Abstract
Toll-like receptors (TLRs) are recognized as fundamental contributors to the immune system function against infections. Hepatitis C virus (HCV) infection represents a global health problem especially in Egypt having the highest HCV prevalence worldwide where HCV infection is a continuing epidemic. The aim of the present study was to investigate the possible association between genetic variation in TLR-3 and TLR-9 and HCV infection and hepatic fibrosis in chronic HCV-positive Egyptian patients. The present study included 100 naïve chronic HCV-positive patients and 100 age- and sex-matched healthy controls. Genotyping of TLR-3 (_7 C/A [rs3775296]), TLR-3 (c.1377C/T [rs3775290]) and TLR-9 (1237T/C [rs5743836]) were done by polymerase chain reaction restriction fragment length polymorphism technique. Frequency of polymorphic genotypes in TLR-3 (_7 C/A), TLR-3 (c.1377C/T) and TLR-9 (1237T/C) were not significantly different between studied HCV-positive patients and controls with P values 0.121, 0.112, and 0.683, respectively. TLR-3 c.1377 T-allele was associated with advanced stage of hepatic fibrosis (P = 0.003).
Introduction
Hepatitis C virus (HCV) infection is a multifactorial disease representing a major health problem in Egypt where the prevalence is almost 10-fold higher than that in other countries.1 The immune system is an essential determinant of viral infection outcome.2 Interactions between the viruses, hepatocytes, and the host immune systems may determine viral persistence and disease progression.3
Toll-like receptors (TLRs) are the fundamental component of the innate immune system and key regulators of acquired immunity. In humans, 10 TLRs proteins (TLR1–10) have been identified,4 that possess different subcellular localization depending on the specific pathogen-associated molecular patterns or damage-associated molecular patterns they recognize,5 thus called pattern recognition receptors. TLRs 1, 2, 4, 5, 6, and 10 are found on the extracellular surface of cells, whereas TLRs 3, 7, 8, and 9 are nucleic acid sensors located within the endoplasmic reticulum and endosomes.6,7 TLRs 3 and 9 recognize microbial nucleic acid molecular patterns,8–11 TLR-3 identifies viral RNA,12 whereas TLR-9 is specific for unmethylated cytosine–phosphate–guanine dinucleotide motifs in bacterial and viral pathogen DNA.8
TLRs have an important role in pathogen recognition and subsequently immune system activation.8,13,14 They trigger inflammatory cytokines production through nuclear factor-κB-dependent or interferon (IFN) regulatory factor (IRF)–dependent signaling pathways. Activation of TLR by pathogen binding stimulates inflammatory cytokines production that triggers induction of type I IFNs.15,16 Type I IFNs (IFN-α, IFN-β) have potent antiviral properties as they interfere with virus replication and possess immunomodulatory activities by promoting multiple immune functions.17–20
Several data support the role of single nucleotide polymorphisms (SNPs) in TLR genes in modulating the risk of viral and bacterial infections. SNPs may alter promoter activity affecting gene expression, messenger RNA (mRNA) conformation and stability, or protein structure and function.21 It has been suggested that promoter polymorphisms in TLR-3 (c.1377C/T, _7 C/A) influence gene expression in response to inflammatory cytokines and causes transcriptional modulation in TLR-3.22 Human TLR-3 promoter region contains a functional IFN-stimulated response element/IRF element.23 TLR-9 1237T/C SNP confers regulatory effects on TLR-9 transcription; higher transcriptional activity was seen in the presence of the CC allelic variant with moderate increase in promoter activity.24 The −1237T/C-C allele generates several regulatory sites, including an IL-6-responsive element and creation of an NF-κB binding site.25,26
HCV infection adds great financial burden on the health sector especially in Egypt, having the highest prevalence of HCV worldwide. HCV-induced end-stage liver disease has become an increasing cause of mortality in Egypt in the last decades.27 “Know your epidemic, know your response” concept necessitates the study of every aspect of the disease that may help in controlling disease dissemination in the community or disease progression which compromises the life quality of chronic HCV-infected patients.
The genetic makeup of the host plays an important role in susceptibility to infections.28,29 In the present work, we studied SNP in TLR-3 (c.1377C/T [rs3775290]), TLR-3 (_7 C/A [rs3775296]), and TLR-9 (1237T/C [rs5743836]) SNPs in 100 naïve chronic HCV-positive Egyptian patients to clarify the role of TLR-3 and TLR-9 polymorphism in HCV infection and the degree of hepatic fibrosis.
Subjects and Methods
Two hundred participants were included in the study; 100 naïve chronic HCV-positive patients and 100 age- and sex-matched normal healthy individuals as control group. Informed consent was obtained from all participants before enrollment, the study was performed in accordance with the Declaration of Helsinki, and the protocols were approved by the Faculty of Medicine, Cairo University Ethics Committee.
HCV infection was diagnosed by anti-HCV antibodies testing and detection of HCV-RNA (Applied Biosystems, Foster City, CA). All patients had no coinfection with hepatitis B virus and no other causes of chronic liver disease.
Liver biopsy, clinical and laboratory evaluation.
Liver biopsy specimens were obtained from all patients included in the study. Histopathologic features of fibrosis and activity were scored according to the Metavir scoring system.30 Fibrosis was staged on a scale of 0–4.
Polymorphism analysis of TLR-3 c.1377C/T (rs3775290), TLR-3 _7 C/A (rs3775296), and TLR-9 1237T/C (rs5743836).
