Abstract
The Rho GTPase proteins Rac1, RhoA and Cdc42 have a central role in regulating the actin cytoskeleton in dendritic spines1, thereby exerting control over the structural and functional plasticity of spines2, 3, 4, 5 and, ultimately, learning and memory6, 7, 8. Although previous work has shown that precise spatiotemporal coordination of these GTPases is crucial for some forms of cell morphogenesis9, the nature of such coordination during structural spine plasticity is unclear. Here we describe a three-molecule model of structural long-term potentiation (sLTP) of murine dendritic spines, implicating the localized, coincident activation of Rac1, RhoA and Cdc42 as a causal signal of sLTP. This model posits that complete tripartite signal overlap in spines confers sLTP, but that partial overlap primes spines for structural plasticity. By monitoring the spatiotemporal activation patterns of these GTPases during sLTP, we find that such spatiotemporal signal complementation simultaneously explains three integral features of plasticity: the facilitation of plasticity by brain-derived neurotrophic factor (BDNF), the postsynaptic source of which activates Cdc42 and Rac1, but not RhoA; heterosynaptic facilitation of sLTP, which is conveyed by diffusive Rac1 and RhoA activity; and input specificity, which is afforded by spine-restricted Cdc42 activity. Thus, we present a form of biochemical computation in dendrites involving the controlled complementation of three molecules that simultaneously ensures signal specificity and primes the system for plasticity.
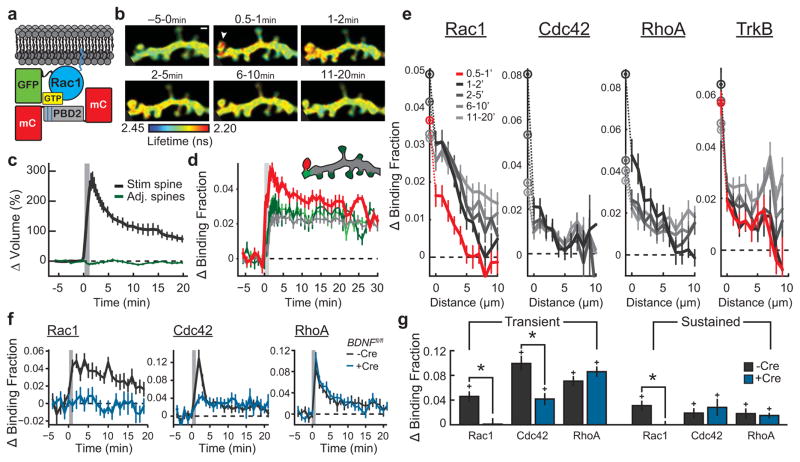
Previous studies using two-photon fluorescence lifetime imaging (2pFLIM) in combination with fluorescence resonance energy transfer (FRET)-based biosensors revealed that the Rho GTPases Cdc42 and RhoA had distinct spatial profiles during sLTP, with Cdc42 showing synapse-restricted activity and RhoA showing a diffuse, heterosynaptic pattern3. To obtain a more complete understanding of the spatiotemporal patterning of Rho GTPase activity during sLTP, we developed a FRET-based sensor for Rac1, following the design of the Cdc42 and RhoA sensors3 (Fig. 1a; validation in Extended Data Figs 1, 2, 3). We transfected rat organotypic hippocampal slices with the sensor using biolistics10, 11 and imaged CA1 pyramidal neurons using 2pFLIM12. When sLTP was induced in single dendritic spines with two-photon glutamate uncaging13, 14 (Fig. 1b, c), Rac1 was rapidly (within ~1 min) activated in the stimulated spine, and remained active for at least 30 min (Fig. 1b, d), notably displaying a more pronounced sustained phase than RhoA or Cdc42 (ref. 3). This activation initially showed limited diffusion, after which it slowly spread over ~10 μm of the parent dendrite until it nearly equalized with activity in the spine (Fig. 1b, d, e, Extended Data Fig. 2a, b). Similar to Cdc42 and RhoA, Rac1 activation was dependent on NMDARs (N-methyl-d-aspartate receptors) and CaMKII (Extended Data Fig. 2d–f), and both pharmacological inhibition of Rac1 and single-cell knockout of Rac1 inhibited sLTP (Extended Data Fig. 4). Thus, like RhoA and Cdc42, Rac1 is a Rho GTPase molecule linking NMDAR–CaMKII signalling to sLTP.
Figure 1. The Rho GTPases Rac1 and Cdc42 convey postsynaptic BDNF–TrkB signalling across both homosynaptic and heterosynaptic domains.
a, Schematic of Rac1 sensor. Monomeric enhanced green fluorescent protein (eGFP) is N-terminally tagged to Rac1 to preserve C-terminal membrane association. GTP binding leads to association with the Pak GTPase binding domain of PAK2R71C,S78A (PBD2; blue lines denote mutations), bringing the mCherry (mCh) fluorophores within the FRET distance of eGFP, decreasing its fluorescence lifetime.
b, Representative 2pFLIM images of Rac1 activation in dendrites during sLTP induced in a single spine with two-photon glutamate uncaging (white arrowhead). Scale bar, 1 μm.
c, Time course of spine volume changes during sLTP induced with two-photon glutamate uncaging (grey window) in the stimulated spine (black) and compared to adjacent spines (green). n = 102 cells/121 spines.
d, Time course of Rac1 activation during sLTP, measured as a change in the fraction of acceptor-bound eGFP–Rac1 in the stimulated spine (black), nearby spines (green), dendrite near the stimulated spine (cyan) and whole dendrite in the image (grey). n = 102/121 (cells/spines).
e, Activations of Rac1 (n = 56/79), Cdc42 (n = 25/38), RhoA (n = 21/23), and TrkB (n = 48/52) (cells/spines) in dendrites as a function of distances from the base of the stimulated spines (lines) and in the stimulated spines (circles).
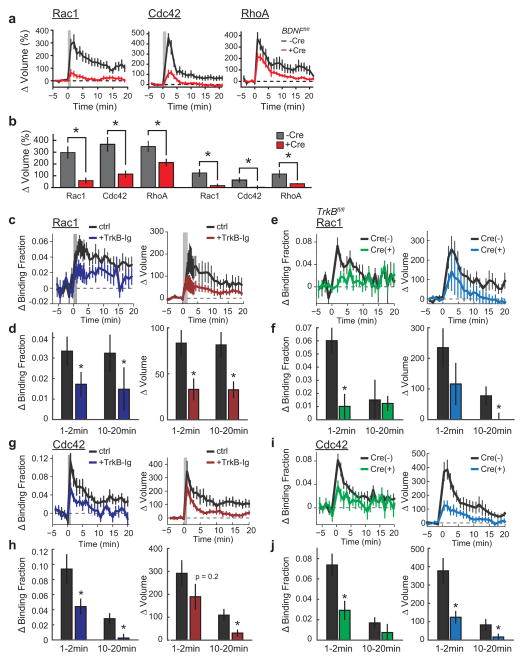
f, Dependence of Rho GTPase activation on postsynaptically synthesized BDNF. Blue indicates co-expression of Cre recombinase in Bdnffl/fl slices along with the Rac1 (left; n = 7/13 Cre−, n = 8/16 Cre+), Cdc42 (middle; n = 5/12 Cre−, n = 4/11 Cre+), or RhoA (right; n = 6/13 Cre−, n = 7/14 Cre+) (cells/spines) sensor. Black represents the corresponding control (Cre−) data.
g, Summary of data from f. Bars represent the average of the activation 1–2 min after stimulation. Error bars represent s.e.m. *P < 0.05 (two-tailed t-test between groups); +P < 0.05 (t-test compared to the baseline).
We next examined whether postsynaptic BDNF15, which also links NMDAR–CaMKII signalling and sLTP, is required for Rho GTPase activation during sLTP. To do this, we used a single-cell knockout technique15, 16 with organotypic hippocampal slices from BDNF conditional knockout mice (Bdnffl/fl)17, 36 coupled with biolistic transfection of Cre recombinase alongside a Rho GTPase sensor. We found that removal of postsynaptic BDNF significantly attenuated Rac1 and Cdc42 activation during sLTP without affecting RhoA (Fig. 1f, g), and reduced the associated expression of sLTP15 (Extended Data Fig. 5a, b). This suggests that Rac1 and Cdc42, but not RhoA, are downstream of postsynaptic BDNF. Similarly, addition of the extracellular BDNF scavenger TrkB-Ig (2 μg ml−1) significantly attenuated the activation of both Rac1 and Cdc42 (Extended Data Fig. 5c, d, g, h). Finally, postsynaptic removal of the BDNF receptor TrkB using Trkbfl/fl (also known as Ntrk2fl/fl) mice also significantly attenuated Rac1 and Cdc42 activation (Extended Data Fig. 5e, f, i, j). These results suggest that an autocrine BDNF–TrkB system controls the activation of Rac1 and Cdc42, thereby instructing two distinct spatial signalling domains relevant to plasticity: a spine-specific domain comprising BDNF–TrkB–Cdc42, and a diffuse domain comprising BDNF–TrkB–Rac1. Since BDNF–TrkB signalling is preferentially enriched in stimulated spines during sLTP (Fig. 1e, Extended Data Fig. 2a), diffusion of Rac1 molecules (Extended Data Fig. 2c) or of Rac1 activators downstream of TrkB7, 18 probably caused the observed spreading of Rac1 activity. By contrast, the spatial spreading of Cdc42 (before its decay) was similar to that of TrkB during the same time period (Extended Data Fig. 2b).
