Abstract
Purpose
Applying CNGA3 gene augmentation therapy to cure a novel causative mutation underlying achromatopsia (ACHM) in sheep.
Methods
Impaired vision that spontaneously appeared in newborn lambs was characterized by behavioral, electroretinographic (ERG), and histologic techniques. Deep-sequencing reads of an affected lamb and an unaffected lamb were compared within conserved genomic regions orthologous to human genes involved in similar visual impairment. Observed nonsynonymous amino acid substitutions were classified by their deleteriousness score. The putative causative mutation was assessed by producing compound CNGA3 heterozygotes and applying gene augmentation therapy using the orthologous human cDNA.
Results
Behavioral assessment revealed day blindness, and subsequent ERG examination showed attenuated photopic responses. Histologic and immunohistochemical examination of affected sheep eyes did not reveal degeneration, and cone photoreceptors expressing CNGA3 were present. Bioinformatics and sequencing analyses suggested a c.1618G>A, p.Gly540Ser substitution in the GMP-binding domain of CNGA3 as the causative mutation. This was confirmed by genetic concordance test and by genetic complementation experiment: All five compound CNGA3 heterozygotes, carrying both p.Arg236* and p.Gly540Ser mutations in CNGA3, were day-blind. Furthermore, subretinal delivery of the intact human CNGA3 gene using an adeno-associated viral vector (AAV) restored photopic vision in two affected p.Gly540Ser homozygous rams.
Conclusions
The c.1618G>A, p.Gly540Ser substitution in CNGA3 was identified as the causative mutation for a novel form of ACHM in Awassi sheep. Gene augmentation therapy restored vision in the affected sheep. This novel mutation provides a large-animal model that is valid for most human CNGA3 ACHM patients; the majority of them carry missense rather than premature-termination mutations.
Keywords: achromatopsia, CNGA3, next-generation sequencing, sheep, Awassi
Congenital achromatopsia (ACHM) is a hereditary vision disorder caused by cone photoreceptor dysfunction.1 In most human cases, ACHM results from mutations in the CNGA3 and CNGB3 genes that code for α and β subunits of the cyclic-nucleotide-gated (CNG) ion channel of the cone photoreceptors, respectively.2,3 Mutations in four additional genes—PDE6C, PDE6H, GNAT2, and ATF6—also cause the disease.4–8
The eye, and particularly the retina, is an ideal target for gene augmentation therapy, as has been demonstrated for several visual impairment illnesses in rodents.9 Ideally, prior to application in humans, safety and efficacy of the gene augmentation therapy should be validated in large-animal models. Indeed, the naturally occurring canine10 and ovine11 models have served to test gene augmentation therapy of ACHM resulting from mutations in CNGB3 and CNGA3, respectively.
Follow-up of gene augmentation therapy in the ovine CNGA3-derived ACHM model reported by our group revealed long-term efficiency in vision restoration for over 4 years (Ezra-Elia R, et al. IOVS 2016;57:ARVO E-Abstract 5149). However, as this model carries a premature stop codon12 that abolishes synthesis of the CNGA3 protein, its validity is questionable for cases of missense CNGA3 mutations, in which the defective subunits may interrupt the function of the restored channel complex. Notably, most human ACHM patients harbor missense rather than stop codon CNGA3 mutations.2,13–15 Missense and amino-acid-deletion mutations of CNGA3 that disrupt cone function have been described in a canine animal model,16 but have not yet been tested in gene augmentation therapy studies.
Recently, we identified a new case of blindness in sheep. Here, we provide evidence that this is a novel model for ACHM and that the causative mutation is c.1618G>A, p.Gly540Ser, located within the GMP-binding domain of the ovine CNGA3 gene. Moreover, we demonstrate that CNGA3 gene augmentation therapy can restore vision in sheep homozygous for this missense ACHM mutation, and that the beneficial effect is retained for at least 20 months.
Materials and Methods
Experimental Design
To characterize the novel ovine blindness, we conducted behavioral maze tests as well as electroretinographic (ERG) recordings. To verify the causative mutation, we applied deep sequencing and bioinformatics analysis, produced heterozygous compound CNGA3 animals, and finally performed gene augmentation therapy by subretinal delivery of an adeno-associated viral vector (AAV)-based vector carrying the intact human CNGA3 gene.
Ethics and Animal Welfare
All experiments were conducted in accordance with the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. Experimental protocols were approved by the Volcani Center Institutional Animal Care and Use Committee, and were conducted in accordance with the principles outlined in the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
For viral injections and electrophysiological recordings, animals were premedicated with acepromazine and pethidine, induced with propofol, and anesthetized and ventilated with isoflurane.
Animals
Birth of blind lambs was reported late in 2012 in a small remote and isolated Local Awassi sheep flock in southern Israel. Shortly thereafter, we visited the flock and took buccal swabs for DNA extraction from all four rams that were present at the time of the visit and from two unaffected ewes and their two affected single lambs. Six affected lambs, three males and three females, were purchased and transferred to the experimental sheep flock of the Volcani Center in Bet Dagan, Israel, for further examination. Not long after that, the flock owner sold all of his sheep, limiting our ability to access additional affected sheep.
