Abstract
Age-related macular degeneration (AMD) is the most common cause of vision loss in elderly individuals throughout the developed world. Inhibitors of vascular endothelial growth factor have been successfully used to treat choroidal neovascularization in late-stage AMD. The pathogenesis of early-stage AMD, however, remains largely unknown, impairing efforts to develop effective therapies that prevent progression to late-stage AMD. To address this, we performed comparative transcriptomics of macular and extramacular retinal pigmented epithelium-choroid (RPE-choroid) tissue from early-stage AMD patients. We found that expression of fatty acid desaturase 1 (FADS1), FADS2, and acetyl-CoA acetyltransferase 2 (ACAT2) is increased in macular but not extramacular tissue, possibly through activation of sterol regulatory element binding factor 1 (SREBF1). Consistent with this, we also found that expression of Fads1 is increased in RPE-choroid in a mouse model of early-stage AMD. In zebrafish, deletion of fads2, which encodes a protein that functions as both Fads1 and Fads2 in other species, enhanced apoptosis in the retina upon exposure to intense light. Similarly, pharmacological inhibition of Srebf1 enhanced apoptosis and reduced fads2 expression in zebrafish exposed to intense light. These results suggest that the SREBF1–FADS1/2 axis may be activated in macular RPE-choroid as a protective response during early-stage AMD and could thus be a therapeutic target for early-stage AMD.
Keywords: Ophthalmology, Pharmaceutical science
1. Introduction
AMD is the most common cause of vision loss in elderly individuals throughout the developed world [1]. AMD can be broadly divided into early and late stages based on clinical features [2]. Early-stage AMD is characterized by pigmentary abnormalities in the macula and accumulation of lipid deposits called drusen between the retinal pigment epithelium (RPE) and Bruch’s membrane [2]. Bruch’s membrane lies over the vascularized choroid, contacting both the vascular endothelium and RPE [3]. Late-stage AMD is usually subdivided into dry (geographic atrophy) and wet (choroidal neovascularization) AMD [2]. Geographic atrophy is not yet treatable [2], whereas choroidal neovascularization can be treated, but not cured, with inhibitors of vascular endothelial growth factor (VEGF) [2]. Therefore, it is important to develop effective therapies that target early-stage AMD to prevent progression to late-stage disease [4]. However, the pathogenesis of early-stage AMD remains largely unknown [2].
Transcriptome analysis can be used to identify gene expression signatures related to the pathogenesis of various diseases, including AMD [5, 6, 7]. Comparison of transcriptome data can also be used to identify robust gene expression changes in a disease with multiple etiologies [8, 9]. In this study, we analyzed two transcriptome datasets for gene expression changes in macular [5, 6] and extramacular [5] RPE-choroid samples from patients with early-stage AMD, and we performed comparative transcriptomics to identify gene expression changes specific to macular RPE-choroid. We found that expression of FADS1, FADS2, and ACAT2 is increased in macular but not extramacular RPE-choroid, possibly through activation of SREBF1. Moreover, expression of Fads1 was elevated in RPE-choroid of a mouse model of early-stage AMD. We also demonstrated protective roles for Srebf1 and Fads2 in a zebrafish model of light-induced retinopathy, suggesting a possible therapeutic approach for early-stage AMD.
2. Materials and methods
2.1. Ethics statement
This study was carried out in strict accordance with Japanese law [The Humane Treatment and Management of Animals (2014), Standards Relating to the Care and Management of Laboratory Animals and Relief of Pain (2013), and the Guidelines for Proper Conduct of Animal Experiments [10, 11, 12]. All efforts were made to minimize animal suffering. Mie University Institutional Animal Care and Use Committee guidelines state that no approval is required for experiments using zebrafish.
2.2. Compounds
Fatostatin and phenylthiourea were obtained from Sigma (St. Louis, MO, USA). Stock solutions were prepared by dissolving in dimethyl sulfoxide (Nacalai Tesque, Kyoto, Japan). 2-Phenoxyethanol was obtained from Wako Chemical (Osaka, Japan).
2.3. Comparative transcriptomics of RPE-choroid from early-stage AMD patients
To identify genes dysregulated in RPE-choroid during early-stage AMD, we downloaded two transcriptome datasets, designated GSE29801 [5] and GSE50195 [6], from Gene Expression Omnibus [13]. The microarray data for GSE29801 were obtained from the macula of AMD patients with Rotterdam grade [14] 2a, 2b, or 3 (n = 16); macula of control subjects (n = 50), extramacula of AMD patients with Rotterdam grade 2a, 2b or 3 (n = 14), and extramacula of control subjects (n = 46). The median ages (range of ages) of the groups for macular of control and AMD patients in GSE29801 are 71 (9–93) years old (yo) and 79 (43–101 yo). The median ages (range of ages) of groups for extramacular of control and AMD patients in GSE29801 are 67.5 (9–93 yo) and 78.5 (43–94 yo), respectively. Data were normalized using the package “limma” [15] in Bioconductor [16]. For GSE50195, the normalized data (macula from AMD patients with RPE changes or drusen, n = 9; macula from control subjects, n = 7) were downloaded from Gene Expression Omnibus. The median ages (range of ages) of groups for macular of control and AMD patients in GSE50195 are 83 (77–93 yo) and 81 (77–92 yo), respectively. Probes with reliable signals were selected and subjected to RankProd [17] analysis to identify differentially expressed genes (DEGs) in each model using a false discovery rate of 20% as the threshold. The list of DEGs in each dataset is shown in Tables S1–S3. DEGs common to macular but not extramacular samples in GSE29801 and GSE50195 are shown in Table S4. DEGs common to both macular and extramacular samples in GSE29801 and GSE50195 are shown in Table S5. Genes with expression changes in the same direction (i.e., upregulated or downregulated) across the datasets are shown in bold.
2.4. Bioinformatic analysis of the genes dysregulated in RPE-choroid from early-stage AMD patients
To identify networks related to genes dysregulated in RPE-choroid during early-stage AMD, we used GeneMANIA in Cytoscape [18]. GeneMANIA uses a database of organism-specific weighted networks to construct a weighted composite functional interaction network between a pair of genes, including co-expression, co-localization, genetic interaction, physical interaction, shared protein domains, and pathway networks [19]. GeneMANIA also analyzes biological pathways significantly enriched in the functional interaction network. The biological pathways enriched in the functional interaction network with adjusted q value <1.0 × 10-10 are shown in Table 1 for the 32 genes dysregulated in macular but not extramacular samples and Table 2 for the 76 genes dysregulated in both macular and extramacular samples. The relationship between two genes and the gene score are shown in Tables S6-S12 for the network related to the 32 genes and Tables S13-S20 for the network related to the 76 genes.
Table 1.
Biological pathways significantly enriched in the functional interaction networks related to 32 genes dysregulated in macular but not extramacular RPE-choroid of early-stage AMD patients.
| GO id | Description | q-value |
|---|---|---|
| GO:0006695 | cholesterol biosynthetic process | 2.7E-22 |
| GO:0016126 | sterol biosynthetic process | 2.7E-22 |
| GO:0008203 | cholesterol metabolic process | 7.1E-18 |
| GO:0016125 | sterol metabolic process | 4.3E-17 |
| GO:0046165 | alcohol biosynthetic process | 8.5E-16 |
| GO:0006694 | steroid biosynthetic process | 1.6E-15 |
| GO:1901617 | organic hydroxy compound biosynthetic process | 1.1E-14 |
| GO:0008202 | steroid metabolic process | 3.7E-12 |
| GO:0006066 | alcohol metabolic process | 1.6E-10 |
Table 2.