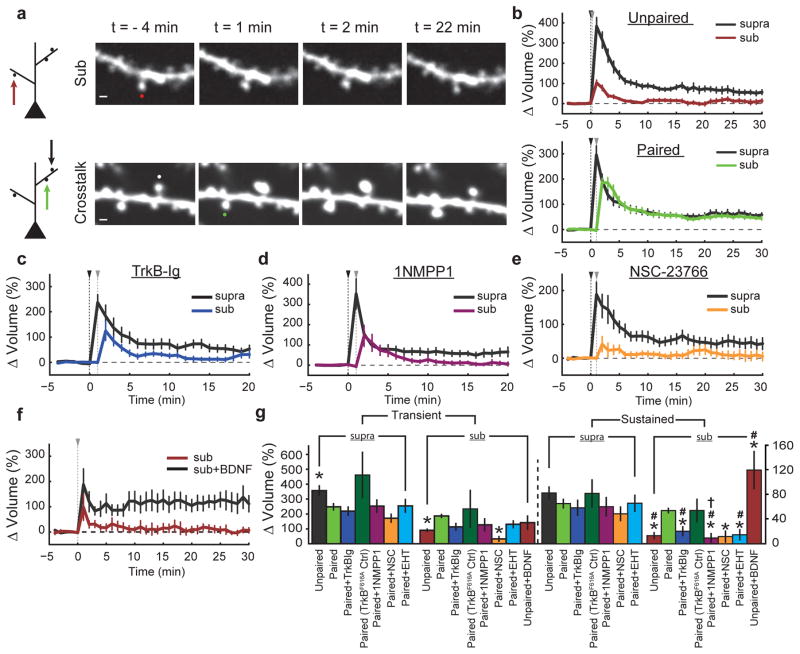
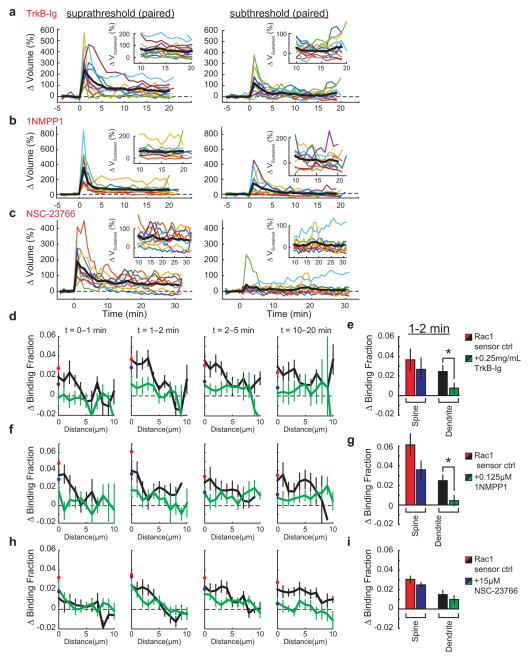
Notably, the length scale of Rac1 spreading is similar to that of synaptic crosstalk, a phenomenon in which sLTP induction briefly facilitates sLTP in nearby (~5–10 μm) spines on the same dendrite19, 20. Thus, we next addressed whether BDNF–TrkB–Rac1 signalling contributes to such crosstalk. Consistent with previous studies19, 20, a suprathreshold sLTP stimulus delivered to one spine allowed a subsequent subthreshold stimulus at a nearby (≤~5 μm) spine to induce sLTP (hereafter referred to as crosstalk) (Fig. 2a, b). To test whether crosstalk requires BDNF–TrkB–Rac1 signalling, we first used partial pharmacological inhibition of this pathway. Although strong inhibition of BDNF signalling has been shown to impair sLTP15, we found that weak inhibition of this pathway with a low concentration (0.25 μg ml−1) of TrkB-Ig preserved sLTP (ΔVsupra sustained = 58 ± 13% (mean ± s.e.m.), in which V denotes spine volume), but attenuated crosstalk (ΔVsub sustained = 20 ± 8%), suggesting that BDNF is required for this process (Fig. 2c, g). To ensure that these effects were due to BDNF signalling through TrkB, we used TrkbF616A mutant mice, which contain a point mutation that renders TrkB uniquely susceptible to inhibition by the small molecule 1NMPP1 (ref. 21). We found that application of a low concentration (0.125 μM) of 1NMPP1 in TrkbF616A slices selectively abolished crosstalk without affecting sLTP (ΔVsupra sustained = 59 ± 16%, ΔVsub sustained = 9 ± 8%) (Fig. 2d, g). Interestingly, weak BDNF–TrkB inhibition also reduced Rac1 activity in dendritic shafts during the time frame of crosstalk induction (1–2 min), without inhibiting the activity in the stimulated spines (Extended Data Fig. 6d–g). Likewise, while a high concentration of the Rac1 inhibitor NSC-23766 significantly reduced sLTP (Extended Data Fig. 4a, c), a small concentration (15 μM) inhibited crosstalk without significantly affecting sLTP (ΔVsupra sustained = 48 ± 12%, ΔVsub sustained = 12 ± 10%) (Fig. 2e, g). This NSC-23766 concentration also trended towards decreasing the Rac1 activation in the shaft (Extended Data Fig. 6h, i). These data suggest that BDNF initiates a signalling cascade capable of lowering the threshold of structural plasticity, consistent with previous reports implicating BDNF in similar phenomena22. Indeed, exogenous BDNF (20 ng ml−1) application for ~10–15 min allowed a subthreshold stimulus alone to induce sLTP (ΔVsub sustained = 119 ± 31%) (Fig. 2f, g). Notably, BDNF application alone was sufficient to activate Rac1 and Cdc42, but had no significant effect on spine volume (Extended Data Fig. 7), suggesting that BDNF is facilitative of, but insufficient for, sLTP. Taken together, these results suggest that the BDNF–TrkB–Rac1 signalling initiated in a single spine during sLTP induction facilitates sLTP in nearby spines, allowing synaptic crosstalk.
Figure 2. BDNF-TrkB-Rac1 signaling is required for synaptic crosstalk.
a, Left, schematic of crosstalk model. Top, in ‘unpaired’ trials, the subthreshold (sub) stimulus was delivered to a single spine. Bottom, in the ‘paired’ (crosstalk) condition, a threshold stimulus was delivered to a spine before the delivery of a subthreshold stimulus to a nearby spine on the same dendrite. Right, representative images of unpaired (top) and paired (bottom) subthreshold stimuli. Scale bars, 1 μm.
b, Quantification of volume change for unpaired (top; n = 24/27 suprathreshold (supra), n = 25/29 sub) (cells/spines) and paired (bottom; n = 35/47) (cells/spine pairs) stimuli. Black and grey triangles indicate suprathreshold and subthreshold stimuli, respectively.
c, Effect of 0.25 μg ml−1 TrkB-Ig on synaptic crosstalk. n = 6/13 (cells/spine pairs).
d, Effect of 0.125 μM 1NMPP1 on synaptic crosstalk in TrkbF616A mice. n = 5/10 (cells/spine pairs).
e, Effect of the Rac1 inhibitor NSC-23766 (NSC; 15 μM) on synaptic crosstalk. n = 6/10 (cells/spine pairs).
f, Effect of 20 ng ml−1 BDNF on spine volume change after a subthreshold stimulus. n = 6/6 sub, n = 6/7 sub plus BDNF.
g, Summary of b–f, with the addition of the Rac1 inhibitor EHT-1864 (EHT; n = 7/12; cells/spine pairs) and TrkbF616A cells in the absence of 1NMPP1 (TrkbF616A ctrl; n = 5/7; cells/spine pairs) showing averages of transient (1–2 min after stimulation) and sustained (>10 min after stimulation) phases of sLTP. Left, spines stimulated with a threshold stimulus. Right, spines stimulated with an unpaired or paired subthreshold stimulus. *P < 0.05 (Dunnet’s test, versus the ‘paired’ subthreshold stimulus standard the crosstalk ‘control’); #P < 0.05 (two-tailed t-test, versus each condition’s paired crosstalk control).
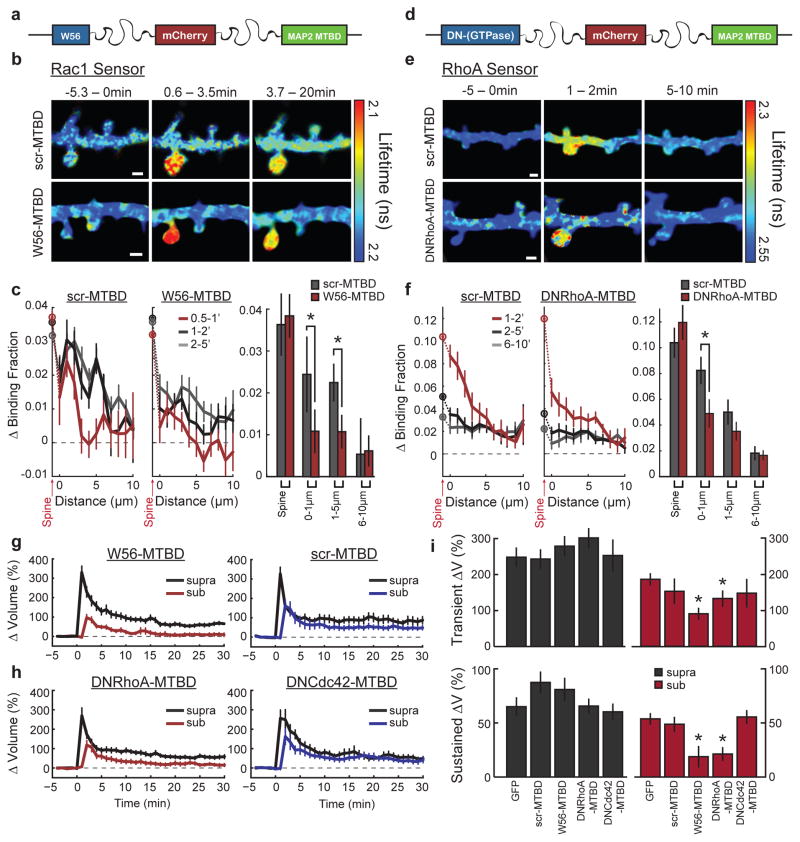
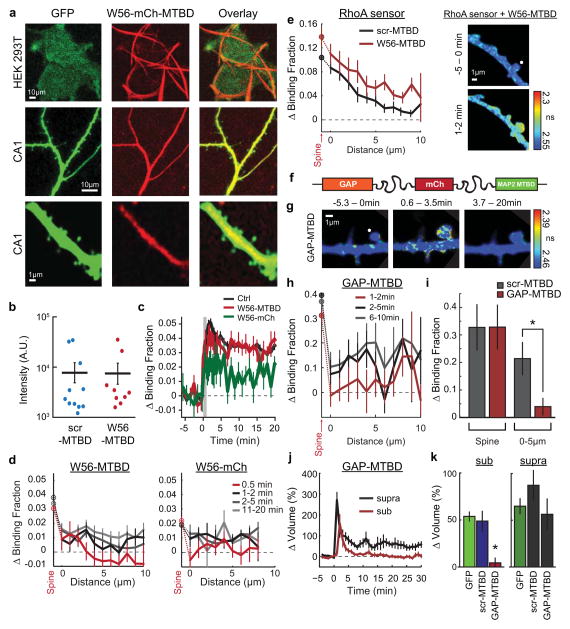
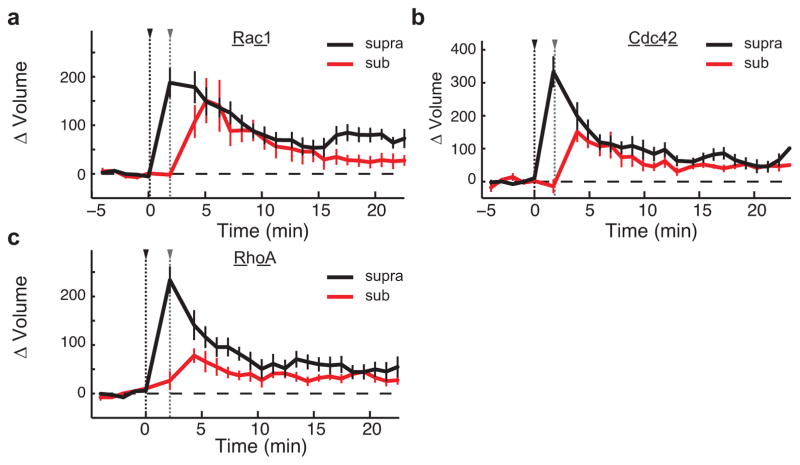
To probe the requirement of heterosynaptic Rac1 activity for synaptic crosstalk more specifically, we devised a strategy to interrupt Rac1 activity spreading out of the stimulated spine during sLTP: we restricted the Rac1 inhibitory peptide W56 (ref. 23) to dendrites by fusing it to the microtubule binding domain (MTBD) of MAP2, concentrating the inhibitor to dendritically enriched microtubules24, 25(Fig. 3a, Extended Data Fig. 8a). This construct, W56–mCh–MTBD (in which mCh denotes mCherry), was concentrated in dendritic shafts and excluded from spines (Extended Data Fig. 8a). We found that this inhibitor significantly reduced Rac1 activation in the dendritic shaft, while largely preserving its activation in the stimulated spine (Fig. 3b, c). By contrast, W56–mCh without MTBD resulted in a more global reduction in Rac1 activation (Extended Data Fig. 8c, d). When the W56 peptide was scrambled (scr–mCh–MTBD), Rac1 activity spreading was normal (Fig. 3b, c). Finally, W56–mCh–MTBD did not change the spatial spreading of RhoA (Extended Data Fig. 8e). These results suggest that W56–mCh–MTBD specifically inhibits the spreading of Rac1 activity into dendrites.
