ABSTRACT
While complex intra- and interspecies microbial community dynamics are apparent during chronic infections and likely alter patient health outcomes, our understanding of these interactions is currently limited. For example, Pseudomonas aeruginosa and Staphylococcus aureus are often found to coinfect the lungs of patients with cystic fibrosis (CF), yet these organisms compete under laboratory conditions. Recent observations that coinfection correlates with decreased health outcomes necessitate we develop a greater understanding of these interbacterial interactions. In this study, we tested the hypothesis that P. aeruginosa and/or S. aureus adopts phenotypes that allow coexistence during infection. We compared competitive interactions of P. aeruginosa and S. aureus isolates from mono- or coinfected CF patients employing in vitro coculture models. P. aeruginosa isolates from monoinfected patients were more competitive toward S. aureus than P. aeruginosa isolates from coinfected patients. We also observed that the least competitive P. aeruginosa isolates possessed a mucoid phenotype. Mucoidy occurs upon constitutive activation of the sigma factor AlgT/U, which regulates synthesis of the polysaccharide alginate and dozens of other secreted factors, including some previously described to kill S. aureus. Here, we show that production of alginate in mucoid strains is sufficient to inhibit anti-S. aureus activity independent of activation of the AlgT regulon. Alginate reduces production of siderophores, 2-heptyl-4-hydroxyquinolone-N-oxide (HQNO), and rhamnolipids—each required for efficient killing of S. aureus. These studies demonstrate alginate overproduction may be an important factor driving P. aeruginosa coinfection with S. aureus.
KEYWORDS: Pseudomonas aeruginosa, Staphylococcus aureus, biofilm, cystic fibrosis, mucoid, polymicrobial
IMPORTANCE
Numerous deep-sequencing studies have revealed the microbial communities present during respiratory infections in cystic fibrosis (CF) patients are diverse, complex, and dynamic. We now face the challenge of determining the influence of these community dynamics on patient health outcomes and identifying candidate targets to modulate these interactions. We make progress toward this goal by determining that the polysaccharide alginate produced by mucoid strains of P. aeruginosa is sufficient to inhibit multiple secreted antimicrobial agents produced by this organism. Importantly, these secreted factors are required to outcompete S. aureus, when the microbes are grown in coculture; thus we propose a mechanism whereby mucoid P. aeruginosa can coexist with S. aureus. Finally, the approach used here can serve as a platform to investigate the interactions among other CF pathogens.
INTRODUCTION
Cystic fibrosis (CF) is the most common fatal, inherited disease among Caucasians. Progressive decline in pulmonary function, resulting from decreased mucociliary clearance, persistent bacterial infections of the airway, and neutrophil-dominated inflammation, is the predominant cause of morbidity and mortality for CF patients (1). CF respiratory infections are notable for their complex polymicrobial nature and recalcitrance to antimicrobial therapeutics. Patients are colonized throughout their lives with a diverse community of pathogens of viral, bacterial, and fungal origins (2). Unfortunately, these infections are rarely eradicated, and patients suffer from frequent exacerbations, hospitalization, and often ineffective treatments with intravenous and inhaled antibiotics. While the importance of interspecies interactions during infection is increasingly appreciated, most studies are still performed with single microbial species in culture, and our knowledge of polymicrobial interactions is limited.
Pseudomonas aeruginosa and Staphylococcus aureus are two of the most prevalent and often the most problematic pathogens in CF. Both S. aureus and P. aeruginosa exhibit intrinsic and acquired antibiotic resistance, making these infections difficult to treat (3, 4). S. aureus is among one of the earliest pathogens to infect pediatric CF patients, whereas P. aeruginosa infections are intermittent early on until a dominant clone emerges and P. aeruginosa becomes the predominant pathogen later in life (5). This inverse pattern of infection has led many investigators to speculate that P. aeruginosa eliminates S. aureus during infection—perhaps outcompeting S. aureus for limited nutrients in the lung and/or producing antimicrobial factors to kill S. aureus directly (recently reviewed in reference 6). These hypotheses are supported by several in vitro studies that demonstrate P. aeruginosa can inhibit the growth or reduce the viability of S. aureus through multiple mechanisms, including sequestration of iron and inhibition of S. aureus respiration via production of the secondary metabolite 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO) (7–16).
Importantly, despite evidence that P. aeruginosa outcompetes S. aureus in vitro, CF patients acquire coinciding pulmonary infections with P. aeruginosa and S. aureus. We and others have observed a correlation between coinfection and poor patient outcome, whereby infection with both P. aeruginosa and S. aureus correlates with increased pulmonary exacerbations and lower baseline forced expiratory volumes of the lung in 1 s (FEV1) compared to patients who were monoinfected with only S. aureus or P. aeruginosa (17–19). Examination of 234 CF patients at Emory+Children's Center for Cystic Fibrosis and Airways Disease Research revealed that 73 patients (31%) were coinfected with P. aeruginosa and S. aureus (17). Moreover, recent data demonstrate higher rates of infection with S. aureus in patients later in life than previously appreciated (20). These observations suggest our currently held model that P. aeruginosa outcompetes S. aureus during infection is oversimplified. We therefore seek to gain a greater fundamental understanding of how coinciding infections with P. aeruginosa and S. aureus occur in CF patients in an effort to more effectively eliminate these infections.
One hypothesis to explain coinciding P. aeruginosa-S. aureus infections in CF patients is spatial segregation of these species during infection, such that P. aeruginosa antimicrobials are unable to access and thereby kill S. aureus during infection. For example, utilizing a wound-like model of P. aeruginosa-S. aureus coinfection, DeLeon et al. established species coexistence for up to 7 days (21). The authors attribute S. aureus survival to the spatial separation of these species established during biofilm formation in this model. Moreover, physical separation between P. aeruginosa and S. aureus microcolonies has been observed in human wound biopsy specimens (22). For CF pulmonary infections, recent studies suggest that P. aeruginosa and S. aureus may occupy the same airspace during infection. Hogan and colleagues identified P. aeruginosa and S. aureus in the same lobes of the lung by examining protected brush samples (23), and Wakeman et al. have presented histological evidence of bacteria with the morphological characteristics of Pseudomonas and Staphylococcus infecting the same niche in explanted lungs from a CF patient (24). However, due to current technical constraints studying in vivo infections, if and to what extent meaningful physical interactions occur between these pathogens in the context of CF infections is completely unknown.
We therefore sought to design a study to gain insight into whether P. aeruginosa and S. aureus may interact during infection and, if so, how such coexistence can occur in light of the observation that P. aeruginosa can kill S. aureus in vitro. We reasoned that P. aeruginosa may adopt phenotypes that limit its antagonism toward S. aureus. To test this hypothesis, we first asked if P. aeruginosa isolates from CF patients were able to outcompete S. aureus in vitro, as previously observed, and importantly if strains from coinfected patients were less competitive than P. aeruginosa isolates obtained from patients who were not infected with S. aureus. Consistent with this idea, we observed that P. aeruginosa isolates from coinfected patients were more permissive to growth with S. aureus than P. aeruginosa isolates from monoinfected patients. Investigation into the nature of coexistence revealed that the presence of a mucoid phenotype limited P. aeruginosa antimicrobial action toward S. aureus. We found that mucoidy reduces the production of siderophores, HQNO, and rhamnolipids; each is required for robust killing of S. aureus (7–16). Moreover, overproduction of the polysaccharide alginate was sufficient to decrease the production of these key compounds. Together these studies suggest that genotypic and phenotypic modifications of P. aeruginosa during infection may contribute to coinfection with S. aureus.
RESULTS
P. aeruginosa isolates from coinfected CF patients are less antagonistic toward S. aureus than isolates from monoinfected CF patients.
In a previous study, we investigated the clinical outcome of CF patients during coinfection with P. aeruginosa and S. aureus. We observed that coinfection correlated with poor patient outcome, including a decline in lung function, compared to monoinfected patients (17). To investigate if there are differences in the isolates from these patients that may contribute to establishing coinfection, we obtained 28 P. aeruginosa strains and 20 S. aureus strains from patients who were either coinfected or monoinfected from the CF Biospecimen Registry (CFBR) at Emory+Children's Center for Cystic Fibrosis and Airways Disease Research. Note that monoinfection refers to the absence of either P. aeruginosa or S. aureus during the review year, but other pathogens may be present. Positive cultures for Burkholderia, Stenotrophomonas, Achromobacter, Acinetobacter, Chryseobacterium, Klebsiella, Streptococcus spp., Haemophilus influenzae, and Escherichia coli were recorded, but the frequencies of these other microbes were not sufficiently high to establish significant correlations between the presence of individual microbes and clinical outcomes.
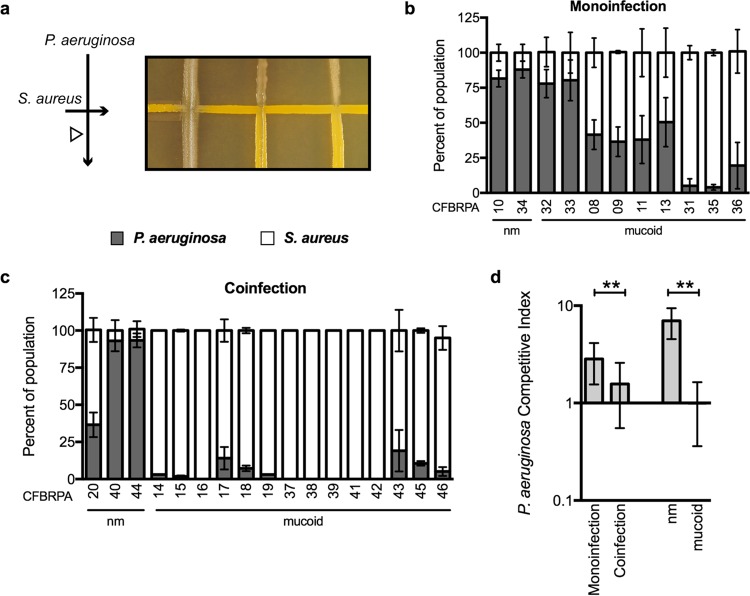
Based on our previous findings showing worse clinical outcomes for coinfected patients, we then asked the following question: do P. aeruginosa and S. aureus strains from coinfected patients grow better in coculture than strains from monoinfected patients? To address this question, each P. aeruginosa clinical isolate was challenged with a laboratory strain of S. aureus (USA300 LAC, JE2) and S. aureus clinical isolates with a laboratory strain of P. aeruginosa (PAO1) in an agar plate-based cross-streak assay. Single colonies of S. aureus and P. aeruginosa were cross-streaked according the schematic in Fig. 1a. Note that results were similar regardless of the order in which the pathogens were cross-streaked: i.e., P. aeruginosa followed by S. aureus, or vice versa. The interactions between the two species was examined by visually inspecting growth at the intersection of the cross-streak and by determining the percentage of the population of P. aeruginosa and S. aureus by recovering bacterial growth in the poststreak (white arrowhead in Fig. 1a) and enumerating the number of CFU for each P. aeruginosa and S. aureus strain on selective media (Pseudomonas isolation agar [PIA] and mannitol salts agar [MSA], respectively). The P. aeruginosa clinical isolates shown in Fig. 1a are representative of the spectrum of P. aeruginosa inhibition of S. aureus, ranging from most competitive on the left to least competitive on the right.