Genomic DNA extraction was done from peripheral blood samples using Gene JET Whole Blood Genomic DNA purification Mini kit (Thermo Scientific, Waltham, MA) according to the manufacturer's instructions. Isolated DNA was stored at −70°C until used for polymerase chain reaction (PCR) amplification. Polymorphism analysis was done by PCR–restriction fragment length polymorphism technique. All PCR reactions were performed in a total volume of 25 μL containing 150 ng genomic DNA, 2X Taq Green PCR Master Mix, 25 pM each of forward and reverse primers (Biosearch Technologies, Novata, CA). PCR amplification was carried out in the DNA thermal cycler (PTC programmable thermal controller; MJ Research, Watertown, MA). Amplification conditions were initial denaturation at 95°C for 5 minutes followed by 35 cycles of 95°C for 45 seconds, * for 45 seconds, and 72°C for 30 seconds, with final extension for 7 minutes at 72°C (* 55°C for TLR-3 c.1377C/T and TLR-9 1237T/C and 59°C for TLR-3 _7 C/A). The amplified PCR products were visualized by 4% agarose gel electrophoresis under ultraviolet light. The primers sequences that were used and amplified product size are shown in Table 1.
Table 1.
Summary of polymerase chain reaction restriction fragment length polymorphism assay
| Polymorphism | Primer sequence (5′–3′) | Length (bp) | Restriction enzyme | Fragments size (bp) | Reference |
|---|---|---|---|---|---|
| TLR-3 _7 C/A (rs3775296) | GCATTTGAAAGCCATCTGCT | 279 | MboII | AA: 257, 17 | 31 |
| AAGTTGGCGGCTGGTAATCT | AC: 279, 257, 17 | ||||
| CC: 279 | |||||
| TLR-3 c.1377C/T (rs3775290) | CCAGGCATAAAAAGCAATATG | 337 | TaqI | CC: 274, 63 | 31 |
| GGACCAAGGCAAAGGAGTTC | CT: 337, 274, 63 | ||||
| TT: 337 | |||||
| TLR-9 1237T/C (rs5743836) | ATGGGAGCAGAGACATAATGGA | 135 | BstNI | TT: 108, 27 | 21 |
| CTGCTTGCAGTTGACTGTGT | TC:108, 60, 48, 27 | ||||
| CC: 60, 48, 27 |
Digestion of the amplified product by specific restriction enzyme for each polymorphism was done as follows: 10 μL of the amplified product was mixed with 1 μL restriction enzyme (Thermo Scientific, Waltham, MA) and the mixture was incubated for 10 minutes (at 37°C for MboII and BstNI and at 65°C for TaqI). The product was analyzed by gel electrophoresis using 4% agarose gel (Promega, Madison, WI). The separated fragments were stained with ethidium bromide and visualized along with a 50-base pair (bp) ladder (MBI Fermentas, Vilnius, Lithuania) as a size marker using transilluminator (Bio-Rad). The restriction enzyme used and the resulting bp length are shown in Table 1, Figures 1–3
Figure 2.
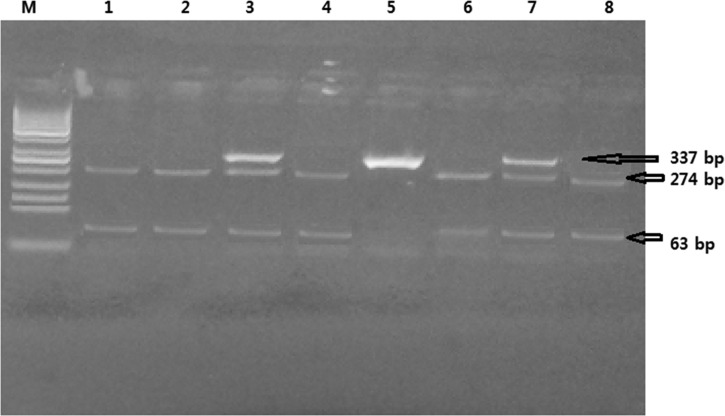
Characterization of the TLR-3 c.1377C/T polymorphism using Taq I restriction enzyme. Ethidium bromide–stained 4% agarose gel. Cases 1, 2, 4, 6, and 8 show homozygous CC (wild genotype): two bands—274 and 63 bp were detected. Cases 3 and 7 show heterozygous CT genotype: three bands—337, 274, and 63 were detected. Case 5 shows homozygous TT genotype: one band—337 was detected. M: polymerase chain reaction marker (50-100-150-200-250-300-350-400 bp, etc.).
.
Figure 1.
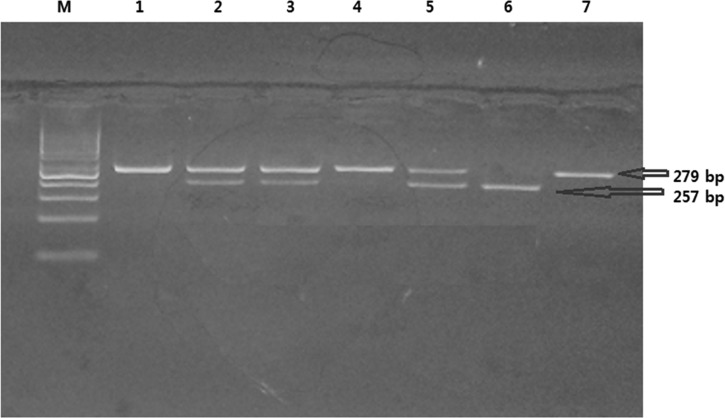
Characterization of the TLR-3 _7 C/A polymorphism using MboII restriction enzyme. Ethidium bromide–stained 4% agarose gel. Cases 1, 4, and 7 show homozygous CC (wild genotype): one band—279 was detected. Cases 2, 3, and 5 show heterozygous AC genotype: three bands—279, 257, and 17 were detected. Case 6 shows homozygous AA genotype: two bands—257, 17 were detected. M: polymerase chain reaction marker (50-100-150-200-250-300-350-400 bp, etc.). The 17-bp band cannot be visualized in the horizontal gel electrophoresis.