Figure 3. Inhibition of signal spreading of Rac1 and RhoA prevents synaptic crosstalk.
a, Schematic of the dendritic Rac1 inhibitor construct. W56 is fused to mCh and the microtubule binding domain (MTBD) of human MAP2 via intermediate linker sequences (wavy lines).
b, Representative 2pFLIM images of the effect of W56–mCh–MTBD (W56–MTBD; bottom) or scrambled control (scr–MTBD; top) on the activation profile of the Rac1 sensor. Scale bars, 1 μm.
c, Quantification of the spreading of Rac1 in scrambled control (left; n = 10/17) versus W56–mCh–MTBD (middle; n = 22/36) (cells/dendrite). Data represent the change in binding fraction in the dendrite as a function of distance from the stimulated spine (lines) and in the stimulated spines (circles) in the indicated time epochs. Right, Rac1 signal spreading in the presence of W56–mCh–MTBD across the indicated spatial windows at 1–2 min after stimulation. *P < 0.05, two-tailed t-test.
d, Schematic of the dendritic RhoA inhibitor, dominant-negative (DN) RhoA–mCh–MTBD.
e, Representative 2pFLIM images of the effect of DNRhoA–mCh–MTBD expression on the activation profile of the RhoA sensor (bottom) versus scrambled control (top). Scale bars, 1 μm.
f, Quantification of the spreading of RhoA in control (n = 8/17) versus DNRhoA–mCh–MTBD (n = 11/18) (cells/dendrite). Right, summary of RhoA spreading data. *P < 0.05, two-tailed t-test.
g, Effect of W56–mCh–MTBD (left; n = 30/47) versus scr–mCh–MTBD (right; n = 14/17) (cells/spine pairs). expression on synaptic crosstalk.
h, Effect of DNRhoA–mCh–MTBD (left; n = 14/26) and DNCdc42–mCh–MTBD (right; n = 10/15) (cells/spine pairs) on synaptic crosstalk. i, Summary of crosstalk experiments. Right, averages for the crosstalk spine. Left, averages of the first (‘LTP’) spine. *P < 0.05, Dunnet’s test, compared to eGFP control.
Consistent with the hypothesis that Rac1 activity spreading is required for synaptic crosstalk, the expression of W56–mCh–MTBD reduced crosstalk without significantly affecting sLTP (Fig. 3g). By contrast, a similar expression level of scr–mCh–MTBD affected neither sLTP nor crosstalk (Fig. 3g, Extended Data Fig. 8b). To test the specificity of this manipulation further, we tethered the Rac1-specific GTPase-activating protein ARHGAP15 (ref. 26) to MTBD. This construct also reduced Rac1 activity spreading without significantly affecting activation in the spine, and inhibited crosstalk without affecting sLTP (Extended Data Fig. 8f–k). Thus, activation of Rac1 in the dendritic shaft is necessary for synaptic crosstalk.
Since RhoA activation also spreads into nearby spines during sLTP, we next tested whether this BDNF-independent signal is also required for synaptic crosstalk. To do this, we tethered dominant-negative RhoA (DNRhoA) to mCh–MTBD (Fig. 3d). Expressing DNRhoA–mCh–MTBD significantly reduced a portion of RhoA activity spreading (Fig. 3e, f), and also blocked crosstalk without significantly affecting sLTP (ΔVsupra sustained = 66 ± 6%, ΔVsub sustained = 21 ± 6%) (Fig. 3h, i). Importantly, using this same strategy against Cdc42 (the activity of which is compartmentalized in spines) had no effect on either sLTP or crosstalk (ΔVsupra sustained = 60 ± 7%, ΔVsub sustained = 56 ± 6%) (Fig. 3h, i), suggesting that this approach is only effective when targeting proteins with diffusive activation profiles. Taken together, these data suggest that the convergence of BDNF-dependent Rac1 signalling and BDNF-independent RhoA signalling at nearby spines primes these regions for facilitated structural plasticity.
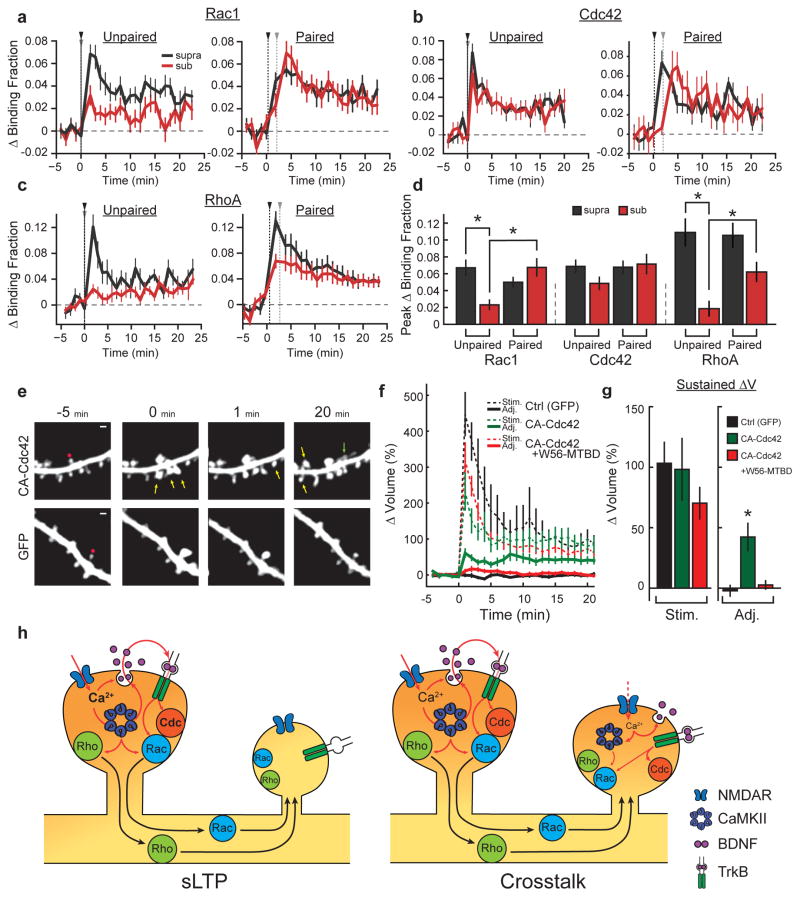
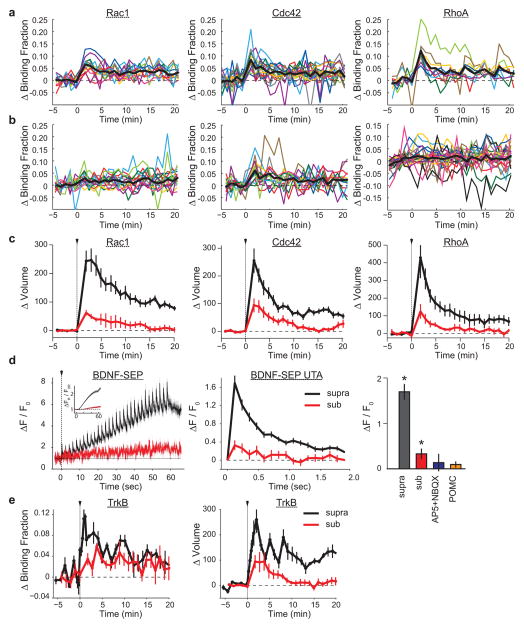
To examine how crosstalk is achieved despite signal spreading conferring activation of only a subset of GTPases, we measured the activation of Rho GTPases in response to both unpaired and paired suprathreshold and subthreshold stimuli (Fig. 4a–d; see Extended Data Fig. 9a, b for response variability and Extended Data Figs 9c and 10 for volume curves of subthreshold stimuli). We found that both RhoA and Rac1, which show diffusive activity profiles, were only weakly activated by unpaired subthreshold stimuli (Fig. 4a, c). However, when paired with nearby suprathreshold stimuli, signal spreading supplied the crosstalk spines with additional Rac1 and RhoA activation (Fig. 4a, c, d). By contrast, Cdc42 was strongly activated by a subthreshold stimulus in both the unpaired and paired conditions, thus explaining how nearby spines achieve the required levels of Cdc42 activation during crosstalk (Fig. 4b, d). Thus, through a combination of low-threshold, spine-specific Cdc42 activation and signal spreading of RhoA/Rac1 provided by nearby sLTP, Rho GTPase signalling complementation facilitates detection of weak synaptic activity proximal to sites of sLTP. Notably, a weak stimulus also caused a modest level of BDNF release (as measured by BDNF fused to the pH-sensitive fluorophore superecliptic pHluorin; BDNF–SEP) and TrkB activation, supporting the notion that this pathway is still functional during a weak stimulus (Extended Data Fig. 9d, e).