FIG 1 .
P. aeruginosa isolates from coinfected CF patients are less competitive with S. aureus. (a) Schematic of the method for cross-streak assay. S. aureus (USA300 LAC, JE2) was streaked onto tryptic soy agar followed by cross-streaking with P. aeruginosa isolates (CFBRPA) from the CF Biospecimen Registry (CFBR) at Emory+Children's Center for Cystic Fibrosis and Airways Disease Research. Coculture assays were performed by cross-streaking P. aeruginosa CF isolates and S. aureus on an agar surface, and the percentage of the total population of P. aeruginosa (gray bars) and S. aureus (white bars) recovered post-cross-streak (white arrowhead in panel a) were enumerated by plating on selective media and dividing the number of P. aeruginosa or S. aureus CFU by the total CFU (S. aureus plus P. aeruginosa). P. aeruginosa strains from panel b were isolated from CF patients who were infected with only P. aeruginosa (monoinfection), and strains in panel c were isolated from patients who were coinfected with P. aeruginosa and S. aureus. Mucoid and nonmucoid (nm) phenotypes are indicated. In panel d, a summary of the competitive index (CI) of all P. aeruginosa strains is indicated according the patient group from which they were isolated or their mucoid phenotype. CI was calculated by dividing the percentage of P. aeruginosa by the percentage of S. aureus recovered in the post-cross-streak. Error bars indicate standard deviations from three biological replicates performed in triplicate. Statistical significance was determined by performing an unpaired two-tailed t test. **, P < 0.001.
We found that P. aeruginosa clinical strains recovered from patients who were monoinfected with P. aeruginosa on average outcompeted S. aureus strain JE2 (Fig. 1b), and P. aeruginosa strains had a higher competitive index (CI [P. aeruginosa/S. aureus]) (Fig. 1d, leftmost bar). This pattern of competition was significantly different from that observed for P. aeruginosa isolates from patients that were coinfected with S. aureus (Fig. 1c), where the CI was on average 2-fold lower (Fig. 1d, second bar). However, when S. aureus isolates from CF patients were screened by cross-streaking with P. aeruginosa strain PAO1, no difference in the CI for S. aureus isolates was observed between mono- and coinfected patients (see Fig. S1 in the supplemental material).
S. aureus cystic fibrosis clinical isolates are not competitive with nonmucoid P. aeruginosa PAO1. Cross-streak coculture assays of S. aureus isolates (CFBRSA) from the CF Biospecimen Registry (CFBR) with P. aeruginosa strain PAO1 were performed by cross-streaking the two species on an agar surface, and the percentages of the populations of P. aeruginosa (gray bars) and S. aureus (white bars) recovered post-cross-streak were enumerated by plating on selective media and counting the number of CFU, according to the schematic in Fig. 1a. S. aureus strains in panel a were isolated from CF patients who were infected with only S. aureus (monoinfected), and strains in panel b were isolated from patients who were coinfected with P. aeruginosa and S. aureus. Error bars indicate the standard deviations of four biological replicates performed in triplicate. Competitive indices were calculated by dividing the percentage of S. aureus by the percentage of P. aeruginosa recovered post-cross-streak, and a statistically significant difference by an unpaired two-tailed t test was not observed between isolates from patients who were monoinfected versus those coinfected. Download FIG S1, TIF file, 0.2 MB (205.6KB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mucoid conversion reduces P. aeruginosa inhibition of S. aureus.
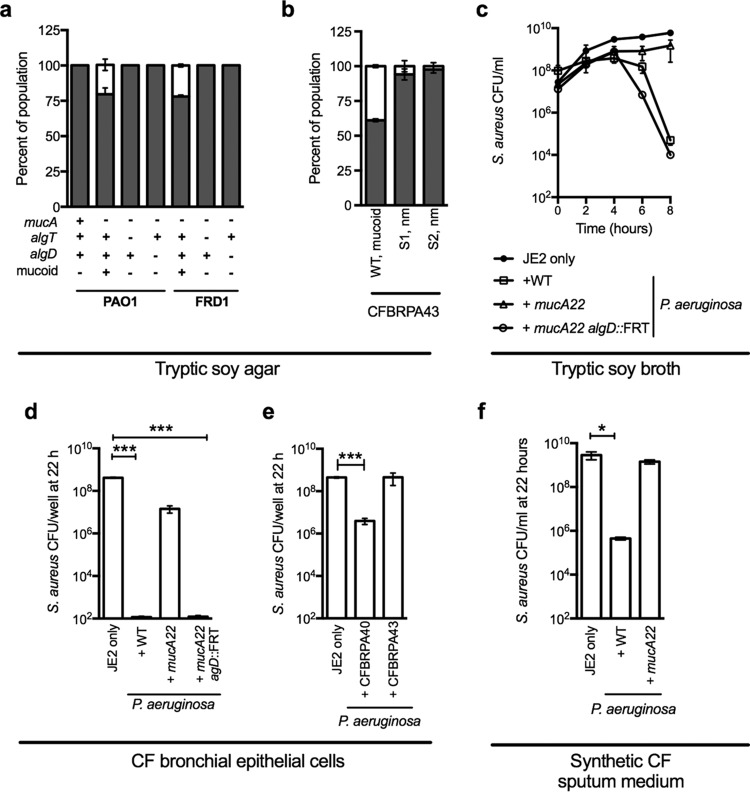
An additional correlation was revealed during this screen, whereby P. aeruginosa isolates that exhibited a mucoid phenotype displayed a CI 6-fold lower on average than that of nonmucoid isolates (Fig. 1d, right 2 bars). Mucoid conversion is generally characterized by overproduction of the polysaccharide alginate, and the designations as nonmucoid and mucoid in Fig. 1 were based on visual observation of alginate production of colonies grown on agar. To test the hypothesis that conversion to the mucoid phenotype promotes P. aeruginosa coexistence with S. aureus, isogenic P. aeruginosa mutants in genes responsible for regulation of alginate synthesis were examined. Mucoid conversion occurs most frequently in P. aeruginosa clinical strains via acquisition of mutations in the anti-sigma factor gene mucA. In nonmucoid strains, intact MucA sequesters the alternative sigma factor, σ22 (AlgT/U), which is the primary regulator of alginate production (25). In mucoid strains, inactivation of MucA (most frequently via the mucA22 mutation, ΔG430, frameshift) results in release of AlgT, which is now free to activate transcription of the alginate biosynthetic operon initiated at the algD promoter (PalgD) (26). We therefore competed isogenic mutants of the primary regulators of alginate synthesis in P. aeruginosa PAO1 with S. aureus JE2 to examine the contribution of mucoid conversion to coexistence with S. aureus in the cross-streak assay. Consistent with the correlations observed with P. aeruginosa clinical isolates, nonmucoid P. aeruginosa PAO1 outcompeted S. aureus. In contrast, introduction of the mucA22 mutation in PAO1, which confers a mucoid phenotype, reduced P. aeruginosa competition with S. aureus (Fig. 2a). Next, to determine if constitutive activation of the sigma factor AlgT as a result of MucA inactivation is responsible for the decreased competition of P. aeruginosa, we disrupted the algT gene in the mucA22 mutant and observed restored competition to wild-type (WT) levels. While alginate overproduction is the most visually apparent phenotype in P. aeruginosa strains when the mucA gene is disrupted, AlgT regulates a number of other factors that have been shown to contribute to competition with S. aureus, including genes required for the synthesis of pyoverdine, pyochelin, phenazines, and hydrogen cyanide (27–30). We would therefore hypothesize that AlgT regulates antistaphylococcal factors independent of alginate overproduction. However, when alginate production was selectively removed by disruption of algD (the first gene in the alginate biosynthetic operon) in the mucA22 mutant, P. aeruginosa was now able to outcompete S. aureus. This result suggests that alginate production may modulate P. aeruginosa competition with S. aureus. These phenotypes were also reproduced in the mucoid CF isolate FRD1 (mucA22) and its nonmucoid derivatives, FRD mucA22 algT33::TnS1 and FRD mucA22 algD::xylE (Fig. 2a).
FIG 2 .
Mucoid conversion prevents P. aeruginosa killing of S. aureus. In vitro P. aeruginosa-S. aureus coculture assays on tryptic soy agar (a and b) and broth (c), in biofilm growth on human CF bronchial epithelial cells (CFBE) (d and e), and in synthetic CF sputum medium (f). In panels a and b, the percentages of the populations of P. aeruginosa (gray) and S. aureus (white) recovered post-cross-streak are indicated. Isogenic P. aeruginosa PAO1 and FRD1 variants, with the mucA, algT, and algD genotypes and the mucoid phenotype indicated below (a), and mucoid P. aeruginosa CFBRPA43 and nonmucoid suppressors (S1, nm and S2, nm) of P. aeruginosa CFRBPA43 (b) were cross-streaked with S. aureus strain JE2. Panel c shows the viable count of S. aureus JE2 over time for the indicated strains. In panels d, e, and f, the viability of S. aureus JE2 after 22 h of competition with the indicated P. aeruginosa strains is indicated. Error bars indicate standard deviations from at least three biological replicates performed in triplicate. Statistical significance was determined by performing an unpaired two-tailed t test. ***, P < 0.0001.
To determine if mutations in mucA are responsible for the mucoid phenotype in a representative mucoid isolate from a coinfected patient in this study, the genomic sequence of the mucA gene from the clinical P. aeruginosa isolate CFBRPA43 was examined. Indeed, strain CFBRPA43 possessed a mucA22 mutation, which would be predicted to disrupt mucA function. To investigate the contribution of mucoid conversion in isolate CFBRPA43, we isolated nonmucoid suppressors by serial passage in broth culture and identified colonies displaying a nonmucoid phenotype. In CF, the most frequent suppressor of mucoid mutations occur within the algT gene (31). We therefore sequenced algT, confirmed the presence of a mutation in algT (S1, 8-nucleotide insertion at position 138; S2, C245A), and compared the competition of two suppressors to the mucoid parental strain in the cross-streak assay. As expected, the nonmucoid variants outcompeted S. aureus compared to the mucoid parental strains (Fig. 2b). These data demonstrate that alginate overproduction correlates with the loss of P. aeruginosa-mediated inhibition of S. aureus and suggest that mucoid conversion may be a factor promoting P. aeruginosa and S. aureus coexistence during CF respiratory infections.
To determine if P. aeruginosa outcompetes S. aureus by inhibiting growth or reducing S. aureus viability, P. aeruginosa and S. aureus competition was monitored in shaking broth culture over the course of 8 h by enumerating P. aeruginosa and S. aureus CFU on selective media (PIA and MSA, respectively) every 2 h. Similar to previous studies, P. aeruginosa viability was not altered by the presence of S. aureus under any of the conditions examined (10) (see Fig. S2a and b in the supplemental material). For S. aureus, coculture with either mucoid or nonmucoid P. aeruginosa did not alter viability or growth rate during the initial stages of growth. However, after approximately 4 h of competition, the viability of S. aureus drastically decreased in the presence of either nonmucoid strains, PAO1 or PAO1 mucA22 algD::FRT (FLP recombination target) (with representative kinetic analysis shown in Fig. 2c). These data indicate that nonmucoid P. aeruginosa is capable of killing S. aureus during competition and is not simply inhibiting growth. On the other hand, no decrease in viability of S. aureus was observed in the presence of the mucA22 mutant through the duration of the experiment, indicating that alginate-overproducing strains have reduced ability to kill S. aureus. Identical results were observed when mucoid and nonmucoid P. aeruginosa strains were grown in competition with S. aureus methicillin-sensitive strain Newman (Fig. S2c).
