Abstract
Background:
Algal cells produce neutral lipid when stressed and this can be used to generate biodiesel.
Objective:
Salt stressed cells of the model microalgal species Chlamydomonas reinhardtii were tested for their suitability to produce lipid for biodiesel.
Methods:
The starchless mutant of C. reinhardtii (CC-4325) was subjected to salt stress (0.1, 0.2 and 0.3 M NaCl) and transesterification and GC analysis were used to determine fatty acid methyl ester (FAME) content and profile.
Results:
Fatty acid profile was found to vary under salt stress conditions, with a clear distinction between 0.1 M NaCl, which the algae could tolerate, and the higher levels of NaCl (0.2 and 0.3 M), which caused cell death. Lipid content was increased under salt conditions, either through long-term exposure to 0.1 M NaCl, or short-term exposure to 0.2 and 0.3 M NaCl. Palmitic acid (C16:0) and linolenic acid (C18:3n3) were found to increase significantly at the higher salinities.
Conclusion:
Salt increase can act as a lipid trigger for C. reinhardtii.
Keywords: Microalgae, biofuel, FAME, salt, lipid, stress, biodiesel
1. INTRODUCTION
As fossil fuels continue to deplete, the search for ways to utilise the energy in natural resources to meet the electricity and fuel demands of our world continues. One seemingly unlimited source of energy is light from the sun. Photosynthetic organisms provide a natural and abundant mechanism for converting sunlight into chemical energy, and therefore provide an obvious choice in the search for new energy sources. Due to the problems associated with first and second generation biofuels, research is now focussing on the possibilities of third generation biofuels based on microalgae [1].
Microalgae are highly efficient at converting sunlight into useful products for biofuels. Most notably, they can produce triacylglycerols (TAGs) in abundance under certain conditions, and this is highly desirable because TAGs can be converted by transesterification to biodiesel [2]. No biofuel has yet achieved price parity with petroleum derived fuels, including algal biodiesel [3, 4].
For algal biodiesel to become a successful replacement fuel for oil-derived liquid fuels, several advances need to be made in algal biofuels research. Firstly, TAG must be made on a large enough scale to be able to meet the demand for liquid fuels. Secondly, biodiesel prices must be able to compete with fossil fuel prices [5]. TAG yields from algal cultures are currently limited by the oil contents of the algae, which has a large impact on price; Jorquera et al. [6] show that algal biodiesel would be economically competitive with petroleum oil if the cellular oil percentage content was increased from 30% to 60%.
Algae use TAG as an energy storage product when growth is curtailed by the absence of a key nutrient, such as nitrogen [7, 8] or phosphorus [9], or by environmental stress such as high salinities [10]. Under stress conditions, the photosynthetically fixed carbon supply exceeds the ability of the cell to multiply, causing the build up of carbon in storage molecules [11]. These stressors causing the onset of lipid production are therefore known as “lipid triggers”. The formation, pathways, and composition of different lipid types within algae are thoroughly researched and documented [12, 13], and now the challenge lies in comprehensively linking these to the genes, proteins, metabolic pathways, and environmental conditions that trigger their synthesis.
Chlamydomonas reinhardtii is often used as a model algal species and is highly tractable for genetic transformations [14], and therefore invaluable for knowledge of lipid metabolism that could subsequently be used in genetic engineering of a high TAG-producing alga. Nitrogen deprivation has been the main focus in lipid manipulation in C. reinhardtii, but sulphur stress has also been investigated and shown to be a lipid trigger [15]. Sharma et al. [16] provide a full description of different stresses used to investigate the lipid profile in different species.
Salt stress (normally increased salt levels above optimum requirements) has been shown to cause TAG increase in Nannochloropsis salina [17], Desmodesmus abundans [18], and Botryococcus braunii [19]. Venkata Mohan and Devi [20] also found a higher lipid productivity in their mixed microalgal culture when 17 mM NaCl was added. Dunaliella salina has also shown increases in lipid content in response to high salt concentrations of 1000 mM [21]. Whilst salt has been shown to induce lipid changes in microalgae, as demonstrated in these studies, only one study has investigated the effect of salt stress on lipid content in Chlamydomonas reinhardtii [10]. This study used up to 100 mM NaCl to induce increased TAG content, but no information was given on any salt induced changes to the lipid profile.
Other Chlamydomonas species have shown to have their lipids increased by salt stress. An Arctic Chlamydomonas strain (species not identified) showed varying lipid yield and profiles in response to increased salinity levels (up to 1000 mM), with polyunsaturated fatty acids induced by salt stress [22]. Another Antarctic ice alga “Chlamydomonas sp. ICE-L” showed the highest lipid content of 23% W/W at 16% NaCl [23]. Chlamydomonas nivalis has also been explored for the effect of salt on lipid biomarkers [24-26], as well as one Nile Red investigation to find the highest lipid expression under different salt concentrations. This was found using 1% and 1.25% NaCl [24].
The freshwater species Chlamydomonas mexicana has been grown under salt stress and found to have a significant increase in lipid content under 0.05 M NaCl [27], demonstrating that the salt stress method of lipid trigger can be used in freshwater species.
The lack of full lipid profile investigation into C. reinhardtii under salt stress provides an opportunity to investigate this model species under a new stress condition that could provide further understanding of lipid metabolism changes under stress. Unlike nitrogen stress, salt stress does not necessarily have the same limits on protein synthesis and cell division, and therefore might address the issues that affect overall lipid productivity in an algal culture.
2. MATERIALS AND METHODS
2.1. Algal Species
Algal species Chlamydomonas reinhardtii strain CC-4325 sta1-1 mt- [Ball I7] was obtained from the Chlamydomonas Resource Centre (www.chlamycollection.org).