Figure 4. Signal spreading provides additive activation of high-threshold signals during synaptic crosstalk.
a, Comparison of Rac1 activation in response to unpaired (left; suprathreshold (black) n = 12/12, subthreshold (red) n = 15/15; cells/spines) and paired crosstalk (right; n = 5/9; cells/spine pairs) stimuli. Black and grey arrows indicate supra/subthreshold stimulus onset.
b, Same as a for Cdc42 activity (left: unpaired; suprathreshold n = 14/18, subthreshold n = 14/20; right: paired; n = 9/10).
c, Same as a and b for RhoA activity (left: unpaired; suprathreshold n = 9/9, subthreshold n = 17/22; right: paired; n = 13/24).
d, Summary of data from a–c. Bars represent average peak activity. For unpaired conditions, the peak is the average of the first 2 min following uncaging. For paired conditions, the peak for suprathreshold spines corresponds to the first 2 min after the stimulus, while the peak of the crosstalk spines was based on the timing of maximal response for each GTPase (Rac1 and Cdc42: 1–2 min after the second stimulus; RhoA: 1–2 min after the first stimulus) to account for signal spreading. *P < 0.05, analysis of variance (ANOVA) with Tukey–Kramer’s post-hoc test.
e, Top, representative two-photon images of the effect of expressing CA-Cdc42 on the spine-specificity of sLTP. Yellow arrows mark non-targeted spines that increased in intensity after uncaging at the target (red circle) spine. The green arrow represents a new spine. Bottom, representative images of a typical sLTP experiment in which there is no heterosynaptic effect of uncaging. Image size, 12 × 12 μm.
f, Quantification of the average change in nearby (<~5 μm) spine volume after glutamate uncaging in control (black curve; n = 13 cells/13 targeted spines/112 neighbouring spines), CA-Cdc42-expressing (green curve; n = 9 cells/14 targeted spines/102 neighbouring spines), and CA-Cdc42- plus W56–mCh–MTBD-expressing (red curve; n = 5 cells/11 targeted spines/78 neighbouring spines) cells.
g, Summary of data in f. *P < 0.05, ANOVA with Tukey–Kramer’s test.
h, Proposed model. Calcium through NDMARs from a strong stimulus activates CaMKII, supporting autocrine BDNF–TrkB signalling, and conferring actin cytoskeleton remodelling through Rac1 and Cdc42. In parallel, CaMKII-dependent RhoA activation also acts on the actin cytoskeleton. The combination of the three GTPases is necessary to produce sLTP. Rac1 and RhoA activity spread out of the stimulated spine into the dendrite and surrounding spines, which is insufficient to produce sLTP. However, even a weak stimulus (red dotted line), can cause sufficient BDNF release to activate Cdc42, complementing the activity of Rac1 and RhoA and allowing synaptic crosstalk.
The compartmentalized nature of Cdc42 activity in spines probably serves to prevent nonspecific structural plasticity at nearby inactive synapses. Consistent with this model, stimulation of a single spine on cells expressing constitutively active (CA)-Cdc42 caused significant enlargement of surrounding, unstimulated spines (ΔVnearby sustained = 42 ± 11%) (Fig. 4e–g). This is in sharp contrast to control conditions, in which nearby spines show no average change in volume (ΔVnearby sustained = −2 ± 4%) (Fig. 4e–g). Importantly, sLTP in the stimulated spines was similar to that of control (Fig. 4e–g). Furthermore, when CA-Cdc42 was co-expressed with W56–mCh–MTBD, sLTP in nearby spines was suppressed without affecting sLTP at the stimulated spines (ΔVnearby sustained = 3 ± 3%; ΔVstim sustained = 70 ± 13%) (Fig. 4e–g), suggesting that the heterosynaptic effects imparted by CA-Cdc42 depend on Rac1 signal spreading. Thus, removing the compartmentalization of Cdc42 activation degrades the input-specificity of sLTP, likely by complementing the typical priming effects of diffusive Rac1 and RhoA signals.
Collectively, our data suggest that simultaneous activation of Rac1, Cdc42 and RhoA predicts the occurrence of sLTP, while activation of a subset of these proteins primes spines for sLTP (Fig. 4h). The activation of both Cdc42 and Rac1 required postsynaptic, autocrine BDNF, and conveyed the occurrence of sLTP over both spine-specific (Cdc42) and heterosynaptic (Rac1) domains. The spreading of BDNF–TrkB-mediated Rac1 signalling out of the stimulated spine was necessary for facilitating sLTP in nearby spines, consistent with the known pro-plasticity properties of BDNF22, 27, 28, 29. The spreading of BDNF-independent RhoA activation was also necessary for synaptic crosstalk, suggesting that both BDNF-dependent and -independent signalling pathways must be coordinated to achieve this phenomenon. The combination of spine-specific, low-threshold Cdc42 activation together with diffusive, high-threshold RhoA and Rac1 activation is perfectly positioned to simultaneously achieve both spine-specific homosynaptic sLTP and facilitation of sLTP in surrounding spines. Thus, our model, based on the coincident activation of three small GTPase molecules Rac, Rho and Cdc42, together with the autocrine BDNF signalling, provides a unified theory for both homo- and heterosynaptic plasticity.
Methods
Reagents
Human recombinant BDNF was purchased from Millipore; d-2-amino-5-phosphonovalerate (d-AP5) and NSC-23766 were from Tocris; 1′-naphthylmethyl-4-amino-1-tert-butyl-3-(p-methylphenyl)pyrazolo[3,4-d] pyrimidine (1NMPP1) was from Santa Cruz and Shanghai Institute of Materia Medica, Chinese Academy of Sciences; and TrkB-Ig was a gift from Regeneron. The tat-CN21 peptide (YGRKKRRQRRRKRPPKLGQIGRSKRVVIEDDR) was synthesized by GenScript.
Animals
All animal procedures were approved by the Duke Univeristy School of Medicine Animal Care and Use Committee. Both male and female rats and mice were used. TrkbF616A mutant mice were provided by D. Ginty21. Bdnffl/fl and Trkbfl/fl were provided by L. Parada17, 36. Rac1fl/fl animals were acquired from C. Brakebusch30. The genotype of each animal was verified before and after preparing slices using PCR of genomic DNA isolated from tail DNA before and slice samples after.
Plasmids
Plasmids containing human RAC1 and PAK1(65–118) are gifts from M. Matsuda and S. Soderling, respectively. The Pak GTPase binding domain of PAK2 (PBD2) was prepared by introducing mutations L77P and S115L into PAK1(60–118) using a Site-Directed Mutagenesis kit (Stratagene). W56–mCh–MTBD was prepared by amplifying the Rac1 inhibitory peptide W56 (ref. 23) using overhang PCR with a C-terminal linker (GGGGGGGGGGGGGGGGGGGGGGGGMADQLTEEWHRGTAGPGS) and inserting it into pCAG-mCh-mCh (ref. 3) by removing the first mCh with EcoRI and KpnI restriction digest and replacing it with the W56-linker amplicon, creating pCAG-W56-(linker)-mCh. In parallel, the MTBD of human MAP2 (272-end)25 was isolated from a human cDNA library and PCR amplification. This amplicon was then further amplified with overhang PCR to contain a linker (same as above), and then inserted into pCAG-mCh-mCh using BamHI-NotI restriction digest to produce pCAG-mCh-(Linker)-MTBD. The two constructs were then combined using BamHI plus NotI restriction digest to create pCAG-W56-(linker)-mCh-(linker)-MTBD. The scrambled variant of W56–MTBD was created by randomly re-ordering the residues of W56. ARHGAP15-mCh-MTBD was made by inserting ARHGAP15 (1–723; Addgene plasmid 38903) into the -mCh-MTBD sequence described above by adding EcoRI and KpnI sites at the N and C terminus, respectively.
DNRhoA and DNCdc42 variants of the X-mCh-MTBD construct were prepared by first incorporating an MfeI digestion site on the 3′ end of W56, then removing W56 by digestion with NheI/MfeI and insertion of the dominant-negative construct.
Preparation
Hippocampal slices were prepared from postnatal day 5–7 rats or mice in accordance with the animal care and use guidelines of Duke University Medical Centre. In brief, we deeply anaesthetized the animal with isoflurane, after which the animal was quickly decapitated and the brain removed. The hippocampi were isolated and cut into 350-μm sections using a McIlwain tissue chopper. Hippocampal slices were plated on tissue culture inserts (Millicell) fed by tissue medium (for 2.5 l: 20.95 g MEM, 17.9 g HEPES, 1.1 g NaHCO3, 5.8 g d-glucose, 120 μl 25% ascorbic acid, 12.5 ml l-glutamine, 2.5 ml insulin, 500 ml horse serum, 5 ml 1 M MgSO4, 2.5 ml 1 M CaCl2). Slices were incubated at 35 °C in 3% CO2.
After 1–2 weeks in culture, CA1 pyramidal neurons were transfected with ballistic gene transfer using gold beads (8–12 mg) coated with plasmids containing 30 μg of total cDNA (Rac1 sensor, donor:acceptor = 1:2; eGFP + W56–MTBD, 5:1; Rac1 sensor + W56–MTBD, donor:acceptor:inhibitor = 2:4:1; TrkB sensor, donor:acceptor = 1:1; Cdc42 sensor, donor:acceptor = 1:1; RhoA sensor, donor:acceptor = 1:1). Cells expressing only eGFP were imaged 1–5 days after transfection, cells expressing TrkB were imaged 1–2 days after transfection, and all other plasmid combinations were imaged 2–5 days after transfection.
