Highlights
-
•
Isolation and detailed taxonomical investigation of potential cellulolytic actinobacteria.
-
•
19 morphologically distinct actinobacterial strains were subjected to cellulose degradation assay.
-
•
Proved actinobacteria (MHA15) is an ideal organism for the bioconversion of cellulose.
-
•
If detailed polyphasic taxonomic analysis of the strain MHA15 is made, it could be a new species.
Keywords: Actinobacteria, Actinoalloteichus sp., Cellulase, Havelock island, Systematic characterization
Abstract
Cellulose is the most abounding biopolymer in the world and there is a great interest in using this material as a substrate for various applications and it is the most important renewable resource for bioconversion. Therefore, it is necessary to screen the cellulolytic bioorganisms. In this context, actinobacteria are one of the most efficient prokaryotes, economically and biotechnologically, for their production of about half of the discovered bioactive secondary metabolites and they can metabolize many different compounds. Therefore, the present study was carried out to isolate and screen cellulase enzyme producing marine actinobacterial strains from the sediments of the Havelock island, the Andamans. Totally, 19 morphologically distinct actinobacterial strains were isolated and subjected to cellulose degradation assay. Out of the 19, four strains were found to possess good cellulose degradation activity and the strain MHA15 alone produced higher amount of cellulase enzyme (14.379 1U/ml) than the others. Taxonomical study of the strain MHA15 revealed that it belongs to the genus Actinoalloteichus and the molecular characters showed distinct difference in its phylogenetic relationship (8.4%) with A. cyanogriseus.
1. Introduction
Cellulose constitutes the major form of stocking glucose, obtained through photosynthesis and at the same time, it is the major component of solar energy conversion to biomass. It is also an important constituent of all the plant materials and that is why it is the most abundant organic material in nature, which is renewed every year [1]. Annual production of cellulose has been estimated at 4.0 × 107 t [2]. Proportion of cellulose in plant tissues ranges from 20 to 45% of dry weight and to almost above 90% in cotton fiber [3]. Much of the cellulose in nature exists as waste paper [4].
Because of its highly ordered structure, cellulose is very hard to be degraded and so it is unusable and stocked in nature as waste. Cellulose has been used by man for centuries; however, its enormous potential as a renewable source of energy was recognized only after cellulose degrading enzymes or “cellulases” were identified [5]. Cellulases are the inducible bioactive compounds produced by microorganisms during their growth on cellulosic materials [6]. Due to the increasing knowledge on the mode of action of cellulases, they have been used in enzymatic hydrolysis of cellulosic substances [7]. Although a large number of microorganisms are capable of degrading cellulose, only a few of them produce significant quantities of cell-free bioactive compounds, capable of completely hydrolyzing crystalline cellulose in-vitro. Though numerous investigations have reported the degradation of cellulosic materials, only a few studies have examined which microorganisms meet the industrial requirements [8].
Among the microbes, actinobacteria (actinomycetes) are one of the largest taxonomic units within the Bacterial domain [9], [10] and they are the most efficient prokaryotes to be used economically and biotechnologically for their production of about half of the discovered bioactive secondary metabolites [11]. Actinobacteria can metabolize many different compounds including sugars, alcohols and amino acids. Additionally, many of them (e.g. Streptomyces and Rhodococcus) produce extracellular hydrolytic enzymes to obtain nutrients from cellulose, hemicellulose, proteins and fats [12]. Furthermore, some strains are degrading compounds of macromolecules (lignin, cellulose, chitin, in part starch and aromatic hydrocarbons). Therefore, actinobacteria often occur in materials where organic matter is degraded [13], such as soils, organic compost heaps and building materials. Their metabolic diversity is due to their extremely large genome which codes for a variety of transcription factors that in turn control gene expression, allowing them to respond to specific needs [12]. Having these in mind, present study was aimed at to isolate and screen cellulase enzyme producing marine actinobacterial strains from the Havelock island sediments.
2. Materials and methods
2.1. Collection of samples and isolation of actinobacterial strains
Sediment samples were collected during the month of November 2011 at a depth of 25 cm from six locations of the Havelock island using a sterile spatula. The samples were placed in sterile polythene covers and brought to the field laboratory immediately and after arrival, necessary dilutions were made to carry out further microbiological analysis.
Isolation of actinobacteria was carried out in Kuster’s agar medium (Glycerol –10 g, Casein – 3 g, KNO3 – 2 g, NaCl –2 g, K2HPO4 – 2 g, MgSO4·7H2O – 0.05 g, CaCO3 – 0.02 g, FeSO4·7H2O –0.01 g, Agar – 15 g, 50% Sea water –1000 ml, pH 7 ± 0.1). The autoclaved Kuster’s agar medium containing petriplates were prepared aseptically. To minimize the fungal and bacterial contaminations, Kuster’s agar medium was supplemented with cycloheximide (10 μg/ml) and nalidixic acid (10 μg/ml) respectively [14]. One gram of pretreated sediment samples was serially diluted using sterile seawater and 0.1 ml of serially diluted samples was added to the petriplates containing Kuster’s agar medium [15] and spread using a ‘L’ shaped glass spreader. The plates were incubated at 37 °C for seven days in an inverted position. The leathery colonies of actinobacteria that appeared on the petriplates were counted from the 5th day onwards upto 28th day. After the incubation period, morphologically distinct colonies were picked up from the petridishes and restraecked in appropriate media and pure cultures were obtained and the slants were maintained at 4 °C for future.