The Ball I7 strain is deficient in the catalytic (small) subunit of ADP-glucose pyrophosphorylase and was obtained by X-ray mutagenesis of the wild type strain CC-4323. A starchless mutant was used as they have the potential for higher lipid production levels under metabolic engineering than wild types [28].
2.2. Algal Culturing
Cultures were grown in a Sanyo Versatile Environmental Test Chamber Model MLR-351H on shakers at 100 rpm. The light intensity was between 130 and 222 µEm-2s-1 recorded across the different shakers. The temperature was 25˚C and light was 24 hour constant cycle. The cultures were grown in 500 mL conical flasks sealed with 55 x 50 mm polyurethane foam sponges (Fisherbrand Scientific, UK, product code 12904301) to allow some air, and therefore carbon dioxide, to enter the flask. They were grown for 48 hours in 200 mL normal TAP medium, for mixotrophic growth, and then centrifuged at 3000 g for 5 minutes and re-suspended at an early-to-mid log growth phase (in this case measured as 0.44 OD at 750 nm) in either normal TAP medium or TAP medium with NaCl salt added to the desired concentration.
C. reinhardtii has been reported to tolerate salt concentrations of up to 200 mM NaCl [29]. A range of salt concentrations (0, 100, 200 and 300 mM NaCl) was used, with each culture condition grown in triplicate.
2.3. Growth Measurements
2.3.1. Optical Density and Dry Cell Weight
To measure growth, optical density (OD) at 750 nm was taken as a measure of culture density. OD was measured on an Ultraspec 2100 Pro spectrophotometer (Serial no. 88446, Biochrom Ltd., Cambridge, UK), using 1 mL cuvettes, and using deionised water as a blank.
The other indication of culture growth used was dry cell weight (DCW). This is a normalising measure of culture growth and volume. Dry cell weight is important to know, since all other aspects of the culture can then be measured against it in terms of an overall percentage of the culture yield. This can be used as a normalising basis for lipid content. DCW was measured by centrifuging a sample from the culture using a Hermle Z400K centrifuge (Labnet International Inc., New Jersey, USA) at 1,200 g for 10 minutes at 4˚C, and transferring the pellet to a pre-weighed 1.5 mL Eppendorf tube. The sample was frozen, first at -20˚C, then at -80˚C, and then freeze-dried. The new tube and sample weight was used to obtain the DCW of the culture. DCW data is not shown here.
2.3.2. Microscopy and Cell Morphology
Cells were photographed under x1000 magnification using an Olympus BX51 microscope (Smcs Limited), with ProgRes® C5 camera attachment (Jenoptik, Germany) and ProgRes® CapturePro 2.6 imaging software. Photographs were used as a visual reference to examine cells.
2.4. Direct Transesterification and Gas Chromatography Protocol for FAME Analysis of Algal Lipids
2.4.1. Sample Preparation
Samples were harvested, washed in 1 x phosphate buffered saline (PBS) buffer and pelleted. They were stored in -20˚C freezer, and freeze dried. The DCW was calculated and dilution of 10 mg mL-1 was made using distilled water. 100 µL sample was taken and added to 2 mL Eppendorf tube. 500 µL glass beads (425-600 µm i.d., acid washed, from Sigma) were added to the sample, with 1.2 mL of methanol:chloroform (1:2) solution. The samples were bead beaten using a Disruptor Genie bead beater (Scientific Industries, New York, USA, Serial No. D68-10198 and D48-1014) on a 2 minute disruption and 2 minute interval on ice cycle. There were 10 cycles. The samples were centrifuged at 16,000 g at 4˚C for 5 minutes. The supernatant was transferred to a new Eppendorf tube. 400 µL chloroform (Fisher Scientific UK) and 400 µL HPLC grade ultrapure water (Fisher Scientific UK, code: W/0106/17) were added to each sample. Samples were centrifuged for 15 minutes at 7000 g at 4˚C. The organic phase, or bottom layer, was transferred to a glass vial and evaporated to dryness under inert nitrogen gas. 250 µL chloroform:methanol (1:1) solution and 100 µL 10% BF3:methanol solution were added to each sample. Samples were incubated at 80˚C for 90 minutes, then cooled at room temperature for 10 minutes. To each sample 300 µL ultrapure water and 600 µL hexane (Fisher Scientific UK) were added and samples were transferred to a fresh Eppendorf tube. They were vortexed for 1 minute and centrifuged at 7000 g for 5 minutes at 4˚C. The top organic layer was transferred to a fresh glass vial and evaporated to dryness under inert nitrogen gas before re-suspending in 100 µL hexane ready for injection into the gas chromatograph.
2.4.2. Gas Chromatography
Analysis was carried out on a Thermo Finnigan TRACE 1310 Gas Chromatograph with FID detector and autosampler (Thermo Scientific, Hertfordshire, UK), and a TRACE TR-FAME capillary column (obtained from Thermo) (dimensions 25 m x 0.32 mm x 0.25 µm). The sample volume of 1 µL was injected in split injection mode at 250˚C. The oven ramp was held at 150˚C for 1 minute, and increased by 10˚C min-1 up to 250˚C, then held at 250˚C for 1 minute. Split injection was carried out at split ratio 50, split flow 75 mL min-1 and carrier flow 1.5 mL min-1. The FID detector was set to 250˚C, air flow 350 mL min-1 and hydrogen flow 35 mL min-1. The GC was calibrated using Supelco 37 Component FAME mix (Supelco, Bellefonte, PA).
2.4.3. Data Analysis
This data contained analytical replicates (3 injections of the same sample) and three biological replicates. In some cases, analytical replicates showed the presence of FAMEs in one but not in another. Therefore the average of the available replicates was taken, but if a value was missing it was discounted and not recorded as a “0”.