For structural plasticity experiments, conditional knockout slices (Bdnffl/fl and Rac1fl/fl) were transfected with either eGFP alone or eGFP and tdTomato-Cre (1:1) for 3–7 days before imaging. For sensor experiments in these slices, the sensors were used in the ratios listed above with an amount of Cre recombinase equal to the amount of donor DNA. The presence of Cre was confirmed by nuclear-localized tdTomato signal.
HEK293T cells (ATCC) were cultured in DMEM supplemented with 10% fetal calf serum at 37 °C in 5% CO2. Transfection was performed at ~50–90% cell confluency using Lipofectamine (Invitrogen) and 2 μg ml−1 of total cDNA/35 mm dish, following the ratios listed above. Cells were used as an expression platform only, and were thus not rigorously tested for potential contamination from other cell lines.
2pFLIM
FRET imaging using a custom-built two-photon fluorescence lifetime imaging microscope was performed as previously described3, 31, 32. Two-photon imaging was performed using a Ti-sapphire laser (MaiTai, Spectraphysics) tuned to a wavelength of 920 nm, allowing simultaneous excitation of eGFP and mCh. All samples were imaged using <2 mW laser power measured at the objective. Fluorescence emission was collected using an immersion objective (60×, numerical aperture 0.9, Olympus), divided with a dichroic mirror (565 nm), and detected with two separate photoelectron multiplier tubes (PMTs) placed downstream of two wavelength filters (Chroma, HQ510-2p to select for green and HQ620/90-2p to select for red). The green channel was fitted with a PMT having a low transfer time spread (H7422-40p; Hamamatsu) to allow for fluorescence lifetime imaging, while the red channel was fitted with a wide-aperture PMT (R3896; Hamamatsu). Photon counting for fluorescence lifetime imaging was performed using a time-correlated single photon counting board (SPC-150; Becker and Hickl) controlled with custom software31, while the red channel signal was acquired using a separate data acquisition board (PCI-6110) controlled with Scanimage software33.
Two-photon glutamate uncaging
A second Ti-sapphire laser tuned at a wavelength of 720 nm was used to uncage 4-methoxy-7-nitroindolinyl-caged-l-glutamate (MNI-caged glutamate) in extracellular solution with a train of 4–6 ms, 4–5 mW pulses (30 times at 0.5 Hz) near a spine of interest (‘sLTP stimulus’). Experiments were performed in Mg2+ fee artificial cerebral spinal fluid (ACSF; 127 mM NaCl, 2.5 mM KCl, 4 mM CaCl2, 25 mM NaHCO3, 1.25 mM NaH2PO4 and 25 mM glucose) containing 1 μM tetrodotoxin (TTX) and 4 mM MNI-caged l-glutamate aerated with 95% O2 and 5% CO2 at 30 °C, as described previously. Subthreshold stimuli were delivered using a train of 1 ms, 4–5 mW pulses (30 times at 0.5 Hz). Crosstalk experiments were performed by first delivering an sLTP stimulus (4–6 ms), then delivering a subthreshold stimulus to a nearby (~2–5 μm) spine on the same dendrite ~90 s later, as previously described19. Anywhere from 1–5 spines were stimulated per cell, and a maximum of 3 crosstalk experiments were performed on a single cell.
Spine volume analysis
Spine volume was calculated as the background-subtracted integrated fluorescence intensity over a region of interest around the dendritic spine head (fluorescence, F). Change in spine volume was measured as F/F0, in which F0 is the average fluorescence intensity before stimulation. Analysis of two-photon images outside of the context of 2pFLIM was performed in ImageJ.
All experiments involving dendritic inhibitor constructs were performed in a blinded fashion until the experiments were complete (when the groups significantly diverged, or until 15–20 individual experiments across at least three cells were complete, whichever came first).
2pFLIM data analyses
To measure the fraction of donor bound to acceptor, we fit a fluorescence lifetime curve summing all pixels over a whole image with a double exponential function convolved with the Gaussian pulse response function:
| (1) |
where τAD is the fluorescence lifetime of donor bound with acceptor, PD and PAD are the fraction of free donor and donor bound with acceptor, respectively, and H(t) is a fluorescence lifetime curve with a single exponential function convolved with the Gaussian pulse response function:
in which τD is the fluorescence lifetime of the free donor, τG is the width of the Guassian pulse response function, F0 is the peak fluorescence before convolution and t0 is the time offset, and erfc is the error function.
We fixed τD to the fluorescence lifetime obtained from free eGFP (2.6 ns). To generate the fluorescence lifetime image, we calculated the mean photon arrival time, 〈t〉, in each pixel as:
then, the mean photon arrival time is related to the mean fluorescence lifetime, 〈τ〉, by an offset arrival time, t0, which is obtained by fitting the whole image:
For small regions-of-interest (ROIs) in an image (spines or dendrites), we calculated the binding fraction (PAD) as:
Measurements of the affinity between Rho GTPases and RBDs
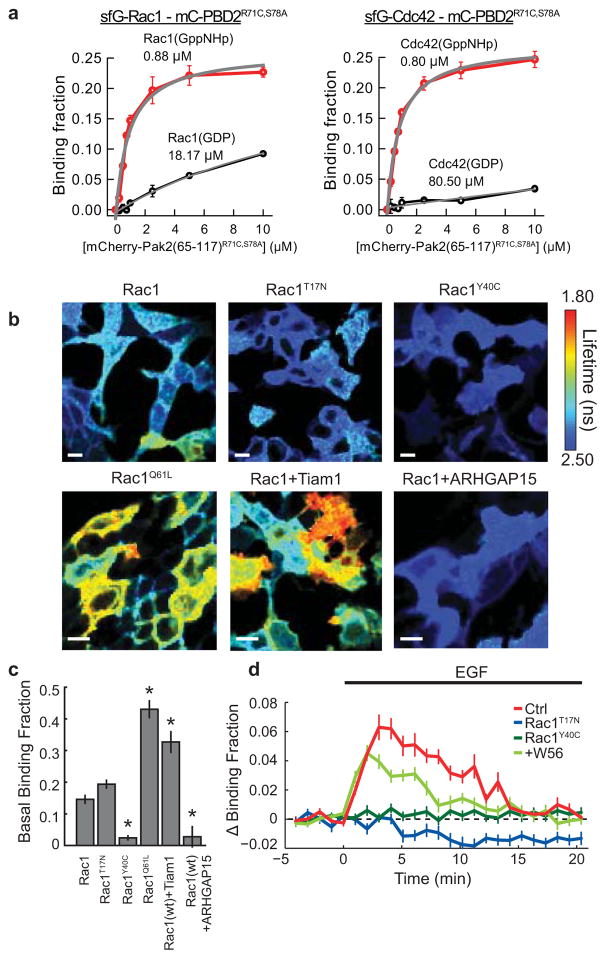
Polyhistidine-tagged super-folder GFP (sfGFP)–Rac1, mCh-PBD2 and their mutants were cloned into the pRSET bacterial expression vector (Invitrogen). Proteins were overexpressed in Escherichia coli (DH5α), purified with a Ni+-nitrilotriacetate (NTA) column (HiTrap, GE Healthcare), and desalted with a desalting column (PD10, GE Healthcare) equilibrated with PBS. The concentration of the purified protein was measured by the absorbance of the fluorophore (sfGFP, A489 nm = 83,000 cm−1 M−1 (ref. 34); mCh, A587 nm = 72,000 cm−1 M−1 (ref. 35)).
Purified sfGFP–Rac1 was loaded with GppNHp (2′,3′-O-N-methyl anthraniloyl-GppNHp) and GDP by incubating in the presence of tenfold molar excess of GppNHp and GDP in MgCl2-free PBS containing 1 mM EDTA for 10 min, respectively. The reaction was terminated by adding 10 mM MgCl2. sfGFP–Rac1 and mCh–PBD2 were mixed and incubated at room temperature for 20 min. FRET between sfGFP and mCh was measured under 2pFLIM, and the fraction of sfGFP–Rac1 bound to mCh–PDB2 was calculated by fitting the fluorescence lifetime curve with a double exponential function (equation (1)). The dissociation constant was obtained by fitting the relationship between the binding fraction and the concentration of mCh–PDB2 ([mCh–PDB2]) with a Michaelis–Menten function.
Statistical methods
Sample sizes for all experiments were chosen based on signal-to-noise ratios identified in pilot experiments.
The variances of all data were estimated and compared using Bartlett’s test or Levene’s test before further statistical analysis.
The distribution patterns of Rho GTPase sensor activity was determined by performing a Shapiro–Wilk test for normality on the peak response (the same points used for statistical comparisons). All of the sensors tested adhered to the null hypothesis, and thus are considered normally distributed. As such, parametric statistics were used to compare values of Rho GTPases response.
For multiple comparisons of sensor activity, data were first subjected to ANOVA, followed by a post-hoc test to determine statistical significance, according to the structure of the comparison being made. In cases where each condition is being compared to all other conditions in the group, the Tukey–Kramer method was used. In cases where each condition is being compared to a single control, Dunnet’s test was used instead.
To compare values of non-normally distribution changes in spine volume, data were log-transformed to resolve skewness, then subjected to normal parametric statistics, as indicated in the figure legends. To support these statistical claims, non-parametric statistics were also applied to the original, non-transformed data using a Wilcoxon rank-sum test in place of t-tests, and the Kruskal–Wallis procedure in place of ANOVA, followed by a post-hoc analysis using Dunn’s test. All of the data tested were significant by both of these approaches.
Data were only excluded if obvious signs of poor cellular health (for example, dendritic blebbing, spine collapse) were apparent.
Crosstalk experiments comparing different genetic perturbations were performed in a blinded fashion. Experimenters were unblinded when either statistical significance was reached, or when experimental number was comparable to similar experiments that had reached statistical significance.
