Abstract
Reduction of preventable hospital readmissions that result from chronic or acute conditions like stroke, heart failure, myocardial infarction and pneumonia remains a significant challenge for improving the outcomes and decreasing the cost of healthcare delivery in the United States. Patient readmission rates are relatively high for conditions like heart failure (HF) despite the implementation of high-quality healthcare delivery operation guidelines created by regulatory authorities. Multiple predictive models are currently available to evaluate potential 30-day readmission rates of patients. Most of these models are hypothesis driven and repetitively assess the predictive abilities of the same set of biomarkers as predictive features. In this manuscript, we discuss our attempt to develop a data-driven, electronic-medical record-wide (EMR-wide) feature selection approach and subsequent machine learning to predict readmission probabilities. We have assessed a large repertoire of variables from electronic medical records of heart failure patients in a single center. The cohort included 1,068 patients with 178 patients were readmitted within a 30-day interval (16.66% readmission rate). A total of 4,205 variables were extracted from EMR including diagnosis codes (n=1,763), medications (n=1,028), laboratory measurements (n=846), surgical procedures (n=564) and vital signs (n=4). We designed a multistep modeling strategy using the Naïve Bayes algorithm. In the first step, we created individual models to classify the cases (readmitted) and controls (non-readmitted). In the second step, features contributing to predictive risk from independent models were combined into a composite model using a correlation-based feature selection (CFS) method. All models were trained and tested using a 5-fold cross-validation method, with 70% of the cohort used for training and the remaining 30% for testing. Compared to existing predictive models for HF readmission rates (AUCs in the range of 0.6–0.7), results from our EMR-wide predictive model (AUC=0.78; Accuracy=83.19%) and phenome-wide feature selection strategies are encouraging and reveal the utility of such data-driven machine learning. Fine tuning of the model, replication using multi-center cohorts and prospective clinical trial to evaluate the clinical utility would help the adoption of the model as a clinical decision system for evaluating readmission status.
1. Introduction
1.1. Hospital readmission rates – a bottleneck in delivering high value-high volume precision healthcare
Precision healthcare aims to ensure every patient receive optimal care throughout the onset, maintenance or recovery phases of a disease. Close coordination between different players in the health system is required to integrate and deliver high-quality care. Patients, providers and the care management team play a pivotal role in delivering low-cost, high value and high volume care for patients with diverse healthcare requirements. Improving the quality of healthcare delivery is a challenging task for providers and an important priority for regulatory agencies. As an attempt to reduce healthcare cost, lower healthcare disparities and increase overall quality of care, healthcare regulatory agencies including Centers for Medicaid and Medicare Services (CMS, https://www.cms.gov/) have proposed the Hospital Readmission Reduction Program (HRRP; See: https://www.cms.gov/medicare/medicare-fee-for-service-payment/acuteinpatientpps/readmissions-reduction-program.html). Depending on the performance of a given provider (or hospital) with respect to the regional, state and federal performance rankings, penalties are levied on healthcare providers. In response, in order to reduce readmissions providers have used commercial or in-house readmission assessment tools to predict 30-day readmission rates, but the overall readmission rates still remain high in various provider sites. In 2015, 2,592 U. S hospitals out of 5,627 registered hospitals in the country received penalties from the CMS (http://khn.org/news/half-of-nations-hospitals-fail-again-to-escape-medicares-readmission-penalties/) for not effectively tackling readmission rates. Despite decades of research, interventions, operational improvements and systems engineering methods, readmission remains a major challenge for patients, providers and payers alike.
