Introduction
In the United States, the incidence and mortality rates for Merkel cell carcinoma (MCC) have more than tripled in the last 2 decades.1 The main risk factors involved in MCC pathogenesis include ultraviolet light exposure, immunosuppression, and Merkel cell polyomavirus (MCpV).2 The most common sites include the head and neck (53%) and extremities (34%–35%), with trunk and mucosal surfaces representing less than 10% of cases.3
MCC has a high propensity for local recurrence, with a median time between diagnosis and recurrence of 6 months.4 We present a case of a 79-year-old woman who initially presented with 2 MCCs of the nasal bridge and left arm, followed by a third MCC on the right nose 8 years later. We used array comparative genomic hybridization (aCGH) testing to determine whether the most recent tumor was the result of metastasis or represented a delayed primary recurrence of MCC. Through this case, we suggest that using aCGH to distinguish between primary and metastatic disease can play an important role in future MCC management.
Case history
A 79 year-old woman first presented in 2004 with 2 small red papules involving the nasal bridge and the left arm. Biopsy found MCC (Fig 1). Each MCC was treated by wide local excision and postoperative radiation therapy. No lymph nodes were removed at that time. Positron emission tomography/computed tomography (PET/CT) scan showed no evidence of metastasis.
Fig 1.
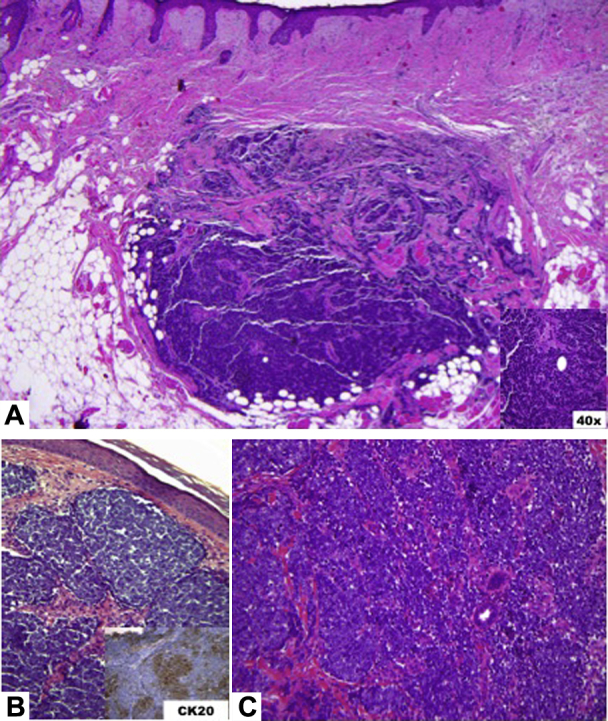
A, Wide local excision of the right side of the nose from 2012 found thin islands and cords and small nests of atypical cells with finely distributed basophilic nuclei without prominent nucleoli and mild pink cytoplasm. Mitotic figures and occasional necrotic cells are identified. Biopsy of the nose from 2004 (B) and the left arm (C) stained positive with CK20.
The patient's course was uneventful until June 2012 when she presented with an 8-mm × 6-mm atrophic lesion on the right side of the nose. Biopsy found MCC, and the patient underwent wide local excision with a negative sentinel lymph node biopsy result (SLNB). Histopathology findings showed MCC and nodular basal cell carcinoma (Fig 1). There was no evidence of systemic metastasis on PET/CT.
The patient is currently closely followed up with total skin examinations every 4 to 6 months. To date, there has been no evidence of recurrence at previous sites or development of new primary lesions. aCGH analysis was performed on tissue from the 2004 left arm and 2012 right side of the nose, and both samples were found to contain multiple chromosomal aberrations, but no significant overlap was seen (Figs 2 and 3). It was determined that the 2 samples most likely represented 2 distinct tumors rather than a reflection of metastasis. The tissue block from the 2004 nose MCC had been destroyed.
