To the Editor
Information on research funding is important to various groups, including investigators, policy analysts, advocacy organizations and, of course, the funding agencies themselves. But informatics resources devoted to research funding are currently limited. In particular, there is a need for information on grants from the US National Institutes of Health (NIH), the world’s largest single source of biomedical research funding, because of its large number of awards (~80,000 each year) and its complex organizational structure. NIH’s 25 grant-awarding Institutes and Centers have distinct but overlapping missions, and the relationship between these missions and the research they fund is multifaceted. Because there is no comprehensive scheme that characterizes NIH research, navigating the NIH funding landscape can be challenging.
At present, NIH offers information on awarded grants via the RePORTER website (http://projectreporter.nih.gov). For each award, RePORTER provides keyword tags, plus ~215 categorical designations assigned to grants via a partially automated system known as the NIH research, condition and disease categorization (RCDC) process (http://report.nih.gov/rcdc/categories). But keyword searches are not optimal for various information needs and analyses, and the RCDC categories are only intended to meet specific NIH reporting requirements, rather than to comprehensively characterize the entire NIH research portfolio.
To facilitate navigation and discovery of NIH-funded research, we created a database (https://app.nihmaps.org/) in which we use text mining to extract latent categories and clusters from NIH grant titles and abstracts. This categorical information is discovered using two unsupervised machine-learning techniques. The first is topic modeling, a Bayesian statistical method that discerns meaningful categories from unstructured text (see Supplementary Methods for references). The second is a graph-based clustering method that produces a two-dimensional visualized output, in which grants are grouped based on their overall topic-and word-based similarity to one another. The database allows specific queries within a contextual framework that is based on scientific research rather than NIH administrative and categorical designations.
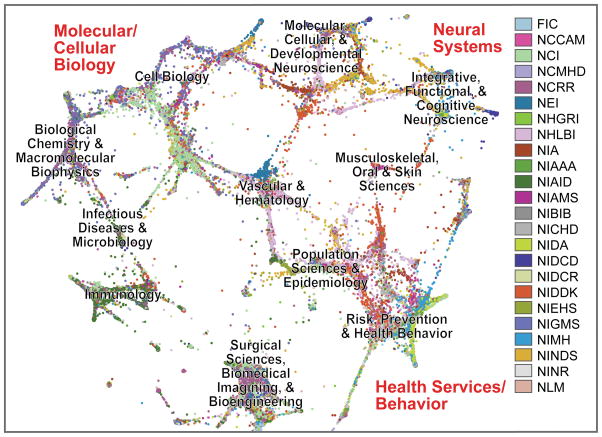
We found that topic-based categories are not strictly associated with the missions of individual Institutes but instead cut across the NIH, albeit in varying proportions consistent with each Institute’s distinct mission (Supplementary Table 1). The graphical map layout (Fig. 1) shows a global research structure that is logically coherent but only loosely related to Institute organization (Supplementary Table 1).
Figure 1. Graphically clustered NIH grants, as rendered from a screenshot of the NIHMaps user interface.
NIH awards (here showing grants from 2010; ~80,000 documents) were scored for their overall topic and word similarity, and the resulting document distance calculations were used to seed a graphing algorithm. Grants are represented as dots, color-coded by NIH Institute and are clustered based on shared thematic content. For acronyms and separate views with each Institute highlighted, see the legend for Supplementary Table 1. Labels in black were automatically derived from review assignments of the underlying documents. Labels in red indicate a global structure that was reproducible using multiple different algorithm settings.
We describe four example use cases (Supplementary Data). First, we show a query using an algorithm-derived category relevant to angiogenesis (Supplementary Fig. 1). Unlike standard keyword-based searches, this type of query allows retrieval of grants that are truly focused on a particular research area. In addition, the resulting graphical clusters reveal clear patterns in the relationships between the retrieved grants and the multiple Institutes funding this research. Second, we examine an NIH peer review study section. The database categories and clusters clarify the complex relationship between the NIH Institutes and the centralized NIH peer review system, which is distinct and independent from the Institutes. Third, we show an analysis of the NIH RCDC category ‘sleep research’ in conjunction with the database topics, the latter providing salient categorical information in greater detail than the officially reported category. Finally, we show how the database can be used for unbiased discovery of research trends, and we document the remarkable increase in funding for research on microRNA biology from 2007 to 2009. Changes in topics associated with this burgeoning area demonstrate a transition in the nature of the research, from basic cellular and molecular biology to investigations of complex physiological processes and disease diagnoses.