2.2. Enzyme screening and preparation of crude enzyme cellulase
Morphologically distinct actinobacterial strains MHA1to MHA19 were analyzed by the spot inoculation method using two different enzymatic agar media such as Cellulose Congo-Red agar medium (Cellulose powder −1.88 g, KH2PO4 −0.5 g, MgSO4·7H2O −0.25 g, Congo red −0.2 g, Gelatin −2.0 g, Agar −16 g, 50% Seawater −1000 ml) and Carboxy Methyl Cellulose agar medium (Carboxy methyl cellulose −10 g, NaNO3–2 g, KH2PO4–1 g, MgSO4·7H2O −0.05 g, FeSO4·7H2O − 0.01 g, Yeast extract −0.1 g, Agar −16 g, 50% Seawater −1000 ml, pH 7 ± 0.1) for the preliminary screening for cellulase enzyme activity [16], [17], [44]. The clear zone diameter was measured in order to select the highest cellulase activity producer.
A loopful of culture or the strain was incubated into carboxy methyl cellulose (CMC) medium in 100 ml (Carboxy methyl cellulose −10 g, NaNO3 − 2 g, KH2PO4–1 g, MgSO4·7H2O −0.05 g, FeSO4·7H2O −0.01 g, Yeast extract −0.1 g, 50% Seawater −1000 ml, pH 7 ± 0.1) in Erlenmeyer flasks (250 ml) in triplicate. Flasks were incubated at 37 °C for 5–7 days in shaker incubator at 150 rpm. Part of samples (5 ml) was withdrawn from each flask after 7 days of fermentation and centrifuged at 2000 rpm for 10 min. The clear supernatant broth was collected aseptically to determine the enzyme yield.
2.3. Cellulase enzyme assay
Assay for the cellulolytic activities of cellulase was done by DNS (3,5-dinitrosalicyclic acid) method [18]. Cellulase activity was determined by incubating the crude enzyme (0.5 ml); 1% (w/v) CMC (0.5 ml) in 110 mM potassium phosphate buffer (pH 7) was mixed and incubated for 20 min at 70 °C; 1% DNS reagent (3 ml) (Dinitrosalicyclic acid −10 g, Phenol −2 g, Sodium sulphate −0.05 g, NaOH −10 g, Distilled water −1000 ml) was added in a capped test tube and the mixture was heated at 90 °C for 5–15 min to develop red brown colour. Then, 1 ml of potassium sodium tartrate (Rochelle salt solution) was added to stabilize the colour. After cooling to room temperature in a cold water bath, absorbance was read in a UV–vis spectrophotometer (UV-2450, Shimadzu, Japan) at 575 nm. The reducing sugar formed was quantified as glucose using dinitrosalicyclic acid.
2.4. Standard glucose solution (0.1 M)
A stock solution was prepared dissolving glucose (1.8 g) in potassium phosphate buffer (100 ml). Different concentrations of glucose standard solution were taken and diluted to final value of 1 ml. One ml of diluted standard solution and 1 ml of potassium phosphate buffer (pH 7) were mixed and incubated for 20 min at 70 °C; 3 ml of DNS reagent (1%) was added in a capped test tube and the mixture was heated at 90 °C for 5–15 min to develop red brown colour. Then, 1 ml of potassium sodium tartrate (Rochelle salt solution) was added to stabilize the colour. After cooling to room temperature in a cold water bath, absorbance was read in a UV–vis spectrophotometer (UV-2450, Shimadzu, Japan) at 575 nm.
A standard curve was obtained with standard glucose solution without the enzyme. One unit (U) of CMCase activity is the amount of enzyme required to produce 1 μmol reducing sugar (glucose) per min in reaction mixture under specified conditions.
2.5. Taxonomical investigation
2.5.1. Genus level identification
Characterization and subsequent identification of the strain MHA15 to the genus level were made based on the chemical composition of the cell wall and micro-morphological studies [19], [20], [21]. Characterization of the strain MHA15 was made by following the method, described by Shirling and Gottlieb [22], using the standard ISP medium.
2.5.2. Species level identification
Species level identification of the strain was made based on the keys and by using the Bergey’s Manual of Determinative Bacteriology. The status of the strain was also confirmed based on its molecular characters (16S rDNA gene sequences), with the following methodology.