The biological replicates also sometimes showed FAMEs in one replicate but not another. In this case, missing values were recorded as “0”, therefore there were always 3 biological replicates for each sample.
3. RESULTS AND DISCUSSION
3.1. Optical Microscopy of C. reinhardtii Cells
3.1.1. Effect of Adding 0.1 M NaCl on Cell Morphology
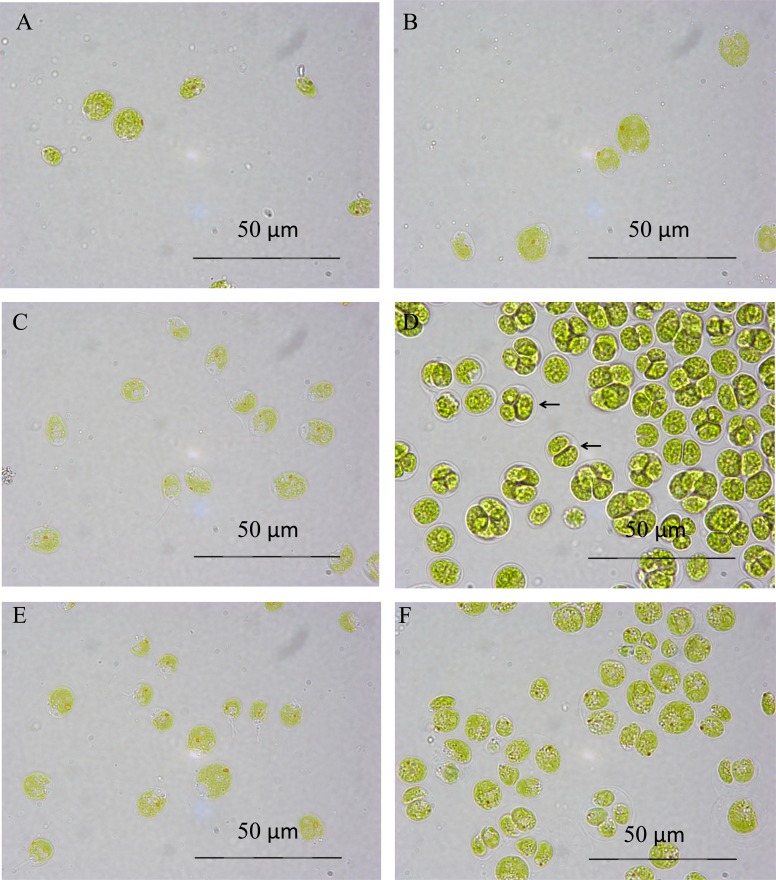
Fig. (1) shows the morphology of the cells under control and 0.1 M salt conditions over time. The cells grown in 0.1 M salt medium (B, D, F) show signs of being in abnormal cell division stages, clustered together in groups of four, rather than individual cells as seen in the control TAP medium (A, C, E). This occurs less in the latter stages (at 119 hours) of the culture growth, and does not show any sign at the initial time point of three hours of being different to the control conditions. The colour of the cells does not appear to be adversely affected by the presence of salt. Both sets of conditions show healthy pigmentation and intact cells. The main difference here is the 0.1 M NaCl stressed culture being in the process of abnormal cell division and having cells clustered together in a membrane known as a coenobium (indicated by arrows in Fig. (1D)).
Fig. (1).
Optical microscopy of C. reinhardtii under TAP conditions (A, C, E) and 0.1 M salt conditions (B, D, F) at 3 (A, B), 47 (C, D), and 119 (E, F) hours. All pictures taken at x1000 magnification.
3.1.2. Effect of Adding 0.2 and 0.3 M NaCl on Cell Morphology
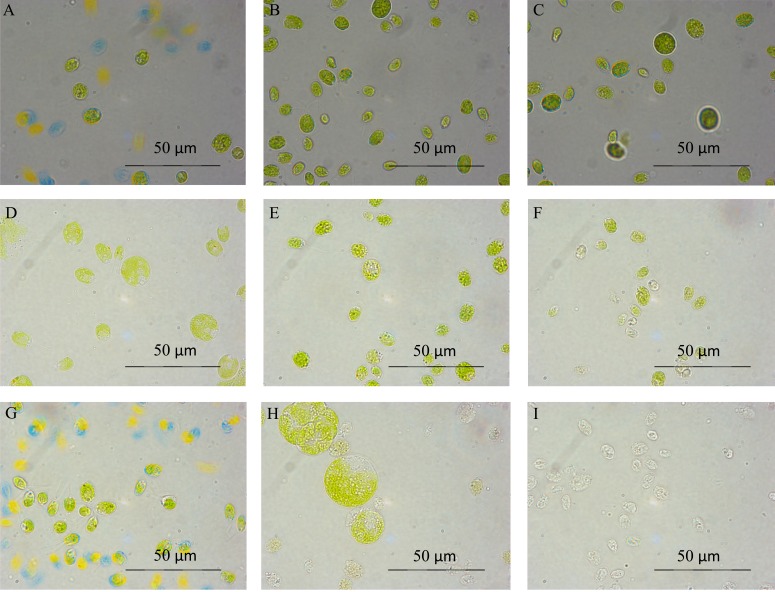
Fig. (2) shows the effect of 0.2 and 0.3 M salt on cultures in comparison to control TAP medium conditions. 0.2 M salt conditions retain a significant amount of green pigmentation, but towards the end of the growth period some cells start to lyse and become “ghosts” and those which are still green become much bigger and change drastically in morphology Fig. (2H). 0.3 M NaCl, in comparison, appears to cause the cells to shrink after the initial time point Fig. (2F), and after this the cells start to degrade and become “ghosts” Fig. (2I). 0.3 M salt is clearly a condition under which this strain cannot survive.