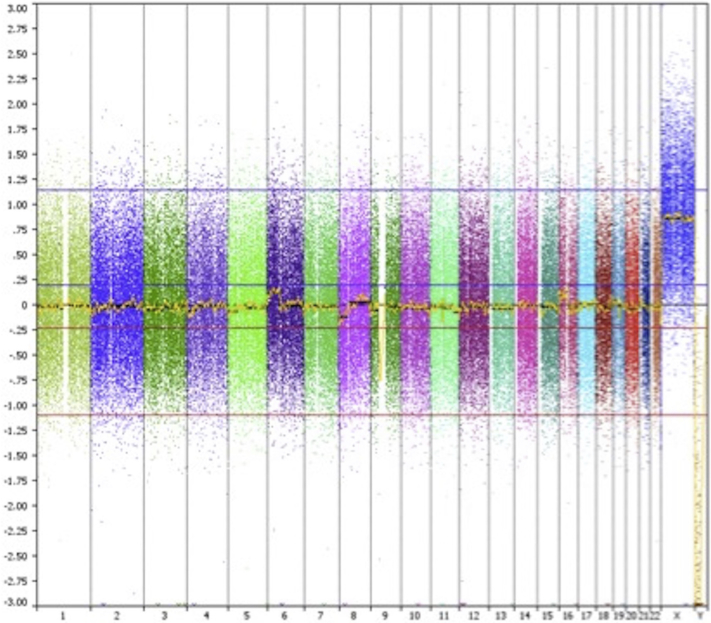
Fig 2.
aCGH results from 2004 left arm shows multiple gains and losses.
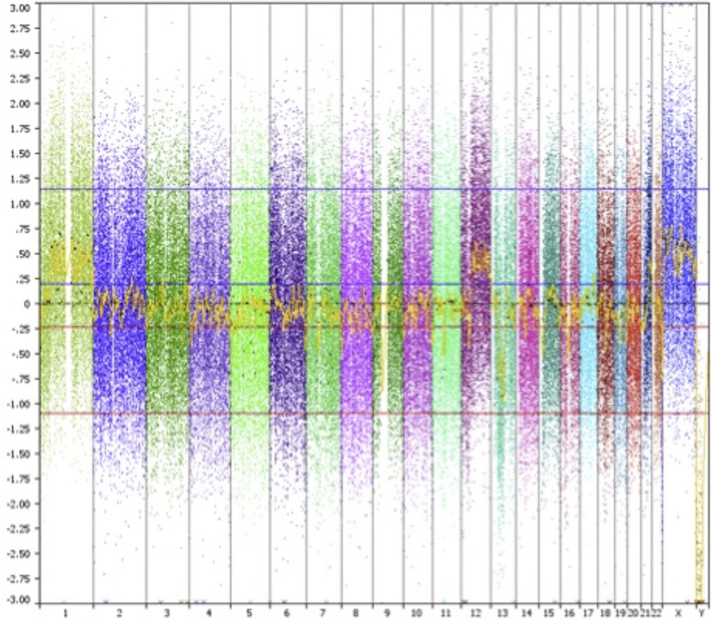
Fig 3.
aCGH results from 2012 specimen from the right side of the nose show multiple gains and losses. There was not significant overlap between the 2004 and 2012 data, suggesting they are separate primary lesions.
Discussion
MCC is a rare, cutaneous malignancy associated with a propensity for rapid local recurrence and regional lymph node metastasis.5 The overall 1-year survival rate is estimated to be 57.3% in the United States.3
We used aCGH to analyze copy number variations in our case, attempting to distinguish between primary versus recurrent/metastatic disease. Our aCGH analysis of available formalin-fixed paraffin-embedded (FFPE) tissue from the 2004 left arm MCC and 2012 right side of the nose MCC showed multiple copy number aberrations with no shared aberrations between the 2 tumors. Given the aCGH evidence, the distinct locations of the 3 lesions (2004 nasal bridge, 2004 left arm, 2012 right side of the nose) and the absence of evidence for metastasis based on SLNB and PET/CT scans, the 2012 lesion from the right side of the nose is not derived from the 2004 left arm tumor. Our case is limited by the lack of comparison of the 2004 nasal bridge MCC, and no definite conclusion can be drawn on whether the 2012 tumor from the right side of the nose is a delayed recurrence of the 2004 nasal bridge MCC.
To our knowledge, there have only been 3 studies to date that have used genomic analysis to investigate the relationship between multiple cutaneous MCCs.6, 7, 8 Nagy et al7 used aCGH to show that although MCCs of the lip and palatine tonsil, separated in time by 7 years, showed 31 distinct copy number alterations, they did not arise independently, as they shared copy number changes at 45 other chromosomal loci.7 Ahronowitz et al6 used aCGH testing of an MCC of the right cheek and left ankle that presented within 4 months of one another with negative SLNB and PET/CT scan on presentation, to show identical copy number variation profiles.8 These studies indicate that physically disparate cutaneous MCCs may represent metastasis rather than separate primary tumors, even after prolonged time intervals and in the absence of lymph node or distant organ involvement. Although aCGH does have the ability to compare chromosomal aberrations between tumors, one of the limitations that we also faced in this study is the propensity for high background noise, especially in FFPE samples. Despite this finding, there were no aberrations that reached threshold levels, and therefore significance, that were shared between the 2 tumors.