2.5.3. Genomic DNA isolation and amplification of 16S rDNA
Total genomic DNA was extracted from the actinobacterial broth by phenol- chloroform isoamyl alcohol method, which removes the protein and their cellular components from the nucleic acid to obtain the pure DNA [23]. DNA sample (10 μl) was mixed with 2 μl of loading (6×) dye and loaded in 1% agarose gel. The separated DNA was visualized by UV transilluminator.
Each 50 μl amplification reaction contained 1 μl template DNA (50–200 ng), 5 μl 10× PCR buffer, 1 μl both forward [27F (5′-AGAGTTTGATCMTGGCTCAG-3′)] and reverse [1492R (5′-GGYTACCTTGTTACGACTT-3′)] primers, 1 μl dNTP mix (10 mM), 6 μl MgCl2 (25 mM), 2.5 U Taq DNA polymerase, 2.5 μl DMSO and 31.5 μl sterile water. The reaction conditions were initial denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 90 s. A final extension was performed at 72 °C for 10 min [24]. Reaction products were electrophoresed on a 1% agarose gel with 2 μl ethidium bromide and visualized under UV transilluminator and then purified.
2.5.4. 16S rDNA sequencing and phylogenetic analysis
The purified fragment was directly sequenced using a Ampli Tag FS DNA sequencing Kit (Applied Biosystem). The data were analyzed using applied Biosystem DNA editing and assembly software and sequences comparisons were obtained using the Micro Seq Software.
Sequence similarity search was made for the 16S rDNA sequence of MHA15 by applying its sequence to BLAST search of the NCBI (National Centre for Biotechnological Information, USA). Final editing of sequence alignment was done using BioEdit [25] and phylogenetic analysis was performed with version 5 of the MEGA (Molecular Evolutionary Genetics Analysis) software package [26]. A phylogenetic tree was constructed by using the Neighbour-joining tree-making algorithum [27]. The topology of the phylogenetic tree was evaluated by using the booststrap resampling method of Felsenstein [28] with 1000 replicates.
3. Results
3.1. Isolation and cellulase enzyme screening of the actinobacteria
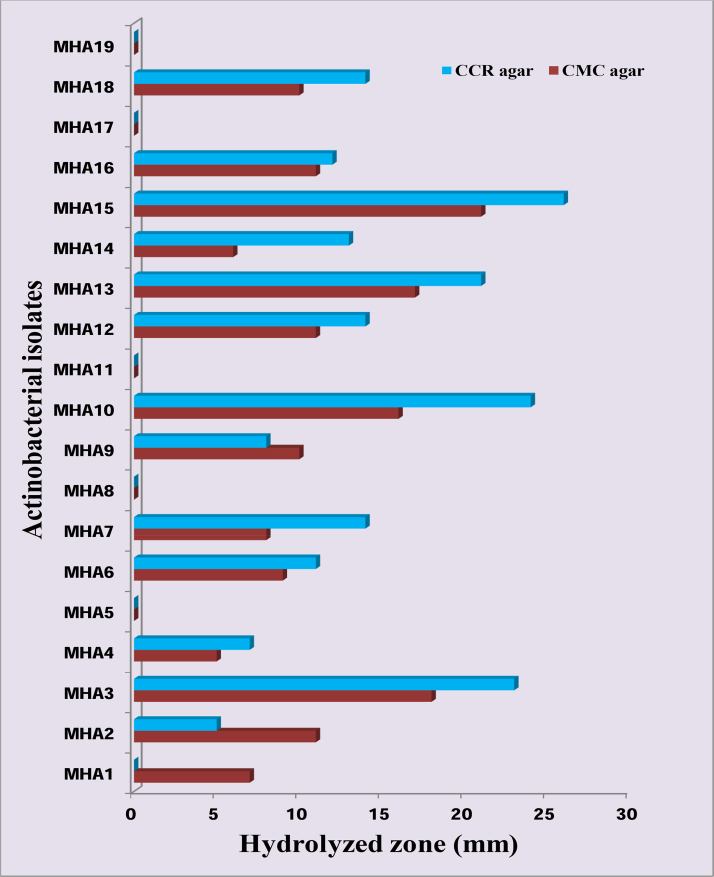
Actinobacterial colonies were isolated from the sediment samples of the six stations of the Havelock island, using Kuster’s agar medium and a total of 19 morphologically distinct actinobacterial strains were obtained. All these morphologically distinct strains were subjected to enzyme screening using two different media. In both the Cellulose Congo-Red agar medium and Carboxy methyl cellulose agar medium, higher cellulose degradation zone, the maximum cellulose degradation characteristic, was found in MHA3, MHA10, MHA13 and MHA15 (Fig. 1, Fig. 2).
Fig. 1.
Screening for the cellulase enzyme activity in CCR agar and CMC agar media.