Fig. (2).
Optical microscopy of C. reinhardtii grown in TAP (A, D, G), 0.2 (B, E, H) and 0.3 (C, F, I) M salt TAP medium at time points 3 (A, B, C), 23 (D, E, F), 76 (G, H, I) hours at x1000 magnification.
3.2. Growth Measured Through Optical Density
Cultures were grown to mid log phase and then re-suspended to 0.44 OD750 with the new appropriate media, rather than growing the cultures in the salt medium from the start. This has the advantage of having a high level of initial biomass at the start, and then introducing a shock stress to the cultures. This is based on the idea, found in the literature [21], of introducing stress part way through the growth phase in order to maximise the lipid yield of a culture. This has the disadvantage of killing the cells if the salt molarity is too high, and therefore the application of salt stress needs to be carefully considered to allow the culture to continue healthy growth without killing the cells and creating dead biomass, whilst still introducing a significant level of stress into the cells. The advantage is that the culture will be at a high level of biomass when the stress is applied, and therefore testing of the culture can take place at early as well as later stages of stress application.
3.2.1. Effect of Adding 0.1 M NaCl on Culture Growth
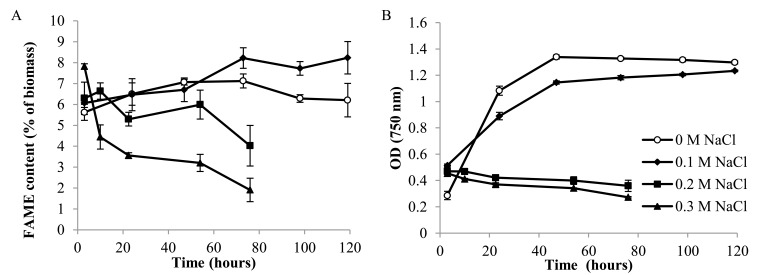
Fig. (3B) shows the optical density, taken as a measure of culture density and productivity, of this strain under low (0.1 M) NaCl conditions compared to control (0 M NaCl) growth conditions. The first (3 hour) time point optical density was at a higher value in the salt conditions than it was in the 0 M control conditions. However, the growth curve in 0.1 M NaCl medium then becomes shallower compared to the control (no added salt) cultures, demonstrating that the cells do not divide as fast as in 0.1 M NaCl TAP medium. The final optical density, as both curves plateau, is similar at 119 hours, demonstrating that at this concentration, 0.1 M NaCl does not have a severe impact on the biomass productivity of the culture.
Fig. (3).
Total FAME content as a percentage of dry cell weight (A) and OD750 (B) in 0.1, 0.2 and 0.3 M NaCl cultures and a control (0 M NaCl) culture (n=3).
This experiment provides conditions under which salt is having an impact on the algal physiology and biological responses, but does not severely hamper the culture's ability to grow and survive. This demonstrates that a low level of salt can be tolerated by this strain, but that no added salt in the medium is a more suitable growth condition for reaching high cell density quickly.
3.2.2. Effect of Adding 0.2 and 0.3 M NaCl on Culture Growth
Fig. (3B) demonstrates that, compared to the control growth conditions of no added salt TAP medium, the 0.2 and 0.3 M salt conditions cause a complete arrest of culture growth, as shown by the steady and slight decline in culture OD. Between 0.1 M and 0.2 M NaCl, there must be a critical level of salt tolerance beyond which the cells cease to be able to carry out their normal biological functions. As with the 0.1 M NaCl, there is an initial increase in optical density at the first time point of 3 hours under both salt conditions. Salt conditions therefore must have an immediate effect on the culture density. This seems unlikely to be due to a spike in cell division at this point, therefore it may be due to an effect on cell morphology that makes the culture appear denser. These cultures were not washed with PBS buffer when taking the OD, therefore we must consider that there may be a slight diffference due to medium composition, however, the dissolved salts should give a negligible effect on optical density. When comparing the OD750 of 0 M NaCl TAP and 0.2 M NaCl TAP sterile medium, both gave an OD of 0.000.
3.3. Discussion of Growth and Cell Morphology Data
The OD and algal microscopy provide information on how this strain grows under varying salt conditions.
The 0.1 M salt condition has some effect on how fast the culture grows and the log phase photograph Fig. (1D) shows the presence of sacs around groups of cells (indicated by arrows) which are not present in the control TAP conditions. This salt condition does not hamper the ability of the culture to grow and reach a similar level of biomass to the normal TAP culture, but it clearly changes the mechanism of cell division.
0.2 M salt arrests the growth of the culture, as shown by OD data, but photographs of microscopy show that the culture keeps its pigmentation for the majority of the culture growth cycle Fig. (2H). This suggests that the cells may be surviving without growth of the culture. In contrast, 0.3 M salt shows the same growth arrest, but also demonstrates cell lysis through loss of pigmentation and cell contents, leaving the remaining biomass as ghost cells and cell fragments Fig. (2I). This may be autophagic cell death, which has been shown to occur in other algae species under salt stress [30]. A loss of pigmentation is an expected response in green algae under high salt stress, although they may also respond with an increase in cell size rather than a decrease [31], as seen in Fig. (2H).
3.4. Lipid Results
Fig. (3A) 1 shows the overall FAME content of the GC tests under the three different salt concentrations as well as a control. These figures are obtained by adding together all the detected FAME weights in a run and calculating it as a percentage weight of the total biomass. The total FAME content increases in 0.1 M NaCl after 50 hours in the salt stressed cultures, whereas they decrease slightly in the control conditions after this point. For the 0.1 M NaCl culture, this is also the point at which stationary phase was reached (see Fig. (3B)), showing that the increase in FAMEs coincided with the culture ceasing to proliferate. The increase in FAME may therefore be due to diverting of carbon that is no longer being used for growth (a similar rise is seen at the start of the control culture, at a lower level). This increase under moderate salt stress of 0.1 M NaCl is, however, important, as it shows that salt can increase lipid content in this species.