Nonmucoid P. aeruginosa outcompetes S. aureus. P. aeruginosa-S. aureus coculture assays during planktonic growth (a, b, and c) and during biofilm growth on CF bronchial epithelial cells (CFBE [d]). In panels a and b, the viability of P. aeruginosa strain PAO1 (WT) and isogenic mutants was measured as log10 CFU/well over the course of 8 h in monoculture (a) and in coculture with S. aureus strain JE2 (b). In panels c and d, the viability of S. aureus JE2 was measured as log10 CFU/ml after 8 h of coinfection in panel c and CFU/well over 22 h in panel d. Data in panels a, b, and d are representative of three biological replicates, and in panel c, error bars indicate standard deviations from three biological replicates performed in triplicate. Statistical significance was determined by performing an unpaired two-tailed t test. ***, P < 0.0001. Download FIG S2, TIF file, 0.2 MB (237KB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mucoid conversion prevents P. aeruginosa killing of S. aureus during competition in models of CF respiratory infection.
During CF pulmonary infections, both P. aeruginosa and S. aureus exist in complex and dynamic communities that influence their intra- and interspecies interactions (32, 33). Recently, we have established an in vitro human airway cell—dual bacterial biofilm infection model, whereby P. aeruginosa and S. aureus form mixed-species biofilms on monolayers of human bronchial epithelial cells homozygous for the ΔF508 cystic fibrosis transmembrane conductance regulator mutation (CFBE) (10, 34). To determine how alginate overproduction influences the dynamics of interspecies interactions in this model, wild-type (WT) PAO1, mucA22, and mucA22 algD::FRT strains were cocultured with S. aureus JE2 on CFBE cells for 22 h, and CFU were enumerated at 6, 18, and 22 h. Similar to the observation in Fig. 2c for broth culture, nonmucoid P. aeruginosa (both WT and mucA22 algD::FRT strains) coexists with S. aureus early during culture, followed by a sharp decrease in S. aureus viability by 18 h (representative kinetic CFU illustrated in Fig. S2d in the supplemental material and at the 22-h time point in Fig. 2d). This finding is consistent with a previous report from our group using nonmucoid P. aeruginosa strains PAO1 and PA14 cocultured with S. aureus strain 8325-4 (10). In contrast, S. aureus viability is unaltered for the duration of the experiment when cocultured with the mucA22 mutant. Additionally, representative nonmucoid (CFBRPA40) and mucoid (CFBRPA43) P. aeruginosa clinical isolates, identified to be competitive and noncompetitive with S. aureus in the cross-streak assay, respectively (Fig. 1c), were selected for analysis in this model. As predicted, nonmucoid P. aeruginosa strain CFRBPA40 reduced the viability of S. aureus, compared to mucoid P. aeruginosa strain CFBRPA43 (Fig. 2e). Finally, to investigate if nutrient availability in the CF respiratory environment influences competitive dynamics between P. aeruginosa and S. aureus, coculture growth was monitored in synthetic CF sputum medium (35). In accordance with prior observations, S. aureus viability was reduced during growth with nonmucoid P. aeruginosa (PAO1), but remained unaltered in the presence of mucoid P. aeruginosa (PAO1 mucA22) (Fig. 2f). These data demonstrate that mucoid conversion supports P. aeruginosa-S. aureus coexistence in mixed-species communities during growth in models that mimic the CF respiratory environment.
Induction of alginate reduces P. aeruginosa antimicrobial activity independent of AlgT activation.
The data presented thus far demonstrate that mucoid conversion limits the ability of P. aeruginosa to reduce S. aureus viability; however, whether overproduction of alginate by P. aeruginosa is sufficient for this interaction is unclear. Therefore, to study this behavior in more detail, we sought to engineer a P. aeruginosa strain whereby alginate production can be modulated in a nonmucoid strain with intact mucA. Such a strategy would allow us to assess the influence of alginate overproduction on P. aeruginosa physiology and gene expression, without the activation of the entire AlgT regulon. Alginate is a high-molecular-weight acidic polysaccharide composed of nonrepeating subunits of selectively O-acetylated d-mannuronic acid and its C5′ epimer, l-guluronic acid (36). Genes required for alginate biosynthesis are primarily organized within a single operon (algD, -8, -44, -K, -E, -G, -X, -L, -I, -J, -F, and -A), with the exception of the algC gene (see the schematic in Fig. 3a). algC encodes a phosphomannomutase required to convert mannose-1-phosphate to mannose-6-phosphate (37). To engineer a strain that bypasses the need for AlgT activation of these genes (schematic in Fig. 3b), we replaced the promoter of the alginate biosynthetic operon (PalgD) with an arabinose-inducible promoter (araC-ParaBAD) on the chromosome of nonmucoid strain PAO1. Additional regulation of alginate synthesis occurs at the posttranscriptional level, whereby polymerization by the inner membrane proteins Alg8 and Alg44 requires binding of the second messenger bis-(3′,5′)-cyclic dimeric GMP (c-di-GMP) to Alg44 (38). In mucA mutants, the requisite c-di-GMP would be provided through the action of diguanylate cyclases activated by AlgT. To circumvent the need for AlgT activation, PalgD::araC-ParaBAD was constructed in a mutant with constitutively high levels of c-di-GMP (PAO1 ΔwspF) (39).
FIG 3 .
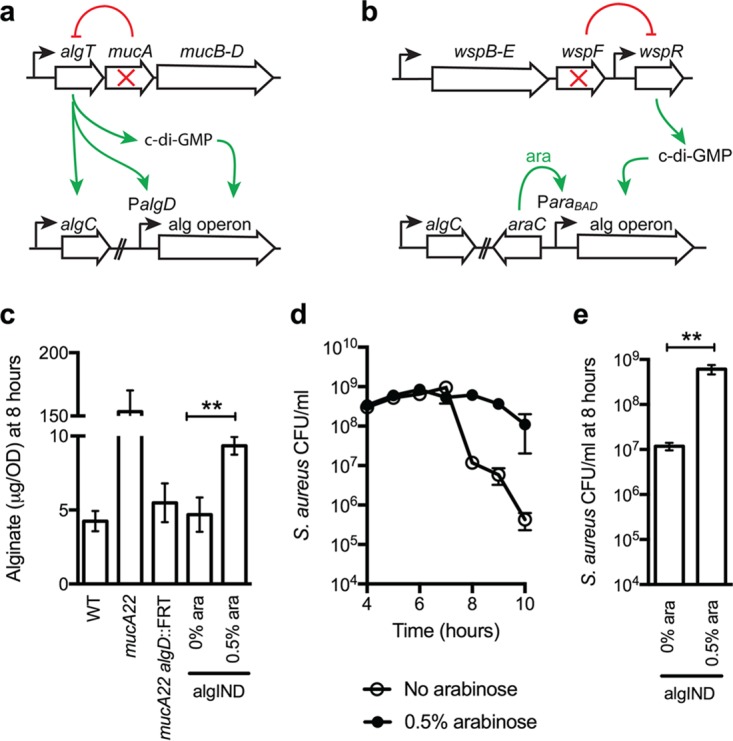
P. aeruginosa alginate production promotes coexistence with S. aureus. (a) Abbreviated schematic of alginate regulation in P. aeruginosa. Disruption of MucA results in release of AlgT, activating transcription of the algC gene, genes encoded in the alginate biosynthetic operon (alg operon, under control of the algD promoter PalgD), and bis-(3′,5′)-cyclic dimeric GMP (c-di-GMP)—all required for alginate synthesis. (b) Schematic of alginate regulation in P. aeruginosa PAO1 ΔwspF PalgD::araC-ParaBAD (PAO1algIND), whereby PalgD was replaced by araC-ParaBAD, to place the alg operon under inducible control of arabinose. Deletion of wspF results in inhibition of WspR and production of c-di-GMP. In panel c, alginate was extracted from nonmucoid wild-type PAO1 (WT) and the PAO1 mucA22, PAO1 mucA22 algD::FRT, and PAO1algIND mutants grown without arabinose (0% ara) and with 0.5% arabinose (0.5% ara) and quantified by a standard carbazole assay. In panels d and e, the log10 CFU/ml for S. aureus JE2 are indicated when grown in the presence of PAO1algIND without (open circles) and with (solid circles) 0.5% arabinose for 10 h (d); S. aureus log10 CFU/ml at 8 h are shown only in panel e. Error bars indicate standard deviations from four biological replicates performed in triplicate. Statistical significance was determined by performing an unpaired two-tailed t test. **, P < 0.01.
To determine if PAO1 ΔwspF PalgD::araC-ParaBAD (abbreviated “PAO1algIND,” for “PAO1 alginate inducible,” from this point forward) produces alginate only in the presence of arabinose under the conditions utilized in the coculture assays, PAO1algIND was grown with and without 0.5% arabinose for 8 h in a shaking broth culture, and the amount of alginate produced was determined by a standard carbazole assay. (Growth rates for both P. aeruginosa and S. aureus were identical with and without arabinose.) Alginate production increased by approximately 2-fold in the presence of 0.5% arabinose (9.3 µg/optical density at 600 nm [OD600]) compared to without arabinose, demonstrating alginate synthesis can be induced using this strategy (Fig. 3c). The increase in alginate production was modest however compared to PAO1 mucA22 (153.4 µg/OD600) (Fig. 3c; note y axis). To determine if this level of alginate production is sufficient to prevent P. aeruginosa-mediated reduction of S. aureus viability, PAO1algIND was grown with and without arabinose in the presence of S. aureus. Without arabinose, PAO1algIND reduced the viability of S. aureus by ~1,000-fold by 8 h, whereas in the presence of 0.5% arabinose, S. aureus viability was maintained for the 10 h of the assay period (Fig. 3d and e). The time required for uninduced PAO1algIND to initiate killing of S. aureus was longer than observed for PAO1 (8 h compared to 4 h [Fig. 2c]). Overall, however, the sustained viability of S. aureus observed when competed with PAO1algIND plus arabinose supports the previous observation that alginate prevents P. aeruginosa killing of S. aureus. Furthermore, our analysis of the PAO1algIND strain suggests that the amount of alginate produced by PAO1 mucA22 is in excess of the amount required to reduce killing of S. aureus.
Alginate overproduction reduces the expression of a subset of P. aeruginosa virulence genes.