Biological pathways significantly enriched in the functional interaction networks related to 76 genes dysregulated in both macular and extramacular RPE-choroid of early-stage AMD patients.
| GO id | Description | q-value |
|---|---|---|
| GO:0016056 | rhodopsin mediated signaling pathway | 2.8E-28 |
| GO:0007603 | phototransduction, visible light | 6.5E-27 |
| GO:0022400 | regulation of rhodopsin mediated signaling pathway | 1.3E-26 |
| GO:0009584 | detection of visible light | 2.0E-26 |
| GO:0007602 | phototransduction | 3.3E-26 |
| GO:0009583 | detection of light stimulus | 8.9E-26 |
| GO:0009581 | detection of external stimulus | 2.1E-24 |
| GO:0009582 | detection of abiotic stimulus | 3.9E-24 |
| GO:0071482 | cellular response to light stimulus | 2.6E-22 |
| GO:0051606 | detection of stimulus | 3.6E-21 |
| GO:0097381 | photoreceptor disc membrane | 8.5E-21 |
| GO:0071478 | cellular response to radiation | 2.6E-19 |
| GO:0009416 | response to light stimulus | 3.5E-19 |
| GO:0008277 | regulation of G-protein coupled receptor protein signaling pathway | 5.8E-18 |
| GO:0071214 | cellular response to abiotic stimulus | 3.2E-17 |
| GO:0009314 | response to radiation | 1.3E-16 |
| GO:0060170 | ciliary membrane | 7.8E-16 |
| GO:0001750 | photoreceptor outer segment | 1.1E-15 |
| GO:0098590 | plasma membrane region | 2.8E-13 |
| GO:0031513 | nonmotile primary cilium | 6.6E-12 |
| GO:0031253 | cell projection membrane | 8.8E-12 |
| GO:0072372 | primary cilium | 1.5E-10 |
| GO:0097458 | neuron part | 5.0E-10 |
To identify transcription factors (TFs) potentially regulating the genes dysregulated in RPE-choroid during early-stage AMD, we used iRegulon [20] in Cytoscape. iRegulon exploits the fact that genes co-regulated by the same TF contain common TF-binding sites, and uses gene sets derived from ENCODE ChIP-seq data (Gerstein et al., 2012). The predicted TFs with normalized enrichment scores (NES) >4 are shown in Table S21 for the 32 genes and Table S22 for the 76 genes.
2.5. Transcriptomics of RPE-choroid and neuroretina from early-stage AMD model mice
To identify genes dysregulated in RPE-choroid and neuroretina during early-stage AMD, we downloaded a transcriptome dataset, designated GSE38671 [21] from Gene Expression Omnibus [13]. The microarray data for GSE38671 were obtained from RPE-choroid or neuroretina from young (7–8 weeks of age) wild type mice (n = 4) or complement factor H (Cfh) null mice (n = 4). Data were normalized using the package “oligo” [22] in Bioconductor [16]. Probes with reliable signals were selected and subjected to RankProd [17] analysis to identify DEGs using a false discovery rate of 30% as the threshold. The gene symbols of the DEGs were converted to those of the human orthologous genes using Life Science Knowledge Bank (World Fusion, Tokyo, Japan). The lists of DEGs in RPE-choroid and neuroretina are shown in Tables S23 and S24, respectively.
2.6. Zebrafish strains
Zebrafish were bred and maintained according to previously described methods [23, 24]. Briefly, zebrafish were raised at 28.5 °C ± 0.5 °C with a 14 h/10 h light/dark cycle. Embryos were obtained and cultured in 0.3 × Danieau’s solution (19.3 mM NaCl, 0.23 mM KCl, 0.13 mM MgSO4, 0.2 mM Ca(NO3)2, 1.7 mM HEPES, pH 7.2) until 7 days post-fertilization (dpf). For modeling light-induced retinopathy, zebrafish were cultured in 0.3 × Danieau’s solution containing 200 μM phenylthiourea.
The effect of fatostatin on light-induced retinopathy was assessed using an AB zebrafish line obtained from Zebrafish International Resource Center (Eugene, OR, USA). To assess the involvement of Fads2 in light-induced retinopathy, we generated fads2 knockout (KO) zebrafish according to the method described previously [25]. Briefly, transcription activator-like effector nucleases (TALENs) targeting exon 6 of the zebrafish fads2 gene were constructed using the Golden Gate TALEN and TAL Effector Kit 2.0 (Addgene #1000000024) [26] and Yamamoto Lab TALEN Accessory Pack (Addgene #1000000030) [27]. Single DNA-binding repeats were assembled into intermediate array vectors, which were subsequently inserted into the final destination vectors, pCS2TAL3-DD and pCS2TAL3-RR (Addgene #37275 and #37276) [28]. The TALEN mRNAs were synthesized using an mMessage mMachine SP6 Kit (Life Technologies, Carlsbad, CA, USA), and 300 ng/μL of each mRNA was then injected into 2–8-cell-stage embryos of the AB zebrafish line. After injection, the embryos were cultured in 0.3 × Danieau’s solution until 5 dpf and reared in the fish farming system with an artificial diet (Meito Suien, Nagoya, Japan) and Artemia (Kitamura, Kyoto, Japan) at 28.5 °C under a 14 h/10 h light/dark cycle. At 4 months post-fertilization, genomic DNA was extracted from the fins of F0 zebrafish according to previous reports [8, 9]. To detect TALEN-induced mutations, a short fragment of the fads2 gene encompassing the target site was amplified from genomic DNA using primers fads2_gF1 and fads2_gR1. The sequences of these primers are shown in Table S25. Three-step PCR was carried out using 40 cycles of 94 °C for 30 s, 60 °C for 30 s, and 68 °C for 30 s. The PCR products were electrophoresed on 10% polyacrylamide gels as described previously [8, 9]. The F0 fish in which the TALEN-induced mutation was present were crossed with the AB strain to obtain F1 progeny. The F1 generation was reared and screened for the presence of the mutation by PCR, as described above. The PCR amplicons were cloned into a pGEM-T Easy vector (Promega, Madison, WI, USA) and the sequences were analyzed using the M13 forward primer. F1 female and F1 male hetero-knockout zebrafish harboring the same mutations in the fads2 gene were crossed to obtain F2 progeny. The F2 generation was reared and screened for the mutation as described above. F2 female and F2 male homo-knockout zebrafish harboring the same mutations in the fads2 gene were crossed to obtain F3 progeny, which were used to assess light-induced retinopathy.
2.7. A larval zebrafish model of light-induced retinopathy
Zebrafish were placed in petri dishes at 3 dpf. The dishes were wrapped in aluminum foil and shielded from light for 48 h. On day 5 dpf, the animals were transferred to 6-well plates (15 larvae/6 ml medium/well) and incubated under normal conditions (28 °C, 14 h [7 am–9 pm] dim light/10 h dark) or placed in a custom-made chamber (Hayashi Factory, Kyoto, Japan) to induce retinopathy as described previously [29].
2.8. TUNEL staining
Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) was performed using an ApopTag Fluorescein In Situ Apoptosis Detection Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s protocol. To assess the effect of Srebf inhibition, albino zebrafish were exposed for 48 h to intense light (13,000 lux) or normal light conditions (14 h/10 h light/dark cycle) in medium with or without 100 nM fatostatin. To assess the effect of Fads2 deficiency, wild-type or fads2-KO zebrafish were treated the same way but in the absence of fatostatin. The zebrafish were then fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS; Nacalai Tesque) at 4 °C overnight. Animals were washed with PBS containing 0.1% Tween 20 (PBST), incubated in water containing 3% H2O2 and 1% KOH at room temperature for 30 min, washed again with PBST, and incubated in 100% methanol at −30 °C overnight. Zebrafish were rehydrated, treated with proteinase K (40 μg/ml) at room temperature for 30 min, washed with PBST, incubated in equilibration buffer at 37 °C for 1 h, and then incubated in working solution containing TdT enzyme and digoxigenin-labeled dNTP at 37 °C for 1 h. The animals were then washed and treated with fluorescein-labeled anti-digoxigenin IgG at 4 °C overnight. Finally, zebrafish were washed once more with PBST and imaged with a SMZ25 stereomicroscope (Nikon, Tokyo, Japan) equipped with a GFP-BP filter. Quantitative analysis of the fluorescent images was performed using Volocity software (PerkinElmer, Waltham, MA, USA). The threshold fluorescence intensity for defining apoptotic areas of the retina was set at five standard deviations above the mean fluorescence intensity of the whole field of view.
2.9. qPCR analysis
Total RNA was extracted from zebrafish exposed to intense light or normal light conditions in the presence or absence of fatostatin using an RNAqueous Micro Kit (Takara, Kyoto, Japan) according to the manufacturer’s protocol. RNA concentrations were determined using a NanoDrop spectrophotometer (Thermo Scientific, Waltham, MA, USA), and cDNAs were generated using a ReverTra Ace qPCR RT Kit (Toyobo). qPCR was performed using an ABI Prism 7300 (Life Technologies Carlsbad, CA, USA) with THUNDERBIRD SYBR qPCR Mix (Toyobo). The thermal cycling conditions were: 95 °C for 1 min followed by 40 cycles of 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 45 s. We measured the expression of fads2 and eukaryotic translation elongation factor 1 alpha 1 like 1 (eef1a1l1) mRNA. The fads2 mRNA levels were normalized to eef1a1l1 mRNA levels to correct for variability in the initial template concentration and the conversion efficiency of the reverse transcription reaction. The primer sequences are shown in Table S25.