These cultures were exposed to a salt condition (0.1 M NaCl) that they were able to tolerate, therefore although the cultures had a slightly reduced growth rate at the start of the experiment the cells can be assumed to have functioned well in these conditions.
In the higher salt concentrations of 0.2 and 0.3 M NaCl, a different effect is seen. Normal TAP conditions show a steady and slight increase in FAME content overall. When comparing to time point 3 hours, the 0.2 and 0.3 M NaCl salt conditions show an increase in overall FAME content compared with the control conditions, and this increase is especially marked in the 0.3 M salt cultures. This time point (t=3) is therefore of particular interest as it demonstrates a distinct (although small) increase in lipid content under high salt stress conditions which can then be further investigated. It may be that the cell is responding to the shock of the salt environment by directing resources into fatty acids at a fast rate, however this must be investigated properly by looking at the individual FAME data. The FAME content then decreases dramatically over time in the salt conditions, although much more markedly in the 0.3 M NaCl cultures than in the 0.2 M NaCl cultures. This can be explained by the fatty acids in the cells degrading as the cells start to lyse and break down. There is some slight fluctuation in the 0.2 M salt conditions rather than a continuous decline, suggesting that some of the cells are still functioning and altering their fatty acid content even though the culture is not dividing. This culture then shows a sharp decrease at the last time point, when the cells are starting to lose their shape, as demonstrated in Fig. (2), suggesting the cells are breaking down in both structure and fatty acid content at this stage.
To more fully analyse the GC results, the individual components of the FAME data were analysed. The individual FAME components of the algal biomass can be analysed in several ways. In this research, a 1 mg sample was analysed to compare the total amounts of FAME detected therein. The relative percentage contributions towards the biomass, or the relative make up of the FAMEs in the total FAME, could also be examined. In this case, the percentage of the biomass that was FAME was calculated and compared, since this was a good way to compare the patterns of the different fatty acids.
The FAMEs that are present (from the 37 FAME mix used as a standard) in lower quantities are C6:0, C8:0, C10:0, C11:0, C12:0, C13:0, C14:0, C14:1, C15:0, C16:1 and C17:1. The FAMEs present in higher quantities are C16:0, C17:0, C18:0, C18:1cis, C18:2cis, and C18:3n3. Occasionally C20:5, C21:0 and C20:3n6 are seen, but not consistently throughout samples. Since the chain lengths were dominated by C16, C17 and C18, these chain lengths were examined in more detail.
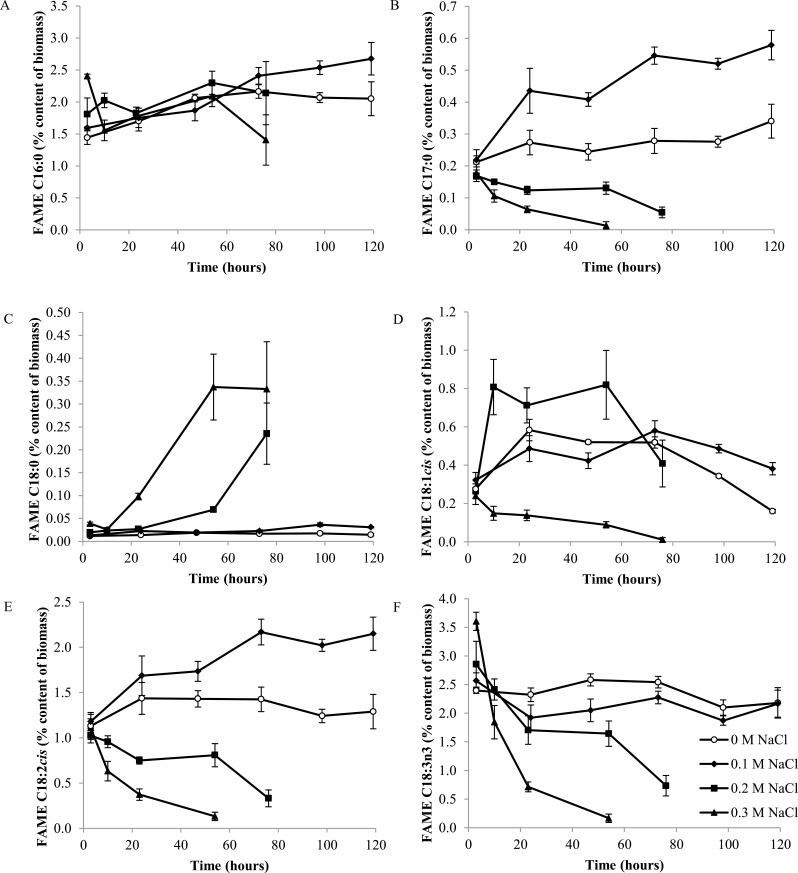
Fig. (4) shows the response of the main FAMEs to salt stress. The levels of C16:0 in the 0.1 M NaCl cultures continue to rise beyond the range of the control cultures, and appear to be a major contributor to the overall rise in FAMEs observed Fig. (4A). This happens after stationary phase has been reached. Similar patterns are seen in C18:1cis (or oleic acid) and C18:2cis; in these major FAME elements, the 0.1 M salt stressed cultures had higher quantities of the FAME element in stationary phase than the cultures grown in control TAP medium Figs. (4D, E). Only C18:2cis and C17:0 showed higher quantities in 0.1 M salt stressed cells throughout the sampling points Figs. (4B, E). C18:3n3 decreased in the 0.1 M salt stress cultures, although by the last sampling point the quantities were at similar levels again Fig. (4F). This suggests a move away from C18:3n3 (alpha-linolenic acid) towards the other main FAME groups in response to salt stress.