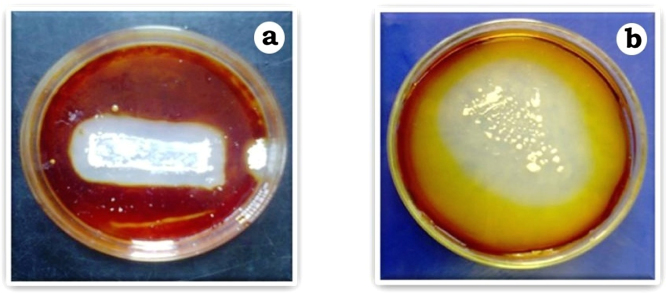
Fig. 2.
Cellulolytic activity of strain MHA15 in CCR agar (a) and CMC agar (b) media.
3.2. Cellulase enzyme assay
Based on the performance, MHA3, MHA10, MHA13 and MHA15 were selected as potential cellulase enzyme producers and these strains were subjected to cellulase enzyme screening using the DNS method and glucose was used as the standard. By comparing the glucose standard curve, cellulase enzyme production of the selected actinobacterial strains was measured. Higher amount of cellulase enzyme production was 14.379 1U/ml in MHA15, followed by MHA3 (14.213 1U/ml), MHA13 (13.356 1U/ml) and MHA10 (13.119 1U/ml). Among them, MHA15 alone produced more quantity of cellulase enzyme.
3.3. Taxonomical investigation of strain MHA15
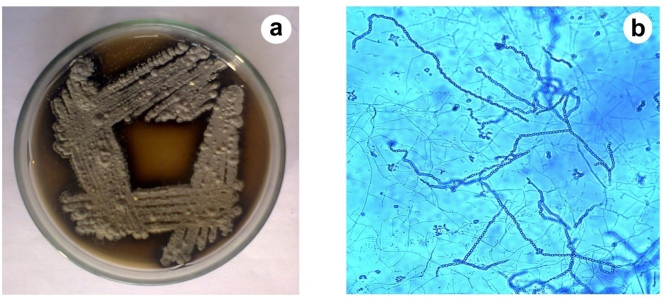
Strain MHA15 formed extensively branched substrate mycelium and aerial hyphae that produced long spore chains (Fig. 3). In ISP2 agar, black with gray coloured aerial spores were formed (Fig. 3). In Peptone yeast extract iron agar, reverse side and soluble pigments were produced. Melanin pigment was also produced on ISP7 agar. The culture grew well when it was supplemented with the carbon sources viz. arabinose, xylose, inositol, mannitol, fructose, rhamnose and sucrose. No growth was observed in raffinose. In the strain MHA15, cell wall characteristics showed the presence of glucose, mannose, galactose and rhamnose as whole-cell sugars and meso-diaminopimilic acid as the amino acid in the cell wall and glycine was absent, indicating that this strain belongs to the cell wall chemo type III (Table 1).
Fig. 3.
Morphological characters (a-aerial mass colour; b- long spore chain) of strain MHA15.
Table 1.
Comparative characteristics of the strain MHA15 with the A. cyanogriseus.
| Characters studied | Strain MHA15 | A. cyanogriseus |
|---|---|---|
| I. Cell wall amino acids | ||
| LL-DAP | – | – |
| Meso- DAP | + | + |
| Glycine | – | – |
| II. Whole cell sugars | ||
| Arabinose | – | – |
| Galactose | + | + |
| Rhamnose | + | + |
| Glucose | + | + |
| Mannose | + | + |
| III. Cell wall chemotype | III | III |
|
IV. Characters studied (as per Tamura et al., 2000) |
||
| Colour of aerial mycelium | Blue-gray | Blue-gray |
| Melanoid pigment | Black | Black |
| Reverse side pigment | Black | Black |
| Soluble pigment | Black | Black |
| Spore chain | Long spore chain | Long spore chain |
| Carbon source assimilation | ||
| Arabinose | + | ± |
| Xylose | + | + |
| Inositol | + | – |
| Mannitol | + | + |
| Fructose | + | – |
| Rhamnose | + | + |
| Sucrose | + | ± |
| Raffinose | – | – |
| V. Molecular characters | ||
| Sequenced gene | 16S rDNA | 16S rDNA |
| NCBI Accession number | KF668663 | NR_024650 |
| No of bp. | 873 | 1468 |
| Similarity level with closest neighbour strain (%) | 91.6 | – |
Positive (+); Negative (−); weakly utilized (±).