The affected animals at Bet Dagan were kept as an isolated group, and with time, one of the females became pregnant from the affected males and lambed three single male lambs in three successive parities, all of which were day-blind. Thus, the newly characterized day blindness in Local Awassi was found to be hereditary. Two of the six blind sheep were killed for histologic and immunohistochemical studies. Genotyping of the day-blind Local Awassi sheep revealed that none of them carried the known c.706C>T mutation12 (data not shown).
In addition to the novel model reported here, we included in the study another model of ovine ACHM that we previously identified,11,12 namely day-blind Improved Awassi ewes homozygous for the c.706C>T stop codon mutation; unaffected Afec-Assaf sheep from the Volcani Center experimental flock at Bet Dagan were used as well.
Behavioral Assessment
Maze-navigation testing of affected and control unaffected sheep was conducted as previously described.11 Briefly, animals were directed to pass through a 9-m-long maze with two barrier obstacles. A group of sheep positioned at the end of the maze attracted the tested animal to pass through the maze as quickly as possible. Passage time and number of collisions with the obstacles or maze walls were recorded for each trial. A trial lasted up to 30 seconds, and a passage time of 30 seconds was ascribed to animals that failed to successfully navigate the maze. For each sheep, a test consisted of two successive trials, with the barriers randomly rearranged in their right or left orientation between trials to avoid a learning effect.
Two of the affected animals underwent gene augmentation therapy in their right eye. Postoperative sessions of maze testing for those animals were conducted with alternate patching of the treated and untreated eyes.
ERG Analysis
Cone function was evaluated by full-field flash ERG, using a handheld mini Ganzfeld stimulator (HMsERG; Ocuscience, Henderson, NV, USA) with a band pass of 0.3 to 300 Hz as previously described.17 Briefly, following 10 minutes of light adaptation (30 cd/m2), responses were recorded to four increasing intensities of flicker stimuli (1, 2.5, 5, and 10 cd·s/m2). At each of the four stimulus intensities, 32 flashes, presented at 1 Hz, were averaged to generate the single photopic flash response; this was followed by a series of cone flicker responses to eight increasing frequencies (flashes presented at 10–80 Hz, with 128 responses averaged at each frequency). Flicker-response amplitudes were measured between peak and trough. Where no flicker response was detected, a value of zero was inserted as the amplitude. The critical flicker fusion frequency (CFFF), or the highest frequency at which the animal could resolve flicker, was determined for each of the four intensities. Recordings were conducted on five experimental animals: four of the original Local Awassi and one of their progeny at a mean ± SD age of 23.4 ± 11.4 months (range, 10–38 months, median 20 months), as well as on two of the treated sheep 2 months after gene augmentation therapy.
Histology and Immunohistochemistry (IHC) of Affected Lambs' Eyes
Two of the six original Local Awassi sheep were euthanized with 20% embutramide, 5% mebezonium iodide, and 0.5% tetracaine hydrochloride (0.1 mL/kg; T-61; Intervet Canada Corp., Kirkland, QC, Canada), and both eyes from each animal were then enucleated and fixed in Davidson solution (4 parts glacial acetic acid, 12 parts 95% ethyl alcohol, 5 parts 16% paraformaldehyde solution, 15 parts distilled water). Procedures for section preparation, histology, and IHC were as previously described.11 Briefly, deparaffinized and dehydrated sections were incubated in a decloaking chamber with 10 mM citrate buffer (pH 6.0) at 110°C for 4 minutes. After blocking with PBS containing 1% bovine serum albumin, 0.1% Triton X-100, and 3% normal donkey serum, sections were incubated overnight at 4°C with one of the following primary antibodies: anti-blue opsin (goat polyclonal, 1:75; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), anti-red/green opsin (rabbit polyclonal, 1:100; Chemicon International, Inc., Billerica, MA, USA), or anti-CNGA3 (goat polyclonal, 1:50; Santa Cruz Biotechnology). After a wash, the appropriate secondary antibody was applied for 1 hour: Cy2-conjugated donkey anti-rabbit IgG (1:200) or Rhodamine Red-X-conjugated donkey anti-goat IgG (1:200; both from Jackson ImmunoResearch Laboratories, West Grove, PA, USA). Peanut agglutinin (PNA) conjugated to fluorescein was used for general identification of cone photoreceptors (Vector Laboratories, Burlingame, CA, USA). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI)-containing mounting medium (Vector Laboratories). To determine the specificity of the antigen–antibody reaction, corresponding negative controls were performed.