Extended Data
Extended Data Fig. 1. Design and characterization of the Rac1 sensor.
a, Measurements of the affinity between sfGFP-Rac1 or sfGFP-Cdc42 and the p21-activated-kinase-derived acceptor construct, Pak2(65-117)R71C,S78A. The binding fraction was measured using 2pFLIM (see Methods) across several concentrations of the acceptor construct. The dissociation constant was obtained by fitting the data (red) with a Michaelis-Menten function (gray).
b. Representative fluorescence lifetime images of Rac1 sensor variants in HEK293T cells. Cells were transfected with a 1:2 donor:acceptor ratio of wild-type, dominant negative (Rac1T17N), substrate-binding dead (Rac1Y40C), or constitutively active (Rac1Q61L) variants of Rac1 with mCherry-PBD2R71C,S78A-mCherry. Some experiments included the addition of an additional construct (a Rac1 GEF, Tiam1, or GAP, ARHGAP15) in a 1:2:1 donor:acceptor:GEF/GAP ratio. Cells were imaged 12–36 hours after transfection in a warmed solution containing 30mM Na_HEPES(pH 7.3), 130mM NaCl, 2.5 mM KCl, 1mM CaCl2, 1mM MgCl2, 2mM NaHCO3, 1.25mM NaH2PO4, and 25mM glucose. Warmer colors indicate a lower fluorescence lifetime value/higher binding fraction of mEGFP-Rac1 to the acceptor construct. Scale bars represent 50μm.
c. Basal binding fraction of Rac1-PBD2R71C, S78A for the conditions listed in (b). Error bars represent s.e.m. Asterisks represent a statistically significant difference from the Rac1(wt) control condition as determined by ANOVA followed by post-hoc tests using the least significant difference (p < 0.0001).
d. Time course of Rac1 activation in HEK293T cells upon application of 100 μg/mL epidermal growth factor (EGF). Control (Ctrl) experiment corresponds to mEGFP-Rac1 (donor)+mCherry-PBD2R71C,S78A-mCherry (acceptor) expression alone (n = 39 cells/8plates), Rac1T17N (n = 33 cells/4 plates) and Rac1Y40C (n = 40 cells/5 plates) corresponds to the expression of the donor variant with the acceptor in the same ratio as controls, and Rac1+W56 corresponds to the expression of the wild-type donor with the acceptor and the Rac1 inhibitory peptide W56.
Extended Data Fig. 2. Characterization of the Rac1 sensor during single spine structural plasticity in hippocampal slices.
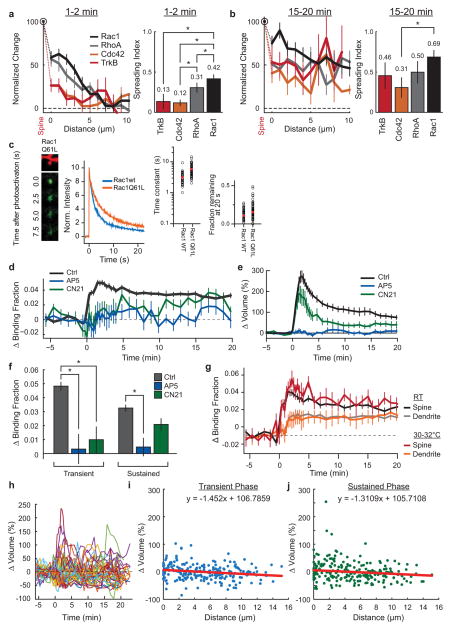
a. (left) Comparison of the signal spreading of Rac1, RhoA, Cdc42, and TrkB, normalized to spine activity from 1–2 minutes post stimulus onset. Error bars represent s.e.m. Data corresponds to that shown in Fig. 1 (Main Text). (right) Comparison of the spreading indices of the four sensors (Spreading Index = Activity in dendrite (1–5 μm) from 1–2 minutes post-stimulation/maximum spine activation). Error was estimated by bootstrapping. Asterisks indicate a statistically significant difference as deteremined by an independent-samples t-test (p<0.05).
b. Same as (a), but for 15–20 min post stimulation.
c. Analysis of the diffusion time constants of constitutively active (Q61L) and wild-type Rac1 using photo-activatable GFP (paGFP). (left) Representative 2p images of paGFP-Rac1Q61L in a single dendritic spine after photo-activation. While active Rac1 shows a slightly slower diffusion that Rac1(wt) (middle), both Rac1Q61L and Rac1(wt) diffuse away from the spine with approximately equal diffusion time constants (right two plots; wt: n = 41 spines/4 cells; Rac1Q61L: 67 spines/7 cells).
d. Pharmacological characterization of Rac1 signal during sLTP. Rac1 activation in control (ctrl; n = 102 cells/121 spines) conditions during 2p-glutamate uncaging, in the presence of the NMDA receptor blocker APV (APV; 100μM; n = 6 cells/13 spines), and in the presence of the cell-permeable CaMKII inhibitory peptide tatCN21 (CN21; 10μM; n = 5 cells/11 spines). Error bars represent s.e.m.
e. Time course of spine volume change for experiments (d). Error bars represent s.e.m.
f. Summary of effect of AP5 and CN21 on the transient (1–2min post stimulation; Ctrl ΔBF = 0.049±0.003; +AP5 ΔBF = 0.00±0.01; +CN21 ΔBF = 0.01±0.01) and sustained (10+minutes post stimulation; Ctrl ΔBF = 0.033±0.002; +AP5 ΔBF = 0.006±0.007; +CN21 ΔBF = 0.026±0.05) phases of Rac1 activation during sLTP. Error bars represent s.e.m. Asterisks indicate a statistically significant difference as determined by an independent samples t-test (p<0.05).
g. Effects of near-physiological temperature of Rac1 sensor activation. Perfusion was warmed with a heating block holding the ACSF container, and the temperature measured at the back of the perfusion chamber. (RT = room temperature, n = 102 cells/121 spines; for 30–32°C, n = 11 cells/13 spines). Error bars represent s.e.m.
h. Variability of unstimulated spine volume changes after the induction of sLTP in a nearby spine in Rac1 sensor-overexpressing neurons. Data shown are timecourses of unstimulated spines close to the site of sLTP induction. While there is no average change in nearby spine volume with this stimulus (Fig. 1c, Main Text), there is occasional enlargement or shrinkage. Data corresponds to 100 randomly selected spines from the total of 777 nearby spines measured for the average depicted in Fig. 1c.
i. Transient (1–2min) change in volume of nearby spines as a function of distance from the stimulated spine. Data correspond to all nearby spines measured for the experiments in which the spatial profile of Rac1 was measured (n = 56 cells/79 experiments/218 nearby spines; Fig 1e, Main Text). Inset equation corresponds to the linear model of best fit.
j. Same as (i), but for the sustained (10–20 min) change in volume of nearby spines.
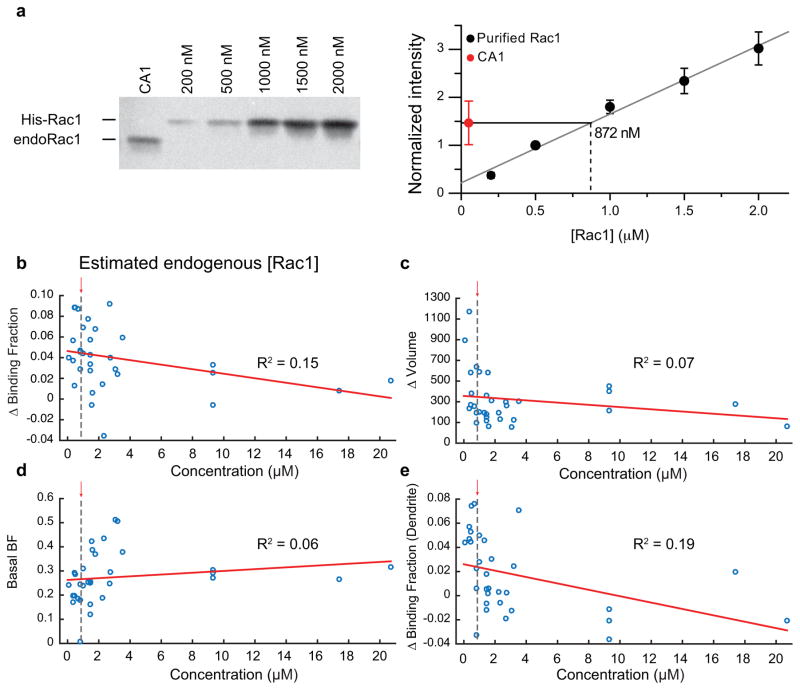
Extended Data Fig. 3. Estimation of endogenous Rac1 concentration and effects of Rac1 sensor overexpression.
a. (left) Representative Western blot used to analyze endogenous Rac1 expression (endo-Rac1) from the CA1 region of hippocampal slices cultures compared to known concentrations of purified polyhistidine-tagged Rac1 (His-Rac1). (right) Quantification of protein expression level for His-Rac1 from western blot shown in (a) averaged over three experiments. The concentration of endogenous Rac1 was estimated by measuring the intersection of the intensity of Rac1 protein from CA1 on the established calibration curve.
b–e. Effect of concentration of the Rac1 sensor on the observed change in binding fraction or volume of the spine with glutamate uncaging (c and d, respectively), the basal binding fraction of the sensor (e), or the change in binding fraction in the dendrite in response to uncaging (f). Rac1 sensor concentration was estimated by making a standard curve of the intensity values known concentrations of mEGFP at a range of imaging powers. The estimated endo-Rac1 concentration is plotted with a dashed line for comparison.