In each case, the machine-learned topics are robustly correlated with funding by specific NIH Institutes, highlighting the importance of the underlying categories to the NIH. The patterns elucidated in this framework are consistent with Institute policies, but obtaining similar information in the absence of the current database would require extensive exploration of Institute websites, followed by time-consuming research on appropriate keywords for queries of specific categories. Our database offers an alternative approach that enables rapid and reproducible retrieval of meaningful categorical information.
To ensure transparent and accurate representations of the algorithm-derived topics, we provide extensive contextual information derived from the documents associated with each topic, in a format conducive to spot checks and to detailed examination for cases requiring precise categorical distinctions. Additionally, we implemented a new technique for automatically assessing topic quality using statistics of topic word co-occurrence (Supplementary Methods), which we used for curating the database to identify poor quality topics.
Our use of this graphing algorithm is somewhat different from previous gene expression analyses and scientometric studies based on journal citation linkages (see Supplementary Methods for references). We assessed the information-retrieval capabilities of the graphs and found that they performed well relative to the document similarity measures that served as inputs. Notably, rather than forming isolated clusters, in this case the algorithm produced a lattice-like structure, in which clusters are linked by strings of aligned documents whose topical content is jointly relevant to the clusters at either end of each string (Supplementary Fig. 1). In addition to providing extra ‘subcluster’ resolution of content that falls between clusters, this lattice-like framework formed a logical organizational structure, merging the local, intermediate and global levels of the graph.
The categories and clusters represented in this database are comprehensive and thus provide reference points from which various information requirements can be addressed by users with divergent interests and needs. Perhaps more importantly, they provide a basis for discovery of interrelationships among concepts and documents that otherwise would be obscure.
Supplementary Material
The word “angiogenesis” was typed into the topic query field in the user interface, which activated an auto-populate function that displayed the top ten words for the corresponding topic. The word list is shown in the upper left corner of the panel. A topic threshold setting of 10% was applied in the user interface (see Supplementary Data for a description of topic-based query design). This setting resulted in a retrieval of ~350 documents from twelve different NIH Institutes, with the top seven Institutes shown in the histogram at the lower left. The retrieved grants are represented as push-pin markers color-coded by Institute; >75% were located in the region of the graph demarcated by the red dashes, whereas the remaining 25% were located in other clusters appropriate to their topical focus. The grants in the demarcated region were organized as a string between clusters focused on cancer mechanisms/therapeutics in the upper left, and vascularization of specific tissues in the lower right. This organization highlights the lattice-like structure of the graph layout, in which major clusters are connected by strings of documents with joint focus between the clusters at either end of the string, providing “between-cluster” resolution and elucidating the layout organization.
Each topic has a dedicated page that provides extended topic related data, for the purpose of spot checks, or for detailed investigations in cases where precise categorical distinctions are needed. A: The top window includes topic words, title words, and extracted phrases, displayed with font grayscale in proportion to per-topic probability. B: Co-occurring topics and semantically similar topics, with links to the corresponding topic pages, enable accurate determination of the categorical distinctions inferred by the algorithm. C: Left panel shows associated thesaurus data from the three different portions of the modeled corpus: NIH RCDC Concepts (for grants starting in 2007), NIH CRISP (for grants through 2006), and PubMed MeSH. A listing of associated PubMed journals is also included (not shown). Right panel shows NIH grants data, including Institutes, grant mechanisms, Review Panels, and Program Announcements. D: The bottom panel contains a full list of associated documents in descending order of per-document proportional allocation (only a partial list is shown here), which allows determination of the threshold above which documents are relevant to a given information need.