Deep Sequencing and Bioinformatics Analysis
DNA extracted from one day-blind and one unaffected Awassi sheep was deep-sequenced using the Illumina (San Diego, CA, USA) HiSeq2000 platform according to the manufacturer's pair-end protocol (ENA accession no. PRJEB12018). Average fragment length was 580 bp, and 100-bp sequence reads were obtained from both ends. Each DNA sample was applied to two lanes yielding ∼11-fold coverage for the sheep genome.
Sequence reads were mapped to 5020 conserved-sequence stretches of genes involved in hereditary visual impairment using the Fast Alignment Search Tool suite (mrsFAST-ultra-3.2.0; http://sourceforge.net/projects/mrsfast/; in the public domain). These conserved sequences were obtained by aligning 242 human genes listed in the Retinal Information Network (https://sph.uth.edu/Retnet/disease.htm; in the public domain) (RetNet) to the sheep genome (v3.1) using the UCSC genome browser and the Galaxy Web-based platform (https://usegalaxy.org/; in the public domain). The obtained BAM-formatted files were analyzed using Genome Analysis Toolkit or GATK (GenomeAnalysisTK-2.7-2; https://www.broadinstitute.org/gatk/; in the public domain) for variant discovery. For each polymorphic site, homozygous variations for the day-blind sheep and heterozygosity or homozygosity for the alternative allele in the normal Awassi sheep were further evaluated by scoring amino acid substitutions using Grantham scores18 and PROVEAN analysis v1.1 (http://provean.jcvi.org/seq_submit.php; in the public domain),19 which scores the effect of protein-sequence variation on protein function.
Production of Compound Heterozygotes
Three homozygous Improved Awassi ewes carrying the stop codon c.706C>T mutation were hand mated with one of the affected Local Awassi rams. Following lambing, buccal swabs were collected from the newborn lambs for DNA extraction. At 5 months of age, behavioral assessment of the visual ability of the compound heterozygous lambs was performed.
CNGA3 Genotyping
DNA was extracted from buccal swabs using standard DNA-extraction methods, and genotyping was performed by Sanger sequencing. Polymerase chain reaction primers (Supplementary Table S1) were designed according to the ovine gene sequence (GenBank accession no. FN377574). Polymerase chain reaction products spanning ovine CNGA3 exons 8 or 9, where the previous (c.706C>T) or novel (c.1618G>A) mutation is located, respectively, were amplified and sequenced as previously described.12
Gene Augmentation Therapy
To confirm that a mutation at the CNGA3 gene is the causative mutation for the novel sheep day blindness, we carried out a gene augmentation therapy study in which we treated affected sheep with AAV5 vector containing intact human CNGA3 cDNA under the control of the red/green opsin promoter.11 Sequence homology of human CNGA3 gene (NCBI Reference Sequence: XP_005252850.1) to the ovine orthologue gene (NCBI Reference Sequence: FN377575) was 93%. Treating sheep carrying a stop codon mutation at CNGA3 with this vector resulted in long-term recovery of their visual function. The viral vector used in this study for gene augmentation therapy, the surgical procedures for subretinal vector delivery, and the postoperational ophthalmologic examinations were recently described.11 Briefly, two 2-year-old affected Local Awassi rams, homozygous for the novel missense CNGA3 mutation, were treated unilaterally with a subretinal injection of AAV5 vector containing the normal human CNGA3 cDNA under control of the PR2.1 red/green opsin promoter; 4.5 to 5 × 1011 VG/mL was delivered in ∼500 μL balanced salt solution. The visual function of the treated rams was evaluated by behavioral assessment at 20, 199, 330, 620, and 759 days after the operation and by ERG recording (as described above), carried out 2 months after vector delivery.
Statistical Analysis
For the behavioral examination, raw and log-transformed values of passage time and number of collisions were subjected to analysis of variance (ANOVA) using the General Linear Model procedure of the Jump IN computer package (SAS Institute, Inc., Cary, NC, USA). The statistical model for comparing performance of control unaffected and affected sheep under scotopic or photopic conditions included the effects of genotype (unaffected or affected) and trial (n = 2).
The statistical model for comparing photopic passage time and number of collisions before and 20, 199, 330, 620, and 759 days after gene augmentation treatment for two rams included the effects of animal (n = 2) and status, which is defined by the time of the test (before treatment or 20, 199, 330, 620, or 759 days after treatment), and the eye covering condition—uncovered, right eye (treated) covered, left eye (untreated) covered. Trial effect (n = 2) was included in the model within status. P < 0.05 was considered significant. Results are expressed as least square means ± standard error (SE).
For ERG, to test for differences in cone function between the Improved Awassi sheep with the previously described mutation,17 Local Awassi sheep with the newly found mutation, and normal control sheep, the Kruskal-Wallis 1-way ANOVA was applied; results with P ≤ 0.05 were considered statistically significant. In cases of significance, the Mann-Whitney nonparametric test was used for pairwise comparisons, and criteria for significance for these post hoc tests were set according to the Bonferroni correction at P ≤ 0.017.