Figure 3.
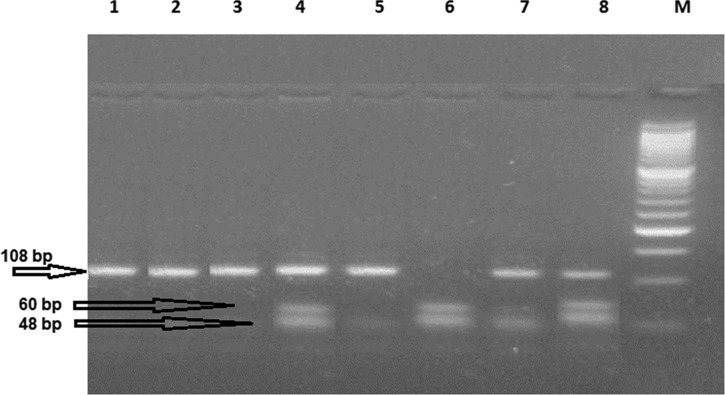
Characterization of the TLR-9 (−1237 T/C) polymorphism using BstNI restriction enzyme. Ethidium bromide–stained 4% agarose gel. Cases 1, 2, 3, and 5 show homozygous TT (wild genotype): two bands—108 and 27 bp were detected. Case 6 shows homozygous CC genotype: three bands—60, 48, and 27 bp were detected. Cases 4, 7, and 8 show heterozygous TC genotype: four bands—108, 60, 48, and 27 bp were detected. M: polymerase chain reaction marker (50-100-150-200-250-300-350-400 bp, etc.). The 27-bp band cannot be visualized in the horizontal gel electrophoresis.
Statistical analysis.
Data were analyzed using SPSS (Statistical Package for Social Science) program for statistical analysis, (version 20; SPSS Inc., Chicago, IL). χ2 or Fisher's exact test (when expected counted in 25% of the cell or more is five) was used to compare qualitative variables. Logistic regression analysis was used to calculate odds ratios and 95% confidence intervals for risk estimation. P values less than 0.05 were considered significant.
Results
Baseline demographic, clinical, and laboratory data of the studied patients are shown in Table 2.
Table 2.
Baseline clinical and laboratory data of the patients and controls
| Patients (N = 100) | Controls (N = 100) | |
|---|---|---|
| Age (years) | 41.1 ± 9.8 | 40.30 ± 10.01 |
| BMI | 26.72 ± 3.5 | 27.21 ± 4.0 |
| Sex | ||
| Male (%) | 65 (65) | 70 (70) |
| Female (%) | 35 (35) | 30 (30) |
| ALT (U/L) | 53.74 ± 39.5 | 19.5 ± 7.9 |
| AST (U/L) | 47.37 ± 32.2 | 20.9 ± 4.5 |
| T. Bil (mg/dL) | 0.81 ± 0.46 | 0.31 ± 0.11 |
| Alkaline phosphatase (mg/dL) | 103.4 ± 62.7 | – |
| Albumin (gm/dL) | 4.2 ± 0.48 | 4.5 ± 0.34 |
| Creatinine (mg/dL) | 0.99 ± 0.17 | 0.97 ± 0.18 |
| Glucose (mg/dL) | 97.7 ± 21.0 | 92.7 ± 11.3 |
| WBC (109/L) | 6.5 ± 1.8 | 6.5 ± 1.9 |
| Hb (g/dL) | 13.6 ± 1.5 | 13.9 ± 1.1 |
| Platelets (109/L) | 225 ± 59.5 | 221.4 ± 47.2 |
| PC (%) | 92.2 ± 9.7 | – |
| AFP (ng/mL) | 4.1 ± 4.2 | – |
| HCV viral load (IU/mL × 105) | 45.8 ± 32.5 | – |
| Stage of fibrosis | ||
| Stage 1 | 65 (65%) | |
| Stage 2 | 15 (15%) | |
| Stage 3 | 20 (20%) | |
AFP = alpha fetoprotein; ALT = alanine aminotransferase; AST = aspartate transaminase; BMI = body mass index; Hb = hemoglobin; HCV = hepatitis C virus; PC = prothrombin concentration; T. Bil = total bilirubin; WBC = white blood count. Data are presented as mean ± standard deviation and number (%).
The frequency of the studied genetic polymorphisms in HCV patients and controls is presented in Table 3. There was no statistically significant difference noticed in the distribution of TLR-3 _7 C/A, TLR-3 c.1377C/T, and TLR-9 1237T/C genotypes between HCV patients and controls. Also, combined genotype analysis of the studied genetic polymorphisms showed that coinheritance of the genetic polymorphism in the three studied genes did not confer increased risk to HCV infection.
Table 3.