1.2. Readmission rate assessment directive by CMS
The CMS (https://www.medicare.gov/hospitalcompare/Data/30-day-measures.html) directive on unplanned readmission grades the results of five diseases, two surgical procedures and a quantitative estimate of hospital-wide readmission rates. The conditions that CMS evaluates for readmission rates include three specific cardiovascular diseases (heart attack, heart failure, and stroke), one respiratory disease (chronic obstructive pulmonary disease) and an infectious disease (pneumonia). The hospital-wide readmission rates assess the readmission status of patients admitted to internal medicine, surgery/gynecology, pulmonary, cardiovascular, and neurology services. Further, the 30-day mortality measures determine death rates associated these services. Implementing data-driven methods that consider all available clinical variables in a hypothesis-free approach could identify new features driving clinical outcomes. Such an approach could also provide insights into mechanistic or operational factors that could improve clinical outcomes 1–4. Heart failure is one of the first core measures by The Joint Commission to assess hospital quality initiatives as part of National Hospital Inpatient Quality Measures. Achieving the lowest readmission rates possible is thus critical to provide high-quality care and improve quality assessments (See: https://www.jointcommission.org/core_measure_sets.aspx).
1.3. Improving quality of healthcare delivery and outcomes using EMR-wide phenomic data
Implementation of precision phenotyping algorithms and development of prescriptive prediction models models using phenomic data could aid in the discovery of new knowledge from biomedical and healthcare big data generated in the hospital setting5,6. Mining of phenomic big data enables the identification of new or unknown features or combinatorial features driving clinical outcomes. Electronic medical records (EMR) provide access to clinical phenome data and enable better understanding of various clinical phenotypes and the associated outcomes in a systematic manner. Design, development, and deployment of predictive and prescriptive models using EMR-based methods could help to accelerate stratification of patients at risk for improved care. Deploying validated predictive patterns in a clinical setting could improve the quality of healthcare delivery and may have a positive impact on patient outcomes. Phenomics7 is a relatively new omics term used to define collectively the measurement of phenotypic characteristics of biological entities that include the physical and biochemical traits of organisms including humans. Human phenomics can benefit by leveraging EMRs as a longitudinal data source for the collection of clinical and health traits. While the data currently available within EMR for building a complete picture of a human phenomic state is limited, it is rapidly improving with the integration of genomic data, sensor data and other non-clinical data elements3,4. Phenome-wide association studies (PheWAS) studies aim to understand the role of a genetic variant identified from genome-wide association studies (GWAS) in increasing or decreasing the likelihood of observing other diseases in a case-control cohort. PheWAS studies are now revealing the molecular architecture of the pleiotropic nature of genetic variants in mediating multiple diseases1,8.
1.4. Predictive modeling of readmission rates in heart failure and need for improvement
Heart failure is a heterogeneous condition characterized by progressive inability of the heart to supply sufficient blood to the organs of the body. HF is associated with high degree of morbidity and mortality, and 50% of patients with HF die within five years of diagnosis. Heart failure accounts for 43% of Medicare spending even though this patient population only makes up 14% of all Medicare beneficiaries. Heart failure is the top cause of readmission for the Medicare fee-for-service patient population and costs approximately 38 billion dollars annually. Several attempts have reported on the utility, accuracy and actionability of predictive models to model and predict potential readmission associated with heart failure hospitalization. Previously reported models have been built using clinical variables and covariates such as age, sex, race, socioeconomic factors, body mass index, laboratory measures, biomarkers (e.g. B-type natriuretic peptide levels), comorbidities (e.g. neurological disorders, type II diabetes mellitus, etc.), behavioral factors, functional phenotyping of cardiovascular systems (e.g. left ventricular ejection fraction), discharge follow-ups and medications 9–12. Some models have used billing and procedural codes extracted from EMR or other hospital administration databases. Continuous hemodynamic monitoring devices have also been used to predict readmission rates 13–15. The predictive power of such HF readmission models remains weak, with Area Under Curve (AUC) values generally in the range of 0.6–0.7. Such models provide only modest utility for predicting which patients may return to the hospital for readmission. There is an immediate need for tools that may be used at the bedside or as part of discharge disposition planning to assess and minimize risk for readmission. Studies led by Hosseinzadeh et.al16 leverage claims data to predict all-cause readmissions, and Duggal et.al17 used EMR-derived clinical and administrative data to predict readmission in the setting of a diabetes cohort. To the best of our knowledge, our study is one of the first attempts to use phenome-wide data to identify novel factors driving readmissions related to congestive heart failure and develop EMR-wide prediction models with orthogonal validation to predict the readmission event.