The concordance test was performed following probability calculations for concordance between the observed phenotypes and genotypes as previously described.20
Results
Behavioral Assessment of Affected Sheep Under Scotopic and Photopic Conditions
Results of maze-navigation tests conducted at 1 year of age on the four Local Awassi blind sheep revealed day blindness in the affected sheep. Under rod-mediated scotopic conditions, the affected and unaffected sheep navigated through the maze similarly (average passing time of 5.5 ± 0.4 seconds with practically no collisions). However, under cone-mediated photopic conditions, while control unaffected animals navigated the maze in 5.6 ± 0.5 seconds with practically no collisions, affected sheep came close to failing, with an average passing time of 28 ± 4 seconds and 6.5 ± 3.0 collisions.
ERG Recording in Control and Affected Day-Blind Sheep
Results of the ERG recordings demonstrated residual cone function that was significantly attenuated compared to normal control animals, and was similar to the ERG records of Improved Awassi day-blind sheep that carry the stop codon mutation21 (Supplementary Figs. S1, S2).
When single-flash parameters (a- and b-wave amplitudes and implicit times) and flicker parameters (CFFF and amplitudes at 20 Hz) at all four intensities (1, 2.5, 5, and 10 cd·s/m2) were compared, we found significant differences between the Improved Awassi sheep with the previously described mutation17 and the Local Awassi sheep with the newly found mutation on the one hand, and normal control sheep on the other, in all 24 of these parameters (P ≤ 0.05). In an intragroup comparison, all 24 parameters in the Local Awassi sheep with the newly found mutation were significantly different from those in normal control sheep (P ≤ 0.017), but not significantly different (P > 0.05) from those in the Improved Awassi sheep with the previously described mutation.
Histology and IHC of Retinas of Control and Affected Sheep
No differences were noted between affected and unaffected Local Awassi sheep in histopathological examination of hematoxylin and eosin (H&E)-stained retinal sections (Supplementary Fig. S3A). Immunohistochemical evaluation of affected retinas revealed a generally preserved retinal structure with a large number of cone photoreceptors (Supplementary Fig. S3B) expressing both red/green and blue cone opsins (Supplementary Fig. S3C).
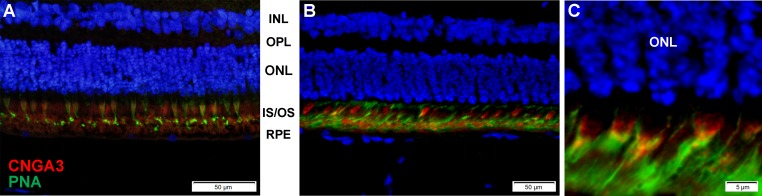
Staining of the affected sheep retinas with anti-CNGA3 antibody demonstrated CNGA3 protein expression in the inner and outer segments of the photoreceptors (Fig. 1).
Figure 1.
Immunohistochemical staining shows CNGA3 protein expression (red fluorescence) in cone photoreceptors in nonaffected (A) as well as affected mutant CNGA3 retinas (B, C). Costaining with anti-peanut agglutinin (PNA) was performed to identify cones (green fluorescence). INL, inner nuclear layer; OPL, outer plexiform layer; ONL, outer nuclear layer, containing nuclei of photoreceptors; IS/OS, inner + outer segments of the photoreceptors; RPE, retinal pigment epithelium.
Bioinformatics Analysis to Locate the Causative Mutation
Sequence reads obtained from whole-genome sequencing of day-blind and unaffected Local Awassi sheep were compared. Narrowing the comparison to conserved regions of genes involved in visual impairment in humans revealed 111 single-nucleotide polymorphisms (SNPs) and one indel (data not shown). None of the nucleotide substitutions were capable of premature termination; 42 of the SNPs in 30 genes encoded nonsynonymous amino acid substitutions (Supplementary Table S2), and 10 of these in 9 genes were deleterious according to PROVAN score.19 In the day-blind Local Awassi, only one of these mutations, CNGA3 p.Gly540Ser, was previously associated with ACHM in humans.2 This mutation was novel in sheep and in a position orthologous to the p.Gly525Asp mutation in the GDP-binding site of the human CNGA3. We excluded as causative another CNGA3 mutation (p.Ser17Leu) with a deleterious score, which was located in the signal-peptide region and has been previously found to segregate in normal Local and Improved Awassi populations.12
Since the CNGA3 p.Gly540Ser substitution was mapped to an evolutionarily conserved site among vertebrates (Supplementary Fig. S4), we further investigated its relevant SNP c.1618G>A in DNA samples collected from the affected Local Awassi sheep. Genotyping (Supplementary Fig. S5) showed that (1) one of the four rams of the Local Awassi flock was heterozygous for the c.1618 G>A mutation, while the three other rams were noncarriers (homozygous GG); (2) the two affected lambs in the flock were both homozygous for the mutation (AA), and their two mothers were heterozygous (GA); (3) all six day-blind sheep brought to the Volcani Center were homozygous for the c.1618G>A mutation. Thus, concordance between the genotype and phenotype data supported the hypothesis that the novel day blindness is inherited in an autosomal recessive mode, and that c.1618G>A is its causative mutation. Assuming an allelic frequency of 0.125 for this mutation based on its prevalence in the rams, the probability of observing such concordance by chance is 3.2 × 10−15.