The distribution of TLR-3 _7 C/A, TLR-3 c.1377C/T, and TLR-9 1237T/C genotypes in HCV patients and controls
| Genotype | Patients (100) | Control (100) | P value | OR (95% CI) | |||
|---|---|---|---|---|---|---|---|
| Count | % | Count | % | ||||
| TLR-3_ 7C/A | AA | 2 | 2.0 | 3 | 3.0 | 1.000 | 0.748 (0.122–4.599) |
| AC | 24 | 24.0 | 14 | 14.0 | 0.076 | 1.923 (0.927–3.989) | |
| CC | 74 | 74.0 | 83 | 83.0 | Reference | ||
| AA + AC | 26 | 26.0 | 17 | 17.0 | 0.121 | 1.715 (0.863–3.410) | |
| Allele A | 28 | 14 | 20 | 10 | 0.218 | 1.468 (0.796–2.698) | |
| Allele C | 172 | 86 | 180 | 90 | Reference | ||
| TLR-3-1377 C/T | TT | 6 | 6.0 | 6 | 6.0 | 0.763 | 0.833 (0.254–2.731) |
| CT | 28 | 28.0 | 39 | 39.0 | 0.094 | 0.598 (0.327–1.094) | |
| CC | 66 | 66.0 | 55 | 55.0 | Reference | ||
| TT + CT | 34 | 34.0 | 45 | 45.0 | 0.112 | 0.630 (0.356–1.115) | |
| Allele T | 40 | 20 | 51 | 25.5 | 0.190 | 0.730 (0.456–1.169) | |
| Allele C | 160 | 80 | 149 | 74.5 | Reference | ||
| TLR-9 1237T/C | CC | 3 | 3.0 | 6 | 6.0 | 0.396 | 0.474 (0.019–2.464) |
| TC | 19 | 19.0 | 20 | 20.0 | 0.813 | 0.901 (0.381–2.130) | |
| TT | 78 | 78.0 | 74 | 74.0 | Reference | ||
| CC + TC | 22 | 22.0 | 26 | 26.0 | 0.683 | 0.803 (0.365–1.768) | |
| Allele C | 25 | 12.5 | 32 | 16 | 0.405 | 0.750 (0.380–1.479) | |
| Allele T | 175 | 87.5 | 168 | 84 | Reference | ||
CI = confidence Interval; OR = odds ratio. P value < 0.05 is considered significant.
HCV patients were stratified according to hepatic fibrosis stage, one group with mild hepatic fibrosis (F1) and another group with advanced hepatic fibrosis (F2, F3). Different parameters that may relate to fibrosis progression were studied. Patients with advanced hepatic fibrosis have significantly elevated alpha-fetoprotein (AFP) serum levels; 5.82 ± 6.07 ng/mL versus 3.29 ± 2.39 ng/mL in mild hepatic fibrosis (P = 0.004). Otherwise, there were no statistically significant differences noticed between the two patient groups regarding age, gender, and baseline laboratory data (data not shown). No statistically significant difference was found with regard to the distribution of TLR-3 _7 C/A, TLR-9 1237T/C across the two patient groups, but with regard to TLR-3 c.1377C/T, the T-allele was found to be associated with advanced stage of hepatic fibrosis (Table 4).
Table 4.
Association of gene polymorphism and degree of liver fibrosis
| Genotype | Degree of liver fibrosis | P value | OR (95% CI) | ||||
|---|---|---|---|---|---|---|---|
| Mild hepatic fibrosis F1 (N = 65) | Advanced hepatic fibrosis F2, F3 (N = 35) | ||||||
| Count | % | Count | % | ||||
| TLR-3_ 7C/A | AA | 0 | 0.0 | 2 | 5.7 | 0.99 | NA |
| AC | 17 | 26.2 | 7 | 20.0 | 0.59 | 0.76 (0.27–2.06) | |
| CC | 48 | 73.8 | 26 | 74.3 | Reference | ||
| AA + AC | 17 | 26.2 | 9 | 25.7 | 0.962 | 0.97 (0.38–2.49) | |
| Allele A | 17 | 13 | 11 | 16 | 0.608 | 1.23 (0.54–2.81) | |
| Allele C | 113 | 87 | 59 | 84 | Reference | ||
| TLR-3-1377 C/T | TT | 4 | 6.2 | 2 | 5.7 | 0.562 | 1.70 (0.28–10.20) |
| CT | 10 | 15.4 | 18 | 51.4 | < 0.001 | 6.12 (2.33–16.04) | |
| CC | 51 | 78.5 | 15 | 42.9 | Reference | ||
| TT + CT | 14 | 21.5 | 20 | 57.1 | < 0.001 | 4.85 (1.98–11.86) | |
| Allele T | 18 | 14 | 22 | 31.4 | 0.003 | 2.85 (1.40–5.79) | |
| Allele C | 112 | 86 | 48 | 68.6 | Reference | ||
| TLR-9 1237T/C | CC | 1 | 1.5 | 2 | 5.7 | 0.22 | 4.5 (0.38–52.05) |
| TC | 10 | 15.4 | 9 | 25.7 | 0.17 | 2.02 (0.73–5.62) | |
| TT | 54 | 83.1 | 24 | 68.6 | Reference | ||
| CC + TC | 11 | 16.9 | 11 | 31.4 | 0.095 | 2.25 (0.85–5.90) | |
| Allele C | 12 | 9 | 13 | 18.6 | 0.057 | 2.24 (0.96–5.22) | |
| Allele T | 118 | 91 | 57 | 81.4 | Reference | ||
CI = confidence interval; OR = odds ratio. P value < 0.05 is considered significant.
Discussion
Knowing that genetic variations in the TLR-signaling pathway contributed in either susceptibility or resistance to several infectious diseases,32,33 we studied the impact of TLR-3 (_7 C/A [rs3775296], c.1377C/T [rs3775290]) and TLR-9 (1237T/C [rs5743836]) SNPs on HCV infection.