2.10. Statistical analysis
Statistical analysis was performed using Prism 6 (GraphPad, La Jolla, CA, USA). Group means were compared by analysis of variance. Alpha was set at 0.05. Tukey’s multiple comparisons test was used for post hoc analysis when significant effects were found by analysis of variance. Data are shown as the mean ± standard error (SEM).
3. Results
3.1. Identification of genes dysregulated in RPE-choroid of early-stage AMD patients
To identify genes dysregulated in RPE-choroid during early-stage AMD, we downloaded two transcriptome datasets, GSE29801 [5] and GSE50195 [6], from a public database [13]. GSE29801 included transcriptome data from macular and extramacular RPE-choroid tissue isolated from AMD patients with Rotterdam grade [14] 2a, 2b, or 3 and from individuals with no features of AMD (controls). GSE50195 included transcriptome data from macular RPE-choroid tissue isolated from AMD patients with RPE changes or drusen or from control subjects. Using a false discovery rate of 20% as the threshold, we identified 1,133 and 1,083 genes in GSE29801 GSE50195, respectively, that were dysregulated in macular RPE-choroid of AMD patients compared with controls (Tables S1 and S2). We also identified 1,214 genes dysregulated in extramacular RPE-choroid of AMD patients (Table S3). In the AMD patient datasets, we identified 55 genes dysregulated in macular but not extramacular RPE-choroid and 128 genes dysregulated in both macular and extramacular RPE-choroid (Fig. 1). Among these 55 and 128 genes, we found that 32 and 76 genes, respectively, had expression changes in the same direction in both datasets (Tables S4 and S5, shown in bold). These results suggest that the 32 genes dysregulated in the macular RPE-choroid may be related to the pathogenesis of AMD.
Fig. 1.
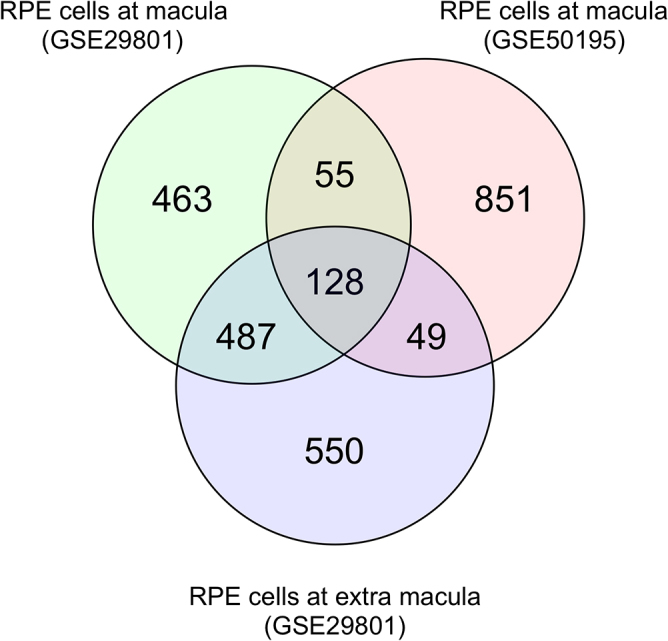
Venn diagrams of the number of genes dysregulated in RPE-choroid of early-stage AMD patients. Transcriptome data of RPE-choroid samples from patients with early-stage AMD (GSE29801 and GSE50195) were downloaded from a public database. Genes differentially expressed in RPE-choroid from AMD patients versus healthy controls were identified using a false discovery rate of 20% as the threshold. The number of differentially expressed genes in each group and the overlap between groups are shown.
3.2. Cholesterol biosynthesis pathways is dysregulated in macular RPE-choroid of early-stage AMD patients
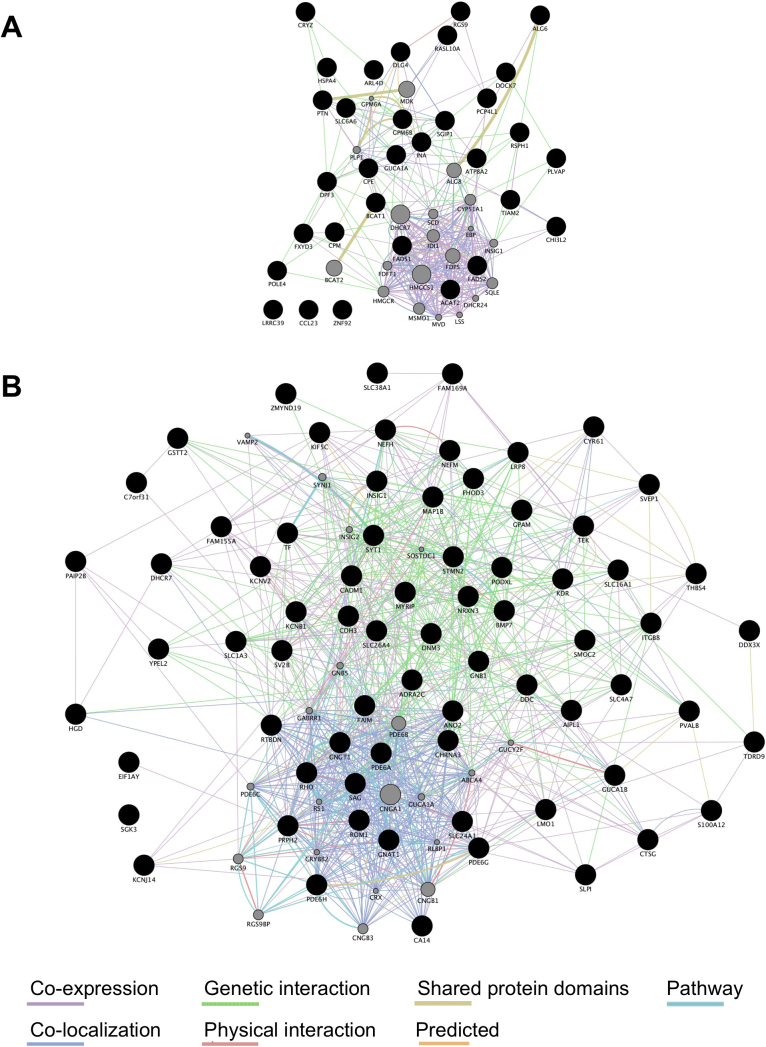
We next analyzed the functional interaction networks related to the genes dysregulated in the RPE-choroid tissues of AMD patients using GeneMANIA [19]. Fig. 2A and Table S6-S12 show the functional interaction networks related to the 32 genes dysregulated in macular but not extramacular RPE-choroid of AMD patients. Fig. 2B and Table S13-S20 show the functional interaction networks related to the 76 genes dysregulated in both macular and extramacular RPE-choroid of AMD patients. Pathways related to cholesterol biosynthesis were significantly enriched in the functional interaction network for genes dysregulated in macular but not extramacular RPE-choroid (Table 1), suggesting that dysregulation of cholesterol biosynthesis in macular RPE-choroid may be involved in the pathogenesis of AMD. Pathways related to rhodopsin-mediated signaling were significantly enriched in the functional interaction network for genes dysregulated in both macular and extramacular RPE-choroid (Table 2), consistent with previous reports demonstrating that rod photoreceptor function is impaired in early-stage AMD [4, 30].
Fig. 2.
Functional interaction networks related to genes dysregulated in RPE-choroid of early-stage AMD patients. (A) The 32 genes dysregulated in macular but not extramacular RPE-choroid (shown as black circles) were subjected to GeneMANIA searches to identify functional interaction networks. (B) The 76 genes dysregulated in both macular and extramacular RPE-choroid (shown as black circles) were subjected to GeneMANIA searches to identify functional interaction networks. The size of the gray circles denotes the score in the functional network (Tables S3 and S4).