Production of Compound Heterozygotes Carrying the c.706C>T and c.1618G>A Mutations
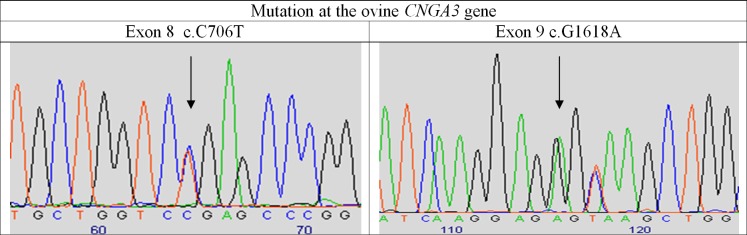
To validate c.1618G>A as the causative mutation for the new form of ovine day blindness, we generated five compound heterozygous lambs by mating a Local Awassi ram homozygous for the CNGA3 c.1618G>A mutation and three ewes homozygous for the previously identified c.706C>T CNGA3 mutation. All five lambs that were heterozygous at both of the mutation sites (Fig. 2) were day-blind, as revealed by behavioral assessment at 5 months of age.
Figure 2.
Chromatograms following sequencing compound heterozygous No. 3909 for mutation sites at the ovine CNGA3 gene that are associated with ACHM. Arrow denotes the position of the mutations at exons 8 and 9.
CNGA3 Augmentation Therapy
An AAV5 vector containing the 2.1-kb human red/green opsin promoter and human CNGA3 cDNA was injected into the subretinal space of the right eye of each of two 2-year-old day-blind Local Awassi rams. Using a green fluorescent protein (GFP) reporter gene, we were able to prove cone-specific expression of the AAV5 vector in the affected eyes (Supplementary Figs. S3D, S3E).
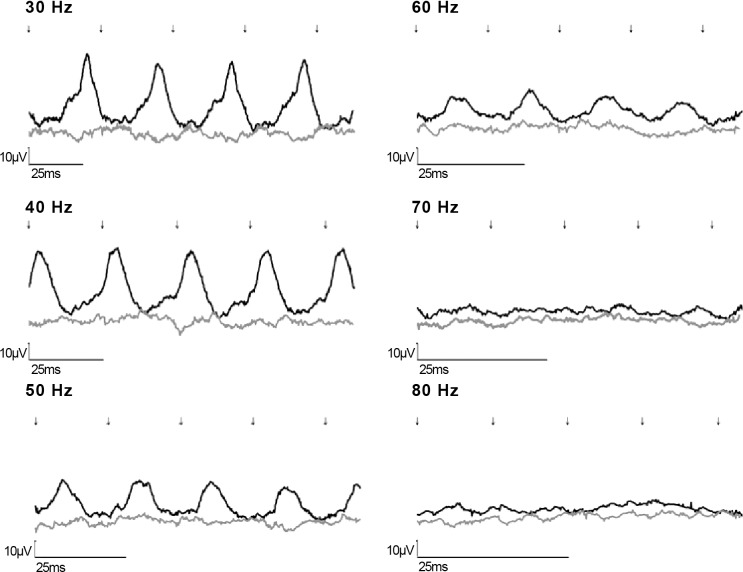
Photopic ERG recordings conducted 2 months after the operation showed similar major improvement of cone function in the treated eyes but not in the untreated fellow eyes of both rams (Fig. 3), similar to the improvement seen following gene augmentation therapy of sheep carrying the p.Arg236* mutation.11
Figure 3.
Cone flicker tracings (30–80 Hz [A–F], respectively) at the highest intensity (10 cd·s/m2) following gene augmentation treatment of a day-blind sheep affected with the new missense mutation. Traces of the treated eye (black) and fellow untreated eye (dark gray line) are shown. Flash onset indicated by arrows. Note that the x-axis scale differs in each part of the figure.