P. aeruginosa secretes several antistaphylococcal effectors which can inhibit respiration (hydrogen cyanide, quinolones, and phenazines) and sequester iron (siderophores) (7–16). To determine how alginate overproduction prevents P. aeruginosa from killing S. aureus, we asked if alginate overproduction alters the expression profile of genes associated with physiological pathways important for S. aureus interactions and infection in the CF airway. We utilized NanoString digital multiplexed gene expression technology (40, 41) to quantify the expression level of P. aeruginosa mRNA transcripts in mucoid PAO1 mucA22 compared to nonmucoid PAO1 mucA22 algD::FRT, as well as PAO1algIND grown with and without arabinose. We utilized gene-specific probes described previously (NanoString codeset PAV2) that monitor the expression of 75 transcripts associated with biofilm formation, polysaccharide production, iron acquisition, quorum sensing, and virulence (41). In brief, transcripts were monitored using a set of two hybridization probes complementary to each transcript of interest, with one probe enabling the capture of the transcript and the other containing a unique fluorescent barcode for direct transcript enumeration that reflects abundance in the sample.
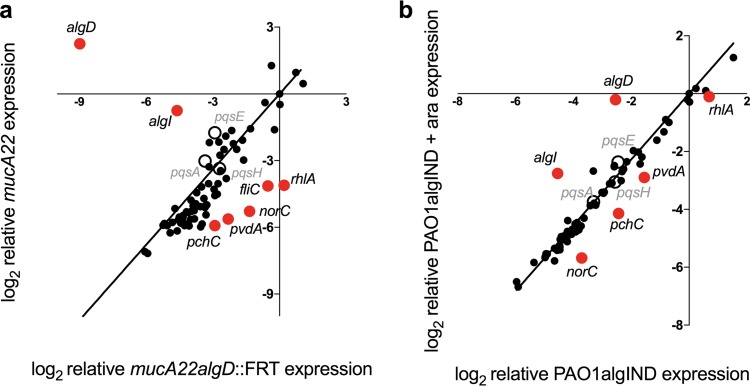
To assess changes in P. aeruginosa transcript profiles during alginate production, the relative levels of expression for each gene in the PAV2 NanoString codeset were compared between PAO1 mucA22 and PAO1 mucA22 algD::FRT (Fig. 4a), as well as PAO1algIND with and without arabinose to induce alginate production (Fig. 4b). Differences in raw transcript levels were also analyzed by their ranked abundance (heat map shown in Fig. S3a and raw data in Table S1 in the supplemental material). The relative transcript levels of five genes in the codeset were significantly lower (rhlA, norC, fliC, pvdA, and pchC) in the mucA22 mutant compared to the mucA22 algD::FRT mutant (Fig. 4a). As expected, we observed significantly higher levels of algD transcript in the mucA22 compared to the mucA22 algD::FRT strain. However, a larger amount of algI was also observed. This observation could be a result of inactivation of downstream genes in the alginate biosynthetic operon in the mucA22 algD::FRT strain or from the acquisition of a secondary mutation in algT during the construction of the algD::FRT strain. To confirm that algT has not been altered in this strain, the sequences of algT in PAO1 mucA22 and PAO1 mucA22 algD::FRT were compared and confirmed to be identical. The expression of algT and the subset of AlgT-regulated genes examined here were also not significantly different between these strains, confirming that alginate production is reduced in PAO1 mucA22 algD::FRT, but it retains constitutive expression of algT (see Table S2 in the supplemental material). In concordance with the comparison of the mucoid mucA22 mutant and its nonmucoid derivative PAO1 mucA22 algD::FRT, transcript levels of rhlA, norC, pvdA, and pchC were lower in PAO1algIND when grown in the presence of arabinose (and thus producing alginate), compared to without this inducer (Fig. 4b). While transcript levels of fliC were also lower, the difference was not statistically significant. The relative expression of algD in PAO1algIND in the presence of arabinose was less than the expression in PAO1 mucA22, which corresponds to the modest increase in alginate production observed in Fig. 3c for the alginate-inducible strain. Accordingly, a smaller change in the relative expression of the rhlA, pvdA, pchC, and norC genes was also observed. Finally, gene expression profiles for P. aeruginosa when grown in competition with S. aureus were examined, and no significant difference in gene expression was observed (Table S1), as we reported previously using transcriptome sequencing (RNA-Seq) analysis (10).
FIG 4 .
Alginate overproduction decreases the expression of P. aeruginosa genes required to reduce S. aureus viability. Relative expression of a subset of P. aeruginosa virulence genes was compared in PAO1 mucA22 (mucoid) and PAO1 mucA22 algD::FRT (nonmucoid) strains (a) and PAO1algIND strains grown with (mucoid) and without 0.5% arabinose (ara) (nonmucoid) (b) by the NanoString nCounter analysis system. The abundance of 75 transcripts was examined with a custom-designed codeset. Transcripts were log2 transformed and normalized to two P. aeruginosa housekeeping genes (rpoD and ppiD). Genes determined to be significantly differentially regulated when alginate is produced by an unpaired t test followed by the two-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli (with q = 1% for false discovery) are indicated in red. Genes involved in the PQS pathway are indicated with open circles.
Expression analysis of P. aeruginosa virulence genes during alginate overproduction. mRNA transcript abundance of a subset of P. aeruginosa virulence genes was compared in PAO1 mucA22 (mucoid) and PAO1 mucA22 algD::FRT (nonmucoid) strains by the NanoString nCounter analysis system. (a) A heat map was generated based on the ranked abundance (from 1 to 75) of the raw transcript values for each of the three biological replicates for each strain. Each cluster of genes is labeled with their known or predicted functions. The color gradient (white to purple) indicates the relative expression level of each gene with purple indicating the most highly expressed genes. (b to e) The expression of pvdA (b), pqsL (c and d), and rhlA (e) relative to the housekeeping gene rpoD was determined by qRT-PCR for the indicated P. aeruginosa strains. The means of biological triplicates (b, c, and e) and quadruplicates (d) and standard deviations are indicated. Statistical significance was determined by one-way ANOVA followed by a Dunnett’s multiple comparison test. *, P < 0.05; **, P < 0.01. Download FIG S3, TIF file, 78 MB (79.9MB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw NanoString data. Download TABLE S1, XLSX file, 0.1 MB (132KB, xlsx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative expression of AlgT-regulated genes in PAV2 between PAO1 mucA22 and PAO1 mucA22 algD::FRT. Download TABLE S2, DOCX file, 0.1 MB (58.6KB, docx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alginate overproduction reduces pyoverdine production.
mRNA analysis revealed that expression of genes required for siderophore production (pvdA and pchC, for the synthesis of pyoverdine and pyochelin, respectively) were lower when alginate was overproduced. These siderophores specifically chelate iron (Fe3+), a role previously demonstrated to be involved in P. aeruginosa inhibition of S. aureus growth (8, 10). We therefore determined if pyoverdine (the predominant P. aeruginosa siderophore) plays a role in inhibiting growth of S. aureus under the conditions utilized in this study. We first confirmed reduced expression of pvdA in the mucA22 mutant compared to the WT and mucA22 algD::FRT strains by quantitative real-time PCR (qRT-PCR) (Fig. S3b).
We then competed P. aeruginosa deficient in pvdA with S. aureus and observed a significant but modest restoration of S. aureus viability compared to the WT (Fig. 5a), as previously reported (10). We then asked if the observed decrease in pvdA expression when alginate is overproduced, results in decreased pyoverdine production. Pyoverdine is fluorescent when excited at 400 nm, and we utilized this property to determine the amount of pyoverdine produced as measured in relative fluorescent units (RFU) per OD600 of P. aeruginosa cultures grown for 8 h under the conditions utilized in the coculture assays. The mucA22 mutant produced approximately 40% less pyoverdine than nonmucoid, WT P. aeruginosa. When algD is deleted in the mucA22 mutant, pyoverdine production is restored (Fig. 5b), demonstrating that pyoverdine levels are lower in alginate-producing P. aeruginosa strains.
FIG 5 .
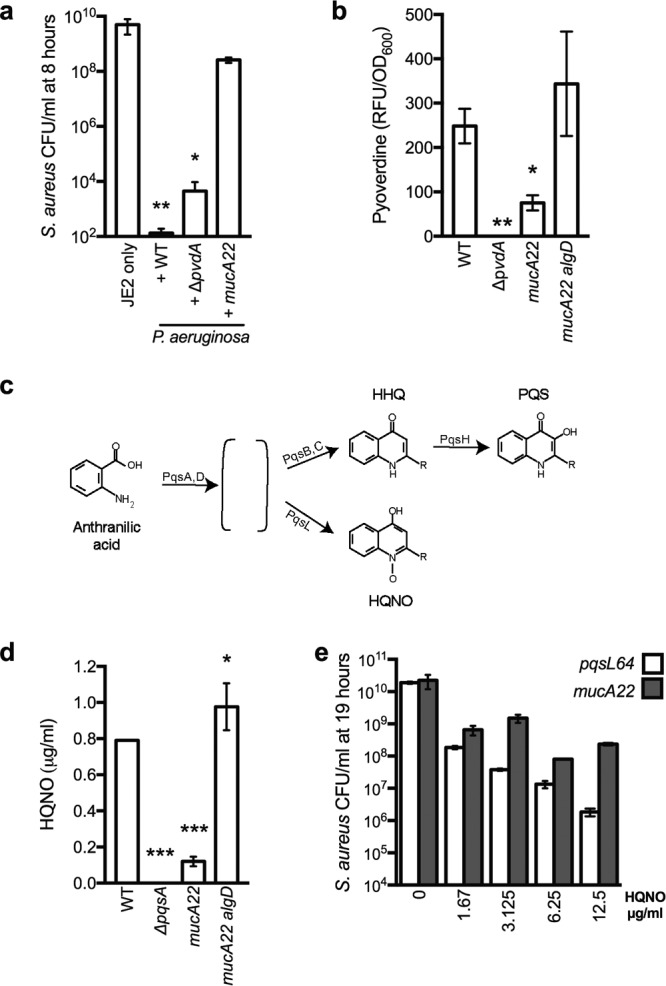
Alginate overproduction inhibits antistaphylococcal exoproducts. (a) In vitro P. aeruginosa-S. aureus coculture assays in planktonic culture with the indicated strains. Log10 CFU/ml for S. aureus JE2 are indicated after 8 h of incubation. (b) Pyoverdine was quantified as relative fluorescence units (RFU)/OD600 produced by planktonic P. aeruginosa strains grown for 8 h. (c) Schematic of 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO) synthesis. The PqsA to -E, -L, and -H enzymes catalyze the synthesis of a series of 4-hydroxy-2-alkylquinolones (HAQs). The conversion of anthranilic acid to uncharacterized intermediates (indicated by the brackets) is catalyzed by PqsA and -D, followed by conversion to either 4-hydroxy-2-heptylquinoline (HHQ) by PqsB and -C (which can be converted to 3,4-dihydroxy-2-heptylquinoline [PQS] by PqsH) or converted to HQNO by PqsL. (d) HQNO quantification by LC-MS from supernatants derived from the indicated strains following 8 h of incubation in planktonic culture and (e) in vitro P. aeruginosa-S. aureus coculture assays in planktonic culture with P. aeruginosa PAO1 pqsL64 (white) and PAO1 mucA22 (gray) in the presence of the indicated concentrations of HQNO. Log10 CFU/ml for S. aureus JE2 are indicated after 19 h of incubation. Error bars indicate standard deviations from three biological replicates performed in triplicate. In panels a and b, statistical significance was determined by performing an analysis of variance (ANOVA) followed by a Dunnett’s multiple comparison test comparing each condition to the WT in panels a and d (*, P < 0.05; **, P < 0.01; ***, P < 0.0001) and to JE2 only in panel b. In panel e, statistical significance was determined by performing independent ANOVA analyses for PAO1 pqsL64 and PAO1 mucA22 followed by a Dunnett’s multiple comparison test comparing the viability of S. aureus JE2 in the presence of each HQNO concentration to the condition without HQNO, and the viability was significantly decreased at each concentration: for PAO1 pqsL64; P ≤ 0.0001, and for PAO1 mucA22, P ≤ 0.01.