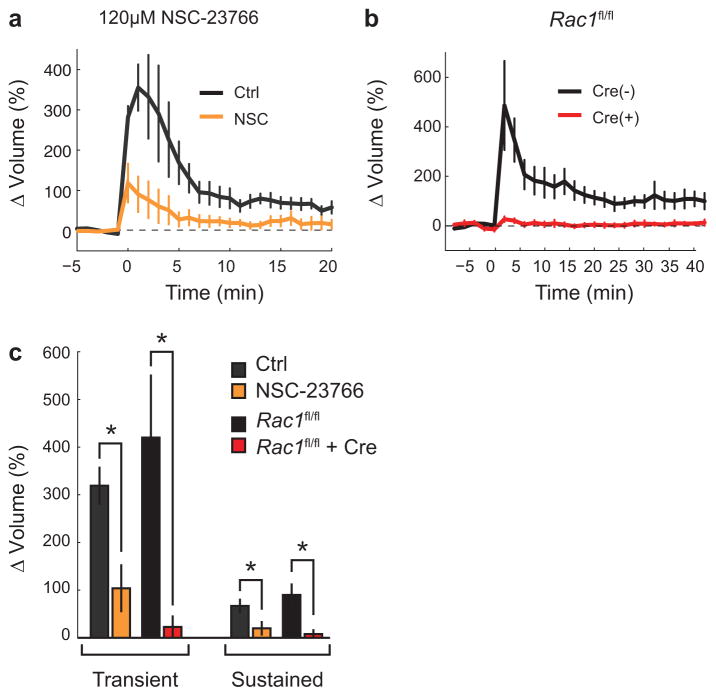
Extended Data Fig. 4. Effect of Rac1 inhibition on sLTP.
a. The effect of a high concentration of NSC-23766 (120 μM) on uncaging-evoked spine volume change. Experiments were performed in mEGFP-expressing rat hippocampal CA1 pyramidal neurons. Error bars represent s.e.m. n = 5 cells/9 ctrl spines; n = 5 cells/15 +NSC-23766 spines.
b. Effect of single-cell Rac1 KO on spine sLTP. Rac1fl/fl slices were transfected with either GFP alone (‘Cre (−)’; black curve, n = 5 cells/5 spines) or GFP+tdTomato-Cre recombinase (‘Cre (+)’; red curve; n = 7 cells/7 spines). Error bars represent s.e.m.
c. Summary of data shown in (a) and (b). For NSC-23766 experiments, Ctrl ΔVTransient = 319 ± 38%; Ctrl ΔVSustained = 67 ± 13%; +NSC-23766 ΔVTransient = 104 ± 48%; +NSC-23766 ΔVSustained = 20 ± 13%. For Rac1 KO experiments, Cre(−) ΔVTransient = 420 ± 130%; Cre(−) ΔVSustained = 90 ± 22%; Cre(+) ΔVTransient = 23 ± 9%; Cre(+) ΔVSustained = 8 ± 8%. Error bars represent s.e.m. Asterisks represent a statistically significant difference as determined by an independent samples t-test (p<0.05). All data is depicted as mean ± s.e.m.
Extended Data Fig. 5. Characterization of Rho-GTPase dependence on BDNF-TrkB signaling.
a–b. (a) Dependence of sLTP (ΔVolume) in Rho GTPase sensor–expressing neurons on postsynaptic BDNF. Data correspond to the volume data from experiments shown in 1f. Black = Creneg data, and red = Crepos data. (b) Summary of data from (a). Bars represent the average spine volume change from 10min to the end of the experiment. Error bars represent s.e.m. *p<0.05 (two-tailed t-test).
c–d. (c) Dependence of Rac1 on extracellular BDNF. (left) Rac1 (n = 7 cells/11 ctrl spines, 8 cells/14 TrkB-Ig spines), activity in the presence of 2 mg/mL TrkB-Ig. (right) Spine volume change in Rac1 sensor-overexpressing cells in the two conditions. Gray bars indicate duration of uncaging bout. (d) Summary of data shown in (c). All data is depicted as mean ± sem. Asterisks indicate a statistically significant difference as indicated by an independent-samples t-test.
e–f. (e) Dependence of Rac1 on postsynaptic TrkB. (left) Rac1 (n = 3 cells/8 ctrl/Cre(−) spines, 3 cells/8 Cre(+) spines), activity in the presence or absence of Cre recombinase in TrkBfl/fl mouse slices. (right) Spine volume change in Rac1 sensor-overexpressing cells in the two conditions. Gray bars indicate duration of uncaging bout. (f) Summary of data shown in (e). All data is depicted as mean ± sem. Asterisks indicate a statistically significant difference as indicated by an independent-samples t-test.
g–h. (g) Dependence of Cdc42 on extracellular BDNF. (left) Cdc42 (n = 7 cells/12 spines, 5 cells, 12 TrkB-Ig spines), activity in the presence of 2 mg/mL TrkB-Ig. (right) Spine volume change in Rac1 sensor-overexpressing cells in the two conditions. Gray bars indicate duration of uncaging bout. (h) Summary of data shown in (g). All data is depicted as mean ± sem. Asterisks indicate a statistically significant difference as indicated by an independent-samples t-test.
i–j. (g) Dependence of Cdc42 on postsynaptic TrkB. (left) Cdc42 (n = 5 cells/12 ctrl/Cre(−) spines, 6 cells/16 Cre(+) spines), activity in the presence or absence of Cre recombinase in TrkBfl/fl mouse slices. (right) Spine volume change in Cdc42 sensor-overexpressing cells in the two conditions. Gray bars indicate duration of uncaging bout. (j) Summary of data shown in (i). All data is depicted as mean ± sem. Asterisks indicate a statistically significant difference as indicated by an independent-samples t-test.
Extended Data Fig. 6. Characterization of the effects of weak pharmacological inhibition of BDNF-TrkB signaling on sLTP and Rac1 activity spreading.
a. Individual data points for crosstalk experiments under low [TrkB-Ig] exposure. Each plot contains the average values of the experiments (thick, black curve) along with the corresponding individual experiments for the suprathreshold spine (left) and the subthreshold spine (right). Inset figures correspond to a close-up view of the data distributions for 10+ minutes (the values used to calculate the sustained volume changes shown in Fig. 2 of the main text).
b. Same as (a), but for crosstalk experiments using low [1NMPP1].
c. Same as (a) and (b), but for crosstalk experiments using low [NSC-23766].
d. Effect of 0.125μg/mL TrkB-Ig on Rac1 signal spreading. Each plot represents a specific time epoch after glutamate uncaging onset. Curves represent control (red = spine; black = dendrite) and +TrkB-Ig (blue = spine; green = dendrite) conditions plotted as a function of distance from the stimulated spine. n = 5 cells/6 ctrl spines; n = 5 cells/9 +TrkB-Ig spines.
e. Summary of data presented in (d). Bars represent averages of the indicated temporal window across 1–5μm of the dendrite. Asterisks indicate a statistically significant difference as determined by a t-test.
f. Same as (d), but with the absence (red/black) and presence (blue/green) of 0.125μM 1NMPP1 in TrkBF616A slices. n = 5 cells/8 ctrl spines; n = 5 cells/11 +1NMPP1 spines.
g. Summary of data presented in (f). Bars represent averages of the indicated temporal window across 1–5μm of the dendrite. Asterisks indicate a statistically significant difference as determined by a t-test.
h. Same as (d) and (f), but in the absence (red/black) and presence (blue/green) of the Rac1 inhibitor, 15 μM NSC-23766. n = 6 cells/8 ctrl spines; n = 8 cells/13 +NSC-23766 spines)
i. Summary of data presented in (h). Bars represent averages of the indicated temporal window across 1–5μm of the dendrite. Asterisks indicate a statistically significant difference as determined by a t-test.
Extended Data Fig. 7. Effect of BDNF application on Rho GTPase signaling.
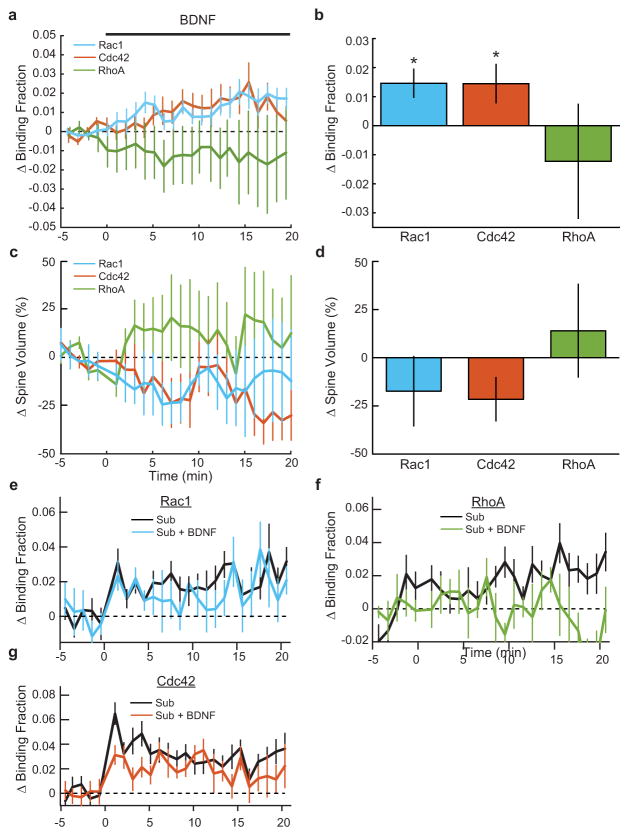
a. Effect of bath application of 20ng/mL exogenous BDNF on Rac1 (n = 8 cells), Cdc42 (n = 10 cells), and RhoA (n = 5 cells) sensors. Asterisks indicate a statistically significant difference from zero as determined by a one-sample t-test.
b. Summary of data shown in (a).
c. Effects of BDNF application on spine volume in Rac1 (blue), Cdc42 (orange), and RhoA (green) sensor-expressing cells. Data is from the experiments presented in (a,b)
d. Summary of data presented in (c). ANOVA determined that there was no significant difference between the sensor conditions. None of the conditions showed a significant difference from zero as determined by a one-sample t-test.
e. Response of Rac1 to a subthreshold stimulus in the presence of 20ng/mL BDNF. (ctrl n = 15 cells/15 spines; +BDNF n = 6 cells/6 spines)
f. Same as (e), but for RhoA. (ctrl n = 14 cells/18 spines; +BDNF = 7 cells/8 spines)
g. Same as (e,f), but for Cdc42. (ctrl n = 17 cells/20 spines; +BDNF = 7 cells/7 spines)
Extended Data Fig. 8. Characterization of the dendritic Rac1 inhibitor approach.
a. Representative two-photon images illustrating the filamentous distribution HEK293T cells (top row), consistent with microtubule localization, and the largely dendrite-specific localization in CA1 neurons (bottom two rows) of W56-mCherry-MTBD in comparison to an mEGFP cell fill (first column). HEK293T cells were imaged at 1050nm to increase specificity of excitation for mCherry vs. mEGFP. All images were acquired 2–5 days after transfection.
b. Comparison of the expression levels of scr-MTBD and W56-MTBD from a subset of cells used for synaptic crosstalk experiments. Expression was estimated by acquiring intensity values in the red channel of an ~10 μm section of a secondary dendrite using ImageJ. Each point represents a single cell.
c. Average time course of Rac1 activation for cells expressing only the Rac1 sensor (Ctrl, black curve; n = 105 spines), the Rac1 sensor + W56-mCherry-MTBD (W56-MTBD, red curve; n = 21 cells/33 spines), and the Rac1 sensor + W56-mCherry (W56-mCh, green curve; n = 6 cells/13 spines). Error bars represent s.e.m.
