Abstract
Pre-mRNA processing is an important step in gene expression and its regulation leads to the expansion of the gene product repertoire. Serine/arginine-rich (SR) proteins are key players in intron recognition and spliceosome assembly and contribute significantly to the alternative splicing process. Due to several duplication events, at least 19 SR proteins are present in the Arabidopsis genome which is almost twice as much as in humans. They fall into seven different subfamilies, three of them homologous to metazoan splicing factors whereas the other four seem to be specific for plants. The current data show that most duplicated genes have different spatio-temporal expression patterns indicating functional diversification. Interestingly, the majority of SR protein genes are alternatively spliced and in some cases this process was shown to be under developmental and/or environmental control. This might greatly influence gene expression of target genes as also exemplified by ectopic expression studies of particular SR proteins.
Keywords: pre-mRNA processing, splicing factor, SR protein, Arabidopsis, genome, duplication
Introduction
The family of serine-arginine (SR) splicing factors includes proteins highly conserved in metazoa and plants. They are involved in multiple steps of the splicing reaction and play an important role both in constitutive and alternative splicing. Typically, they contain one or two RNA recognition motifs (RRM) and a C-terminal domain rich in serine and arginine residues (RS domain). The basic mechanism of splicing is similar in metazoa and plants. However, there are some variations in the conservation of the branch point sequence and polypyrimidine tract [1]. Intron recognition might be different in plants, because animal introns are not processed correctly in plants [2, 3].
A plenitude of SR proteins in Arabidopsis
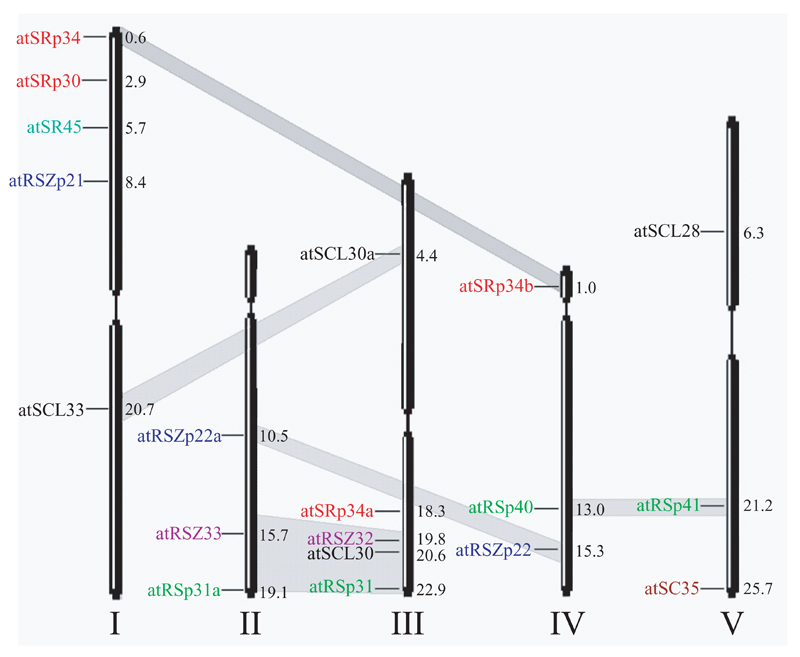
Studies of splicing in plants were directed mostly to the isolation of purified proteins important for intron recognition, namely SR proteins (Fig. 1). The existence of SR proteins in plants was shown by screening plant proteins with antibodies specific for a phosphoepitope present in the animal SR proteins [4, 5]. It has been demonstrated that plants possess at least two homologues of human SF2/ASF (atSRp34/SR1 and atSRp30) [4, 5]. Two additional members of this subfamily (atSRp34a and atSRp34b) have been identified by in silico genome analysis [6], and data base search of Arabidopsis thaliana EST libraries confirmed that both of them are expressed (Fig. 1). Three homologues of human splicing factor 9G8 with a characteristic Zn knuckle motif have been found in Arabidopsis [7, 8]. Furthermore, it was shown that plants not only have more paralogues of human orthologues, but also possess plant specific SR proteins with a unique domain organization [9–11]. A systematic survey of the complete Arabidopsis genome has revealed that at least 18 genes encoding SR proteins are present in A. thaliana [6] (Fig. 1), in contrast to eleven genes in humans [12, 13] and seven in C. elegans [14] . It is believed that gene families