2. Methods
The Mount Sinai Institutional Review Board approved the study. An author (JJ) act as the honest data broker to ensure PHI and HIPAA adherence during the data management, analytics and machine learning. Data scientists and research scientists in the project received a deidentified database from the Mount Sinai Data Warehouse. All analyses were performed using the deidentified data.
2.1. Mount Sinai Heart cohort and characteristics of heart failure cohort
The study cohort consists of a database of 1,068 individuals admitted to Mount Sinai Heart service during the year 2014. The principal diagnosis of heart failure using the CMS directive was used to compile HF patients. Each patient readmitted to any service of Mount Sinai within 30-days after the discharge of an HF primary encounter is defined as a “case”. The remainder of patients who did not return to the hospital within 30-days were defined as “controls”. Patients admitted to other locations of Mount Sinai Health System or other hospitals within New York city/state or other states in country were not captured. An author (DR) manually phenotyped the cohort and classified the patients as part of a quality control initiative at Mount Sinai Hospital. As an exploratory study with low case rate, no patient exclusion criteria were applied to the dataset.
2.2. Clinical data analytics and EMR-wide machine learning
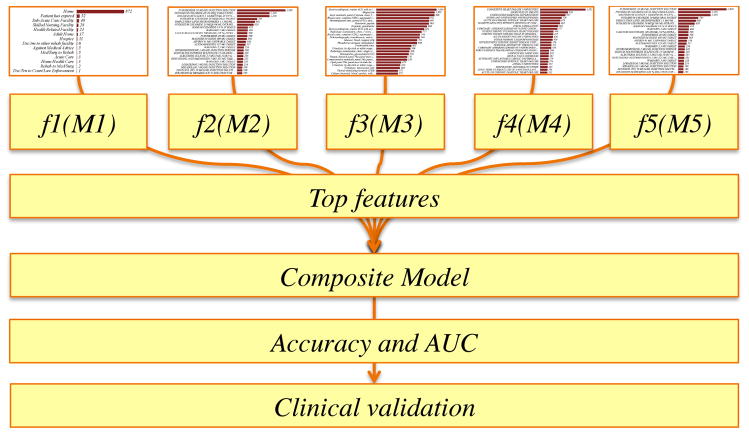
Data was stored in a MySQL database indexed using a unique hexadecimal identifier associated with the data for the visit about HF. Only data about the primary encounter (admission with HF as primary diagnosis) is employed in the analysis. All figures were generated using Wizard for Mac (http://www.wizardmac.com/) and Weka 18–21. A Naïve Bayes model is used for machine learning. Exploratory data analyses were performed using Elasticsearch and Kibana (https://github.com/elastic/kibana). All models were independently created using 70% of the dataset for training and 30% of the dataset for testing. Bayesian models were created using features unique to each data element and feature selection was performed using correlation based feature subset selection across two classes. Orthogonal validation of machine learning models was performed with logistic regression. Principal component analyses to understand the variability of features were performed using the Python-based scikit-learn package (http://scikit-learn.org/) and visualized using matplotlib (http://matplotlib.org/). Testing accuracies were estimated using the 5-fold cross validation approach. We define the classification task as a binary classification problem, where RA=“Readmitted” patient and NonRA=“Not readmitted patient”. Weka provides a suite of state-of-the-art machine learning algorithms using a programmatic interface in Java. We used the native Naïve Bayesian classifier in Weka without modification in this exploratory analysis. The algorithm was selected as a rational choice based on prior studies on modeling of readmission prediction16 Feature ranking and selection22,23 was performed using a correlation-based feature selection (CFS) method. CFS is a widely used feature selection strategy that aims to find subset of features with significant discriminatory power to perform the classification but which are uncorrelated in feature space. Feature selection is implemented using the “CfsSubsetEval” method in Weka (http://weka.sourceforge.net/doc.dev/weka/attributeSelection/CfsSubsetEval.html). Orthogonal class-specific statistical significance was estimated using Kolmogorov-Smirnov test (distribution estimates), t-test (differences across class-labels), Z-score or Mann-Whitney (median estimates) depending on the data type tested (lab-test, medication, procedure etc.) across the groups (RA and NonRA). An overview of the study design is provided in Figure 1.