3.3. Identification of potential TFs for genes dysregulated in RPE-choroid samples from early-stage AMD patients
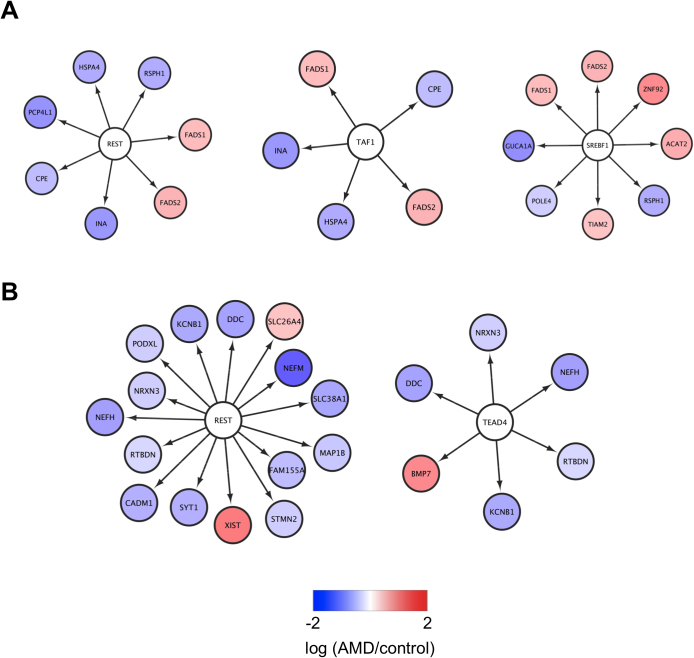
We next used iRegulon to identify potential TFs for genes dysregulated in RPE-choroid samples from AMD patients [20]. TATA box binding protein associated factor 1 (TAF1) and SREBF1 were identified as potential TFs for genes dysregulated in macular but not extramacular RPE-choroid (Fig. 3A and Table S21). TEA domain family member 4 (TEAD4) and RE1-silencing transcription factor (REST), which has previously been associated with early-stage AMD [31], were identified as a potential TFs for genes dysregulated in both macular and extramacular RPE-choroid (Fig. 3B and Table S22). Lastly, FADS1 and FADS2, which stimulate synthesis of polyunsaturated fatty acids [32] and have also been associated with AMD [33, 34, 35, 36, 37], were identified as common targets of TAF1, REST, and SREBF1 (Fig. 3A). These findings suggest that dysregulation of FADS1 and FASD2 in macular RPE-choroid may play a critical role in early-stage AMD.
Fig. 3.
Identification of key transcription factors for genes dysregulated in RPE-choroid of early-stage AMD patients. (A) The 32 genes dysregulated in macular but not extramacular RPE-choroid of early-stage AMD patients were subjected to iRegulon. (B) The 76 genes dysregulated in both macular and extramacular RPE-choroid of early-stage AMD patients were subjected to iRegulon. The transcription factors and their targets identified by GeneMANIA are shown.
3.4. Fads1 is increased in RPE-choroid of a mouse model of early-stage AMD
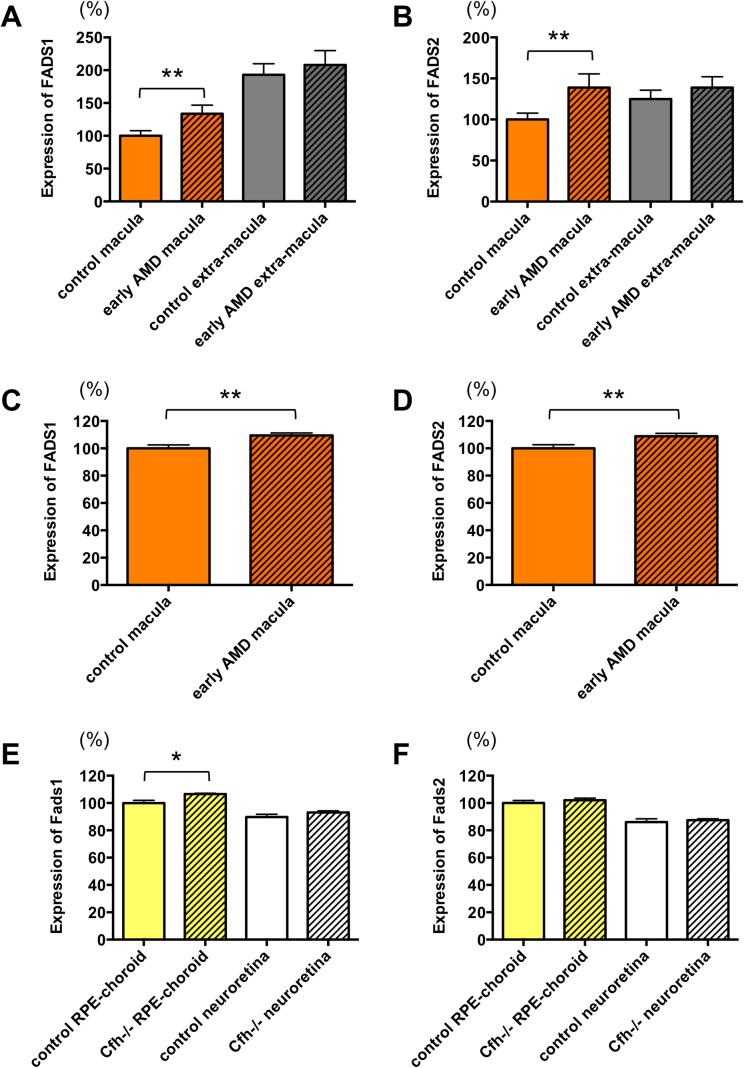
To examine whether Fads1 and Fads2 might also be dysregulated in a mouse model of early-stage AMD, we searched a public database [13] and obtained a transcriptome dataset from RPE-choroid and neuroretina of young complement factor H KO (Cfh−/−) mice [21]. In humans, a single nucleotide polymorphism of the CFH gene results in a loss-of-function Y402H mutation in the protein and confers an increased risk for AMD [38]. After analyzing the transcriptome datasets for genes differentially expressed in young wild-type and Cfh null mice, we identified 132 DEGs in RPE-choroid (Table S23) and 31 in neuroretina (Table S24). Fads1 expression was significantly increased in RPE-choroid, but not in neuroretina, of young Cfh null mice (Fig. 4E), which is consistent with the increase in FADS1 expression in macular RPE-choroid of early-stage AMD patients (Fig. 4A, C). However, Fads2 expression was not significantly different between young wild-type and Cfh null mice (Fig. 4F), which contrasts with the increased levels of FADS2 in macular RPE-choroid of early-stage AMD patients (Fig. 4B, D).
Fig. 4.
Expression of FADS1 and FADS2 in early-stage AMD patients and mice.