3.4. Molecular identification
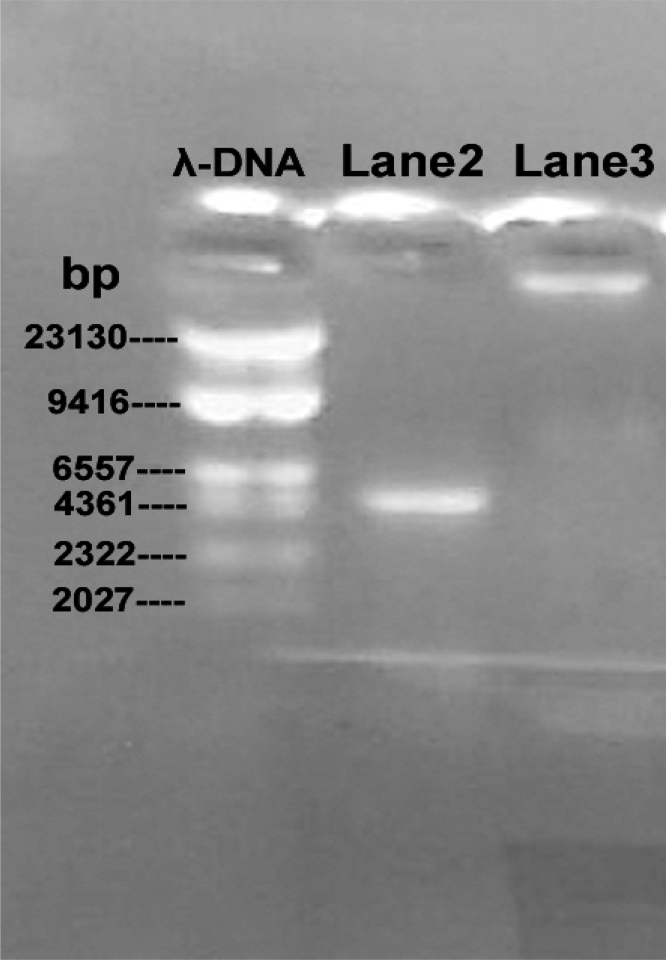
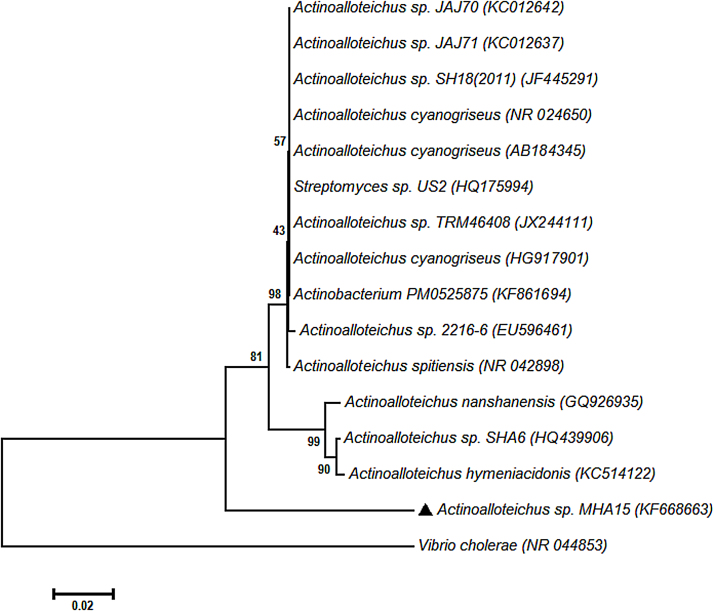
Further, for molecular identification, the strain MHA15 genomic DNA was extracted and purified and genomic DNA’s 16S rDNA gene was amplified using Universal primers. Amplified product and genomic DNA were loaded in lanes 2 and 3 respectively and amplified 16S rDNA gene showed a distinct band pattern with a molecular base pairs approximately near to 4361 bp (Fig. 4). A 873 bp of 16S rDNA sequences was determined for the strain MHA15, which was submitted to the GenBank (National Center for Biotechnology Information, USA) and an accession number (KF668663) was obtained. Based on the molecular data (16S rDNA sequences), phylogenetic tree was constructed on the comparison of 16S rRNA gene (873 bp) of the strain MHA15 with the previously reported sequence of Actinoalloteichus species (Accession number: NR_024650) deposited in the GenBank and closest species molecular data were also obtained from the GenBank. Vibrio cholerae rooted phylogenetic tree (Fig. 5) indicated that the strain MHA15 is related to the members of the genus, Actinoalloteichus and the strain MHA15 forms a branch with A. cyanogriseus with 91.6% similarity (Table 1).
Fig. 4.
Electrophoresis agarose gel results in strainMHA15.
Fig. 5.
Neighbour-joining tree based on 16S rRNA gene sequences, showing the phylogenetic relationship between the strain MHA15 and related and respective species of the genus Actinoalloteichus. Only bootstrap values (expressed as percentage of 1000 replications) >50% are given at nodes.
Results of the cultural, cell wall chemotypical, morphological, physiological and molecular characters were compared between the strain MHA15 and its closest phylogenic member. The strain MHA15 showed no variation in any of the characters when compared to those of the reference species, A. cyanogriseus. All the conventional characters were similar to those of A. cyanogriseus. But, the molecular characters showed distinct difference in the phylogenetic relationship (8.4%).
4. Discussion
Cellulose is the most important component of plant biomass and it acts as the renewable source for bioconversion to biofuels and by-products. In this respect, actinobacteria, one of the known cellulase producers, have evoked considerable research interest, due to their potential application in the recovery of fermentable sugars from cellulose [29] Further, these microbes are capable of producing an array of different extracellular enzymes including cellulase, chitinase and xylanase. In the present study, 19 actinobacterial isolates were screened for the cellulase activity.