MCpV has been found to clonally integrate in 80% of MCCs with expression of large T and small T antigens.9, 10 Modalities for detecting MCpV in MCC have included PCR amplification of viral DNA and immunohistochemistry with antibodies specific for MCpV large and small T antigens. In addition to detecting the presence of viral DNA, MCpV genome sequencing could potentially be used to distinguish between primary and recurrent or metastatic disease. In 2010, Schrama et al8 found different mutations in the MCpV large T-antigen DNA from 2 MCCs of the contralateral arms, separated in time by 6 years, suggesting 2 primary tumors. In our case, we were unable to perform MCpV genome sequencing or testing because of the age of our specimens and the difficulty in analyzing FFPE samples.
We report the first case, to our knowledge, to use aCGH to show at least 2 distinct primary MCCs in the same patient. Recent advances in the treatment of metastatic MCC through immunotherapies and molecular target therapies underscore the importance of distinguishing between multiple primary tumors versus metastatic MCC. Importantly, the clinical finding of a second lesion at a distant anatomic location, even in the absence of lymph node or distant organ involvement, does not in itself distinguish between diagnosis of a second primary MCC versus distant cutaneous metastasis. Our case highlights that aCGH analysis is a useful diagnostic tool in such diagnostically challenging cases.
Footnotes
Funding sources: None.
Conflicts of interest: None declared.
References
- 1.Fitzgerald T.L., Dennis S., Kachare S.D., Vohra N.A., Wong J.H., Zervos E.E. Dramatic increase in the incidence and mortality from Merkel cell carcinoma in the United States. Am Surg. 2015;81:802–806. doi: 10.1177/000313481508100819. [DOI] [PubMed] [Google Scholar]
- 2.Lebbe C., Becker J.C., Grob J.J. Diagnosis and treatment of Merkel Cell Carcinoma. European consensus-based interdisciplinary guideline. Eur J Cancer. 2015;51:2396–2403. doi: 10.1016/j.ejca.2015.06.131. [DOI] [PubMed] [Google Scholar]
- 3.Albores-Saavedra J., Batich K., Chable-Montero F., Sagy N., Schwartz A.M., Henson D.E. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: a population based study. J Cutan Pathol. 2010;37:20–27. doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 4.Grotz T.E., Tarantola T.I., Otley C.C., Weaver A.L., McGree M.E., Jakub J.W. Natural history of merkel cell carcinoma following locoregional recurrence. Ann Surg Oncol. 2012;19:2556–2562. doi: 10.1245/s10434-011-2161-x. [DOI] [PubMed] [Google Scholar]
- 5.Akhtar S., Oza K.K., Wright J. Merkel cell carcinoma: report of 10 cases and review of the literature. J Am Acad Dermatol. 2000;43:755–767. doi: 10.1067/mjd.2000.106505. [DOI] [PubMed] [Google Scholar]
- 6.Ahronowitz I.Z., Daud A.I., Leong S.P. An isolated Merkel cell carcinoma metastasis at a distant cutaneous site presenting as a second ‘primary’ tumor. J Cutan Pathol. 2011;38:801–807. doi: 10.1111/j.1600-0560.2011.01757.x. [DOI] [PubMed] [Google Scholar]
- 7.Nagy J., Feher L.Z., Sonkodi I., Lesznyak J., Ivanyi B., Puskas L.G. A second field metachronous Merkel cell carcinoma of the lip and the palatine tonsil confirmed by microarray-based comparative genomic hybridisation. Virchows Arch. 2005;446:278–286. doi: 10.1007/s00428-004-1176-0. [DOI] [PubMed] [Google Scholar]
- 8.Schrama D., Thiemann A., Houben R., Kahler K.C., Becker J.C., Hauschild A. Distinction of 2 different primary Merkel cell carcinomas in 1 patient by Merkel cell polyomavirus genome analysis. Arch Dermatol. 2010;146:687–689. doi: 10.1001/archdermatol.2010.121. [DOI] [PubMed] [Google Scholar]
- 9.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendzicki J.A., Moore P.S., Chang Y. Large T and small T antigens of Merkel cell polyomavirus. Curr Opin Virol. 2015;11:38–43. doi: 10.1016/j.coviro.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]