3.5. Fads2 protects against light-induced retinopathy in zebrafish
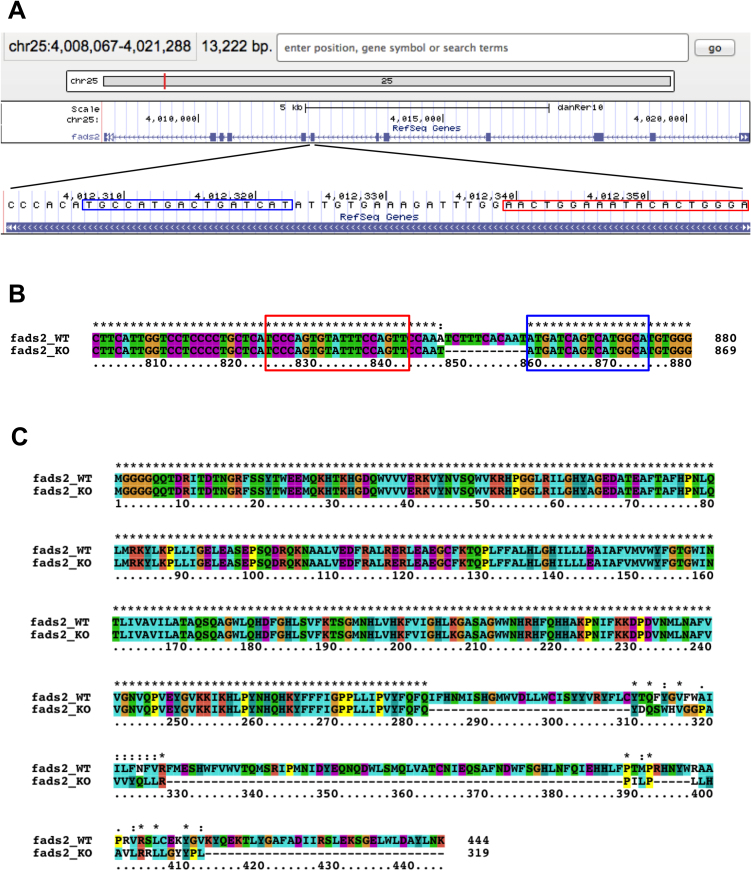
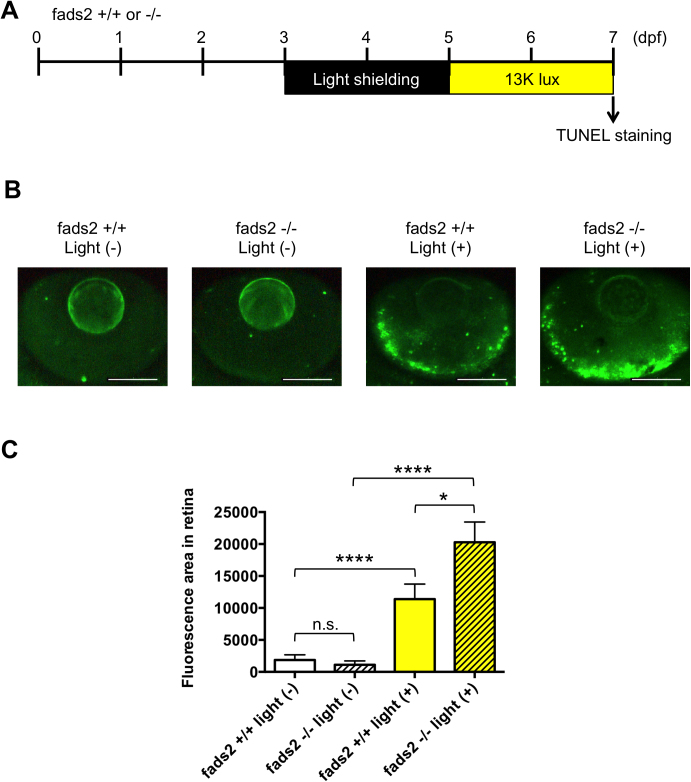
To analyze the functional role of FADS1/2 in retinal pathophysiology, we used a zebrafish model of light-induced retinopathy [29]. Because zebrafish lack a fads1 gene, and the Fads2 protein performs the function of both Fads1 and Fads2 in other species [39], we generated a fads2-KO zebrafish line using TALEN (Fig. 5). As shown in Fig. 6, exposure of zebrafish to intense light significantly increased the level of apoptosis in the retina compared with normal light conditions. The level of apoptosis in the retina of fads2-KO zebrafish exposed to intense light was significantly greater than that of similarly treated control zebrafish. However, knockout of fads2 did not affect retinal apoptosis in zebrafish maintained under normal light conditions. These results suggest that increased expression of FADS1 and FADS2 in macular RPE-choroid may have a protective role in early-stage AMD.
Fig. 5.
Generation of fads2-KO zebrafish. (A) The position of TALEN recognition sites for the zebrafish fads2 gene. (B) Nucleic acid alignment of fads2 from wild-type and fads2-KO zebrafish. (C) Amino acid alignment of Fads2 from wild-type and fads2-KO zebrafish. The target sequences of TALEN are boxed in red and blue.
Fig. 6.
Knockout of fads2 increases retinal apoptosis in a larval zebrafish model of light-induced retinopathy. (A) Protocol for light-induced retinal damage in larval zebrafish. Zebrafish were shielded from light between 3 and 5 days post-fertilization (dpf) and then exposed to normal conditions (14 h 250 lux/10 h dark) or intense light (13,000 lux) for 48 h at 27 °C. After light exposure, whole-mount TUNEL staining was performed. (B) Representative images of TUNEL staining in the retina of control or fads2 knockout zebrafish exposed to intense light (indicated as light +) or normal light conditions (light −). Scale bars, 100 μm. (C) Quantitative analysis of retinal apoptosis in zebrafish exposed to the conditions shown in (B). *p < 0.05, ****p < 0.0001. Data are the mean ± SEM of 11–14 eyes/group.
3.6. Srebf1 protects against light-induced retinopathy in zebrafish
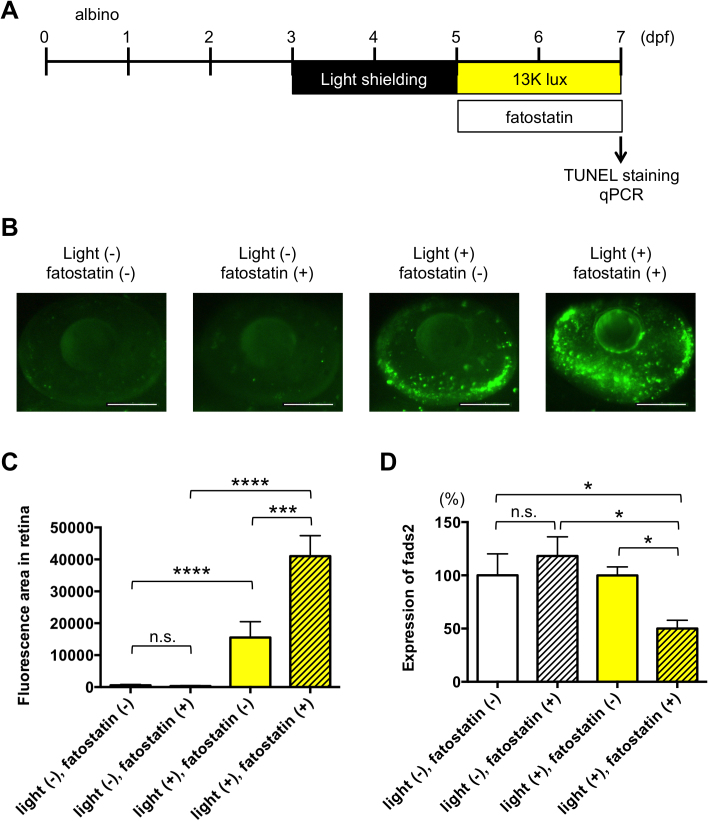
Bioinformatics analysis identified REST, TAF1, and SREBF1 as potential TFs for FADS1/2 (Fig. 3). It has been reported that SREBF1 may be activated in response to reduced levels of docosahexaenoic acid (DHA), thereby increasing the expression of FADS1/2 and synthesis of DHA [40]. To examine whether SREBF1 may be involved in regulating FADS expression and retinal pathophysiology, we tested the effect of a specific inhibitor of Srebf, fatostatin [41, 42], in the larval zebrafish model of light-induced retinopathy [29] (Fig. 7A). Zebrafish treated with fatostatin showed significantly increased retinal apoptosis (Fig. 7B, C) and decreased fads2 expression (Fig. 7D) after exposure to intense, but not normal, light conditions. These results suggest that exposure of zebrafish to intense light may activate retinal Srebf1, possibly secondary to reduced DHA levels, and thus increase fads2 expression and DHA synthesis in an attempt to protect the retina from light-induced damage.
Fig. 7.
Inhibition of Srebfs increases retinal apoptosis in a larval zebrafish model of light-induced retinopathy. (A) Protocol for light-induced retinal damage in larval zebrafish. Zebrafish were shielded from light between 3 and 5 days post-fertilization (dpf) and then exposed to normal conditions (14 h 250 lux/10 h dark) or intense light (13,000 lux) in the presence or absence of 100 nM fatostatin for 48 h at 27 °C. After light exposure, whole-mount TUNEL staining was performed. (B) Representative images of TUNEL staining in the retina of zebrafish exposed to intense light (indicated as light +) or normal light conditions (light −) in the presence or absence of fatostatin. Scale bars, 100 μm. (C) Quantitative analysis of retinal apoptosis in zebrafish exposed to the conditions shown in (B). ***p < 0.001, ****p < 0.0001. n.s., not significant. Data are the mean ± SEM of 10–15 eyes/group. (D) qPCR analysis of fads2 mRNA levels in zebrafish exposed to intense light (indicated as light +) or normal light conditions (light −) in the presence or absence of fatostatin. Expression levels are relative to those in normal light conditions in the absence of fatostatin. Data are the mean ± SEM of 3–6 zebrafish/group. *p < 0.05.