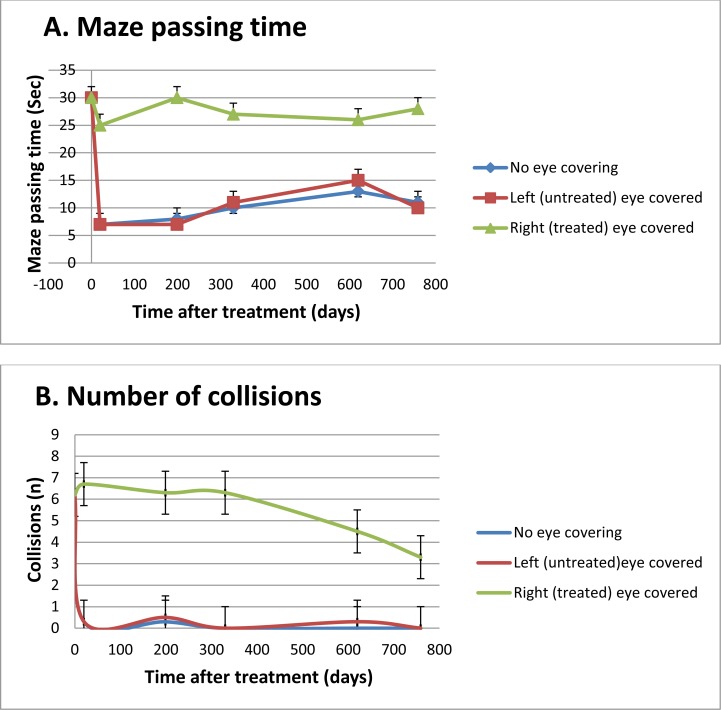
Behavioral assessments conducted 20, 199, 330, 620, and 759 days after gene augmentation therapy (Supplementary Video S1) on two affected rams confirmed long-term restoration of photopic vision (Fig. 4). Before treatment, the two affected rams practically failed the maze test. Up to 25 months after CNGA3 gene augmentation therapy, the rams succeeded in navigating the maze with no eye covering or with the left (untreated) eye covered. However, they failed in all cases to pass through the maze when the treated (right eye) was covered.
Figure 4.
Maze passage time (A) and number of collisions (B) (mean ± SE) of affected rams (n = 2) before and 20, 199, 330, 620, and 759 days after CNGA3 gene augmentation therapy.
Discussion
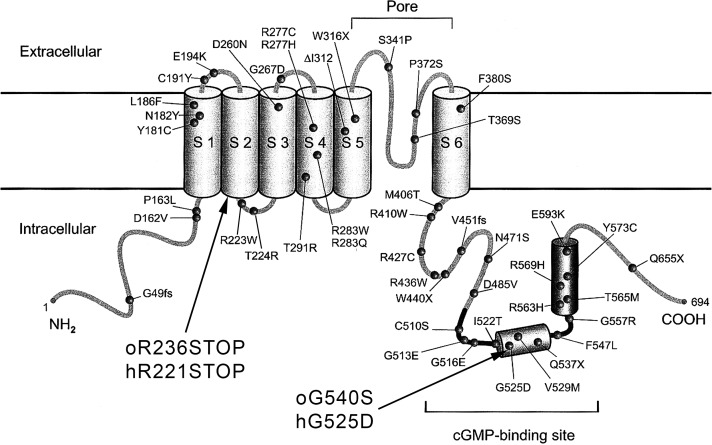
While genome-editing tools have been suggested as an efficient way to generate, in the future, large-animal models for human diseases,22 spontaneously occurring mutations in canines and sheep still play a pivotal role in current evaluations of the efficiency of gene augmentation therapy for CNGB3- and CNGA3-related disorders.10,11 To date, only three CNGA3 mutations have been reported in large animals: the ovine CNGA3 p.Arg236* mutation and two canine CNGA3-associated mutations, p.Arg424Trp and p.Val644del.16 In the present study, we characterized a novel missense mutation at the cGMP-binding domain of the ovine CNGA3 gene—p.Gly540Ser (Fig. 5)—which causes day blindness, as shown by behavioral and ERG tests. We further demonstrated that CNGA3 gene augmentation therapy can restore cone vision in the affected animals. A limited number of affected animals and the absence of specific antibodies that differentiate between ovine and human CNGA3 protein molecules (93% similarity) prevented further investigation of the expression of the transgenic gene in the treated animals.
Figure 5.
Location of the ovine day-blindness mutations with respect to topological model of the human CNGA3 gene. Reprinted with permission from Wissinger B, Gamer D, Jägle H, et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001;69:722–737. © 2001 The American Society of Human Genetics. Published by Elsevier, Inc.
In 2008, Komaromy and al.23 noted that dogs represent a valuable model for the development of cone-directed gene therapy, due to the existence of two canine ACHM lines with either a genomic deletion (i.e., functional null) or missense mutation in the CNGB3 gene. Both models were then used to evaluate the efficacy of gene-replacement therapy with a recombinant AAV.10 The robustness and stability of the observed treatment were mutation independent, but dependent on promoter and age at treatment.