Panels A and B show two high-resolution views of a single region of the graph layout (red dashed box), labeled by (A) topic words, and (B) the predominant study sections from the underlying grants. Five of the six study sections in B are from the “Integrative, Functional, and Cognitive Neuroscience” Integrated Review Group (described at http://cms.csr.nih.gov/PeerReviewMeetings/CSRIRGDescriptionNew/IFCNIRG/). Note that the two predominant clusters (covering pain and reward/addiction, respectively) are connected in the layout’s lattice-like structure by grants devoted to research on opiates, a research topic relevant to both clusters. Panel C shows a larger region of the graph (blue dashed box) with results from a query for grants reviewed by one of the panels, the Somatosensory & Chemosensory Systems Study Section. Restricting this query to the region marked by the red dashed box (corresponding to the region shown in Panels A and B), using a bounding box feature in the graphical user interface, reveals topics and clusters that are not readily apparent from divisions between study sections, but are highly relevant to NIH Institute funding patterns. For acronyms and descriptions of NIH Institutes, see the Legend following Supplementary Table 1.
We queried the topic database for the NIH RCDC Category “Sleep Research,” and restricted the query, in succession, to each of the four indicated topics. We used two different topic threshold cutoffs, 5% and 10%, to compile an estimated range of grants for each topic. In this analysis the topic assignments to grants are not mutually exclusive, and therefore for each Institute, the sum of the four topics is usually somewhat larger than the total number of awards. Note that the table shows clear divergences in funding patterns among the Institutes (for acronyms and Institute descriptions see Supplementary Table 1).
We queried the database for awards from 2009 classified under the NIH RCDC Category “Sleep Research.” Panel A shows the layout of the retrieved grant set, with >90% of the awards falling on the right side of the graph. The lower left inset shows the top four topics associated with this category, while the two insets on the right show detail of two predominant clusters, which together account for ~56% of the awards. The two selected clusters were analyzed using a bounding box feature of the user interface, revealing distinct Institute representations and research emphases. As seen in Panel B, the top cluster is focused on circadian biology, with prominent funding from NIGMS, whereas the bottom cluster (Panel C) is focused on sleep disorders, neurobiology of sleep/arousal, and sleep-disordered breathing, with prominent funding from NHLBI. Relative differences in NIH Institute representations between these clusters reflect divergences in funding priorities between these Institutes that are not apparent in the NIH reporting category.
As an initial assessment of research trends represented in the NIH grants topic database, we screened for topics whose relative allocations increased between 2007 and 2009. Shown here is further characterization of the top “hit” from this screen, a topic focused on microRNA biology. Top panels: Non-competing awards from 2007 (i.e., awards based on competing applications from years prior to 2007) are clustered in a region associated with RNA biology (with a smaller cluster focused on cell differentiation), and their topic composition reveals a weighting towards basic science (consistent with prominent funding by NIGMS). Bottom panels: New awards from 2009 are more broadly distributed, with a particular emphasis on cancer biomarkers (consistent with prominent NCI funding). Note that this ~2.5fold increase in retrieved awards contrasts with a markedly opposite proportion of total awards from the two queried sets (~36,000 non-competing vs. ~15,000 new awards), thus indicating an even more substantial proportional increase in the grants on microRNA biology.
In separate experiments we found a strong correlation between topic word co-occurrence within documents and rater evaluations of topic quality. We therefore used this metric for assessment and curation of topics in the current model. Panel A shows a word co-occurrence matrix for a good quality topic. Gray numbers in the diagonal represent document frequency for the corresponding words at the left; off-diagonals represent document co-occurrence frequency for the corresponding word pairs. Word co-occurrence scores were calculated as the unweighted sums of ln(co-occurrence ÷ occurrence) for the top fifteen words in the topic. Panel B shows a corresponding matrix for a poor quality topic; numbers in red reflect low incidence of co-occurrence. Panel C shows all 700 topics plotted for co-occurrence score against topic size (i.e., fraction of the aggregate corpus assigned to a given topic), which we found to correlate inversely with topic quality. Topics in red and yellow were deemed sufficiently poor that they received special indicators in the user interface (see text for details).
We assessed the graph output relative to its input similarity values, using rank order retrieval as a basis for comparison. Panel A shows an example query using the top 100 most similar documents for a single grant. Approximately half of these similar documents were quite proximal on the graph (46 documents in cluster “b”, where the initial grant was located). The other half were dispersed, but note that they were clustered rather than randomly distributed, in groups corresponding to the topic mix of the initiating grant. Scale bars are in arbitrary Graph Units (GU), which relate the example to subsequent analysis. Panel B shows the same analysis for the full grant set, assessing the top 1, 10, 100 and 300 similar grants for their distance on the graph. Median values are labeled, and indicate that half the grants were quite proximal (within 10 graph units for n=100 similar documents), but the other half were widespread, consistent with the example in A. Panel C shows distances for the rank ordered closest grants on the graph, which were used as a basis for comparing graph-based retrieval performance to document similarity-based retrieval (below).