Fig. (4).
Percentage content (of dry cell weight) of major FAME components C16:0 (A), C17:0 (B), C18:0 (C), C18:1cis (D), C18:2cis (E) and C18:3n3 (F) under 0.1, 0.2, 0.3 M NaCl and control conditions (n=3).
In the higher salt stresses of 0.2 and 0.3 M NaCl, different patterns in individual FAMEs are seen. The C16:0 content was increased at the three hour time point in the 0.3 M NaCl culture relative to the control, and then fluctuated. This initial increase is a large part of the overall increase in total FAME content. Since C16:0 is one of the first fatty acids made in fatty acid synthesis, this data suggests that fatty acids are being synthesized at the 3 hour 0.3 M NaCl data point. This may be a response of the cell to salinity stress for which additional lipids are required for maintaining membrane function, or it may be a stress response similar to that seen during nitrogen deprivation where fatty acids start to accumulate because growth is halted. The 0.2 M NaCl culture shows an overall higher level of C16:0, showing higher accumulation without the degradation.
C18:3n3 shows a similar pattern to the C16:0; there is an initial spike in the 0.3 M NaCl stressed culture above that of control conditions, and then a sharp decline as the culture starts to die and the fatty acids degrade or are otherwise lost from the samples.
C18:2cis is linoleic acid, and this is the other biggest contributor (with C16:0) to the FAMEs that also shows an increased quantity under salt stress. Linoleic acid is part of the two-stage desaturation process of oleic acid to linolenic acid [32]. Linolenic acid and the oleic and linoleic acid precursors are found in the photosynthetic tissue of the chloroplast, and linolenic and linoleic acids are the major unsaturated fatty acids in photosynthetic tissue [33]. This suggests that there are some changes taking place in photosynthetic activity under salt stress.
C16:0, or palmitic acid, is commonly found as an ester in triglycerides. It is also the first fatty acid formed during fatty acid synthesis [13]. An increase in this fatty acid suggests that new fatty acids may be being synthesized after the salt stressed culture reaches stationary phase, whilst they are not in the control culture. An alternative hypothesis is that they are being synthesized in both cultures but not used in the salt stressed culture, and that the control culture utilizes these fatty acids in the stationary phase and converts them to other fatty acids. However, if this were the case, we would expect to see the rise of other fatty acids in the later stages of the control cultures as the C16:0 chains are converted. No such effect was observed.
C18:0 rises over time with 0.2 and 0.3 M salt stress, after starting at a similar level to the control conditions. Stearic acid (C18:0) is a saturated fatty acid, from which unsaturated fatty acids can be derived within the algae cell. Since this coincides with the decline of many of the unsaturated C18 chains, it suggests that the conversion from saturated to unsaturated is halted, but that the C18:0 fatty acids are still being made in the cells despite the decaying culture.
C18:1cis shows an interesting pattern, as it does not show the same response in both the high salt conditions. High salt at 0.3 M NaCl causes an overall decline resulting in almost complete degradation of this fatty acid. 0.2 M NaCl salt shows a sharp increase in C18:1cis level after the first time point and continues to be higher than the control culture. It is clear from the growth data that this level of salt halts culture growth, but the microscopy suggests that the cells are still alive throughout this experiment. C18:1cis, therefore, may play a role in salt tolerance and surviving these high salinity conditions.
There was a steady decline of C17:0 and C18:2cis under high salt 0.2 and 0.3 M NaCl conditions, mirroring the overall pattern of total FAME content.
4. CONCLUSION
These data are the first published evidence of the effect of salt stress on the lipid profile of C. reinhardtii. Whilst many studies have investigated the effect of salinity stress on this species, only a single study [10] did so with any information of the effect on lipid content, but this study did not show any information on the lipid profile. Furthermore, their study is limited to 100 mM NaCl stress, and the wild-type strain only. The current study uses a starchless mutant and tests a range of salinities to show the effect of increasing salinity on the lipid profile and quantity in C. reinhardtii.
The ability of C. reinhardtii cultures to grow and function well in the presence of salt reaches a tolerance limit between concentrations 0.1 M and 0.2 M NaCl. 0.1 M NaCl grown cultures show an increase in total FAMEs during the stationary phase of the culture. In the higher salinities of 0.2 and 0.3 M NaCl, increases in the FAMEs occur at the first time point (t=3 hours) only, followed by decline over time. This shows a shock response to the higher salinity, which later changes as the cells are unable to tolerate the osmotic stress. The step up in salt from 0.1 to 0.2 M NaCl shows very different responses in all aspects of the biology investigated here; the difference between slow lipid accumulation in 0.1 M NaCl, versus the sudden steep lipid accumulation in 0.2 and 0.3 M NaCl, is very interesting. This will warrant deeper systems biology analysis to unravel the mechanism(s).
Investigation into FAME composition shows a complex relationship between salinity and fatty acid profile in the starchless mutant of C. reinhardtii. There are two main ways to think about the relevance of the lipid data for current research aims. The first way is how the lipids are responding and providing a biological role in the algal cultures. The second is how these particular cultures are providing information useful for biofuels research. The composition of fatty acids can have a large impact on biodiesel suitability [34], therefore these fluctuations in lipid profile impact the suitability of C. reinhardtii for biodiesel production.
The marked rise in total FAME content at the 3 hour time point in 0.3 M NaCl was identified as a sampling point for future proteomic investigation. This immediate and distinct rise demonstrates the ability of the cells to direct their resources rapidly into lipid components. It has been shown that many different stresses affect lipid content [35] and other species (such as Dunaliella salina) accumulate TAG in response to high salinity. There is some evidence from the data described in this study that lipids accumulate under particular salinity conditions in C. reinhardtii, with the level of salinity being key to this response.