Screening of actinobacterial strains was performed, using the cellulose Congo-Red agar and Carboxy Methyl cellulose agar media. In the cellulose Congo-Red agar medium, 74% of actinobacterial isolates (14 strains) showed the Zones of cellulose hydrolysis whereas, 29% of the actinobacterial isolates (5 strains) did not show the Zones of cellulose hydrolysis. In the Carboxy Methyl cellulose agar medium, 68% of the actinobacterial isolates (13 strains) showed Zones of cellulose hydrolysis whereas, 32% of actinobacterial isolates (6 strains) did not show the Zones of cellulose hydrolysis. In 1983, Veiga et al. [30] isolated 36 strains from marine sediments of about 10 m depth in Za corure Bay (Spain) and only 19 strains (>50%) showed enzymatic degradation of cellulose. Very recently, Gobalakrishnan et al. [44] have reported that the strain AUANI-5 isolated from the Neil island, produced higher cellulase enzyme and which was identified as Streptomyces craterifer.
In the present study, cellulose degradation zone was higher in Carboxy Methyl cellulose agar than that of the cellulose Congo-Red agar. This could be due to the fact that the cellulase is an inducible enzyme and it is affected by the nature of the substrate used for production, as explained by Haung and Monk [31] and due to the less complexity, leading to its easy assimilation by the isolated microbes [32].
Actinobacteria, one of the known cellulase producers have attracted considerable research interest in the recent times due to their potential application [33], [34]. A wide variety of bacteria are known for their production of hydrolytic enzymes with Streptomyces species being the best known enzyme producers [35]. Present study also examined cellulase production in 19 marine antibacterial isolates; among them, four (MHA3, MHA10, MHA13 and MHA15) showed higher cellulase activity and the strain MHA15 was identified as belonging to the genus Actinoalloteichus and it formed a branch with A. cyanogriseus with 91.6% similarity, in the phylogenetic study. Recently, biologically active compounds have been discovered from species belonging to this rare genus. For instance, naturally active melanin and cytotoxic cyclic bipyridine glycosides have been reported from the marine isolated strains, Actinoalloteichus sp. MA-32 and A. cyanogriseus WH1-2216-6 [36], [37]. In fact, Jose et al. [38] reported that the strain JAJ70 (isolated from hypersaline solar saltern) showed significant antimicrobial activity against the different clinical pathogens and the strain was identified as belonging to genus Actinoalloteichus.
Similarly, other researchers have also widely reported cellulase enzyme production by the marine actinobacteria: Veiga et al. [30] isolated 36 Streptomyces strains from 10 m depth of La Corufia Bay, Spain. Among them, 50% of isolates performed cellulolytic activity. Murugan et al. [39] isolated 35 actinobacterial strains from the Vellar estuary, India and examined their cellulose activity. Among them, starin CL-30 (S. actuosus) showed maximum cellulase activity. Stalin et al. [40] reported that 30 isolates (obtained from Tiruchendhur coastal area of Tamil Nadu, India) were screened for cellulase activity and S. clavuligerus alone produced maximum amount of cellulase. Recently, Sirisha et al. [41] reported bioactive compounds obtained from marine actinobacteria isolated from the sediments of the Bay of Bengal, and 24% the strains exhibited cellulase activity. Meena et al. [42] isolated 26 strains of actinobacteria from the marine sediments of the Andaman and Nicobar islands and among them, two Streptomyces species (NIOT-VKKMA02 and NIOT-VKKMA26) showed excellent cellulase activity. Reyad [43] also reported the actinobacterial production of cellulase and ascertained that the halotolerant actinobacteria associated with mangroves are a good source of cellulase.
5. Conclusion
From the present study, it can be stated that the Havelock island coast of the Andamans is a significant source for isolation of potential actinobacteria for biotechnological purpose. Out of the 19 morphologically distinct strains, strain MHA15 showed higher cellulase activity (14.379 1U/ml), suggesting that it is an ideal organism for the bioconversion of cellulose. Detailed investigations revealed that the strain MHA15 belongs to the genus Actinoalloteichus and the molecular characters showed distinct difference in phylogenetic relationship (8.4%) with the closest reference species A. cyanogriseus. If detailed polyphasic taxonomic analysis of the strain is made, it could be a new species.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
Authors thank the Dean, Faculty of Marine Sciences and the authorities of Annamalai University and J.J. College of Arts and Science, for providing with necessary facilities. They also thank Prof. L. Kannan, Former Vice Chancellor, Thiruvalluvar University, Vellore, for encouragement and editorial support.
