [Google Scholar]
- 24.Nadar S.S., Muley A.B., Ladole M.R., Joshi P.U. Macromolecular cross-linked enzyme aggregates (M-CLEAs) of α-amylase. Int. J. Biol. Macromol. 2016;84:69–78. doi: 10.1016/j.ijbiomac.2015.11.082. [DOI] [PubMed] [Google Scholar]
- 25.Delattre C., Pierre G., Gardarin C., Traikia M., Elboutachfaiti R., Isogai A., Michaud P. Antioxidant activities of a polyglucuronic acid sodium salt obtained from TEMPO-mediated oxidation of xanthan. Carbohydr. Polym. 2015;116:34–41. doi: 10.1016/j.carbpol.2014.04.054. [DOI] [PubMed] [Google Scholar]
- 26.Badhe P., Joshi M., Adivarekar R. Optimized production of extracellular proteases by Bacillus subtilis from degraded abattoir waste. J. BioSci. Biotechnol. 2016;5:29–36. [Google Scholar]
- 27.Reza R.T., Pérez C.A.M., González C.A.R., Romero H.M., Casillas P.E.G. Effect of the polymeric coating over Fe3O4 particles used for magnetic separation. Cent. Eur. J. Chem. 2010;8:1041–1046. [Google Scholar]
- 28.Talekar S., Ghodake V., Ghodake T., Rathod P., Deshmukh P., Nadar S., Mulla M., Ladole M. Novel magnetic cross-linked enzyme aggregates (magnetic CLEAs) of alpha amylase. Bioresour. Technol. 2012;123:542–547. doi: 10.1016/j.biortech.2012.07.044. [DOI] [PubMed] [Google Scholar]
- 29.Li B.D., Teoh W.Y., Gooding J.J., Selomulya C., Amal R. Functionalization strategies for protease immobilization on magnetic nanoparticles. Adv. Funct. Mater. 2010;20:1767–1777. [Google Scholar]
- 30.Bradford M.M. A rapid and sensitive method for the quantitation microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 31.Ebrahimi M., Mahboubi F., Naimi-jamal M.R. RSM base study of the effect of deposition temperature and hydrogen flow on the wear behavior of DLC films. Tribol. Int. 2015;91:23–31. [Google Scholar]
- 32.Martins A.B., Graebin N.G., Lorenzoni A.S.G., Fernandez-lafuente R., Ayub M.A.Z., Rodrigues R.C. Rapid and high yields of synthesis of butyl acetate catalyzed by Novozym 435: reaction optimization by response surface methodology. Process Biochem. 2011;46:2311–2316. [Google Scholar]
- 33.Vyas S.K., Shukla S.R. Degumming of eri silk using ionic liquids and optimization through response surface methodology. J. Text. Inst. 2015 doi: 10.1080/00405000.2015.1086196. [DOI] [Google Scholar]
- 34.Meng Y., Wang X., Wu Z., Wang S., Young T.M. Optimization of cellulose nanofibrils carbon aerogel fabrication using response surface methodology. Eur. Polym. J. 2015;73:137–148. [Google Scholar]
- 35.Talekar S., Pandharbale A., Ladole M., Nadar S., Mulla M. Carrier free co-immobilization of alpha amylase, glucoamylase and pullulanase as combined cross-linked enzyme aggregates (combi-CLEAs): a tri-enzyme biocatalyst with one pot starch hydrolytic activity. Bioresour. Technol. 2013;147:269–275. doi: 10.1016/j.biortech.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 36.Seenuvasan M., Malar C.G., Preethi S., Balaji N., Iyyappan J., Kumar M. Anil, Kumar K. Sathis. Immobilization of pectinase on co-precipitated magnetic nanoparticles for enhanced stability and activity. Res. J. Biotchnol. 2013;8 [Google Scholar]
- 37.Swarnalatha V., Esther R.A., Dhamodharan R. Immobilization of α-amylase on gum acacia stabilized magnetite nanoparticles, an easily recoverable and reusable support. J. Mol. Catal. B. 2013;96:6–13. [Google Scholar]
- 38.Ladole M.R., Muley A.B., Patil I.D., Talib M.I., Parate V.R. Immobilization of tropizyme-P on amino-functionalized magnetic nanoparticles for fruit juice clarification. J. Biochem. Technol. 2014;5:838–845. [Google Scholar]
- 39.Soozanipour A., Taheri-kafrani A., Landarani Isfahani A. Covalent attachment of xylanase on functionalized magnetic nanoparticles and determination of its activity and stability. Chem. Eng. J. 2015;270:235–243. [Google Scholar]
- 40.Krishna S.H., Sattur A.P., Karanth N.G. Lipase-catalyzed synthesis of isoamyl isobutyrate-optimization using a central composite rotatable design. Process Biochem. 2001;37:9–16. [Google Scholar]
- 41.Halim S.F.A., Kamaruddin A.H., Fernando W.J.N. Continuous biosynthesis of biodiesel from waste cooking palm oil in a packed bed reactor: optimization using response surface methodology (RSM) and mass transfer studies. Bioresour. Technol. 2009;100:710–716. doi: 10.1016/j.biortech.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Myra F., Manan A., Nurhazwani I., Rahman A., Haziqah Che Marzuki N., Mahat N.A., Abdul Wahab R. Statistical modelling of eugenol benzoate synthesis using Rhizomucor miehei lipase reinforced nanobioconjugates. Process Biochem. 2016;51:249–262. [Google Scholar]
- 43.Marzuki N. Haziqah Che, Huyop F., Aboul-enein H.Y., Mahat A., Wahab R.A. Modelling and optimization of Candida rugosa nanobioconjugates catalysed synthesis of methyl oleate by response surface methodology. Biotechnol. Biotechnol. Equip. 2016;2818(June) [Google Scholar]
- 44.Gumel A.M., Annuar M.S.M., Heidelberg T., Chisti Y. Lipase mediated synthesis of sugar fatty acid esters. Process Biochem. 2011;46:2079–2090. [Google Scholar]
- 45.Bisswanger H. Enzyme assays. Perspect. Sci. 2014;1:41–55. [Google Scholar]
- 46.H Noritomi, Protease-Catalyzed Synthetic Reactions in Ionic Liquids, Ionic Liquids: Applications and Perspectives, Prof. Alexander Kokorin (Ed.), ISBN: 978-953-307-248-7 (2011) InTech.
- 47.Pal A., Khanum F. Efficacy of xylanase purified from Aspergillus niger DFR-5 alone and in combination with pectinase and cellulase to improve yield and clarity of pineapple juice. J. Food Sci. Technol. 2011;48:560–568. doi: 10.1007/s13197-010-0175-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liao L.F., Lien C.F., Shieh D.L., Chen F.C., Lin J.L. FTIR study of adsorption and photochemistry of amide on powdered TiO2: comparison of benzamide with acetamide. Phys. Chem. Chem. Phys. 2002;4:4584–4589. [Google Scholar]
- 49.Garidel P., Schott H. Fourier-transform midinfrared spectroscopy for analysis and screening of liquid protein formulations. Bioprocess Int. 2016:1. (n.d.) [Google Scholar]
- 50.Ren Y., Rivera J.G., He L., Kulkarni H., Lee D.-K., Messersmith P.B. Facile, high efficiency immobilization of lipase enzyme on magnetic iron oxide nanoparticles via a biomimetic coating. BMC Biotechnol. 2011;11:63. doi: 10.1186/1472-6750-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.