As with the previous CNGA3 p.Arg236* mutation, the new p.Gly540Ser mutation was discovered in genetically closed Awassi sheep flocks. No other day blindness–related mutations have been reported in other sheep breeds anywhere in the world. Thus, the possibility that the Awassi have a higher tendency for cone-specific mutations may be put forward, just as ACHM and other inherited retinal diseases have been reported in certain dog breeds but not in others.24
Although the study was performed with a limited number of affected animals, we were able, through behavioral studies and ERG recordings, to confirm inherited day blindness in the affected animals. By a next-generation sequencing strategy and bioinformatics tools, as applied in other studies,25–28 we pinpointed the causative missense mutation, which was located within the cGMP-binding domain of the ovine CNGA3 gene. It is worth noting that in vitro analysis has confirmed impaired function of human CNGA3 polypeptides carrying mutations at the cGMP-binding domain relative to the function of the wild-type polypeptide.29
Finding a new large-animal model for CNGA3 ACHM opens the door for evaluating the translating of gene augmentation therapy into humans in the case of missense mutations, as those mutations are more common than null mutations in human patients.30 We previously showed the long-term effect of CNGA3 gene augmentation therapy for a nonsense mutation leading to a stop codon situation, where the native CNGA3 molecule is absent. In the case of the present missense mutation, the native CNGA3 protein was present (Fig. 1), and during gene augmentation therapy, there could be an interfering effect of the mutated host protein on the delivered normal protein. This might be expected in oligomers such as the CNG channel, for which missense mutations in CNGA3 lead to the formation of abnormal ion channels in combination with normal CNGB3 subunits.13
Our results show the long-term effect of gene augmentation therapy in the case of the p.Gly540Ser mutation, similar to previous results with the CNGA3-null mutation in the ovine model11 (Ezra-Elia R, et al. IOVS 2016;57:ARVO E-Abstract 5149). Therefore, although we have examined only one CNGA3 missense example in sheep thus far, our data support the conclusion that homozygous missense CNGA3 mutations do not prevent successful gene augmentation therapy with the normal wild-type cDNA. Thus, it would appear that the normal CNGA3 protein, when expressed exogenously via an AAV vector, can mediate viable assembly of normal A3 and B3 proteins into functional CNG channels. However, further investigation is required to unravel the precise mechanisms underlying successful therapy in this sheep model of ACHM.
Finally, another possible condition contributing to the success of gene augmentation therapy in our new missense model is that expression of the human CNGA3 in the sheep's eye may induce only a minimal immune response, unlike other therapeutic proteins that are usually recognized by the patient's immune system.31
Supplementary Material
Acknowledgments
This paper is a contribution from the Agricultural Research Organization, Institute of Animal Science, Bet Dagan, Israel.
Supported by The Haim and Esther Koppel Foundation, The Joseph Alexander Foundation, The Chief Scientist of the Israeli Ministry of Health (Grant no. 11892–3-0000), The Israel Science Foundation (Grant no. 1257/15), and the Yedidut Research grant, Israel. WWH received National Institutes of Health Grant P30EY022023 and funds from the Macula Vision Research Foundation, Foundation Fighting Blindness, and Research to Prevent Blindness, Inc., for partial support of this work.
Disclosure: E. Gootwine, None; M. Abu-Siam, None; A. Obolensky, None; A. Rosov, None; H. Honig, None; T. Nitzan, None; A. Shirak, None; R. Ezra-Elia, None; E. Yamin, None; E. Banin, None; E. Averbukh, None; W.W. Hauswirth, AGTC (C, I, R), P; R. Ofri, None; E. Seroussi, None
References
- 1. Roosing S,, Thiadens AA,, Hoyng CB,, Klaver CC,, den Hollander AI,, Cremers FP. Causes and consequences of inherited cone disorders. Prog Retin Eye Res. 2014; 42: 1–26. [DOI] [PubMed] [Google Scholar]
- 2. Wissinger B,, Gamer D,, Jagle H,, et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001; 69: 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kohl S,, Varsanyi B,, Antunes GA,, et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur J Hum Genet. 2005; 13: 302–308. [DOI] [PubMed] [Google Scholar]
- 4. Kohl S,, Baumann B,, Rosenberg T,, et al. Mutations in the cone photoreceptor G-protein alpha-subunit gene GNAT2 in patients with achromatopsia. Am J Hum Genet. 2002; 71: 422–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thiadens AA,, Slingerland NW,, Roosing S,, et al. Genetic etiology and clinical consequences of complete and incomplete achromatopsia. Ophthalmology. 2009; 116: 1984–1989. e1. [DOI] [PubMed] [Google Scholar]