The TLR-9 gene is located on chromosome 3p21.3 and spans approximately 5 kb. The TLR-9 gene has a major coding region exon in one of its two exons.15 TLR-9 has been demonstrated to bind only to DNA virus, so the binding of HCV which is single-stranded RNA to TLR-9 is assumed unlikely.16 However, studies demonstrated that TLR-9 mRNA and protein are downregulated in peripheral blood mononuclear cells of HCV-infected patients compared with normal controls, and are negatively correlated with serum viral copies.34 In addition, TLR-9 stimulation showed antiviral effects in HCV-infected individuals,35 and was found to participate in the early immune response against HCV infection of the central nervous system.36 On the other hand, some other studies showed that TLR-9 level was present at higher levels in HCV patients compared with healthy controls.37 Although data are controversial, it points that TLR-9 plays a role during HCV infection.
In our study, the presence of TLR-9 1237T/C SNP is not associated with susceptibility to HCV infection. Similarly, Wei and others showed no significant association with regard to TLR-9 (rs187084) genotype and allele frequency between chronic HCV patients and subjects who spontaneously cleared the virus.38
In humans, TLR-3-promoter region is responsible for maintenance of promoter integrality and promoter-specific virus responsive element.39 In our study, the TLR-3 (_7 C/A) SNP within the promoter region is not associated with HCV infection, and our results are comparable to results in other ethnicities.31,40,41
We assumed that both TLR-3 and TLR-9 signaling mechanisms work in synergy to establish an antiviral state against HCV infection, so we analyzed whether combined genetic variants in TLRs act as a potential indicator for host susceptibility to HCV infection, but coinheritance of the genetic polymorphism in the three studied genes did not confer increased risk to HCV infection.
Liver fibrosis results from chronic liver injury–mediated inflammation and activation of hepatic stellate cells.42,43 Several studies stated the important role of TLR-3 in the pathophysiology of a variety of liver diseases44–46; TLR-3 is widely expressed on all types of liver cells, including hepatocytes,47 stellate cells,48 sinusoidal endothelial cells,49 Kupffer cells, biliary epithelial cells,50 and immune cells as natural killer cells, natural killer T-cells,51 and liver lymphocytes.49 Animal studies reported that TLR-3 plays a dual role of having a negative or positive effect in liver injury and fibrosis.52–56 Also TLR-9 was found to be associated with hepatic failure, where TLR-9 signals caused hepatic failure by promoting TNF-α production.57
In our study, no symptomatic significance was found with regard to the distribution of TLR-3 _7 C/A and TLR-9 1237T/C between patients with mild hepatic fibrosis and advanced hepatic fibrosis. However, interestingly enough, TLR-3 c.1377 (T)–allele was found to be associated with advanced stage of hepatic fibrosis (P = 0.003). Also, in the present study, patients with advanced hepatic fibrosis had significantly elevated AFP serum levels. This goes with the results of Hu and others, who found that chronic hepatitis C patients had elevated serum AFP that was independently associated with stage III/IV hepatic fibrosis.58
In conclusion, we aimed to demonstrate associations between TLR-3 and TLR-9 SNPs and susceptibility to HCV infection and stage of hepatic fibrosis in HCV-infected Egyptian patients, as recently, TLRs are gaining increased importance due to their role in influencing host immunity, and it has been suggested to use TLRs as biomarkers for HCV pathogenesis and as a novel therapeutic target for improving liver fibrosis.56,59 In our study, frequency of polymorphic genotypes in TLR-3 _7 C/A, TLR-3 c.1377C/T, and TLR-9 (1237T/C) were not significantly different between studied HCV-positive patients and controls, whereas TLR-3 (c.1377 (T)-allele was found to be associated with advanced hepatic fibrosis stage; however, with regard to TLR-9 1237T/C and TLR-3 _7 C/A, no significant association was found.
Our study has certain points of strength as well as certain limitations; the most important issue is that most Egyptian patients are infected with HCV genotype-4 and the predominant subtype is HCV-4a60; thus, the obtained results were not fragmented due to inclusion of various HCV genotypes as the pathogenesis of infection vary according to genotype.61–64 On the other hand, the study encompassed a limited number of cases. Further studies with larger samples of patients are required to further add to the validity of our results, and also studies including other members of TLR family identified in humans are required.
ACKNOWLEDGMENTS
The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Footnotes
Authors' addresses: Rania A. Zayed and Doha A. Mokhtar, Clinical and Chemical Pathology Department, Faculty of Medicine, Cairo University, Cairo, Egypt, E-mails: rania.zayed@kasralainy.edu.eg and dohamokhtar@gmail.com. Dalia Omran and Zinab Zakaria, Department of Endemic Medicine and Hepato-gastroentrology, Faculty of Medicine, Cairo University, Cairo, Egypt, E-mails: daliaomran2007@yahoo.com and zinab.zakaria@yahoo.com. Sameera Ezzat, Community Medicine Department, National Liver Institute, Menofia University, Menofia, Egypt, E-mail: drsekhir@yahoo.com. Mohamed A. Soliman, Specialized Liver Unit, Kasr Alainy Hospital, Cairo University, Cairo, Egypt, E-mail: masoly@yahoo.com. Lamiaa Mobarak, National Hepatology and Tropical Medicine Research Institute, Cairo, Egypt, E-mail: lamiaamobarak@yahoo.com. Hossam El-Sweesy, Tropical Medicine Department, Cairo Fatemic Hospital, Ministry of Health, Cairo, Egypt, E-mail: h_mrcp@yahoo.co.uk. Ghada Emam, Clinical Pathology Department, NINMS, Cairo, Egypt, E-mail: ghadaemam58@gmail.com.