4. Discussion
4.1. Involvement of FADS1/2 in AMD
In this study, we demonstrated that FADS1 and FADS2 expression are significantly increased in macular, but not extramacular, RPE-choroid of early-stage AMD patients. We also showed that FADS1 expression is significantly increased in RPE-choroid, but not neuroretina, of young Cfh null mice, an early-stage AMD model [21]. FADS1 and FADS2 encode delta-5 and delta-6 desaturases, respectively, which are membrane-bound enzymes that catalyze the formation of long-chain polyunsaturated fatty acids such as DHA and eicosapentaenoic acid (EPA) [43]. Both FADS1 and FADS2 are expressed in RPE-choroid [44, 45] and have been associated with AMD [33, 34, 35, 36, 37]. Inhibition of FADS1/2 prevents synthesis of DHA from EPA and blocks EPA-mediated protection of the retina against oxidative stress [32]. Cigarette smoke, one of the most important risk factors for AMD [46], decreases FADS1 and FADS2 activity [47].
After several passages in culture, human RPE cells downregulate expression of FADS1 and FADS2 [48], lose their RPE phenotype, and adopt a mesenchymal phenotype [48]. FADS1 and FADS2 expression is also decreased in chorioretinal endothelial cells after formation of a complement membrane attack complex [49], a major risk factor for AMD [50]. These studies suggest that FADS1 and FADS2 may have important physiological functions in RPE-choroid of AMD. In fact, FADS1 has been identified as a component of the molecular signature of human RPE [45]. In this study, we demonstrated that knockout of zebrafish fads2, which performs the functions of both Fads1 and Fads2 in other species [39], significantly increased retinal apoptosis induced by intense light exposure. Although light-induced retinopathy is generally used as a model of neuroretinal degeneration [51], these results are consistent with a protective role for FADS1 and FADS2 in AMD. To our knowledge, there have been no reports of age-related changes in FADS1 and FADS2 expression in RPE-choroid, suggesting that these molecules are subject to more complex mechanisms of regulation. Further studies using drusen models, such as the chemokine receptor-2 KO mouse [52], will be necessary to directly examine the protective function of FADS1/2 in early-stage AMD.
4.2. Involvement of SREBF1 in AMD
We identified SREBF1 as a potential TF regulating FADS1 and FADS2 expression, consistent with a previous report [40]. We also demonstrated that pharmacological inhibition of Srebf1 in zebrafish significantly increased retinal apoptosis and decreased Fads2 expression in response to intense light exposure. These results, combined with the findings that FADS1 and FADS2 expression is increased in macular RPE-choroid of early-stage AMD patients, support a protective function for SREBF in early-stage AMD by elevating FADS1 and FADS2 production. SREBF1 is known to be expressed in human RPE-choroid [53]. One possible mechanism of SREBF1 activation is via endoplasmic reticulum stress caused by complement activation and oxidative stress [54], both of which have been considered major pathophysiological mechanisms of AMD [55]. Another possibility is that SREBF1 is activated in response to reduced levels of DHA. Interestingly, DHA is present at lower levels in RPE-choroid of AMD patients than of control subjects [56]. Since DHA negatively regulates SREBF1 by stimulating its proteolysis [57], this suggests a mechanism by which SREBF1 activity might be increased in RPE-choroid of AMD patients, thus restoring DHA levels by promoting FADS1/2 transcription [40]. Intense light exposure reduces DHA concentrations in the retina [58, 59]; therefore, it is possible that this mechanism also operates in the zebrafish light-induced retinopathy model to increase Srebf1 activity and elevate fads2 expression. However, further experiments to quantify DHA levels in the zebrafish model will be necessary to validate this hypothesis.
Although Srebf1 appears to have a protective function in the zebrafish model of light-induced retinopathy, it may also have detrimental effects in AMD. SREBF1 is a master regulator of the synthesis of both fatty acid and cholesterol [60] and has been implicated in cholesterol homeostasis in mouse retina [61]. In this study, we found that ACAT2 expression is increased, possibly through SREBF1, in macular RPE-choroid samples from patients with early-stage AMD. Increased ACAT2 expression via activation of SREBF1 has previously been demonstrated in RPE cells [62]. ACAT2 stimulates cholesteryl ester secretion in apoB-containing lipoproteins [63], and a high level of these lipoproteins is a well-established risk factor for AMD [64]. These findings suggest that activation of the SREBF1–ACAT2 axis in macular RPE-choroid may be involved in the pathogenesis of early-stage AMD through stimulation of drusen formation. Further studies using drusen models will help to clarify the functional role of SREBF1 in early-stage AMD.
In conclusion, we performed a comparative transcriptomic analysis of RPE-choroid tissue from early-stage AMD patients and demonstrated that FADS1, FADS2, and ACAT2 are increased in the macula, possibly through activation of SREBF1. We also demonstrated a protective role for Srebf1 and Fads2 in a zebrafish model of light-induced retinopathy. These results suggest that the SREBF1–FADS1/2 axis in macular RPE-choroid may play a protective role in early-stage AMD, although the functional role of the SREBF1–ACAT2 axis remains unknown. It is noteworthy that statins, which activate SREBF1 and increase FADS1 and FADS2 expression while inhibiting cholesterol biosynthesis through HMG-CoA reductase [61, 65], can counteract the detrimental effect of cigarette smoke on FADS1 and FADS2 [66]. Clinical trials of statins for AMD are currently ongoing [67, 68]. Our findings suggest that targeting of the SREBF1–FADS1/2 axis as a therapeutic approach for AMD warrants further investigation.
Declarations
Author contribution statement
Yuhei Nishimura: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Toshio Tanak: Conceived and designed the experiments; Wrote the paper.
Yoshifumi Ashikawa: Performed the experiments; Analyzed and interpreted the data.
Yumi Sato, Mizuki Yuge, Tomoko Tada, Shiko Okabe, Haruka Miyao, Soichiro Murakami, Koki Kawaguchi, Shota Sasagawa, Yasuhito Shimada: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Japan Society for the Promotion of Science KAKENHI (25670127, 15K15051, 24510069, 16K08547), the Long-range Research Initiative of the Japan Chemical Industrial Association (13-PT01-01), and Okasan-Kato Foundation.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study has been deposited at Gene Expression Omnibus under the accession number GSE29801 and GSE50195.
Supplementary content related to this article has been published online at http://dx.doi.org/10.1016/j.heliyon.2017.e00266
Acknowledgments
We are grateful to Drs. Shinsaku Yamane and Hiroshi Yamamoto (Ono Pharmaceutical Co. Ltd.) for insightful discussion about the light-induced retinopathy model of zebrafish. We also thank Junko Koiwa, Yuka Takahashi, Yuka Hayakawa, Chizuru Hirota, and Aiko Sugimura for assistance with the experiments, and Rie Ikeyama and Yuka Mizutani for administrative support.