- 6. Kohl S,, Coppieters F,, Meire F,, et al. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am J Hum Genet. 2012; 91: 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ansar M,, Santos-Cortez RL,, Saqib MA,, et al. Mutation of ATF6 causes autosomal recessive achromatopsia. Hum Genet. 2015; 134: 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kohl S,, Zobor D,, Chiang WC,, et al. Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat Genet. 2015; 47: 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boye SE,, Boye SL,, Lewin AS,, Hauswirth WW. A comprehensive review of retinal gene therapy. Mol Ther. 2013; 21: 509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Komaromy AM,, Alexander JJ,, Rowlan JS,, et al. Gene therapy rescues cone function in congenital achromatopsia. Hum Mol Genet. 2010; 19: 2581–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Banin E,, Gootwine E,, Obolensky A,, et al. Gene augmentation therapy restores retinal function and visual behavior in a sheep model of CNGA3 achromatopsia. Mol Ther. 2015; 23: 1423–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reicher S,, Seroussi E,, Gootwine E. A mutation in gene CNGA3 is associated with day blindness in sheep. Genomics. 2010; 95: 101–104. [DOI] [PubMed] [Google Scholar]
- 13. Reuter P,, Koeppen K,, Ladewig T,, et al. Mutations in CNGA3 impair trafficking or function of cone cyclic nucleotide-gated channels, resulting in achromatopsia. Hum Mutat. 2008; 29: 1228–1236. [DOI] [PubMed] [Google Scholar]
- 14. Nishiguchi KM,, Sandberg MA,, Gorji N,, Berson EL,, Dryja TP. Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum Mutat. 2005; 25: 248–258. [DOI] [PubMed] [Google Scholar]
- 15. Johnson S,, Michaelides M,, Aligianis I,, et al. Achromatopsia caused by novel mutations in both CNGA3 and CNGB3. J Med Genet. 2004; 40: e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tanaka N,, Dutrow EV,, Miyadera K,, et al. Canine CNGA3 gene mutations provide novel insights into human achromatopsia-associated channelopathies and treatment. PLoS One. 2015; 10: e0138943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ezra-Elia R,, Banin E,, Honig H,, et al. Flicker cone function in normal and day blind sheep: a large animal model for human achromatopsia caused by CNGA3 mutation. Doc Ophthalmol. 2014; 129: 141–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grantham R. Amino acid difference formula to help explain protein evolution. Science. 1974; 185: 862–864. [DOI] [PubMed] [Google Scholar]
- 19. Choi Y,, Sims GE,, Murphy S,, Miller JR,, Chan AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012; 7: e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Seroussi E. The concordance test emerges as a powerful tool for identifying quantitative trait nucleotides: lessons from BTA6 milk yield QTL. Anim Genet. 2009; 40: 230–234. [DOI] [PubMed] [Google Scholar]
- 21. Shamir MH,, Ofri R,, Bor A,, et al. A novel day blindness in sheep: epidemiological, behavioural, electrophysiological and histopathological studies. Vet J. 2010; 185: 130–137. [DOI] [PubMed] [Google Scholar]
- 22. Whitelaw CB,, Sheets TP,, Lillico SG,, Telugu BP. Engineering large animal models of human disease. J Pathol. 2016; 238: 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Komaromy AM,, Alexander JJ,, Cooper AE,, et al. Targeting gene expression to cones with human cone opsin promoters in recombinant AAV. Gene Ther. 2008; 15: 1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Narfström K,, Petersen-Jones SM. Diseases of the canine ocular fundus. : Gelatt KN,, Gilger BC,, Kern TJ, Veterinary Ophthalmology. 5th ed Ames, IA: Wiley-Blackwell; 2013: 1303–1392. [Google Scholar]
- 25. Beryozkin A,, Shevah E,, Kimchi A,, et al. Whole exome sequencing reveals mutations in known retinal disease genes in 33 out of 68 Israeli families with inherited retinopathies. Sci Rep. 2015; 5: 13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lazar CH,, Kimchi A,, Namburi P,, et al. Nonsyndromic early-onset cone-rod dystrophy and limb-girdle muscular dystrophy in a consanguineous Israeli family are caused by two independent yet linked mutations in ALMS1 and DYSF. Hum Mutat. 2015; 36: 836–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernandez-San Jose P,, Corton M,, Blanco-Kelly F,, et al. Targeted next-generation sequencing improves the diagnosis of autosomal dominant retinitis pigmentosa in Spanish patients. Invest Ophthalmol Vis Sci. 2015; 56: 2173–2182. [DOI] [PubMed] [Google Scholar]
- 28. Li FF,, Huang XF,, Chen J,, et al. Identification of novel mutations by targeted exome sequencing and the genotype-phenotype assessment of patients with achromatopsia. J Transl Med. 2015; 13: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koeppen K,, Reuter P,, Kohl S,, Baumann B,, Ladewig T,, Wissinger B. Functional analysis of human CNGA3 mutations associated with colour blindness suggests impaired surface expression of channel mutants A3(R427C) and A3(R563C). Eur J Neurosci. 2008; 27: 2391–2401. [DOI] [PubMed] [Google Scholar]
- 30. Zelinger L,, Cideciyan AV,, Kohl S,, et al. Genetics and disease expression in the CNGA3 form of achromatopsia: steps on the path to gene therapy. Ophthalmology. 2015; 122: 997–1007. [DOI] [PubMed] [Google Scholar]
- 31. Cao O,, Hoffman BE,, Moghimi B,, et al. Impact of the underlying mutation and the route of vector administration on immune responses to factor IX in gene therapy for hemophilia B. Mol Ther. 2009; 17: 1733–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.