Figure 1.
EMR-wide machine learning architecture and predictive modeling strategy to find drivers of readmission rates
3. Results
3.1. Cohort characteristics
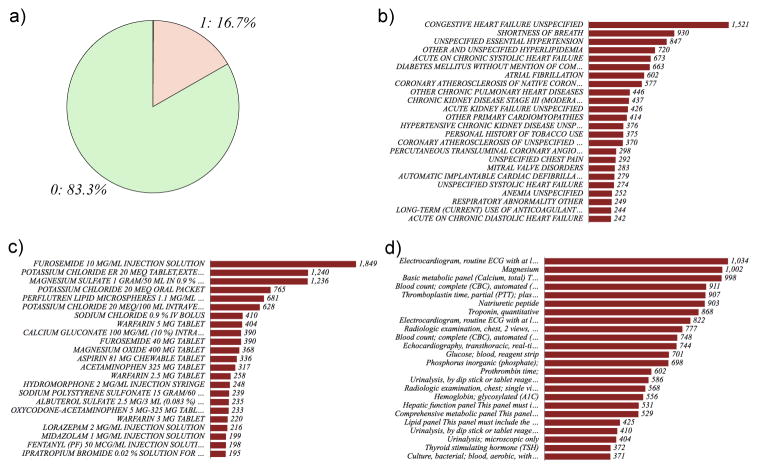
EMR-wide data mining provides a deep view of various data elements in the cohort (Figure 2). A total of 4,205 variables were extracted from EMR. The data from EMR was categorized into five data modalities as diagnosis codes (ICD-9 codes and IMO-codes), procedures (ICD-9, SNOMED-CT and CPT-codes), medications and vital signs. For each patient, the patient encounter specific data is extracted from the EMR. A patient specific filter is used to extract data unique to the visit; the data from the most recent visit of the patients with multiple admissions is incorporated.
Figure 2.
Summary of the study cohort a) case-control ratio: cases are indicated as “1” and controls as “0”. Frequency charts of b) diagnoses c) medications and d) procedures.
Phenomic data extracted from EMR:
Diagnoses codes using ICD-9 (n=1,763): ICD-9 codes (http://www.cdc.gov/nchs/icd/icd9.htm) were extracted from Mount Sinai Data Warehouse. The codes were mapped to ICD-9 or IMO codes (https://www.e-imo.com/problemit-terminology-1); all codes were unified to ICD-9 and normalized using UMLS as the bridge (https://www.nlm.nih.gov/research/umls/mapping_projects/icd9cm_to_snomedct.html).
Medications (n=1,028): Medications prescribed during the hospitalization were compiled using Epic and extracted from Mount Sinai Data warehouse. Medication name, dosage, route of administration was obtained. All medication data was normalized using RxNorm (https://www.nlm.nih.gov/research/umls/rxnorm/).
Laboratory measurements (n=846): Laboratory measures captured in the EMR were compiled; the raw values of the tests without normalization have been used as a matrix of observations with patients as rows and individual tests as columns.
Procedures (n=564): Procedures encoded using SNOMED-CT or ICD-9-CM procedures were used.
Vital signs (n=4): Pulse, respiration rate, systolic blood pressure, heartbeats and temperature were compiled from bedside monitor logs captured in a MySQL database. Vitals were often captured using multiple monitors and approaches. For example temperature was captured at the bedside as axillary temperature, temperature measured via catheter, oral temperature, rectal temperature, or tympanic temperature.