have originated from several rounds of duplication of the ancestral genome and/or its particular regions. Analyses of genome sequence suggested that Arabidopsis had undergone probably three rounds of genome duplications [15]. Exploring duplicated regions using available on-line resourses (http://mips.gsf.de/proj/thal/db/gv/rv/rv_frame.html and http://www.tigr.org/tdb/e2k1/ath1/Arabidopsis_genome_duplication.shtml) revealed that at least twelve genes encoding Arabidopsis SR proteins are located on the duplicated segments (Fig.2). Surprisingly, detailed comparison of two duplicated regions on the bottoms of chromosomes II and III yielded the identification of a previously unknown Arabidopsis SR protein. This nineteenth protein, which we designated atRSp31a, is 72% identical to atRSp31 and is encoded by gene at2g46610. The high degree of gene duplications and the relatively large number of SR proteins in Arabidopsis raises the question about the fate of duplicated genes and redundancy of their functions.
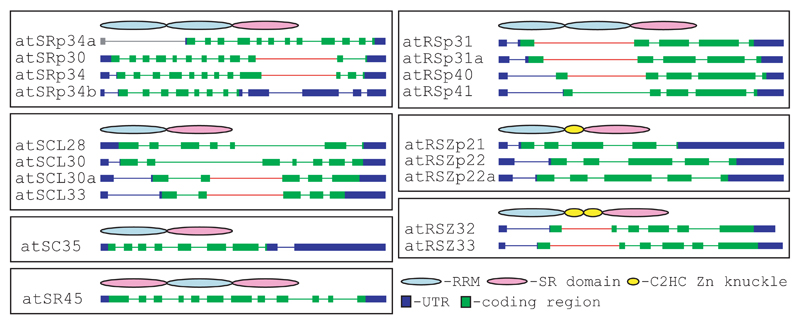
Figure 1.
Family of the Arabidopsis SR protein genes. Genes are grouped in subfamilies [10]. General protein domain organization for each subfamily is shown. Exon-intron organization is based on experimental data, except the first exon and an intron of 5’UTR of atSRp34a. Long introns subjected to alternative splicing are marked with stars.
Figure 2.
Chromosomal distribution of Arabidopsis genes encoding SR proteins. Chromosomes are labelled by roman numerals. Gene positions are indicated in megabases. Grey bands connect duplicated regions, containing genes for SR proteins.
Divergence on the level of transcription
On the protein level the homology is quite high between paralogous pairs of genes. However, diversification of functions in gene families occurs not only on the level of coding sequence, but also by modification of regulatory regions that control spatial and temporal expression. Analyses of paralogous promoter regions of genes encoding SR proteins using different motif search programs and databases (TRANSFAC, PatSearch, and others) suggest that they contain both different and similar cis-acting regulatory elements.
In vivo studies of expression patterns of Arabidopsis SR proteins revealed that they are differentially expressed in certain tissues and under different conditions. A good example is atSRp30 and atSRp34/SR1. According to Northern blot data and RT-PCR analyses, atSRp30 and atSRp34/SR1 are expressed more or less ubiquitously in all plant organs, despite slightly different expression levels [4, 16, 17]. However, detailed histochemical analyses of promoter-reporter gene fusions have shown that they have complementary expression patterns during early development of Arabidopsis seedlings. Interestingly, their expression overlaps during the first stages of lateral root formation, suggesting that both of them are involved in the initiation of lateral roots. During later stages they become separated, and only atSRp34/SR1 is expressed in the cells of the growing root tip, indicating distinct roles of these two proteins in root development [17].