Alginate overproduction reduces HQNO production.
In addition to siderophores, we and others previously reported that 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO) inhibits S. aureus respiration (7, 10, 42), and we further showed that HQNO causes a shift to a fermentative lifestyle and eventual S. aureus cell death during late stage biofilm growth on CFBE cells (10). In our NanoString studies described above, we did not observe a significant change in the expression of genes involved in the Pseudomonas quinolone signal pathway (pqs) in alginate-overproducing strains (open circles in Fig. 4a and b and heat map in Fig. S3a). The PqsA to -E, -L, and -H enzymes catalyze the synthesis of a series of 4-hydroxy-2-alkylquinolones (HAQs [schematic in Fig. 5c]) (15, 43). Anthranilic acid is converted by PqsA and -D to a series of precursors whose precise structures are unknown (indicated by brackets), and these intermediates are ultimately converted to either 4-hydroxy-2-heptylquinoline (HHQ) by PqsB and -C (which can be converted to 3,4-dihydroxy-2-heptylquinoline [PQS] by PqsH) or converted to HQNO by PqsL (15). Because deletion of pqsL reduces the ability of P. aeruginosa to kill S. aureus compared to the wild type (10) (see Fig. S4 in the supplemental material), we would have predicted that alginate overproduction might decrease expression of genes involved in HQNO synthesis, reducing the amount of HQNO produced by mucoid cells. While P. aeruginosa HQNO production peaks in late stationary phase, the expression of the genes encoding enzymes required for HQNO generation peaks in late exponential phase (15). For the NanoString analysis, RNA was extracted from P. aeruginosa cells in late stationary phase. We therefore examined the expression of pqsL by qRT-PCR at various time points in WT PAO1 compared to a P. aeruginosa PAO1 pqsL64 mutant, previously characterized to be deficient in the production of HQNO (44). pqsL expression peaked for WT in late exponential phase (3 h, OD600 of ~1.8) (Fig. S3c), as previously observed (15). Expression of pqsL was then examined in the mucA22 mutant at ~3 h and was found to be modestly but significantly decreased in relative expression compared to mucA22 algD::FRT (Fig. S3d). To determine if HQNO generation is altered, we measure HQNO present in culture supernatants of P. aeruginosa by extracting HQNO from equal cell numbers of P. aeruginosa strains, including the WT and the ΔpqsA, mucA22, and mucA22 algD::FRT mutants, and quantified HQNO by liquid chromatography coupled to mass spectrometry (LC-MS), as previously described (45). Nonmucoid WT P. aeruginosa produced approximately 0.80 μg/ml of HQNO, whereas the mucoid mucA22 mutant produced 0.12 μg/ml HQNO, a significant reduction (Fig. 5d). No HQNO was detected in the ΔpqsA mutant, the negative-control strain. Deletion of algD in the mucA22 mutant restored the production of HQNO to levels that are significantly increased compared to those of the wild type (0.98 μg/ml).
Alginate overproduction inhibits HQNO antimicrobial activity toward S. aureus. (a) In vitro P. aeruginosa-S. aureus coculture assays in planktonic culture with S. aureus JE2 only (a), S. aureus plus P. aeruginosa PAO1 (WT), (b) S. aureus plus P. aeruginosa pqsL64 (c), and P. aeruginosa mucA22 (d) in the presence of either DMSO (vehicle control [open circles]) or 12.5 µg/ml HQNO (solid circles). Log10 CFU/ml for S. aureus JE2 are indicated. Error bars indicate standard deviations from three biological replicates performed in triplicate. Download FIG S4, TIF file, 0.3 MB (324.2KB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
To confirm that reduced HQNO in the mucA22 mutant contributes to decreased killing of S. aureus, we asked if the addition of exogenous HQNO could restore killing of S. aureus. As a control, we first tested if the addition of HQNO could complement a pqsL mutant. As expected, pqsL64 was unable to reduce S. aureus viability (Fig. 5e; Fig. S4C), supporting previous observations that HQNO is required to kill S. aureus (7, 10, 42). We then grew S. aureus in monoculture or in coculture with the P. aeruginosa WT, pqsL64, or mucA22 strain with either 12.5 µg/ml HQNO or dimethyl sulfoxide (DMSO) (vehicle control). The addition of HQNO did not affect the viability of S. aureus alone or in the presence of wild-type P. aeruginosa PAO1 (Fig. S4a and b) but did restore killing mediated by the pqsL64 mutant (Fig. S4c), albeit the amount of time to initiate killing was delayed compared to that with WT P. aeruginosa. Importantly, HQNO partially restored the ability of the P. aeruginosa mucA22 mutant to kill S. aureus (Fig. S4d), suggesting decreased production of HQNO contributes to the reduced antimicrobial activity of the mucoid isolates. The addition of HQNO did not fully restore killing of S. aureus by the PAO1 mucA22 strain, and the concentrations required to restore killing were higher for mucA22 compared to pqsL64 (Fig. 5e).
Rhamnolipids contribute to P. aeruginosa killing of S. aureus.
NanoString analysis of alginate-overproducing strains also revealed lower transcript levels of rhlA compared to those in nonmucoid strains: rhlA is the gene that encodes the first enzyme required for the synthesis of P. aeruginosa surfactants (rhamnolipids) (Fig. 4a and b). RhlA is involved in the synthesis of the fatty acid dimer 3-(3-hydroxyalkanoyloxy) alkanoic acid (HAA) moiety of rhamnolipids from 3-hydroxy fatty acid precursors, which are subsequently converted to monorhamnolipids, and dirhamnolipids, by RhlB and RhlC, respectively (46). HAA, mono-, and dirhamnolipids each have biosurfactant activity, but the type of rhamnolipid produced and the amount depends on the strain and the carbon source (47). Rhamnolipids derived from P. aeruginosa strains isolated from oil-contaminated soil have been shown to have antimicrobial activity against S. aureus (13, 14); therefore, we hypothesized that the production of rhamnolipids may also play a role in P. aeruginosa interactions with S. aureus in our model. We first confirmed reduced expression of rhlA in the mucA22 mutant compared to the WT and mucA22 algD::FRT mutant by qRT-PCR (Fig. S3e). Furthermore, when the rhlA gene was disrupted in P. aeruginosa (PAO1 rhlA::Gm), the viability of S. aureus in coculture was similar to that seen with mucA22 mutant (Fig. 6a).
FIG 6 .
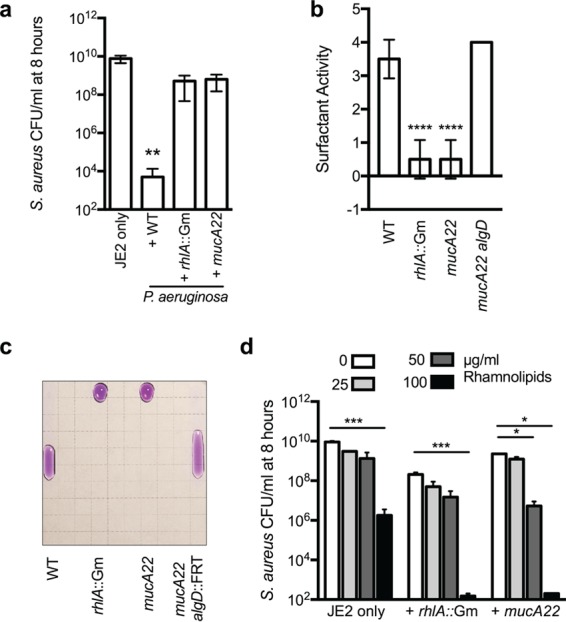
Alginate inhibits the production of rhamnolipids required to kill S. aureus. In vitro P. aeruginosa-S. aureus coculture assays in planktonic culture. In panels a and d, the log10 CFU/ml for S. aureus JE2 are indicated after 8 h of incubation, and in panels b and c, the drop collapse assay was used to measure surfactant activity. Clarified supernatants were serially diluted (1:1) with water plus 0.005% crystal violet for visualization. Twenty microliters of each dilution was spotted onto the underside of the lid of a petri plate and tilted at a 90° angle. As surfactant quantities are reduced by dilution, surface tension increases, resulting in the beading of the droplet. Surfactant scores are equal to the reciprocal of the greatest dilution at which there was surfactant activity (a collapsed drop that migrates down the plate). Quantification is indicated in panel b, and a representative image of all strains at the 1/8 dilution is shown in panel c. In panel d, competitions were performed in the presence of the indicated concentrations of rhamnolipids. Error bars indicate the standard deviation of three biological replicates performed in triplicate (two biological replicates for panel d). In panels a and b, statistical significance was determined by performing an ANOVA followed by a Dunnett’s multiple comparison test comparing each condition to JE2 in panel a and to the WT only in panel b. In panel d, statistical significance was determined by performing an ANOVA followed by a Tukey’s multiple comparison test to compare the mean survival of S. aureus in the presence of each rhamnolipid concentration within strains. *, P ≤ 0.05; **, P < 0.01; ***, P < 0.0001.
To determine if rhamnolipid production is also decreased when alginate is overproduced, we assayed for biosurfactant activity in clarified P. aeruginosa supernatant from the mucoid mucA22 mutant compared to the nonmucoid, WT, and mucA22 algD::FRT strains, utilizing a drop collapse assay, as previously described (48). In brief, the presence of sufficient surfactant disrupts the surface tension of droplets of bacterial supernatant on plastic, which will migrate downward when placed at a 90° angle. Supernatant from each strain (or purified rhamnolipid) was serially diluted 2-fold, and the surfactant activity calculated as 1/dilution at which the drop begins to migrate (see representative images in Fig. S5 and quantification of biological replicates in Fig. 6b). P. aeruginosa WT supernatant, which produces rhamnolipids, exhibits surfactant activity of approximately 3.5, compared to 0.5 when rhlA is disrupted (Fig. 6b). The surfactant activity of PAO1 mucA22 phenocopies the rhlA mutant, and surfactant activity is restored in the mucA22 algD mutant, demonstrating that alginate overproduction reduces surfactant production in P. aeruginosa. A representative image of each strain at the 1/8 dilution in shown in Fig. 6c. We then evaluated the ability of rhamnolipids to kill S. aureus. We found that 100 µg/ml of a 50/50 mixture of mono- and dirhamnolipids reduces the viability of S. aureus from approximately 1 × 1010 CFU/ml to 1 × 106 CFU/ml (Fig. 6d). Whereas the addition of exogenous rhamnolipids to S. aureus in the presence of a rhlA mutant or the mucA22 mutant further reduces S. aureus viability to below 100 CFU/ml, which supports our findings that multiple P. aeruginosa antimicrobials are required to kill S. aureus.