References
- 1.Devi M.D., Kumar M.S. Production, optimization and partial purification of cellulase by Aspergillus niger fermented with paper and timber sawmill industrial wastes. J. Microbiol. Biotechnol. Res. 2012;2(1):120–128. [Google Scholar]
- 2.Avellaneda-Torres L.M., Pulido C.P.G., Rojas E.T. Assessment of cellulolytic microorganisms in soils of Nevados Park: colombia. Braz. J. Microbiol. 2014;45(4):1211–1220. doi: 10.1590/s1517-83822014000400011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Awojobi K.O., Agboola F.K., Arotupin D.J., Oladoy C.O., Olutiola P.O. The screening and selection of Trichoderma species capable of producing extracellular cellulolytic enzymes from soil of decaying plant materials. Ife J. Sci. 2013;15(2):263–271. [Google Scholar]
- 4.Bajaj B.K., Pangotra H., Wani M.A., Sharma P., Sharma A. Partial purification and characterization of a highly thermostable and pH stable endoglucanase from a newly isolated Bacillus strain M-9. Indian J. Chem. Technol. 2009;16:382–387. [Google Scholar]
- 5.Bhat M.K., Bhat S. Cellulose degrading enzymes and their potential industrial applications. Biotechnol. Adv. 1997;15(3–4):583–620. doi: 10.1016/s0734-9750(97)00006-2. [DOI] [PubMed] [Google Scholar]
- 6.Lee S.M., Koo Y.M. Pilot-scale production of cellulase using Trichoderma reesei Rut C-30 in fed-batch mode. J. Microbiol. Biotechnol. 2001;11(2):229–233. [Google Scholar]
- 7.Kubicek C.P., Messner R., Guber F., Mach R.L., Kubicek-Pranz E.M. The Trichoderma cellulase regulatory puzzle: from the interior life of a secretory fungus. Enzyme Microb. Technol. 1993;15(2):90–99. doi: 10.1016/0141-0229(93)90030-6. [DOI] [PubMed] [Google Scholar]
- 8.Bai S., Ravikumar M., Mukeshkumar D.J., Balashanmugam P., Balakumaran M.D., Kalaichelvan P.T. Cellulase production by Bacillus subtilis isolated from cow dung. Arch. Appl. Sci. Res. 2012;4(1):269–279. [Google Scholar]
- 9.Niva M., Hernesmaa A., Haahtela K., Salkinoja-Salonen M., Sivonen K., Haukka K. Actinobacterial communities of boreal forest soil and lake water are rich in mycobacteria. Boreal Environ. Res. 2006;11(1):45–53. [Google Scholar]
- 10.Jiang H., Dong C.Z., Huang Q., Wang G., Fang B., Zhang C., Dong H. Actinobacterial diversity in microbial mats of five hot springs in central and Central-Eastern Tibet, China. Geomicrobiol. J. 2012;29(6):520–527. [Google Scholar]
- 11.Berdy J. Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. 2012;65(8):385–395. doi: 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- 12.Trujillo M.E. eLS John Wiley & Sons Ltd; Chichester: 2008. Actinobacteria.http://www.els.net (10.1002/9780470015902. a0020366) [Google Scholar]
- 13.Schafer J., Jackel U., Kampfer P. Development of a new PCR primer system for selective amplification of Actinobacteria. FEMS Microbiol. Lett. 2010;311(2):103–112. doi: 10.1111/j.1574-6968.2010.02069.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee L.H., Zainal N., Azman A.S., Eng S.K., Goh B.H., Yin W.F., Mutalib N.S.A., Chan K.G. Diversity and antimicrobial activities of actinobacteria isolated from tropical mangrove sediments in Malaysia. Sci. World J. 2014;2014(698178):1–14. doi: 10.1155/2014/698178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuster E., Williams S.T. Production of hydrogen sulphide by Streptomyces and methods for its detection. Appl. Microbiol. 1964;12(1):46–52. [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta P., Samant K., Sahu Isolation of cellulose-degrading bacteria and determination of their cellulolytic potential. Int. J. Microbiol. 2012;2012(578925):1–5. doi: 10.1155/2012/578925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnsen H.R., Krause K. Cellulase activity screening using pure carboxymethylcellulose: application to soluble cellulolytic samples and to plant tissue prints. Int. J. Mol. Sci. 2014;15(1):830–838. doi: 10.3390/ijms15010830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31(3):426–428. [Google Scholar]
- 19.Cummins C.S., Harris H. Studies on the cell-wall composition and taxonomy of Actinomycetales and related groups. Microbiology. 1958;18(1):173–189. doi: 10.1099/00221287-18-1-173. [DOI] [PubMed] [Google Scholar]
- 20.Lechevalier M.P., Lechevalier H. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Bacteriol. 1970;20(4):435–443. [Google Scholar]
- 21.Tamura T., Zhiheng L., Yamei Z., Hatano K. Actinoalloteichus cyanogriseus gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2000;50:1435–1440. doi: 10.1099/00207713-50-4-1435. Pt4. [DOI] [PubMed] [Google Scholar]