The shift in the FA composition seen in these experiments shows an increase in most of the main C16, C17 and C18 chains, with the exception of C18:3n3, for 0.1 M NaCl conditions. For 0.3 M NaCl stress, C16:0 and C18:3n3 showed an initial increase at the elevated FAME time point of 3 hours, whilst C17:0, C18:0, C:18:1cis and C18:2cis do not show these increases. The decline of the health and lipid content of the cultures in 0.2 and 0.3 M NaCl past the initial time point show that beyond 3 hours these conditions are not suitable for algal culture or lipid production. The observed shift towards saturated and polyunsaturated fatty acids is not the desired shift for biodiesel (this would ideally be towards monounsaturated FAs [36, 37]), but there is still some advantage in both long term exposure to 0.1 M NaCl and short term exposure to 0.2 and 0.3 M NaCl causing overall lipid increases.
Using this information to explore lipid metabolism further in the model species could inform future work on manipulation of algae for biofuels research, especially as salinity is a condition that needs to be considered in large scale algal production such as raceway ponds.
ACKNOWLEDGEMENTS
Many thanks to the EPSRC (EP/E036252/1) and the Energy Futures DTC. Thanks to Joseph Longworth for his guidance on algal culturing and sampling. Thanks to Mark Jones for guidance and help in operating the GC.
Footnotes
Figure modified from “Lipid quantification techniques for screening oleaginous species of microalgae for biofuel production” by Hounslow et al. (2016) Eur. J. Lipid Sci. Technol., DOI 10.1002/ejlt.201500469.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
REFERENCES
- 1.Leite G.B., Abdelaziz A.E., Hallenbeck P.C. Algal biofuels: Challenges and opportunities. Bioresour. Technol. 2013;145:134–141. doi: 10.1016/j.biortech.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Fjerbaek L., Christensen K.V., Norddahl B. A review of the current state of biodiesel production using enzymatic transesterification. Biotechnol. Bioeng. 2009;102:1298–1315. doi: 10.1002/bit.22256. [DOI] [PubMed] [Google Scholar]
- 3.Gallagher B.J. The economics of producing biodiesel from algae. Renew. Energy. 2011;36:158–162. [Google Scholar]
- 4.Tercero E.A., Domenicali G., Bertucco A. Autotrophic production of biodiesel from microalgae: An updated process and economic analysis. Energy. 2014;76:807–815. [Google Scholar]
- 5.Kovacevic V., Wesseler J. Cost-effectiveness analysis of algae energy production in the EU. Energy Policy. 2010;38:5749–5757. [Google Scholar]
- 6.Jorquera O., Kiperstok A., Sales E.A., Embirucu M., Ghirardi M.L. Comparative energy life-cycle analyses of microalgal biomass production in open ponds and photobioreactors. Bioresour. Technol. 2010;101:1406–1413. doi: 10.1016/j.biortech.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 7.Longworth J., Noirel J., Pandhal J., Wright P.C., Vaidyanathan S. HILIC- and SCX-based quantitative proteomics of Chlamydomonas reinhardtii during nitrogen starvation induced lipid and carbohydrate accumulation. J. Proteome Res. 2012;11:5959–5971. doi: 10.1021/pr300692t. [DOI] [PubMed] [Google Scholar]
- 8.Chen M., Tang H., Ma H., Holland T.C., Ng K.Y., Salley S.O. Effect of nutrients on growth and lipid accumulation in the green algae Dunaliella tertiolecta. Bioresour. Technol. 2011;102:1649–1655. doi: 10.1016/j.biortech.2010.09.062. [DOI] [PubMed] [Google Scholar]
- 9.Khozin-Goldberg I., Cohen Z. The effect of phosphate starvation on the lipid and fatty acid composition of the fresh water eustigmatophyte Monodus subterraneus. Phytochemistry. 2006;67:696–701. doi: 10.1016/j.phytochem.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Siaut M., Cuine S., Cagnon C., et al. Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 2011;11:1–15. doi: 10.1186/1472-6750-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pal D., Khozin-Goldberg I., Cohen Z., Boussiba S. The effect of light, salinity, and nitrogen availability on lipid production by Nannochloropsis sp. Appl. Microbiol. Biotechnol. 2011;90:1429–1441. doi: 10.1007/s00253-011-3170-1. [DOI] [PubMed] [Google Scholar]
- 12.Guschina I.A., Harwood J.L. Lipids and lipid metabolism in eukaryotic algae. Prog. Lipid Res. 2006;45:160–186. doi: 10.1016/j.plipres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 13.Harwood J.L., Guschina I.A. The versatility of algae and their lipid metabolism. Biochimie. 2009;91:679–684. doi: 10.1016/j.biochi.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 14.Leon-Banares R., Gonzalez-Ballester D., Galvan A., Fernandez E. Transgenic microalgae as green cell-factories. Trends Biotechnol. 2004;22:45–52. doi: 10.1016/j.tibtech.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Cakmak T., Angun P., Demiray Y.E., Ozkan A.D., Elibol Z., Tekinay T. Differential effects of nitrogen and sulfur deprivation on growth and biodiesel feedstock production of Chlamydomonas reinhardtii. Biotechnol. Bioeng. 2012;109:1947–1957. doi: 10.1002/bit.24474. [DOI] [PubMed] [Google Scholar]