Alginate inhibits the production of P. aeruginosa rhamnolipids. The results from a drop collapse assay to measure rhamnolipid-mediated surfactant activity are shown. The clarified supernatants of the indicated strains (a) and purified rhamnolipids (b [50/50 mixture of mono- and dirhamnolipids]) were serially diluted (1:1) with water plus 0.005% crystal violet (for visualization). Surfactant activity was measured by the spread of the droplet down a 90° incline. As surfactant quantities are reduced by dilution, surface tension increases, resulting in the beading of the droplet. Representative images from three biological replicates are shown. Download FIG S5, TIF file, 45.6 MB (46.7MB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
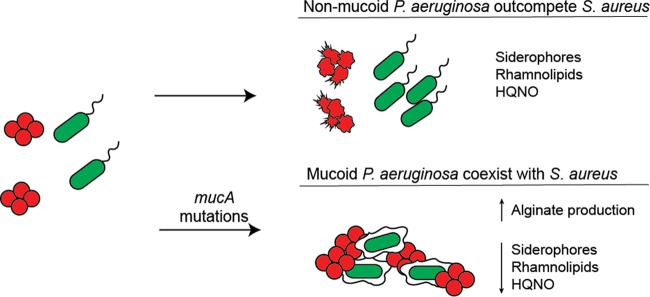
From our studies, we are able to begin to generate a model of how P. aeruginosa and S. aureus may be able to coinfect the same niche in the respiratory tract of CF patients (Fig. 7). During infection P. aeruginosa acquires mutations in the mucA gene, leading to alginate overproduction. Alginate overproduction reduces the generation of siderophores, HQNO, and rhamnolipids by lowering the expression of genes encoding enzymes required for their generation. All three of these factors—HQNO, siderophores, and rhamnolipids—have documented effects in reducing the viability of S. aureus (7–16). These data demonstrate that the production of this polysaccharide is sufficient to block a range of antimicrobials required for reduction of S. aureus viability. The observations that PAO1algIND has reduced ability to kill S. aureus when grown in the presence of arabinose, while only producing a modest level of alginate, suggests that small changes in alginate production during infection could influence polymicrobial dynamics. In agreement with this idea, we observed increased production of both pyoverdine and HQNO compared to the WT when alginate production was completely removed by deletion of algD (Fig. 5b and d). Evidence that even nonmucoid P. aeruginosa strains produce alginate during CF pulmonary infections (41, 49) suggests that the observations in this study may be relevant beyond P. aeruginosa isolates that visibly produce alginate.
FIG 7 .
Proposed model of P. aeruginosa coinfection with S. aureus. Nonmucoid isolates produce a range of antimicrobial agents that can kill S. aureus, including siderophores, rhamnolipids, and HQNO, which allows P. aeruginosa to outcompete S. aureus. If P. aeruginosa acquires mucA mutations during infection leading to overproduction of the polysaccharide alginate, the expression of genes required for siderophore, HQNO, and rhamnolipid synthesis are decreased. These modifications reduce the capacity of P. aeruginosa to outcompete S. aureus, and the two species coexist in the CF lung.
The correlations identified herein through studying P. aeruginosa isolates from coinfected CF patients support the hypothesis that one way P. aeruginosa and S. aureus coexist in the CF lung is through P. aeruginosa reducing antimicrobial generation, driven by alginate overproduction. While we are beginning to advance our knowledge of the distribution of microbial species during CF pulmonary infections, and how and to what extent interspecies interactions occur (50, 51), our understanding remains quite limited. We predict that in certain areas of the lung, P. aeruginosa and S. aureus are spatially segregated, which also likely contributes to S. aureus survival of during coinfection, independent of P. aeruginosa competitive phenotypes. Thus, it is necessary to further investigate the extent of spatial interspecies coexistence in CF, and its influence on microbial physiology and patient outcome. If the former hypothesis is supported, that P. aeruginosa adapts to the presence of competing species, we can explore if such adaptations are specific to S. aureus. Reported studies from the Whiteley group suggest that P. aeruginosa senses and responds to N-acetylglucosamine (GlcNAc) shed from the cell wall of Gram-positive organisms (52); therefore, some specificity of response may be afforded to Gram-positive organisms.
Our expression analysis revealed alginate-overproducing strains decrease expression of genes required for pyoverdine, HQNO, and rhamnolipid synthesis, as well as norC, encoding a nitric oxide reductase, and fliC, encoding flagellin type B. We predict alginate may also exert effects on additional P. aeruginosa genes not examined in this study. Each gene identified here has been reported previously to be directly or indirectly regulated by AlgT (27–30). Our data support a mechanism of indirect regulation, whereby AlgT activates transcription of PalgD, resulting in increased alginate production, which reduces the expression of a subset of genes in the AlgT regulon. In agreement with a model of alginate-dependent regulation independent of AlgT, we did not observe changes in algT gene expression (or additional genes analyzed known to be AlgT regulated) in the mucA22 algD::FRT mutant compared to the mucA22 strain or in PAO1algIND with and without arabinose (Fig. 3; Table S2). However, it remains a formal possibility that limiting alginate production reduces posttranscriptional AlgT activity and alters expression of only a subset of AlgT-regulated genes.
The mechanism by which alginate might reduce expression of genes required for HQNO, siderophore, and rhamnolipid biosynthesis has not been established, yet modulation of relevant P. aeruginosa physiology by this polysaccharide is not unprecedented. Indeed, a recent report demonstrated alginate can interfere with PQS signaling—a feature limited to alginate producers; no such inhibition of signaling was observed for the non-alginate-producing neighbors (53). Alginate has also been shown to restrict the diffusion of oxygen (54–56) and aminoglycoside antibiotics (57), and to bind and sequester reactive intermediates and iron (58). These properties may explain how alginate is able to reduce the production of multiple P. aeruginosa virulence factors. In the case of siderophores, alginate may help concentrate iron locally thus reducing the need to acquire iron via siderophores. Prior studies demonstrating increased alginate production during iron starvation support the hypothesis that alginate aids P. aeruginosa in acquiring iron (59). In this scenario, we may predict that changes in siderophore gene expression occur as part of derepression of the entire Fur regulon. However, in our NanoString studies, we did not observe a significant change in the expression of other Fur-regulated genes within the PaV2 codeset (bfrB, hasR, phuR, and feoB). Alginate might also help to sequester other secreted factors thus increasing their local concentration and feedback inhibiting their production, or making such secreted factors unavailable to the cell. Alternatively, the draw on metabolites to synthesize alginate may deplete intermediates used by other pathways. Such an explanation may inform the reduction in rhamnolipid production, as these surfactants require similar activated sugars for their synthesis. Whether via common or discrete mechanisms, it is clear that production of even a small amount of alginate results in markedly altered cellular physiology.
Our investigation of polymicrobial dynamics in CF isolates reinforces previous observations (10) that in order for P. aeruginosa to kill S. aureus, multiple secreted factors must be generated simultaneously by P. aeruginosa, as deletion of any one of these factors reduces P. aeruginosa ability to kill S. aureus. These antimicrobials may function individually and/or work together to enhance killing. For example, rhamnolipids have been shown to not only have antimicrobial activity, but these surfactants can increase the solubility and activity of other P. aeruginosa metabolites such as PQS (60). A recent report examined S. aureus-P. aeruginosa interactions in a biofilm coculture model in a flow chamber system where the P. aeruginosa exoproducts required for killing would be removed (61). Here the authors observed that nonmucoid P. aeruginosa facilitates S. aureus biofilm formation, whereas mucoid P. aeruginosa tend to outcompete S. aureus. This finding highlights the complexity of interspecies interactions and how environmental conditions may influence such interactions. In CF respiratory infections, microbial communities often persist in thickened, static airway secretions and in close proximity to airway epithelial cells, and the P. aeruginosa antimicrobials described here have not only been detected in the airway of CF patients but may be predictors of airway infection (62, 63). We therefore predict these secreted P. aeruginosa factors are important mediators of infection in CF respiratory disease. Finally, whether the correlation observed between reduced competition by P. aeruginosa and coinfection results from P. aeruginosa adapting in response to the presence of competing species or that reduced competition by P. aeruginosa occurs independently, which then permits growth of other organisms, is currently unknown.
Generation of a complete understanding of the dynamics of respiratory infections is complicated by shifting host pathogens, as well as inter- and intraspecies interactions. Here we revealed that a prevalent P. aeruginosa adaptation during chronic infections, mucoid conversion, correlates with reduced P. aeruginosa competition with S. aureus. Interrogation of this phenotype in vitro informed how mucoid conversion limits P. aeruginosa competition with S. aureus—by reducing production antistaphylococcal factors. Combining these findings with a model of coinfection during biofilm formation on CF bronchial epithelia cells allows us to gain a more complete understanding of how bacteria interact during infection.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table S3 in the supplemental material. P. aeruginosa and Escherichia coli were routinely grown in lysogeny broth (LB) and S. aureus in tryptic soy broth (TSB), with 1.5% agar for solid medium. Synthetic CF sputum medium was made as previously described (35) with 0.5% mucin. Gentamicin at 30 μg/ml and carbenicillin at 250 µg/ml were used for P. aeruginosa, and 100 µg/ml ampicillin was used for E. coli where indicated. Detailed descriptions of the construction of P. aeruginosa mutants can be found in Text S1 in the supplemental material.
Bacterial strains and plasmids used in this study. Download TABLE S3, DOCX file, 0.1 MB (24.3KB, docx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S4, DOCX file, 0.1 MB (18.5KB, docx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download TEXT S1, DOCX file, 0.2 MB (34.1KB, docx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Coculture assays.
P. aeruginosa and S. aureus were cocultured on either agar plates, in shaking broth culture in rich medium (TSB) or synthetic CF sputum medium, or in biofilm growth on CF bronchial epithelial cells. At the indicated time points, aliquots of culture were removed, and P. aeruginosa and S. aureus were enumerated by plating on selective media (Pseudomonas isolation agar [PIA] and mannitol salts agar [MSA], respectively). Detailed methods are provided in Text S1.
Alginate quantification.
Alginate was collected from cultures grown as described above for planktonic coculture assays and isolated as previously described (64), with modifications (65). Detailed protocols can be found in Text S1.
NanoString analysis of P. aeruginosa transcripts.
RNA transcript levels were measured using the NanoString nCounter system (NanoString Technologies, Seattle, WA) and methods described by Geiss et al. (66). We employed a custom-designed codeset containing 75 P. aeruginosa genes (Table S1). The nucleotide sequences were provided to NanoString Technologies, Inc., for codeset design and construction. Each reaction mixture contained 70 ng of RNA, hybridization buffer, reporter probes, capture probes, as well as six positive and eight negative controls. RNA was hybridized with reporter and capture probes overnight at 65°C, and sample preparation ensued at the NanoString preparation station. Finally, targets were counted on the nCounter using 255 fields of view per sample. Data were analyzed using nSolver Analysis software, version 1.1 (NanoString Technologies, Seattle, WA). Raw counts were calibrated to the arithmetic mean of six spiked positive-control transcripts, and count values greater than 2 standard deviations above the average of the eight negative-control probes were considered above background. Detailed protocols for RNA isolation and quantitative real-time PCR can be found in Text S1.