Pair-wise similarity scores (red circles) and graph distances (blue diamonds) were compared at four rank ordered intervals for their ability to retrieve documents with the same top scoring topic (A) or the same NIH Program Code assignment (B). Values are averages over the set of initiating grants. Because of the size of the dataset (~88,000), standard error measurements were too small for representation in the plots.
Acknowledgments
We acknowledge assistance and support from G. LaRowe and N. Skiba at ChalkLabs, and input and feedback from NIH staff during the project. We thank S. Silberberg, C. Cronin, K. Boyack and K. Borner for helpful advice and comments on the manuscript.
This project has been supported through small contracts from the NIH to:
University of Southern California (271200900426P and 271200900244P)
University of Massachusetts (271201000758P, 271200900640P, 271201000704P and 271200900639P)
ChalkLabs LLC (271200900695P and 271201000701P)
TopicSeek LLC (271201000620P and 271200900637P).
Footnotes
Competing Financial Interests
B.W.H. II is employed by Chalklabs, LLC, which was contracted for a portion of the work described in the report. D.N. is owner of TopicSeek, LLC, which was also contracted for a portion of the described work.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The word “angiogenesis” was typed into the topic query field in the user interface, which activated an auto-populate function that displayed the top ten words for the corresponding topic. The word list is shown in the upper left corner of the panel. A topic threshold setting of 10% was applied in the user interface (see Supplementary Data for a description of topic-based query design). This setting resulted in a retrieval of ~350 documents from twelve different NIH Institutes, with the top seven Institutes shown in the histogram at the lower left. The retrieved grants are represented as push-pin markers color-coded by Institute; >75% were located in the region of the graph demarcated by the red dashes, whereas the remaining 25% were located in other clusters appropriate to their topical focus. The grants in the demarcated region were organized as a string between clusters focused on cancer mechanisms/therapeutics in the upper left, and vascularization of specific tissues in the lower right. This organization highlights the lattice-like structure of the graph layout, in which major clusters are connected by strings of documents with joint focus between the clusters at either end of the string, providing “between-cluster” resolution and elucidating the layout organization.
Each topic has a dedicated page that provides extended topic related data, for the purpose of spot checks, or for detailed investigations in cases where precise categorical distinctions are needed. A: The top window includes topic words, title words, and extracted phrases, displayed with font grayscale in proportion to per-topic probability. B: Co-occurring topics and semantically similar topics, with links to the corresponding topic pages, enable accurate determination of the categorical distinctions inferred by the algorithm. C: Left panel shows associated thesaurus data from the three different portions of the modeled corpus: NIH RCDC Concepts (for grants starting in 2007), NIH CRISP (for grants through 2006), and PubMed MeSH. A listing of associated PubMed journals is also included (not shown). Right panel shows NIH grants data, including Institutes, grant mechanisms, Review Panels, and Program Announcements. D: The bottom panel contains a full list of associated documents in descending order of per-document proportional allocation (only a partial list is shown here), which allows determination of the threshold above which documents are relevant to a given information need.
Panels A and B show two high-resolution views of a single region of the graph layout (red dashed box), labeled by (A) topic words, and (B) the predominant study sections from the underlying grants. Five of the six study sections in B are from the “Integrative, Functional, and Cognitive Neuroscience” Integrated Review Group (described at http://cms.csr.nih.gov/PeerReviewMeetings/CSRIRGDescriptionNew/IFCNIRG/). Note that the two predominant clusters (covering pain and reward/addiction, respectively) are connected in the layout’s lattice-like structure by grants devoted to research on opiates, a research topic relevant to both clusters. Panel C shows a larger region of the graph (blue dashed box) with results from a query for grants reviewed by one of the panels, the Somatosensory & Chemosensory Systems Study Section. Restricting this query to the region marked by the red dashed box (corresponding to the region shown in Panels A and B), using a bounding box feature in the graphical user interface, reveals topics and clusters that are not readily apparent from divisions between study sections, but are highly relevant to NIH Institute funding patterns. For acronyms and descriptions of NIH Institutes, see the Legend following Supplementary Table 1.