Appendix A. Supplementary data
References
- 1.Zhang K., Zhang L., Weinreb R.N. Ophthalmic drug discovery: novel targets and mechanisms for retinal diseases and glaucoma. Nat. Rev. Drug Discov. 2012;11:541–559. doi: 10.1038/nrd3745. [DOI] [PubMed] [Google Scholar]
- 2.Fritsche L.G., Fariss R.N., Stambolian D., Abecasis G.R., Curcio C.A., Swaroop A. Age-related macular degeneration: genetics and biology coming together. Annu. Rev. Genomics Hum. Genet. 2014;15:151–171. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kauppinen A., Paterno J.J., Blasiak J., Salminen A., Kaarniranta K. Inflammation and its role in age-related macular degeneration. Cellular and molecular life sciences: CMLS. 2016;73:1765–1786. doi: 10.1007/s00018-016-2147-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller J.W. Age-related macular degeneration revisited-piecing the puzzle: the LXIX Edward Jackson memorial lecture. Am. J. Ophthalmol. 2013;155:1–35. doi: 10.1016/j.ajo.2012.10.018. e13. [DOI] [PubMed] [Google Scholar]
- 5.Newman A.M., Gallo N.B., Hancox L.S., Miller N.J., Radeke C.M., Maloney M.A., Cooper J.B., Hageman G.S., Anderson D.H., Johnson L.V. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 2012;4:16. doi: 10.1186/gm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitmore S.S., Braun T.A., Skeie J.M., Haas C.M., Sohn E.H., Stone E.M., Scheetz T.E., Mullins R.F. Altered gene expression in dry age-related macular degeneration suggests early loss of choroidal endothelial cells. Mol. Vis. 2013;19:2274–2297. [PMC free article] [PubMed] [Google Scholar]
- 7.Tian L., Kazmierkiewicz K.L., Bowman A.S., Li M., Curcio C.A., Stambolian D.E. Transcriptome of the human retina, retinal pigmented epithelium and choroid. Genomics. 2015;105:253–264. doi: 10.1016/j.ygeno.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasagawa S., Nishimura Y., Sawada H., Zhang E., Murakami S., Ashikawa Y., Yuge M., Okabe S., Kawaguchi K., Kawase R. Comparative transcriptome analysis identifies CCDC80 as a novel gene associated with pulmonary arterial hypertension. Front. Pharmacol. 2016;7:142. doi: 10.3389/fphar.2016.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasagawa S., Nishimura Y., Okabe S., Murakami S., Ashikawa Y., Yuge M., Kawaguchi K., Kawase R., Okamoto R., Ito M. Downregulation of GSTK1 Is a Common Mechanism Underlying Hypertrophic Cardiomyopathy. Front. Pharmacol. 2016;7:162. doi: 10.3389/fphar.2016.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Science Council of Japan; 2006. Guidelines for Proper Conduct of Animal Experiments. [Google Scholar]
- 11.Ministry of the Environment; Japan: 28 April 2006. Standards Relating to the Care and Management of Laboratory Animals and Relief of Pain, Notice No.88. revised August 30, 2013. [Google Scholar]
- 12.Ministry of the Environment; Japan: October 1, 1973. The Law for the Humane Treatment and Management of Animals, Law No. 105. revised May 30, 2014. [Google Scholar]
- 13.Barrett T., Troup D.B., Wilhite S.E., Ledoux P., Rudnev D., Evangelista C., Kim I.F., Soboleva A., Tomashevsky M., Marshall K.A. NCBI GEO archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Leeuwen R., Klaver C.C., Vingerling J.R., Hofman A., de Jong P.T. The risk and natural course of age-related maculopathy: follow-up at 6 1/2 years in the Rotterdam study. Archives of ophthalmology (Chicago, Ill.: 1960) 2003;121:519–526. doi: 10.1001/archopht.121.4.519. [DOI] [PubMed] [Google Scholar]
- 15.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitling R., Armengaud P., Amtmann A., Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 18.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montojo J., Zuberi K., Rodriguez H., Bader G.D., Morris Q. GeneMANIA Fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. doi: 10.12688/f1000research.4572.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janky R., Verfaillie A., Imrichova H., Van de Sande B., Standaert L., Christiaens V., Hulselmans G., Herten K., Naval Sanchez M., Potier D. iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS computational biology. 2014;10 doi: 10.1371/journal.pcbi.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Faber C., Williams J., Juel H.B., Greenwood J., Nissen M.H., Moss S.E. Complement factor H deficiency results in decreased neuroretinal expression of Cd59a in aged mice. Invest. Ophthalmol. Vis. Sci. 2012;53:6324–6330. doi: 10.1167/iovs.12-10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carvalho B.S., Irizarry R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westerfield M. University of Oregon Press; Eugene: 2007. A guide for the laboratory use of zebrafish (Danio rerio) [Google Scholar]
- 24.Nishimura Y., Inoue A., Sasagawa S., Koiwa J., Kawaguchi K., Kawase R., Maruyama T., Kim S., Tanaka T. Using zebrafish in systems toxicology for developmental toxicity testing. Congenit. Anom. 2016;56:18–27. doi: 10.1111/cga.12142. [DOI] [PubMed] [Google Scholar]
- 25.Nishimura Y., Okabe S., Sasagawa S., Murakami S., Ashikawa Y., Yuge M., Kawaguchi K., Kawase R., Tanaka T. Pharmacological profiling of zebrafish behavior using chemical and genetic classification of sleep-wake modifiers. Front. Pharmacol. 2015;6:257. doi: 10.3389/fphar.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., Baller J.A., Somia N.V., Bogdanove A.J., Voytas D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39 doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakuma T., Hosoi S., Woltjen K., Suzuki K., Kashiwagi K., Wada H., Ochiai H., Miyamoto T., Kawai N., Sasakura Y. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells. 2013:18. doi: 10.1111/gtc.12037. [DOI] [PubMed] [Google Scholar]
- 28.Dahlem T.J., Hoshijima K., Jurynec M.J., Gunther D., Starker C.G., Locke A.S., Weis A.M., Voytas D.F., Grunwald D.J. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS genetics. 2012;8 doi: 10.1371/journal.pgen.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawase R., Nishimura Y., Ashikawa Y., Sasagawa S., Murakami S., Yuge M., Okabe S., Kawaguchi K., Yamamoto H., Moriyuki K. EP300 protects from light-induced retinopathy in zebrafish. Front. Pharmacol. 2016;7 doi: 10.3389/fphar.2016.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Owsley C., McGwin G., Jr., Jackson G.R., Kallies K., Clark M. Cone- and rod-mediated dark adaptation impairment in age-related maculopathy. Ophthalmology. 2007;114:1728–1735. doi: 10.1016/j.ophtha.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 31.Holliday E.G., Smith A.V., Cornes B.K., Buitendijk G.H., Jensen R.A., Sim X., Aspelund T., Aung T., Baird P.N., Boerwinkle E. Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simon M.V., Agnolazza D.L., German O.L., Garelli A., Politi L.E., Agbaga M.P., Anderson R.E., Rotstein N.P. Synthesis of docosahexaenoic acid from eicosapentaenoic acid in retina neurons protects photoreceptors from oxidative stress. J. Neurochem. 2016;136:931–946. doi: 10.1111/jnc.13487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyers K.J., Johnson E.J., Bernstein P.S., Iyengar S.K., Engelman C.D., Karki C.K., Liu Z., Igo R.P., Jr., Truitt B., Klein M.L. Genetic determinants of macular pigments in women of the Carotenoids in Age-Related Eye Disease Study. Invest. Ophthalmol. Vis. Sci. 2013;54:2333–2345. doi: 10.1167/iovs.12-10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyers K.J., Mares J.A., Igo R.P., Jr., Truitt B., Liu Z., Millen A.E., Klein M., Johnson E.J., Engelman C.D., Karki C.K. Genetic evidence for role of carotenoids in age-related macular degeneration in the Carotenoids in Age-Related Eye Disease Study (CAREDS) Invest. Ophthalmol. Vis. Sci. 2014;55:587–599. doi: 10.1167/iovs.13-13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paun C.C., Ersoy L., Schick T., Groenewoud J.M., Lechanteur Y.T., Fauser S., Hoyng C.B., de Jong E.K., den Hollander A.I. Genetic Variants and Systemic Complement Activation Levels Are Associated With Serum Lipoprotein Levels in Age-Related Macular Degeneration. Invest. Ophthalmol. Vis. Sci. 2015;56:7766–7773. doi: 10.1167/iovs.15-17035. [DOI] [PubMed] [Google Scholar]
- 36.Neale B.M., Fagerness J., Reynolds R., Sobrin L., Parker M., Raychaudhuri S., Tan P.L., Oh E.C., Merriam J.E., Souied E. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc. Natl. Acad. Sci. USA. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fauser S., Smailhodzic D., Caramoy A., van de Ven J.P., Kirchhof B., Hoyng C.B., Klevering B.J., Liakopoulos S., den Hollander A.I. Evaluation of serum lipid concentrations and genetic variants at high-density lipoprotein metabolism loci and TIMP3 in age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2011;52:5525–5528. doi: 10.1167/iovs.10-6827. [DOI] [PubMed] [Google Scholar]