Arabidopsis has a subfamily of SR proteins consisting of atRSp31, atRSp40 (formerly atRSp35), atRSp41 [9] and atRSp31a, with no animal equivalent identified so far. The paralogous pair, atRSp40 and atRSp41, is located on duplicated regions of chromosomes IV and V (Fig. 2). Comparison of expression patterns of their promoters fused to a reporter gene revealed that atRSp41 is strongly expressed in all tissues, while atRSp40 has a similar expression only in the roots and cotyledons and is not detected in young leaves (our unpublished data). This suggests loss of a particular expression domain by one of the paralogues and indicates that these two genes had evolved after duplication and might play distinct roles in plant development.
The Arabidopsis SR proteins subfamily homologous to h9G8 and containing a CCHC zink knuckle motif is represented by three proteins atRSZp21 (SRZ-21), atRSZp22 (SRZ-22) and atRSZp22a [7, 8]. Two of them, atRSZp22a and atRSZp22, are located on the duplicated regions of chromosomes II and IV, respectively (Fig. 2). Transcript distribution of atRSZp21 and atRSZp22 has been shown to vary among tissues, and, in addition, atRSZp21 was more abundant in roots and less abundant in siliques than atRSZp22 [7]. Existing EST clones indicate that atRSZp22a is expressed in flowers and/or siliques and in roots. Histochemical analysis of atRSZp22 promoter activity showed that it is expressed in tapetum, mature pollen, carpel, ovules, developing seeds, and in the elongation zone of the root [10]. Taken together, these data suggest that atRSZp22 and atRSZp22a might have overlapping expression patterns in those organs, however, a more detailed analysis of expression pattern of atRSZp22a is needed .
Plants possess two paralogous SR proteins with a unique domain structure consisting of an N-terminal RNA recognition motif (RRM), two zinc knuckles embedded in a RS region and an acidic C-terminal domain [10]. Genes encoding these proteins are located on the duplicated regions at the bottom arms of chromosomes II and III (Fig. 2). One of those genes, atRSZ33, is expressed during embryogenesis, early seedling formation, and in flower and root development, implicating a role of this protein in these processes [18]. Expression of the second gene, atRSZ32, has not been studied yet, but existing information about EST clones suggests that this gene is also expressed in flowers, siliques and roots, and, additionally, in rosette leaves.
The diversification of spatio-temporal expression patterns of genes encoding Arabidopsis SR proteins implies different target specificity and consequently specialization of their biological functions.
Exon-intron structures and alternative splicing of Arabidopsis SR protein genes
Exon-intron structures of genes encoding Arabidopsis SR proteins are relatively conserved within subfamilies (Fig.1). Members of the atRSp31-41 and atRSZp21-22a subfamilies contain 6 exons, and the ones from atRSZ32-33 subfamily have 7 exons. Paralogous pairs of genes have not only a conserved number of exons, but also show a more similar intron length. The numbers of exons are more variable within the SCL and SF2/ASF subfamilies, comprising either 6-8, or 12-13 exons, respectively. Interestingly, exon-intron structures of atSRp34/SR1 and atSRp34b are almost identical up to exon 10. However, the 3’ end of atSRp34b adopted an organization similar to the SR1B splice variant of atSRp34/SR1 [16] (Fig.1), thus only a truncated protein can be produced. Additionally, the 9th exon of atSRp34b contains a premature stop codon due to a single nucleotide insertion, which results in a protein lacking the SR domain. Therefore, atSRp34b represents a pseudogene which originated from DNA duplication and subsequent disablement through a premature stop codon and alteration of a splice site.