3.2. EMR-wide feature selection and predictive modeling using five different data modalities
The machine learning strategy utilized for our study is outlined in Figure 1. To address the tradeoffs in dealing with a broad range of features using a small number of samples and missing data, we first generated distinct models using different data elements and relevant features were selected. Features were also compared using orthogonal metrics including logistic regression and PCA to understand the variable space and their inherent relationships. Finally, a composite model for performing predictions is generated using features selected from the individual models. As a real-world machine-learning task, we had a small subset of cases (16.7%) compared to the controls (83.3%). We used a random subset of age and sex matched controls to control the bias introduced by imbalanced datasets. We first generated five different NB predictors using individual data elements. Medications were the most predictive with an accuracy of 81% and AUC of 0.615. Procedure codes encoded as binary variable fared poorly with AUCs of <0.50 (ICD-9 procedures) and 0.553 (CPT codes). We did not generate an independent model for feature selection using the four vital signs after accounting for the small number of features. Laboratory values also showed lower AUC (0.535). Exploration of the data using principal component analyses also revealed that procedures had low variance compared to medications. From a healthcare delivery standpoint, this is insightful, as most of the patients have undergone the same type of procedures in the cardiac units. However the medication profiles of patients may vary due to individualized disease comorbidities, side effect profiles, age, and gender. Details of individual models and features identified using feature selection method (See Table 1). Detailed analyses of medications could provide better insights into features driving readmissions (Johnson & Shameer et.al; manuscript in preparation)
Table 1.
Summary of different Bayesian predictors and features compiled using CFS method
| Data-element | Type | Encoding | Accuracy | AUC | Features |
|---|---|---|---|---|---|
| Diagnosis | ICD-9 Diagnosis | Binary | 70.3297% | 0.605 | 34/1763 |
| Procedures | ICD-9-Procedure | Binary | 77.907% | <0.50 | 4/273 |
| Procedure | CPT-codes | Binary | 72.9858% | 0.553 | 8/564 |
| Medications | Medication name and dosage | Binary | 81.9048% | 0.615 | 26/1028 |
| Labs | Non-descriptive lab measurements | Continuous | 73.9336% | 0.535 | 29/846 |
| Composite model | Combined features | Hybrid | 83.9000% | 0.780 | 105 |
3.3. Feature reduction and model refinement
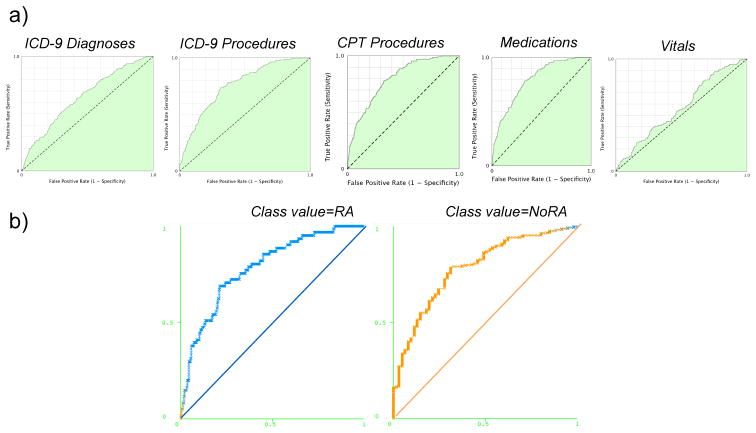
Due to the low percentage of the cases in the cohort under investigation, a high-dimension feature array is prone to overfitting in machine learning of binary classification tasks. To address this, we have used a feature reduction approach. Features were tested to assess predictive value using a classifier based method and regression models. Feature selection approach and an orthogonal validation approach provide insights into a subset of highly predictive variables associated with readmitted subset of patients. The AUCs of regression models were 0.5685, 0.6471, 0.7596 and 0.795 (ICD-9 and CPT) for vitals, diagnoses codes, medications, and procedures respectively (See Figure 4 and 5). The final composite model is developed using 105 features with an AUC=0.78 and cross-validation testing accuracy of 83.19%.