- 38.Gemenetzi M., Lotery A.J. Complement pathway biomarkers and age-related macular degeneration. Eye (London, England) 2016;30:1–14. doi: 10.1038/eye.2015.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monroig O., Rotllant J., Cerda-Reverter J.M., Dick J.R., Figueras A., Tocher D.R. Expression and role of Elovl4 elongases in biosynthesis of very long-chain fatty acids during zebrafish Danio rerio early embryonic development. Biochim. Biophys. Acta. 2010;1801:1145–1154. doi: 10.1016/j.bbalip.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 40.Matsuzaka T., Shimano H., Yahagi N., Amemiya-Kudo M., Yoshikawa T., Hasty A.H., Tamura Y., Osuga J., Okazaki H., Iizuka Y. Dual regulation of mouse Delta(5)- and Delta(6)-desaturase gene expression by SREBP-1 and PPARalpha. J. Lipid Res. 2002;43:107–114. [PubMed] [Google Scholar]
- 41.Kamisuki S., Mao Q., Abu-Elheiga L., Gu Z., Kugimiya A., Kwon Y., Shinohara T., Kawazoe Y., Sato S., Asakura K. A small molecule that blocks fat synthesis by inhibiting the activation of SREBP. Chem. Biol. 2009;16:882–892. doi: 10.1016/j.chembiol.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 42.Ashikawa Y., Nishimura Y., Okabe S., Sasagawa S., Murakami S., Yuge M., Kawaguchi K., Kawase R., Tanaka T. Activation of Sterol Regulatory Element Binding Factors by Fenofibrate and Gemfibrozil Stimulates Myelination in Zebrafish. Front. Pharmacol. 2016;7:206. doi: 10.3389/fphar.2016.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merino D.M., Ma D.W., Mutch D.M. Genetic variation in lipid desaturases and its impact on the development of human disease. Lipids Health Dis. 2010;9:63. doi: 10.1186/1476-511X-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delton-Vandenbroucke I., Grammas P., Anderson R.E. Polyunsaturated fatty acid metabolism in retinal and cerebral microvascular endothelial cells. J. Lipid Res. 1997;38:147–159. [PubMed] [Google Scholar]
- 45.Strunnikova N.V., Maminishkis A., Barb J.J., Wang F., Zhi C., Sergeev Y., Chen W., Edwards A.O., Stambolian D., Abecasis G. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum. Mol. Genet. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pujol-Lereis L.M., Schafer N., Kuhn L.B., Rohrer B., Pauly D. Interrelation Between Oxidative Stress and Complement Activation in Models of Age-Related Macular Degeneration. Adv. Exp. Med. Biol. 2016;854:87–93. doi: 10.1007/978-3-319-17121-0_13. [DOI] [PubMed] [Google Scholar]
- 47.Ghezzi S., Rise P., Ceruti S., Galli C. Effects of cigarette smoke on cell viability, linoleic acid metabolism and cholesterol synthesis, in THP-1 cells. Lipids. 2007;42:629–636. doi: 10.1007/s11745-007-3070-4. [DOI] [PubMed] [Google Scholar]
- 48.Radeke M.J., Radeke C.M., Shih Y.H., Hu J., Bok D., Johnson L.V., Coffey P.J. Restoration of mesenchymal retinal pigmented epithelial cells by TGFbeta pathway inhibitors: implications for age-related macular degeneration. Genome Med. 2015;7:58. doi: 10.1186/s13073-015-0183-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeng S., Whitmore S.S., Sohn E.H., Riker M.J., Wiley L.A., Scheetz T.E., Stone E.M., Tucker B.A., Mullins R.F. Molecular response of chorioretinal endothelial cells to complement injury: implications for macular degeneration. J. Pathol. 2016;238:446–456. doi: 10.1002/path.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitmore S.S., Sohn E.H., Chirco K.R., Drack A.V., Stone E.M., Tucker B.A., Mullins R.F. Complement activation and choriocapillaris loss in early AMD: implications for pathophysiology and therapy. Prog. Retin. Eye Res. 2015;45:1–29. doi: 10.1016/j.preteyeres.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y., Perusek L., Maeda A. Autophagy in light-induced retinal damage. Exp. Eye Res. 2016;144:64–72. doi: 10.1016/j.exer.2015.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ambati J., Anand A., Fernandez S., Sakurai E., Lynn B.C., Kuziel W.A., Rollins B.J., Ambati B.K. An animal model of age-related macular degeneration in senescent Ccl-2- or Ccr-2-deficient mice. Nat. Med. 2003;9:1390–1397. doi: 10.1038/nm950. [DOI] [PubMed] [Google Scholar]
- 53.Zheng W., Reem R.E., Omarova S., Huang S., DiPatre P.L., Charvet C.D., Curcio C.A., Pikuleva I.A. Spatial distribution of the pathways of cholesterol homeostasis in human retina. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kunchithapautham K., Atkinson C., Rohrer B. Smoke exposure causes endoplasmic reticulum stress and lipid accumulation in retinal pigment epithelium through oxidative stress and complement activation. J. Biol. Chem. 2014;289:14534–14546. doi: 10.1074/jbc.M114.564674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holz F.G., Schmitz-Valckenberg S., Fleckenstein M. Recent developments in the treatment of age-related macular degeneration. J. Clin. Invest. 2014;124:1430–1438. doi: 10.1172/JCI71029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu A., Chang J., Lin Y., Shen Z., Bernstein P.S. Long-chain and very long-chain polyunsaturated fatty acids in ocular aging and age-related macular degeneration. J. Lipid Res. 2010;51:3217–3229. doi: 10.1194/jlr.M007518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jump D.B., Depner C.M., Tripathy S. Omega-3 fatty acid supplementation and cardiovascular disease. J. Lipid Res. 2012;53:2525–2545. doi: 10.1194/jlr.R027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.SanGiovanni J.P., Chew E.Y. The role of omega-3 long-chain polyunsaturated fatty acids in health and disease of the retina. Prog. Retin. Eye Res. 2005;24:87–138. doi: 10.1016/j.preteyeres.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 59.Bazan N.G., Molina M.F., Gordon W.C. Docosahexaenoic acid signalolipidomics in nutrition: significance in aging, neuroinflammation, macular degeneration, Alzheimer's, and other neurodegenerative diseases. Annu. Rev. Nutr. 2011;31:321–351. doi: 10.1146/annurev.nutr.012809.104635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye J., DeBose-Boyd R.A. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb. Perspect. Biol. 2011:3. doi: 10.1101/cshperspect.a004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng W., Mast N., Saadane A., Pikuleva I.A. Pathways of cholesterol homeostasis in mouse retina responsive to dietary and pharmacologic treatments. J. Lipid Res. 2015;56:81–97. doi: 10.1194/jlr.M053439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Porstmann T., Griffiths B., Chung Y.L., Delpuech O., Griffiths J.R., Downward J., Schulze A. PKB/Akt induces transcription of enzymes involved in cholesterol and fatty acid biosynthesis via activation of SREBP. Oncogene. 2005;24 doi: 10.1038/sj.onc.1208802. [DOI] [PubMed] [Google Scholar]
- 63.Temel R.E., Hou L., Rudel L.L., Shelness G.S. ACAT2 stimulates cholesteryl ester secretion in apoB-containing lipoproteins. J. Lipid Res. 2007;48:1618–1627. doi: 10.1194/jlr.M700109-JLR200. [DOI] [PubMed] [Google Scholar]
- 64.Curcio C.A., Johnson M., Huang J.D., Rudolf M. Aging, age-related macular degeneration, and the response-to-retention of apolipoprotein B-containing lipoproteins. Prog. Retin. Eye Res. 2009;28:393–422. doi: 10.1016/j.preteyeres.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reardon H.T., Zhang J., Kothapalli K.S., Kim A.J., Park W.J., Brenna J.T. Insertion-deletions in a FADS2 intron 1 conserved regulatory locus control expression of fatty acid desaturases 1 and 2 and modulate response to simvastatin. Prostaglandins Leukot. Essent. Fatty Acids. 2012;87:25–33. doi: 10.1016/j.plefa.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rise P., Ghezzi S., Manzoni C., Colombo C., Galli C. The in vitro effects of cigarette smoke on fatty acid metabolism are partially counteracted by simvastatin. Prostaglandins Leukot. Essent. Fatty Acids. 2009;80:71–75. doi: 10.1016/j.plefa.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Gehlbach P., Li T., Hatef E. Statins for age-related macular degeneration. Cochrane Database Syst. Rev. 2016 doi: 10.1002/14651858.CD006927.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vavvas D.G., Daniels A.B., Kapsala Z.G., Goldfarb J.W., Ganotakis E., Loewenstein J.I., Young L.H., Gragoudas E.S., Eliott D., Kim I.K. Regression of Some High-risk Features of Age-related Macular Degeneration (AMD) in Patients Receiving Intensive Statin Treatment. EBioMedicine. 2016;5:198–203. doi: 10.1016/j.ebiom.2016.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.