P. aeruginosa antimicrobial quantification.
Pyoverdine, HQNO, and rhamnolipids were quantified as previously described (45, 48, 67), respectively. Detailed protocols can be found in Text S1.
Availability of data.
The authors certify that they will comply with mBio’s data policy: data will be made publicly available upon publication and upon request for peer review.
ACKNOWLEDGMENTS
Access to the CF Biospecimen Registry (CFBR) at Emory+Children's Center for Cystic Fibrosis and Airways Disease Research was provided through Children’s Healthcare of Atlanta and the Emory University Pediatric CF Discovery Core. We thank Arlene Stecenko and Maret Maliniak for assistance acquiring bacterial isolates and Jeffery Meisner for insightful discussion and review of the manuscript.
This work was supported by the Cystic Fibrosis Foundation (LIMOLI15F0 to D.H.L. and OTOOLE16GO to G.A.O.), the National Institutes of Health (R37 AI83256-06 to G.A.O., R33 AI105902 to L.G.R., and R01 AI091702 to D.A.H.), the Canadian Institutes of Health Research (Operating Grant no. 13337 to P.L.H.), and the Natural Sciences and Engineering Research Council of Canada (NSERC, graduate scholarship to G.B.W.). P.L.H. is the recipient of a Canada Research Chair. The cell biology studies utilized the resources of the Host Pathogen Interaction Core, supported by the National Institute of General Medical Sciences of the NIH under award P20-GM103413 and the Cystic Fibrosis Research Development Program (STANTO07R0). Clinical strains and associated metadata were obtained from the CF Biospecimen Registry of the CF@LANTA CF Research Center, cosupported by Children’s Healthcare of Atlanta and the Cystic Fibrosis Foundation Research Development Program (MCCART15R0).
Footnotes
Citation Limoli DH, Whitfield GB, Kitao T, Ivey ML, Davis MR, Jr, Grahl N, Hogan DA, Rahme LG, Howell PL, O’Toole GA, Goldberg JB. 2017. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio 8:e00186-17. https://doi.org/10.1128/mBio.00186-17.
REFERENCES
- 1.Taylor-Robinson D, Whitehead M, Diderichsen F, Olesen HV, Pressler T, Smyth RL, Diggle P. 2012. Understanding the natural progression in %FEV1 decline in patients with cystic fibrosis: a longitudinal study. Thorax 67:860–866. doi: 10.1136/thoraxjnl-2011-200953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filkins LM, O’Toole GA. 2015. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog 11:e1005258. doi: 10.1371/journal.ppat.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCallum N, Berger-Bächi B, Senn MM. 2010. Regulation of antibiotic resistance in Staphylococcus aureus. Int J Med Microbiol 300:118–129. doi: 10.1016/j.ijmm.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 5.Talwalkar JS, Murray TS. 2016. The approach to Pseudomonas aeruginosa in cystic fibrosis. Clin Chest Med 37:69–81. doi: 10.1016/j.ccm.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen AT, Oglesby-Sherrouse AG. 2016. Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl Microbiol Biotechnol 100:6141–6148. doi: 10.1007/s00253-016-7596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 1992. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother 30:615–623. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- 8.Mashburn LM, Jett AM, Akins DR, Whiteley M. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol 187:554–566. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffman LR, Déziel E, D’Argenio DA, Lépine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. 2006. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 103:19890–19895. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, O’Toole GA. 2015. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol 197:2252–2264. doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. 2015. Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. J Bacteriol 197:2265–2275. doi: 10.1128/JB.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haba E, Pinazo A, Jauregui O, Espuny MJ, Infante MR, Manresa A. 2003. Physicochemical characterization and antimicrobial properties of rhamnolipids produced by Pseudomonas aeruginosa 47T2 NCBIM 40044. Biotechnol Bioeng 81:316–322. doi: 10.1002/bit.10474. [DOI] [PubMed] [Google Scholar]
- 14.Bharali P, Saikia JP, Ray A, Konwar BK. 2013. Rhamnolipid (RL) from Pseudomonas aeruginosa OBP1: a novel chemotaxis and antibacterial agent. Colloids Surf B Biointerfaces 103:502–509. doi: 10.1016/j.colsurfb.2012.10.064. [DOI] [PubMed] [Google Scholar]
- 15.Déziel E, Lépine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. 2004. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A 101:1339–1344. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hazan R, Que YA, Maura D, Strobel B, Majcherczyk PA, Hopper LR, Wilbur DJ, Hreha TN, Barquera B, Rahme LG. 2016. Auto poisoning of the respiratory chain by a quorum-sensing-regulated molecule favors biofilm formation and antibiotic tolerance. Curr Biol 26:195–206. doi: 10.1016/j.cub.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Limoli DH, Yang J, Khansaheb MK, Helfman B, Peng L, Stecenko AA, Goldberg JB. 2016. Staphylococcus aureus and Pseudomonas aeruginosa co-infection is associated with cystic fibrosis-related diabetes and poor clinical outcomes. Eur J Clin Microbiol Infect Dis 35:947–953. doi: 10.1007/s10096-016-2621-0. [DOI] [PubMed] [Google Scholar]
- 18.Maliniak ML, Stecenko AA, McCarty NA. 2016. A longitudinal analysis of chronic MRSA and Pseudomonas aeruginosa co-infection in cystic fibrosis: a single-center study. J Cyst Fibros 15:350–356. doi: 10.1016/j.jcf.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Hubert D, Réglier-Poupet H, Sermet-Gaudelus I, Ferroni A, Le Bourgeois M, Burgel PR, Serreau R, Dusser D, Poyart C, Coste J. 2013. Association between Staphylococcus aureus alone or combined with Pseudomonas aeruginosa and the clinical condition of patients with cystic fibrosis. J Cyst Fibros 12:497–503. doi: 10.1016/j.jcf.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 20.Cystic Fibrosis Foundation 2016. Cystic Fibrosis Foundation patient registry 2010 annual data report. Cystic Fibrosis Foundation, Bethesda, MD. [Google Scholar]
- 21.DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. 2014. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun 82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fazli M, Bjarnsholt T, Kirketerp-Møller K, Jørgensen B, Andersen AS, Krogfelt KA, Givskov M, Tolker-Nielsen T. 2009. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol 47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogan DA, Willger SD, Dolben EL, Hampton TH, Stanton BA, Morrison HG, Sogin ML, Czum J, Ashare A. 2016. Analysis of lung microbiota in bronchoalveolar lavage, protected brush and sputum samples from subjects with mild-to-moderate cystic fibrosis lung disease. PLoS One 11:e0149998. doi: 10.1371/journal.pone.0149998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakeman CA, Moore JL, Noto MJ, Zhang Y, Singleton MD, Prentice BM, Gilston BA, Doster RS, Gaddy JA, Chazin WJ, Caprioli RM, Skaar EP. 2016. The innate immune protein calprotectin promotes Pseudomonas aeruginosa and Staphylococcus aureus interaction. Nat Commun 7:11951. doi: 10.1038/ncomms11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Govan JR, Deretic V. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60:539–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bragonzi A, Wiehlmann L, Klockgether J, Cramer N, Worlitzsch D, Döring G, Tümmler B. 2006. Sequence diversity of the mucABD locus in Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology 152:3261–3269. doi: 10.1099/mic.0.29175-0. [DOI] [PubMed] [Google Scholar]
- 27.Tart AH, Wolfgang MC, Wozniak DJ. 2005. The alternative sigma factor AlgT represses Pseudomonas aeruginosa flagellum biosynthesis by inhibiting expression of fleQ. J Bacteriol 187:7955–7962. doi: 10.1128/JB.187.23.7955-7962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Firoved AM, Deretic V. 2003. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J Bacteriol 185:1071–1081. doi: 10.1128/JB.185.3.1071-1081.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, Nielsen KF, Jelsbak L, Høiby N, Yang L, Molin S. 2010. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ Microbiol 12:1643–1658. doi: 10.1111/j.1462-2920.2010.02211.x. [DOI] [PubMed] [Google Scholar]
- 30.Wood LF, Ohman DE. 2009. Use of cell wall stress to characterize σ22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol Microbiol 72:183–201. doi: 10.1111/j.1365-2958.2009.06635.x. [DOI] [PubMed] [Google Scholar]
- 31.Ciofu O, Lee B, Johannesson M, Hermansen NO, Meyer P, Høiby N, Scandinavian Cystic Fibrosis Study Consortium . 2008. Investigation of the algT operon sequence in mucoid and non-mucoid Pseudomonas aeruginosa isolates from 115 Scandinavian patients with cystic fibrosis and in 88 in vitro non-mucoid revertants. Microbiology 154:103–113. doi: 10.1099/mic.0.2007/010421-0. [DOI] [PubMed] [Google Scholar]
- 32.Ciofu O, Tolker-Nielsen T, Jensen PØ, Wang H, Høiby N. 2015. Antimicrobial resistance, respiratory tract infections and role of biofilms in lung infections in cystic fibrosis patients. Adv Drug Deliv Rev 85:7–23. doi: 10.1016/j.addr.2014.11.017. [DOI] [PubMed] [Google Scholar]
- 33.Goerke C, Wolz C. 2010. Adaptation of Staphylococcus aureus to the cystic fibrosis lung. Int J Med Microbiol 300:520–525. doi: 10.1016/j.ijmm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Anderson GG, Moreau-Marquis S, Stanton BA, O’Toole GA. 2008. In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells. Infect Immun 76:1423–1433. doi: 10.1128/IAI.01373-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer KL, Aye LM, Whiteley M. 2007. Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J Bacteriol 189:8079–8087. doi: 10.1128/JB.01138-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Linker A, Jones RS. 1966. A new polysaccharide resembling alginic acid isolated from pseudomonads. J Biol Chem 241:3845–3851. [PubMed] [Google Scholar]
- 37.Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front Microbiol 2:167. doi: 10.3389/fmicb.2011.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitney JC, Whitfield GB, Marmont LS, Yip P, Neculai AM, Lobsanov YD, Robinson H, Ohman DE, Howell PL. 2015. Dimeric c-di-GMP is required for post-translational regulation of alginate production in Pseudomonas aeruginosa. J Biol Chem 290:12451–12462. doi: 10.1074/jbc.M115.645051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kulkarni MM. 2011. Digital multiplexed gene expression analysis using the NanoString nCounter system. Curr Protoc Mol Biol Chapter 25:Unit25B.10. doi: 10.1002/0471142727.mb25b10s94. [DOI] [PubMed] [Google Scholar]
- 41.Gifford AH, Willger SD, Dolben EL, Moulton LA, Dorman DB, Bean H, Hill JE, Hampton TH, Ashare A, Hogan DA. 2016. Use of a multiplex transcript method for analysis of Pseudomonas aeruginosa gene expression profiles in the cystic fibrosis lung. Infect Immun 84:2995–3006. doi: 10.1128/IAI.00437-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, Götz F. 2006. Microevolution of cytochrome bd oxidase in staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol 188:8079–8086. doi: 10.1128/JB.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lépine F, Milot S, Déziel E, He J, Rahme LG. 2004. Electrospray/mass spectrometric identification and analysis of 4-hydroxy-2-alkylquinolines (HAQs) produced by Pseudomonas aeruginosa. J Am Soc Mass Spectrom 15:862–869. doi: 10.1016/j.jasms.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 44.D’Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J Bacteriol 184:6481–6489. doi: 10.1128/JB.184.23.6481-6489.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lépine F, Déziel E, Milot S, Rahme LG. 2003. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim Biophys Acta 1622:36–41. doi: 10.1016/S0304-4165(03)00103-X. [DOI] [PubMed] [Google Scholar]