References
- 1.El-Zanaty F, Way A. Egypt Demographic and Health Survey 2008. Cairo, Egypt: Ministry of Health, El Zanaty and Associates, and Macro International; 2009. http://dhsprogram.com/pubs/pdf/FR220/FR220.pdf Available at. Accessed May 17, 2013. [Google Scholar]
- 2.Visvanathan K, Lewin SR. Immunopathogenesis: role of innate and adaptive immune responses. Semin Liver Dis. 2006;26:104–115. doi: 10.1055/s-2006-939755. [DOI] [PubMed] [Google Scholar]
- 3.Bertoletti A, Gehring AJ. The immune response during hepatitis B virus infection. J Gen Virol. 2006;87:1439–1449. doi: 10.1099/vir.0.81920-0. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Takeda K. Toll-like receptor signaling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 5.Isaza-Correa JM, Liang Z, van den Berg A, Diepstra A, Visser L. Toll-like receptors in the pathogenesis of human B cell malignancies. J Hematol Oncol. 2014;7:57. doi: 10.1186/s13045-014-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Neill LA. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 7.Krishnegowda G, Hajjar AM, Zhu J, Douglass EJ, Uematsu S, Akira S, Woods AS, Gowda DC. Induction of proinflammatory responses in macrophages by the glycosylphosphatidylinositols of Plasmodium falciparum: cell signaling receptors, glycosylphosphatidylinositol (GPI) structural requirement, and regulation of GPI activity. J Biol Chem. 2005;280:8606–8616. doi: 10.1074/jbc.M413541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 9.Hemmi H, Kaisho T, Takeuchi O, Sato S, Sanjo H, Hoshino K, Horiuchi T, Tomizawa H, Takeda K, Akira S. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 10.Lee J, Chuang TH, Redecke V, She L, Pitha PM, Carson DA, Raz E, Cottam HB. Molecular basis for the immunostimulatory activity of guanine nucleoside analogs: activation of Toll-like receptor 7. Proc Natl Acad Sci USA. 2003;100:6646–6651. doi: 10.1073/pnas.0631696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heil F, Ahmad-Nejad P, Hemmi H, Hochrein H, Ampenberger F, Gellert T, Dietrich H, Lipford G, Takeda K, Akira S, Wagner H, Bauer S. The Toll-like receptor 7 (TLR7)-specific stimulus loxoribine uncovers a strong relationship within the TLR7, 8 and 9 subfamily. Eur J Immunol. 2003;33:2987–2997. doi: 10.1002/eji.200324238. [DOI] [PubMed] [Google Scholar]
- 12.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 13.Ahmad-Nejad P, Ha¨cker H, Rutz M, Bauer S, Vabulas RM, Wagner H. Bacterial CpG-DNA and lipopolysaccharidesactivate Toll-like receptors at distinct cellular compartments. Eur J Immunol. 2002;32:1958–1968. doi: 10.1002/1521-4141(200207)32:7<1958::AID-IMMU1958>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Kaisho T, Akira S. Toll-like receptors and their signaling mechanism in innate immunity. Acta Odontol Scand. 2001;59:124–130. doi: 10.1080/000163501750266701. [DOI] [PubMed] [Google Scholar]
- 15.Lu KC, Yang HY, Lin YF, Kao SY, Lai CH, Chu CM, Wu CC, Su SL. The T-1237C polymorphism of the Toll-like receptor-9 gene is associated with chronic kidney disease in a Han Chinese population. Tohoku J Exp Med. 2011;225:109–116. doi: 10.1620/tjem.225.109. [DOI] [PubMed] [Google Scholar]
- 16.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 17.Nagano Y, Kojima Y. Immunizing property of vaccinia virus inactivated by ultraviolets rays. C R Seances Soc Biol Fil. 1954;148:1700–1702. [PubMed] [Google Scholar]
- 18.Lindenmann J, Burke DC, Isaacs A. Studies on the production, mode of action and properties of interferon. Br J Exp Pathol. 1957;38:551–562. [PMC free article] [PubMed] [Google Scholar]
- 19.Pestka S, Krause CD, Walter MR. Interferons, interferon like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 20.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207:2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Lu W, Qian Q, Qi W, Hu J, Feng B. Frequency of TLR 2, 4, and 9 gene polymorphisms in Chinese population and their susceptibility to type 2 diabetes and coronary artery disease. J Biomed Biotechnol. 2012;2012:373945. doi: 10.1155/2012/373945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang HY, Lee HS, Lee CH, Fang WH, Chen HC, Salter DM, Su SL. Association of a functional polymorphism in the promoter region of TLR-3 with osteoarthritis: a two-stage case-control study. J Orthop Res. 2013;31:680–685. doi: 10.1002/jor.22291. [DOI] [PubMed] [Google Scholar]
- 23.Heinz S, Haehnel V, Karaghiosoff M, Schwarzfischer L, Müller M, Krause SW, Rehli M. Species specific regulation of Toll-like receptor 3 genes in men and mice. J Biol Chem. 2003;278:21502–21509. doi: 10.1074/jbc.M301476200. [DOI] [PubMed] [Google Scholar]