We queried the topic database for the NIH RCDC Category “Sleep Research,” and restricted the query, in succession, to each of the four indicated topics. We used two different topic threshold cutoffs, 5% and 10%, to compile an estimated range of grants for each topic. In this analysis the topic assignments to grants are not mutually exclusive, and therefore for each Institute, the sum of the four topics is usually somewhat larger than the total number of awards. Note that the table shows clear divergences in funding patterns among the Institutes (for acronyms and Institute descriptions see Supplementary Table 1).
We queried the database for awards from 2009 classified under the NIH RCDC Category “Sleep Research.” Panel A shows the layout of the retrieved grant set, with >90% of the awards falling on the right side of the graph. The lower left inset shows the top four topics associated with this category, while the two insets on the right show detail of two predominant clusters, which together account for ~56% of the awards. The two selected clusters were analyzed using a bounding box feature of the user interface, revealing distinct Institute representations and research emphases. As seen in Panel B, the top cluster is focused on circadian biology, with prominent funding from NIGMS, whereas the bottom cluster (Panel C) is focused on sleep disorders, neurobiology of sleep/arousal, and sleep-disordered breathing, with prominent funding from NHLBI. Relative differences in NIH Institute representations between these clusters reflect divergences in funding priorities between these Institutes that are not apparent in the NIH reporting category.
As an initial assessment of research trends represented in the NIH grants topic database, we screened for topics whose relative allocations increased between 2007 and 2009. Shown here is further characterization of the top “hit” from this screen, a topic focused on microRNA biology. Top panels: Non-competing awards from 2007 (i.e., awards based on competing applications from years prior to 2007) are clustered in a region associated with RNA biology (with a smaller cluster focused on cell differentiation), and their topic composition reveals a weighting towards basic science (consistent with prominent funding by NIGMS). Bottom panels: New awards from 2009 are more broadly distributed, with a particular emphasis on cancer biomarkers (consistent with prominent NCI funding). Note that this ~2.5fold increase in retrieved awards contrasts with a markedly opposite proportion of total awards from the two queried sets (~36,000 non-competing vs. ~15,000 new awards), thus indicating an even more substantial proportional increase in the grants on microRNA biology.
In separate experiments we found a strong correlation between topic word co-occurrence within documents and rater evaluations of topic quality. We therefore used this metric for assessment and curation of topics in the current model. Panel A shows a word co-occurrence matrix for a good quality topic. Gray numbers in the diagonal represent document frequency for the corresponding words at the left; off-diagonals represent document co-occurrence frequency for the corresponding word pairs. Word co-occurrence scores were calculated as the unweighted sums of ln(co-occurrence ÷ occurrence) for the top fifteen words in the topic. Panel B shows a corresponding matrix for a poor quality topic; numbers in red reflect low incidence of co-occurrence. Panel C shows all 700 topics plotted for co-occurrence score against topic size (i.e., fraction of the aggregate corpus assigned to a given topic), which we found to correlate inversely with topic quality. Topics in red and yellow were deemed sufficiently poor that they received special indicators in the user interface (see text for details).
We assessed the graph output relative to its input similarity values, using rank order retrieval as a basis for comparison. Panel A shows an example query using the top 100 most similar documents for a single grant. Approximately half of these similar documents were quite proximal on the graph (46 documents in cluster “b”, where the initial grant was located). The other half were dispersed, but note that they were clustered rather than randomly distributed, in groups corresponding to the topic mix of the initiating grant. Scale bars are in arbitrary Graph Units (GU), which relate the example to subsequent analysis. Panel B shows the same analysis for the full grant set, assessing the top 1, 10, 100 and 300 similar grants for their distance on the graph. Median values are labeled, and indicate that half the grants were quite proximal (within 10 graph units for n=100 similar documents), but the other half were widespread, consistent with the example in A. Panel C shows distances for the rank ordered closest grants on the graph, which were used as a basis for comparing graph-based retrieval performance to document similarity-based retrieval (below).
Pair-wise similarity scores (red circles) and graph distances (blue diamonds) were compared at four rank ordered intervals for their ability to retrieve documents with the same top scoring topic (A) or the same NIH Program Code assignment (B). Values are averages over the set of initiating grants. Because of the size of the dataset (~88,000), standard error measurements were too small for representation in the plots.