- 22.Shirling E.B., Gottlieb D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966;16(3):313–340. [Google Scholar]
- 23.Ahmed O.B., Asghar A.H., Elhassan M.M. Comparison of three DNA extraction methods for polymerase chain reaction (PCR) analysis of bacterial genomic DNA. Afr. J. Microbiol. Res. 2014;8(6):598–602. [Google Scholar]
- 24.Karuppiah V., Aarthi C., Sivakumar K. Enhancement of PCR amplification of actinobacterial 16S rRNA gene using an adjuvant, dimethyl sulphoxide. Curr. Sci. 2011;101(1):22–23. [Google Scholar]
- 25.Hall T.A. BioEdit: a user- friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- 26.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 29.Rathnan R.K., Ambili M. Cellulase enzyme production by Streptomyces sp using fruit waste as substrate. Aust. J. Basic Appl. Sci. 2011;5(12):1114–1118. [Google Scholar]
- 30.Veiga M., Esparis A., Fabregas J. Isolation of cellulolytic actinomycetes from marine sediments. Appl. Environ. Microbiol. 1983;46(1):286–287. doi: 10.1128/aem.46.1.286-287.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haung X.P., Monk C. Purification and characterization of a cellulase from a newly isolated thermophilic aerobic bacterium Caldibacillus cellulovorans gen sp. nov. World J. Microbiol. Biotechnol. 2004;20(1):85–92. [Google Scholar]
- 32.Sadhu S., Saha P., Sen S.K., Mayilraj S., Maiti T.K. Production, purification and characterization of a novel thermotolerant endoglucanase (CMCase) from Bacillus strain isolated from cow dung. SpringerPlus. 2013;2:1–10. doi: 10.1186/2193-1801-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jang H.D., Chen K.S. Production and characterization of thermostable cellulases from Streptomyces transformant T3-1. World J. Microbiol. Biotechnol. 2003;19(3):263–268. [Google Scholar]
- 34.Arunachalam R., Wesely E.G., George J., Annadurai G. Novel Approaches for identification of Streptomyces noboritoensis TBG-V20 with cellulase production. Curr. Res. Bacteriol. 2010;3(1):15–26. [Google Scholar]
- 35.Chellapandi P., Jani H.M. Production of endoglucanase by the native strains of Streptomyces isolates in submerged fermentation. Braz. J. Microbiol. 2008;39(1):122–127. doi: 10.1590/S1517-838220080001000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manivasagan P., Venkatesan J., Senthikumar K., Sivakumar K., Kim S.K. Isolation and characterization of biologically active melanin from Actinoalloteichus sp: MA-32. Int. J. Biol. Macromol. 2013;58:263–274. doi: 10.1016/j.ijbiomac.2013.04.041. [DOI] [PubMed] [Google Scholar]
- 37.Fu P., Liu P., Li X., Wang Y., Wang S., Hong K., Zhu W. Cyclic Bipyridine Glycosides from the marine derived actinomycete Actinoalloteichus cyanogriseus WH1-2216-6. Org. Lett. 2011;13:5948–5951. doi: 10.1021/ol202245s. [DOI] [PubMed] [Google Scholar]
- 38.Jose P.A., Robinson S., Jebakumar D. Taxonomic and antimicrobial profiles of a rare actinomycete isolated from an inland solar saltern (India) Indian J. Geo-Marine Sci. 2015;44(3) [Google Scholar]
- 39.Murugan M., Srinivasan M., Sivakumar K., Sahu M.K., Kannan L. Characterization of an actinomycete isolated from the estuarine finfish, Mugil cephalus Lin: (1758) and its optimization for cellulase production. J. Sci. Ind. Res. 2007;66(5):388–393. [Google Scholar]
- 40.Stalin T., Sathyapriya B., Selvam K. Ecofriendly application of cellulase and xylanase producing marine Streptomyces clavuligerus as enhancer in biogas production from waste. Afr. J. Environ. Sci. Technol. 2012;6(6):258–262. [Google Scholar]
- 41.Sirisha B., Haritha R., Jaganmohan Y.S.Y.V., Sivakumar K., Ramana T. Bioactive compounds from marine actinomycetes isolated from the sediments of Bay of Bengal. Int. J. Pharm. Chem. Biol. Sci. 2013;3(2):257–264. [Google Scholar]
- 42.Meena B., Rajan L.A., Vinithkumar N.V., Kirubagaran R. Novel marine actinobacteria from emerald Andaman & Nicobar Islands: a prospective source for industrial and pharmaceutical byproducts. BMC Microbiol. 2013;13(145):2–17. doi: 10.1186/1471-2180-13-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reyad A.M. Diverse of enzymatically active actinomycetes associated with mangrove rhizospher in Jazan Coast. Ann. Biol. Res. 2013;4(4):100–108. [Google Scholar]
- 44.Gobalakrishnan R., Radha G., Sivakumar K., Naresh Rashmi R.R., Kannan L. Screening of industrially important enzymes of potential marine actinobacteria of the Neil Island: the Andamans, India. J. Bioresour. 2016;3(1):20–30. [Google Scholar]