- 24.Lange NE, Zhou X, Lasky-Su J, Himes BE, Lazarus R, Manuel Soto-Quirós M, Avila L, Celedón JC, Hawrylowicz CM, Raby BA, Litonjua AA. Comprehensive genetic assessment of a functional TLR9 promoter polymorphism: no replicable association with asthma or asthma-related phenotypes. BMC Med Genet. 2011;12:26. doi: 10.1186/1471-2350-12-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carvalho A, Osorio NS, Saraiva M, Cunha C, Almeida AJ, Teixeira-Coelho M, Ludovico P, Pedrosa J, Pitzurra L, Aversa F, Romani L, Castro AG. The C allele of rs5743836 polymorphism in the human TLR9 promoter links IL-6 and TLR9 up-regulation and confers increased B-cell proliferation. PLoS One. 2011;6:e28256. doi: 10.1371/journal.pone.0028256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng MT, Van't Hof R, Crockett JC, Hope ME, Berry S, Thomson J, McLean MH, McColl KE, El-Omar EM, Hold GL. Increase in NF-kappaB binding affinity of the variant C allele of the toll-like receptor 9-1237T/C polymorphism is associated with Helicobacter pylori-induced gastric disease. Infect Immun. 2010;78:1345–1352. doi: 10.1128/IAI.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mostafa I, Abd el Aal M, Syam M. HCV recurrence in adult living related transplantation: initial results of starting population. Liver Transpl. 2005;11:456–462. [Google Scholar]
- 28.Tuite A, Gros P. The impact of genomics on the analysis of host resistance to infectious disease. Microbes Infect. 2006;8:1647–1653. doi: 10.1016/j.micinf.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 29.Clementi M, Di Gianantonio E. Genetic susceptibility to infectious diseases. Reprod Toxicol. 2006;21:345–349. doi: 10.1016/j.reprotox.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Bedossa P, Poynard T, The METAVIR Cooperative Study Group An algorithm for the grading of activity in chronic hepatitis C. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 31.Cheng PL, Eng HL, Chou MH, You HL, Lin TM. Genetic polymorphisms of viral infection-associated Toll-like receptors in Chinese population. Transl Res. 2007;150:311–318. doi: 10.1016/j.trsl.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Schröder NW, Schumann RR. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect Dis. 2005;5:156–164. doi: 10.1016/S1473-3099(05)01308-3. [DOI] [PubMed] [Google Scholar]
- 33.Turvey SE, Hawn TR. Towards subtlety: understanding the role of Toll-like receptor signaling in susceptibility to human infections. Clin Immunol. 2006;120:1–9. doi: 10.1016/j.clim.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 34.Zhou J, Huang Y, Tian D, Xu D, Chen M, Wu H. Expression of Toll-like receptor 9 in peripheral blood mononuclear cells from patients with different hepatitis B and C viral loads. J Huazhong Univ Sci Technol. 2009;29:313–317. doi: 10.1007/s11596-009-0310-2. [DOI] [PubMed] [Google Scholar]
- 35.Broering R, Wu J, Meng Z, Hilgard P, Lu M, Trippler M, Szczeponek A, Gerken G, Schlaak JF. Toll-like receptor-stimulated non-parenchymal liver cells can regulate hepatitis C virus replication. J Hepatol. 2008;48:914–922. doi: 10.1016/j.jhep.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 36.Hu K, Wang GW, Wang ZH. TLR9 mRNA expression and tumor necrosis factor alpha/interleukin-6 secretion in murine microglia by hepatitis C virus stimulation. Zhonghua Yi Xue Za Zhi. 2011;91:1070–1074. [PubMed] [Google Scholar]
- 37.Zhao P, Ma L, Ji H, Yu L, Feng J, Wang J, Liu MY, Jiang YF. The expression of TLR-9, CD86, and CD95 phenotypes in circulating B cells of patients with chronic viral hepatitis B or C before and after antiviral therapy. Mediators Inflamm. 2015;2015:762709. doi: 10.1155/2015/762709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei XS, Wei CD, Tong YQ, Zhu CL, Zhang PA. Single nucleotide polymorphisms of toll-like receptor 7 and toll-like receptor 9 in hepatitis C virus infection patients from central China. Yonsei Med J. 2014;55:428–434. doi: 10.3349/ymj.2014.55.2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanabe M, Kurita-Taniguchi M, Takeuchi K, Takeda M, Ayata M, Oyura H, Matsumoto M, Seya T. Mechanism of up-regulation of human Toll-like receptor receptor 3 secondary to infection of measles virus-attenuated strains. Biochem Biophys Res Commun. 2003;311:39–48. doi: 10.1016/j.bbrc.2003.09.159. [DOI] [PubMed] [Google Scholar]
- 40.Medhi S, Deka M, Deka P, Swargiary SS, Hazam RK, Sharma MP, Gumma PK, Asim M, Kar P. Promoter region polymorphism and expression profile of toll like receptor-3 (TLR-3) gene in chronic hepatitis C virus (HCV) patients from India. Indian J Med Res. 2011;134:200–207. [PMC free article] [PubMed] [Google Scholar]
- 41.Askar E, Bregadze R, Mertens J, Schweyer S, Rosenberger A, Ramadori G, Mihm S. TLR3 gene polymorphisms and liver disease manifestations in chronic hepatitis C. J Med Virol. 2009;81:1204–1211. doi: 10.1002/jmv.21491. [DOI] [PubMed] [Google Scholar]
- 42.Lee YA, Wallace MC, Friedman SL. Pathobiology of liver fibrosis: a translational success story. Gut. 2015;64:830–841. doi: 10.1136/gutjnl-2014-306842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61:1066–1079. doi: 10.1002/hep.27332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seki E, Brenner DA. Toll-like receptors and adaptor molecules in liver disease: update. Hepatology. 2008;48:322–335. doi: 10.1002/hep.22306. [DOI] [PubMed] [Google Scholar]
- 45.Szabo G, Dolganiuc A, Mandrekar P. Pattern recognition receptors: a contemporary view on liver diseases. Hepatology. 2006;44:287–298. doi: 10.1002/hep.21308. [DOI] [PubMed] [Google Scholar]