- 16.Sharma K.K., Schuhmann H., Schenk P.M. High lipid induction in microalgae for biodiesel production. Energies. 2012;5:1532–1553. [Google Scholar]
- 17.Bartley M.L., Boeing W.J., Corcoran A.A., Holguin F.O., Schaub T. Effects of salinity on growth and lipid accumulation of biofuel microalga Nannochloropsis salina and invading organisms. Biomass Bioenergy. 2013;54:83–88. [Google Scholar]
- 18.Xia L., Rong J.F., Yang H.J., He Q.N., Zhang D.L., Hu C.X. NaCl as an effective inducer for lipid accumulation in freshwater microalgae Desmodesmus abundans. Bioresour. Technol. 2014;161:402–409. doi: 10.1016/j.biortech.2014.03.063. [DOI] [PubMed] [Google Scholar]
- 19.Rao A.R., Dayananda C., Sarada R., Shamala T.R., Ravishankar G.A. Effect of salinity on growth of green alga Botryococcus braunii and its constituents. Bioresour. Technol. 2007;98:560–564. doi: 10.1016/j.biortech.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Venkata M.S., Devi M.P. Salinity stress induced lipid synthesis to harness biodiesel during dual mode cultivation of mixotrophic microalgae. Bioresour. Technol. 2014;165:288–294. doi: 10.1016/j.biortech.2014.02.103. [DOI] [PubMed] [Google Scholar]
- 21.Takagi M. Karseno, Yoshida T. Effect of salt concentration on intracellular accumulation of lipids and triacylglyceride in marine microalgae Dunaliella cells. J. Biosci. Bioeng. 2006;101:223–226. doi: 10.1263/jbb.101.223. [DOI] [PubMed] [Google Scholar]
- 22.Ahn J.W., Hwangbo K., Yin C.J., et al. Salinity-dependent changes in growth and fatty acid composition of new Arctic Chlamydomonas species, ArM0029A. Plant Cell Tissue Organ Cult. 2015;120:1015–1021. [Google Scholar]
- 23.An M., Mou S., Zhang X., et al. Expression of fatty acid desaturase genes and fatty acid accumulation in Chlamydomonas sp. ICE-L under salt stress. Bioresour. Technol. 2013;149:77–83. doi: 10.1016/j.biortech.2013.09.027. [DOI] [PubMed] [Google Scholar]
- 24.Lu N., Jiang X.L., Chen F., Yang S.T. Regulation of lipid metabolism in the snow alga Chlamydomonas nivalis in response to NaCl stress: An integrated analysis by cytomic and lipidomic approaches. Process Biochem. 2012;47:1163–1170. [Google Scholar]
- 25.Lu N., Wei D., Jiang X.L., Chen F., Yang S.T. Fatty acids profiling and biomarker identification in snow alga Chlamydomonas nivalis by NaCl stress using GC/MS and multivariate statistical analysis. Anal. Lett. 2012;45:1172–1183. [Google Scholar]
- 26.Lu N., Wei D., Chen F., Yang S.T. Lipidomic profiling and discovery of lipid biomarkers in snow alga Chlamydomonas nivalis under salt stress. Eur. J. Lipid Sci. Technol. 2012;114:253–265. [Google Scholar]
- 27.Salama E.S., Kim H.C., Abou-Shanab R.I., et al. Biomass, lipid content, and fatty acid composition of freshwater Chlamydomonas mexicana and Scenedesmus obliquus grown under salt stress. Bioprocess Biosyst. Eng. 2013;36:827–833. doi: 10.1007/s00449-013-0919-1. [DOI] [PubMed] [Google Scholar]
- 28.James G.O., Hocart C.H., Hillier W., et al. Fatty acid profiling of Chlamydomonas reinhardtii under nitrogen deprivation. Bioresour. Technol. 2011;102:3343–3351. doi: 10.1016/j.biortech.2010.11.051. [DOI] [PubMed] [Google Scholar]
- 29.Leon R., Galvan F. Halotolerance studies on Chlamydomonas reinhardtii glycerol excretion by free and immobilized cells. J. Appl. Phycol. 1994;6:13–20. [Google Scholar]
- 30.Affenzeller M.J., Darehshouri A., Andosch A., Lütz C., Lütz-Meindl U. Salt stress-induced cell death in the unicellular green alga Micrasterias denticulata. J. Exp. Bot. 2009;60:939–954. doi: 10.1093/jxb/ern348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batterto J.C., Vanbaale C. Growth responses of blue-green algae to sodium chloride concentration. Arch. Mikrobiol. 1971;76:151–165. doi: 10.1007/BF00411789. [DOI] [PubMed] [Google Scholar]
- 32.Cherif A., Dubacq J., Mache R., Oursel A., Tremolieres A. Biosynthesis of α-linolenic acid by desaturation of oleic and linoleic acids in several organs of higher and lower plants and in algae. Phytochemistry. 1975;14:703–706. [Google Scholar]
- 33.Harris R.V., James A.T. Linoleic, α-linolenic acid biosynthesis in plant leaves and a green alga. Biochim. Biophys. Acta. 1965;106:456–464. doi: 10.1016/0005-2760(65)90062-7. [DOI] [PubMed] [Google Scholar]
- 34.Islam M.A., Ayoko G.A., Brown R., Stuart D., Heimann K. Influence of fatty acid structure on fuel properties of algae derived biodiesel. Procedia Eng. 2013;56:591–596. [Google Scholar]
- 35.Solovchenko A.E. Physiological role of neutral lipid accumulation in eukaryotic microalgae under stresses. Russ. J. Plant Physiol. 2012;59:167–176. [Google Scholar]
- 36.Stansell G.R., Gray V.M., Sym S.D. Microalgal fatty acid composition: implications for biodiesel quality. J. Appl. Phycol. 2012;24:791–801. [Google Scholar]
- 37.Knothe G. Improving biodiesel fuel properties by modifying fatty ester composition. Energy Environ. Sci. 2009;2:759–766. [Google Scholar]