- 46.Abdel-Mawgoud AM, Lépine F, Déziel E. 2010. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Déziel E, Lépine F, Milot S, Villemur R. 2000. Mass spectrometry monitoring of rhamnolipids from a growing culture of Pseudomonas aeruginosa strain 57RP. Biochim Biophys Acta 1485:145–152. doi: 10.1016/S1388-1981(00)00039-1. [DOI] [PubMed] [Google Scholar]
- 48.Caiazza NC, Shanks RMQ, O’Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bragonzi A, Worlitzsch D, Pier GB, Timpert P, Ulrich M, Hentzer M, Andersen JB, Givskov M, Conese M, Döring G. 2005. Nonmucoid Pseudomonas aeruginosa expresses alginate in the lungs of patients with cystic fibrosis and in a mouse model. J Infect Dis 192:410–419. doi: 10.1086/431516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jorth P, Staudinger BJ, Wu X, Hisert KB, Hayden H, Garudathri J, Harding CL, Radey MC, Rezayat A, Bautista G, Berrington WR, Goddard AF, Zheng C, Angermeyer A, Brittnacher MJ, Kitzman J, Shendure J, Fligner CL, Mittler J, Aitken ML, Manoil C, Bruce JE, Yahr TL, Singh PK. 2015. Regional isolation drives bacterial diversification within cystic fibrosis lungs. Cell Host Microbe 18:307–319. doi: 10.1016/j.chom.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DePas WH, Starwalt-Lee R, Van Sambeek L, Ravindra Kumar S, Gradinaru V, Newman DK. 2016. Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 7:e00796-16. doi: 10.1128/mBio.00796-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korgaonkar AK, Whiteley M. 2011. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol 193:909–917. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang J, Toyofuku M, Sakai R, Nomura N. 2017. Influence of the alginate production on cell-to-cell communication in Pseudomonas aeruginosa PAO1. Environ Microbiol Rep doi: 10.1111/1758-2229.12521. [DOI] [PubMed] [Google Scholar]
- 54.Hassett DJ. 1996. Anaerobic production of alginate by Pseudomonas aeruginosa: alginate restricts diffusion of oxygen. J Bacteriol 178:7322–7325. doi: 10.1128/jb.178.24.7322-7325.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Worlitzsch D, Tarran R, Ulrich M, Schwab U, Cekici A, Meyer KC, Birrer P, Bellon G, Berger J, Weiss T, Botzenhart K, Yankaskas JR, Randell S, Boucher RC, Döring G. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J Clin Invest 109:317–325. doi: 10.1172/JCI13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt A, Hammerbacher AS, Bastian M, Nieken KJ, Klockgether J, Merighi M, Lapouge K, Poschgan C, Kölle J, Acharya KR, Ulrich M, Tümmler B, Unden G, Kaever V, Lory S, Haas D, Schwarz S, Döring G. 2016. Oxygen-dependent regulation of c-di-GMP synthesis by SadC controls alginate production in Pseudomonas aeruginosa. Environ Microbiol 18:3390–3402. doi: 10.1111/1462-2920.13208. [DOI] [PubMed] [Google Scholar]
- 57.Hatch RA, Schiller NL. 1998. Alginate lyase promotes diffusion of aminoglycosides through the extracellular polysaccharide of mucoid Pseudomonas aeruginosa. Antimicrob Agents Chemother 42:974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Learn DB, Brestel EP, Seetharama S. 1987. Hypochlorite scavenging by Pseudomonas aeruginosa alginate. Infect Immun 55:1813–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hassett DJ, Howell ML, Ochsner UA, Vasil ML, Johnson Z, Dean GE. 1997. An operon containing fumC and sodA encoding fumarase C and manganese superoxide dismutase is controlled by the ferric uptake regulator in Pseudomonas aeruginosa: fur mutants produce elevated alginate levels. J Bacteriol 179:1452–1459. doi: 10.1128/jb.179.5.1452-1459.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Calfee MW, Shelton JG, McCubrey JA, Pesci EC. 2005. Solubility and bioactivity of the Pseudomonas quinolone signal are increased by a Pseudomonas aeruginosa-produced surfactant. Infect Immun 73:878–882. doi: 10.1128/IAI.73.2.878-882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang L, Liu Y, Markussen T, Høiby N, Tolker-Nielsen T, Molin S. 2011. Pattern differentiation in co-culture biofilms formed by Staphylococcus aureus and Pseudomonas aeruginosa. FEMS Immunol Med Microbiol 62:339–347. doi: 10.1111/j.1574-695X.2011.00820.x. [DOI] [PubMed] [Google Scholar]
- 62.Barr HL, Halliday N, Barrett DA, Williams P, Forrester DL, Peckham D, Williams K, Smyth AR, Honeybourne D, Whitehouse JL, Nash EF, Dewar J, Clayton A, Knox AJ, Cámara M, Fogarty AW 20 October 2016. Diagnostic and prognostic significance of systemic alkyl quinolones for P. aeruginosa in cystic fibrosis: a longitudinal study. J Cyst Fibros doi: 10.1016/j.jcf.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 63.Martin LW, Reid DW, Sharples KJ, Lamont IL. 2011. Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals 24:1059–1067. doi: 10.1007/s10534-011-9464-z. [DOI] [PubMed] [Google Scholar]
- 64.Knutson CA, Jeanes A. 1968. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem 24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 65.Cesaretti M, Luppi E, Maccari F, Volpi N. 2003. A 96-well assay for uronic acid carbazole reaction. Carbohydr Polym 54:59–61. doi: 10.1016/S0144-8617(03)00144-9. [DOI] [Google Scholar]
- 66.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K. 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 67.Imperi F, Tiburzi F, Visca P. 2009. Molecular basis of pyoverdine siderophore recycling in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 106:20440–20445. doi: 10.1073/pnas.0908760106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S. aureus cystic fibrosis clinical isolates are not competitive with nonmucoid P. aeruginosa PAO1. Cross-streak coculture assays of S. aureus isolates (CFBRSA) from the CF Biospecimen Registry (CFBR) with P. aeruginosa strain PAO1 were performed by cross-streaking the two species on an agar surface, and the percentages of the populations of P. aeruginosa (gray bars) and S. aureus (white bars) recovered post-cross-streak were enumerated by plating on selective media and counting the number of CFU, according to the schematic in Fig. 1a. S. aureus strains in panel a were isolated from CF patients who were infected with only S. aureus (monoinfected), and strains in panel b were isolated from patients who were coinfected with P. aeruginosa and S. aureus. Error bars indicate the standard deviations of four biological replicates performed in triplicate. Competitive indices were calculated by dividing the percentage of S. aureus by the percentage of P. aeruginosa recovered post-cross-streak, and a statistically significant difference by an unpaired two-tailed t test was not observed between isolates from patients who were monoinfected versus those coinfected. Download FIG S1, TIF file, 0.2 MB (205.6KB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nonmucoid P. aeruginosa outcompetes S. aureus. P. aeruginosa-S. aureus coculture assays during planktonic growth (a, b, and c) and during biofilm growth on CF bronchial epithelial cells (CFBE [d]). In panels a and b, the viability of P. aeruginosa strain PAO1 (WT) and isogenic mutants was measured as log10 CFU/well over the course of 8 h in monoculture (a) and in coculture with S. aureus strain JE2 (b). In panels c and d, the viability of S. aureus JE2 was measured as log10 CFU/ml after 8 h of coinfection in panel c and CFU/well over 22 h in panel d. Data in panels a, b, and d are representative of three biological replicates, and in panel c, error bars indicate standard deviations from three biological replicates performed in triplicate. Statistical significance was determined by performing an unpaired two-tailed t test. ***, P < 0.0001. Download FIG S2, TIF file, 0.2 MB (237KB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression analysis of P. aeruginosa virulence genes during alginate overproduction. mRNA transcript abundance of a subset of P. aeruginosa virulence genes was compared in PAO1 mucA22 (mucoid) and PAO1 mucA22 algD::FRT (nonmucoid) strains by the NanoString nCounter analysis system. (a) A heat map was generated based on the ranked abundance (from 1 to 75) of the raw transcript values for each of the three biological replicates for each strain. Each cluster of genes is labeled with their known or predicted functions. The color gradient (white to purple) indicates the relative expression level of each gene with purple indicating the most highly expressed genes. (b to e) The expression of pvdA (b), pqsL (c and d), and rhlA (e) relative to the housekeeping gene rpoD was determined by qRT-PCR for the indicated P. aeruginosa strains. The means of biological triplicates (b, c, and e) and quadruplicates (d) and standard deviations are indicated. Statistical significance was determined by one-way ANOVA followed by a Dunnett’s multiple comparison test. *, P < 0.05; **, P < 0.01. Download FIG S3, TIF file, 78 MB (79.9MB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Raw NanoString data. Download TABLE S1, XLSX file, 0.1 MB (132KB, xlsx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Relative expression of AlgT-regulated genes in PAV2 between PAO1 mucA22 and PAO1 mucA22 algD::FRT. Download TABLE S2, DOCX file, 0.1 MB (58.6KB, docx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alginate overproduction inhibits HQNO antimicrobial activity toward S. aureus. (a) In vitro P. aeruginosa-S. aureus coculture assays in planktonic culture with S. aureus JE2 only (a), S. aureus plus P. aeruginosa PAO1 (WT), (b) S. aureus plus P. aeruginosa pqsL64 (c), and P. aeruginosa mucA22 (d) in the presence of either DMSO (vehicle control [open circles]) or 12.5 µg/ml HQNO (solid circles). Log10 CFU/ml for S. aureus JE2 are indicated. Error bars indicate standard deviations from three biological replicates performed in triplicate. Download FIG S4, TIF file, 0.3 MB (324.2KB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Alginate inhibits the production of P. aeruginosa rhamnolipids. The results from a drop collapse assay to measure rhamnolipid-mediated surfactant activity are shown. The clarified supernatants of the indicated strains (a) and purified rhamnolipids (b [50/50 mixture of mono- and dirhamnolipids]) were serially diluted (1:1) with water plus 0.005% crystal violet (for visualization). Surfactant activity was measured by the spread of the droplet down a 90° incline. As surfactant quantities are reduced by dilution, surface tension increases, resulting in the beading of the droplet. Representative images from three biological replicates are shown. Download FIG S5, TIF file, 45.6 MB (46.7MB, tif) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains and plasmids used in this study. Download TABLE S3, DOCX file, 0.1 MB (24.3KB, docx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S4, DOCX file, 0.1 MB (18.5KB, docx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download TEXT S1, DOCX file, 0.2 MB (34.1KB, docx) .
Copyright © 2017 Limoli et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.