A remarkable feature in the majority of Arabidopsis genes encoding SR proteins is the presence of one or two relatively long introns ranging in size from 417 to 1099 nt (i.e. 12 out of 19 genes have introns > 400 nt). Such introns are not common in plants. Typically, two thirds of the plant introns are shorter than 150 nt [1]. The positions of these long introns tend to be conserved between Arabidopsis paralogues (Fig.1). Interestingly, published evidence [10, 11, 16–18] and analysis of the Arabidopsis EST data base reveal that in nine of these genes the long introns are alternatively spliced. These splice variants encode truncated proteins due to stop codons generated by included intron sequences. Long introns in atSCL33/SR33 and in atRSZ33 are located in their RRMs , thus alternative splicing results in the production of shortened proteins containing only a part of the RRM [10, 11]. In contrast, location of a long intron in the 3’ part of genes encoding atSRp30 and atSRp34/SR1 results in protein isoforms differing in their RS domains [16, 17]. Interestingly, in addition to a more diverse exon-intron structure, members of the SF2/ASF and SCL subfamilies have the highest occurrence of alternative splicing events in the long and adjacent introns. For example, atSRp34/SR1 has at least seven different splice variants [16, 17], and four splice forms were detected for atSRp30 [17]. RT-PCR data have shown multiple transcripts for atSCL33/SR33, and the nature of four of them was resolved [11].
Alternative splicing creates an important potential for regulation of gene expression. Splicing patterns of Arabidopsis SR protein genes are under tight spatio-temporal control, leading to a different abundance of splice variants in different tissues and at developmental stages [4, 9, 11, 17]. Environmental conditions can also modulate the splicing pattern of a gene, as it was shown by the temperature control of the ratio of atSRp34/SR1 alternative forms [16]. Consequently, production of the full length protein is not only controled by transcription but also by alternative splicing.
In vivo modulation of SR protein expression levels
As individual members of Arabidopsis SR proteins possess distinct expression patterns it is plausible to speculate that they are likely to have non-redundant functions at least under some conditions. In metazoa, SR proteins were shown to have both essential and redundant functions [14, 19, 20]. Gene knockout is a powerful tool to dissect the function of an individual gene. However, in case of multigene families, simultaneous disruption of several genes might be required to reveal their biological functions. The overexpression approach proved to be successful in studies of both biochemical and physiological roles of SR proteins in plants.
It was shown that Arabidopsis SR proteins can modulate splicing patterns of other genes encoding SR proteins. For example, overexpression of atSRp30 in transgenic plants resulted in the down-regulation of mRNA encoding full-length atSRp34/SR1 protein, which together with their distinct expression patterns suggest antagonistic functions [17]. Overexpression of atRSZ33 in transgenic plants led to alteration of splicing patterns of atSRp30 and atSRp34/SR1. Moreover, atRSZ33 regulates its own expression, as splicing of its endogenous pre-mRNA was changed in transgenic plants [18]. In case of atSRp34/SR1, overexpression of the full-length protein or its variants does not influence its own splicing profile [16].
Limited evidence exists on the functions of particular SR proteins in plant development. It was shown that increased protein level of atSRp30 in transgenic plants led to delay in transition from vegetative to reproductive phase, prolongation of life cycle, and to increased size of plant [17]. Overexpression of atRSZ33 caused changes in fertilization, embryogenesis, and development of shoot apical and root meristems, which resulted from impaired cell expansion and division. In addition, levels and distribution of the plant hormone auxin was affected in transgenic plants [18]. Since ectopic expression can possibly mimic function of a paralogous gene, it would be interesting to compare the impact of overexpression of paralogous pairs of genes on plant development.
Our analyses of the Arabidopsis SR protein family show the importance of gene duplication and alternative splicing as a source of functional protein diversity.
Arabidopsis SR proteins and corresponding gene names
atSRp30 - at1g09140; atSRp34/SR1 - at1g02840; atSRp34a - at3g49430; atSRp34b - at4g02430; atRSp31a - at2g46610; atRSp31 - at3g61860; atRSp40 - at4g25500; atRSp41 - at5g52040; atRSZp21 - at1g23860; atRSZp22 - at4g31580; atRSZp22a - at2g24590; atRSZ32 - at3g53500; atRSZ33 - at2g37340; atSCL28 - at5g18810; atSCL30 - at3g55460; atSCL30a - at3g13570; atSCL33/SR33 - at1g55310; atSC35 - at5g64200; atSR45 - at1g16610.