d. Comparison of the effect of W56-mCherry-MTBD (left; n = 21 cells/33 spines) to untargeted W56-mCherry (right; n = 6 cells/13 spines) on the spatial profile of Rac1 activation. Data is represented as mean ± s.e.m. ‘Ctrl’ data corresponds to the data shown in Fig. 1d (Main Text).
e. Effect of expression of W56-MTBD on RhoA sensor activation. (left) Spatial profile of RhoA activation during sTLP induction in the presence of scr-MTBD ctrl (black curve) and W56-MTBD (red curve). (right) Representative 2pFLIM images of RhoA activation in the presence of W56-MTBD. White circle indicates the targeted spine.
f. Schematic of the design of the GAP-based Rac1 dendritic inhibitor. A Rac1 GAP (ARHGAP15) replaced W56 in the general dendritic inhibitor construct (see Fig. 3a, Main Text).
g. Representative 2pFLIM images of the effect of GAP-mCh-MTBD on Rac1 signal spreading for the indicated time windows. White circle indicates the targeted spine.
h. Quantification of the effects of expression of GAP-mCherry-MTBD on Rac1 signal spreading after glutamate uncaging. Data is depicted as the change in binding fraction in the dendrite as a function of distance from the stimulated spine (change in binding fraction in spine plotted on y axis). n = 8 cells/11 spines
i. Summary of the data depicted in (g,h). Also shown is data for Rac1 spreading from scr-MTBD (see Fig. 3d in main text) for comparison. Asterisk indicates statistically significant difference as determined by an independent-samples t-test (p<0.05).
j. Effects of GAP-mCherry-MTBD expression on synaptic crosstalk. n = 4 cells/8 crosstalk experiments
k. Summary of the data presented in (j). Asterisk denotes a statistically significant difference as determined by an ANOVA and post-hoc test using the Tukey-Kramer method.
Extended Data Fig. 9. Effect of a subthreshold stimulus on single spines of sensor-expressing CA1 neurons.
a. Plots showing the variability of the response of the Rac1 (left), Cdc42 (middle), and RhoA (right) sensors to a suprathreshold stimulus. Thick black curves correspond to the averages depicted in Fig. 4a–c (‘unpaired’ condition) of the main text. The peak responses (1–2 min post stimulation, the same points used for statistical claims in Fig. 4) were subjected to a Shapiro-Wilk test to confirm the normality of the data. All data supported the null-hypothesis, illustrating that the data are Guassian distributed and justifying the use of parametric statistics.
b. Plots showing the variability of the response of the Rac1 (left), Cdc42 (middle), and RhoA (right) sensors to a subtreshold stimulus. Thick black curves correspond to the averages depicted in Fig. 4a–c (‘unpaired’ condition) of the main text.
b. Change in spine volume with glutamate uncaging (arrow and dotted line) during an unpaired threshold (black) and subthreshold (red) stimuli for CA1 pyramidal cells expressing the Rac1 (left), Cdc42 (center), or RhoA (right) sensors. Shaded region represents s.e.m. Data corresponds to the volume curves for the data presented in Fig. 4 (Main Text).
c. 8Hz two-photon imaging of BDNF-SEP intensity during an unpaired suprathreshold (black) or subthreshold (red) stimulus. The left panel corresponds to the full time course. Glutamate uncaging stimuli are delivered at 0.5Hz beginning at t = 0, indicated by the black arrow and dashed line. Inset figure shows the associated volume change (as measured from mCh cell fill) of the two conditions. The middle panel shows the uncaging-triggered average of 30 16-frame bins (corresponding to each uncaging pulse) and thus shows the average response to individual glutamate uncaging events. The right panel shows the average of the first point after uncaging for the indicated conditions. Both suprathreshold and subthreshold conditions show a statistically significant difference from zero, while the presence of AP5+NBQX or the POMC peptide eliminate this signal. Error bars represent s.e.m. n = 28 cells/217 spines (LTP), 5/84 (Sub), 2/46 AP5+NBQX), and 2/29 (POMC).
d. (Left) Activation of the TrkB sensor in response to an unpaired suprathreshold (black; n = 4 cells/5 spines) or subthreshold (red; n = 6 cells/8 spines) stimulus. (Right) Change in spine volume in a cell expressing the TrkB sensor in response to an unpaired threshold or subthreshold stimulus. Error bars represent s.e.m
Extended Data Fig. 10. Change in volume of paired spines during synaptic crosstalk in Rho GTPase-expressing CA1 neurons.
a. Spine volume change in response to a suprathreshold (black curve; black arrow indicates stimulus initiation) and a paired subthreshold (red curve, gray arrow indicates stimulus initiation) stimulus in spines from cells expressing the Rac1 sensor. n = 6 cells/12 crosstalk experiments. Error bars represent s.e.m.
b. Same as (a), but for Cdc42-expressing cells. n = 9 cells/10 crosstalk experiments.
c. Same as (a and b), but for RhoA-expressing cells. n = 8 cells/12 crosstalk experiments.
Acknowledgments
This work was supported by grants from NIMH (R01MH080047 (RY), R01NS068410 (RY)), NINDS (F31NS078847 (SH) R01NS05621 (JOM), DP1NS096787(RY)), JSPS KAKENHI (HM), JST PRESTO (HM), the Wakeman Fellowship at Duke University (SH) and Max Planck Florida Institute (RY). We thank Drs. A. E. West, S. Soderling, and F. Wang for guidance and valuable discussion, and Yasuda and McNamara lab members for discussion. We also thank Dr. R. Puranam for isolating MTBD. Finally, we thank D. Kloetzer for managing the lab.
Footnotes
Author Contributions
N.G.H, S.C.H, J.O.M. and R.Y. designed the experiments. N.G.H, S.C.H, and H.M. developed the sensor and inhibitors; N.G.H. and S.C.H collected the data with assistance from C.E.H; N.G.H, S.C.H, C.E.H, and R.Y. analysed the data; N.G.H., S.C.H., J.M.O. and R.Y. wrote the paper. All authors discussed the results and comments on this manuscript. The authors declare no competing financial interests.
References
- 1.Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10:927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- 2.Dietz DM, et al. Rac1 is essential in cocaine-induced structural plasticity of nucleus accumbens neurons. Nat Neurosci. 2012;15:891–896. doi: 10.1038/nn.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamoto K, Nagai T, Miyawaki A, Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat Neurosci. 2004;7:1104–1112. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- 5.Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haditsch U, et al. A central role for the small GTPase Rac1 in hippocampal plasticity and spatial learning and memory. Mol Cell Neurosci. 2009;41:409–419. doi: 10.1016/j.mcn.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai KOO, et al. TrkB phosphorylation by Cdk5 is required for activity-dependent structural plasticity and spatial memory. Nat Neurosci. 2012;15:1506–1515. doi: 10.1038/nn.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Il Hwan K, Hong W, Scott HS, Ryohei Y. Loss of Cdc42 leads to defects in synaptic plasticity and remote memory recall. eLife. 2014;3:e02839. doi: 10.7554/eLife.02839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McAllister AK. Biolistic transfection of neurons. Sci STKE. 2000;2000:pl1. doi: 10.1126/stke.2000.51.pl1. [DOI] [PubMed] [Google Scholar]
- 11.Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 12.Yasuda R, et al. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 13.Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Honkura N, Matsuzaki M, Noguchi J, Ellis-Davies GC, Kasai H. The subspine organization of actin fibers regulates the structure and plasticity of dendritic spines. Neuron. 2008;57:719–729. doi: 10.1016/j.neuron.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Harward SC, et al. Autocrine BDNF–TrkB signalling within a single dendritic spine. Nature. 2016;538:99–103. doi: 10.1038/nature19766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu W, et al. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luikart BW, et al. TrkB has a cell-autonomous role in the establishment of hippocampal Schaffer collateral synapses. J Neurosci. 2005;25:3774–3786. doi: 10.1523/JNEUROSCI.0041-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamoto Y, Yamauchi J, Tanoue A, Wu C, Mobley WC. TrkB binds and tyrosine-phosphorylates Tiam1, leading to activation of Rac1 and induction of changes in cellular morphology. Proc Natl Acad Sci USA. 2006;103:10444–10449. doi: 10.1073/pnas.0603914103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harvey CD, Svoboda K. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 2007;450:1195–1200. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harvey CD, Yasuda R, Zhong H, Svoboda K. The spread of Ras activity triggered by activation of a single dendritic spine. Science. 2008;321:136–140. doi: 10.1126/science.1159675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, et al. A chemical-genetic approach to studying neurotrophin signaling. Neuron. 2005;46:13–21. doi: 10.1016/j.neuron.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Huber KM, Sawtell NB, Bear MF. Brain-derived neurotrophic factor alters the synaptic modification threshold in visual cortex. Neuropharmacology. 1998;37:571–579. doi: 10.1016/s0028-3908(98)00050-1. [DOI] [PubMed] [Google Scholar]
- 23.Gao Y, Xing J, Streuli M, Leto TL, Zheng Y. Trp(56) of rac1 specifies interaction with a subset of guanine nucleotide exchange factors. J Biol Chem. 2001;276:47530–47541. doi: 10.1074/jbc.M108865200. [DOI] [PubMed] [Google Scholar]
- 24.Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong H, et al. Subcellular dynamics of type II PKA in neurons. Neuron. 2009;62:363–374. doi: 10.1016/j.neuron.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seoh ML, Ng CH, Yong J, Lim L, Leung T. ArhGAP15, a novel human RacGAP protein with GTPase binding property. FEBS Lett. 2003;539:131–137. doi: 10.1016/s0014-5793(03)00213-8. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka J, et al. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 2008;319:1683–1687. doi: 10.1126/science.1152864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korte M, et al. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci USA. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosch M, et al. Structural and molecular remodeling of dendritic spine substructures during long-term potentiation. Neuron. 2014;82:444–459. doi: 10.1016/j.neuron.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chrostek A, et al. Rac1 is crucial for hair follicle integrity but is not essential for maintenance of the epidermis. Mol Cell Biol. 2006;26:6957–6970. doi: 10.1128/MCB.00075-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yasuda R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr Opin Neurobiol. 2006;16:551–561. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 32.Murakoshi H, Lee SJ, Yasuda R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat Biotech. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 35.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotech. 2005;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 36.He XP, et al. Conditional deletion of TrkB but not BDNF prevents epileptogenesis in the kindling model. Neuron. 2004;43:31–42. doi: 10.1016/j.neuron.2004.06.019. [DOI] [PubMed] [Google Scholar]