- 46.Mencin A, Kluwe J, Schwabe RF. Toll-like receptors as targets in chronic liver diseases. Gut. 2009;58:704–720. doi: 10.1136/gut.2008.156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khvalevsky E, Rivkin L, Rachmilewitz J, Galun E, Giladi H. TLR3 signaling in a hepatoma cell line is skewed towards apoptosis. J Cell Biochem. 2007;100:1301–1312. doi: 10.1002/jcb.21119. [DOI] [PubMed] [Google Scholar]
- 48.Wang B, Trippler M, Pei R, Lu M, Broering R, Gerken G, Schlaak JF. Toll-like receptor activated human and murine hepatic stellate cells are potent regulators of hepatitis C virus replication. J Hepatol. 2009;51:1037–1045. doi: 10.1016/j.jhep.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 49.Xiao X, Zhao P, Rodriguez-Pinto D, Qi D, Henegariu O, Alexopoulou L, Flavell RA, Wong FS, Wen L. Inflammatory regulation by TLR3 in acute hepatitis. J Immunol. 2009;183:3712–3719. doi: 10.4049/jimmunol.0901221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura M, Funami K, Komori A, Yokoyama T, Aiba Y, Araki A, Takii Y, Ito M, Matsuyama M, Koyabu M, Migita K, Taniguchi K, Fujioka H, Yatsuhashi H, Matsumoto M, Ishibashi H, Seya T. Increased expression of Toll-like receptor 3 in intrahepatic biliary epithelial cells at sites of ductular reaction in diseased livers. Hepatol Int. 2008;2:222–230. doi: 10.1007/s12072-008-9055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner T, Chen Q, Jin Y, Ajuber M. Toll-like receptor 3 ligand dampens liver inflammation by stimulating Valpha14 invariant natural killer T cells to negatively regulate gamma/delta T cells. Am J Pathol. 2010;176:1779–1789. doi: 10.2353/ajpath.2010.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin S, Gao B. Toll-like receptor 3 in liver diseases. Gastroenterol Res Pract. 2010;2010:971270. doi: 10.1155/2010/750904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong Z, Wei H, Sun R, Hu Z, Gao B, Tian Z. Involvement of natural killer cells in PolyI:C-induced liver injury. J Hepatolology. 2004;41:966–973. doi: 10.1016/j.jhep.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 54.Jeong WI, Park O, Radaeva S, Gao B. STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating NK cell cytotoxicity. Hepatology. 2006;44:1441–1451. doi: 10.1002/hep.21419. [DOI] [PubMed] [Google Scholar]
- 55.Byun JS, Suh YG, Yi HS, Lee YS, Jeong WI. Activation of toll-like receptor 3 attenuates alcoholic liver injury by stimulating Kupffer cells and stellate cells to produce interleukin-10 in mice. J Hepatolology. 2013;58:342–349. doi: 10.1016/j.jhep.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 56.Seo W, Eun HS, Kim SY, Yi HS, Lee YS, Park SH, Jang MJ, Jo E, Kim SC, Han YM, Park KG, Jeong WI. Exosome-mediated activation of toll-like receptor 3 in stellate cells stimulates interleukin-17 production by γδ T cells in liver fibrosis. Hepatology. 2016;64:616–631. doi: 10.1002/hep.28644. [DOI] [PubMed] [Google Scholar]
- 57.Dejager L, Libert C. Tumor necrosis factor alpha mediates the lethal hepatotoxic effects of poly(I:C) in d-galactosamine-sensitized mice. Cytokine. 2008;42:55–61. doi: 10.1016/j.cyto.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 58.Hu KQ, Kyulo NL, Lim N, Elhazin B, Hillebrand DJ, Bock T. Clinical significance of elevated alpha-fetoprotein (AFP) in patients with chronic hepatitis C, but not hepatocellular carcinoma. Am J Gastroenterol. 2004;99:860–865. doi: 10.1111/j.1572-0241.2004.04152.x. [DOI] [PubMed] [Google Scholar]
- 59.Firdaus R, Biswas A, Saha K, Mukherjee A, Pal F, Chaudhuri S, Chandra A, Konar A, Sadhukhan PC. Modulation of TLR 3, 7 and 8 expressions in HCV genotype 3 infected individuals: potential correlations of pathogenesis and spontaneous clearance. Biomed Res Int. 2014;2014:491064. doi: 10.1155/2014/491064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ray SC, Arthur RR, Carella A, Bukh J, Thomas DL. Genetic epidemiology of hepatitis C virus throughout Egypt. J Infect Dis. 2000;182:698–707. doi: 10.1086/315786. [DOI] [PubMed] [Google Scholar]
- 61.Dusheiko GM, Schmilovitz-Weiss H, Brown D, McOmish F, Yap PL, Sherlock S, McIntyre N, Simmonds P. Hepatitis C virus genotypes: an investigation of type-specific differences in geographic origin and disease. Hepatology. 1994;19:13–18. [PubMed] [Google Scholar]
- 62.McHutchison JG, Gordon SC, Schiff ER, Shiffman ML, Lee WM, Rustgi VK, Goodman ZD, Ling MH, Cort S, Albrecht JK. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 63.Yoshioka K, Kakumu S, Wakita T, Ishikawa T, Itoh Y, Takayanagi M, Higashi Y, Shibata M, Morishima T. Detection of hepatitis C virus by polymerase chain reaction and response to interferon alpha therapy: relationship to genotypes of hepatitis C virus. Hepatology. 1992;16:293–299. doi: 10.1002/hep.1840160203. [DOI] [PubMed] [Google Scholar]
- 64.Zein NN, Rakela J, Krawitt EL, Reddy KR, Tominaga T, Persing DH. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Ann Intern Med. 1996;125:634–639. doi: 10.7326/0003-4819-125-8-199610150-00002. [DOI] [PubMed] [Google Scholar]