Acknowledgements
This work was supported by grants from the Österreichischer Fonds zur Förderung der wissenschaftlichen Forschung (FWF) to AB (SFB F017/10/11).
Abbreviations used
- RRM
RNA recognition motif
- SR
serine-arginine
- RS
arginine-serine
- EST
expressed sequence tag
- UTR
untranslated region
References
- 1.Lorkovic ZJ, Wieczorek Kirk DA, Lambermon MH, Filipowicz W. Trends Plant Sci. 2000;5:160–7. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- 2.Barta A, Sommergruber K, Thompson D, Hartmuth K, Matzke MA, Matzke AJM. Plant Mol Biol. 1986;6:347–357. doi: 10.1007/BF00034942. [DOI] [PubMed] [Google Scholar]
- 3.Van Santen VL, Spritz RA. Gene. 1987;56:253–265. doi: 10.1016/0378-1119(87)90142-9. [DOI] [PubMed] [Google Scholar]
- 4.Lazar G, Schaal T, Maniatis T, Goodman HM. Proc Natl Acad Sci U S A. 1995;92:7672–6. doi: 10.1073/pnas.92.17.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopato S, Mayeda A, Krainer AR, Barta A. Proc Natl Acad Sci U S A. 1996;93:3074–9. doi: 10.1073/pnas.93.7.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lorkovic ZJ, Barta A. Nucleic Acids Res. 2002;30:623–35. doi: 10.1093/nar/30.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Golovkin M, Reddy AS. Plant Cell. 1998;10:1637–48. doi: 10.1105/tpc.10.10.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopato S, Gattoni R, Fabini G, Stevenin J, Barta A. Plant Mol Biol. 1999;39:761–73. doi: 10.1023/a:1006129615846. [DOI] [PubMed] [Google Scholar]
- 9.Lopato S, Waigmann E, Barta A. Plant Cell. 1996;8:2255–2264. doi: 10.1105/tpc.8.12.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopato S, Forstner C, Kalyna M, Hilscher J, Langhammer U, Indrapichate K, Lorkovic ZJ, Barta A. J Biol Chem. 2002;277:39989–98. doi: 10.1074/jbc.M206455200. [DOI] [PubMed] [Google Scholar]
- 11.Golovkin M, Reddy AS. J Biol Chem. 1999;274:36428–38. doi: 10.1074/jbc.274.51.36428. [DOI] [PubMed] [Google Scholar]
- 12.Graveley BR. RNA. 2000;6:1197–211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin C, Manley JL. Cell. 2002;111:407–17. doi: 10.1016/s0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 14.Longman D, Johnstone IL, Caceres JF. EMBO J. 2000;19:1625–37. doi: 10.1093/emboj/19.7.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. Proc Natl Acad Sci U S A. 2002;99:13627–32. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lazar G, Goodman HM. Plant Mol Biol. 2000;42:571–81. doi: 10.1023/a:1006394207479. [DOI] [PubMed] [Google Scholar]
- 17.Lopato S, Kalyna M, Dorner S, Kobayashi R, Krainer AR, Barta A. Genes Dev. 1999;13:987–1001. doi: 10.1101/gad.13.8.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalyna M, Lopato S, Barta A. Mol Biol Cell. 2003;14:3565–77. doi: 10.1091/mbc.E03-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ring HZ, Lis JT. Mol Cell Biol. 1994;14:7499–506. doi: 10.1128/mcb.14.11.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang J, Takagaki Y, Manley JL. Genes Dev. 1996;10:2588–99. doi: 10.1101/gad.10.20.2588. [DOI] [PubMed] [Google Scholar]