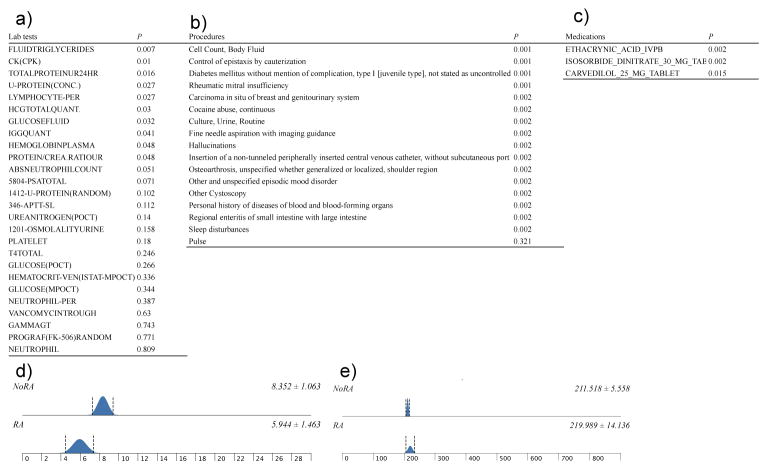
Figure 4.
Orthogonal validation of discriminating features a) laboratory tests b) procedures and diagnoses c) medications d) absolute neutrophil count (P=0.051) e) platelet count (P=0.180)
A brief summary of features significant in feature selection method and the orthogonal validation approach is provided below (also see Figure 5):
a) Procedures: out of 12 procedures, codes for invasive procedures including fine needle aspirations with imaging guidance, intravenous catheterization, routine culture and cell count were significant procedures. As procedures were counted as individual events, the subset of readmitted patients has higher frequency of these procedures compared to patients not readmitted. Repetitive tests for culture and cell count could also indicate potential infection or other complications. b) Medications: amongst the 1,028 medications, our analyses indicate 28 medications as features with discriminatory power. Three medications (carvedilol 25 mg tablet, ethacrynic acid IVPB and isosorbide dinitrate 30 mg tablet) were validated using logistic regression approach. However, we noted that only 2.7% of the cohort received carvedilol 25 mg, and all of them were part of the readmission subset. Previous work has potentially indicated that increasing in carvedilol dosage may lead to better a outcome on readmission rate24. c) Diagnosis: chronic conditions like type 1 diabetes (ICD-9 code 250.01), osteoarthritis; manifestations of cancer (ICD-9 code 233); neurological or psychiatric conditions (mood disorders, hallucinations, sleep disturbances cocaine abuse); cardiovascular structural conditions like rheumatic mitral insufficiency and gastrointestinal conditions such as enteritis were conditions significantly associated with readmission rates. Onco-cardiology assessment of patients may also help in reducing the readmission rates in high-risk patients. Assessment of cardiovascular patients for psychosocial aspects and careful evaluation of individual comorbidities could help to reduce the readmission rates and adherence to the medications 25–28. d) Laboratory values: laboratory values were least predictive in the individual modeling stage. During the orthogonal validation step, creatinine kinase, glucose-fluid, fluid triglycerides and lymphocytes were significant. Optimal glycemic control is a key factor in determining positive outcomes in heart failure patients, especially in those with diabetes mellitus 29. We noted that features identified using our feature selection method are concordant with earlier findings. For example, we have identified glucose-fluid and type-1 diabetes as predictive factors. We have also identified psychiatric illness, a known factor that influences readmission ratesin the setting of complex diseases.
3.4. Comparison with current heart failure readmission models
In this work we use EMR-wide feature selection and machine learning to discover novel features and develop new predictors to predict readmission rates. One of the first predictive modeling of hospital readmissions using healthcare data from Quebec, Canada by Hosseinzadeh et.al16 showed that Naïve Bayes models (0.65) performed better than Random Forest models (0.64). Using a diabetes cohort from a hospital in India, Duggal et.al17 showed that Naïve Bayes (0.67) showed higher readmission associated savings compared to logistic regression (0.67), Random Forests (0.68), Adaboost (0.67) and Neural Networks (0.62). Futoma et.al30 showed that Random Forests (0.68) and deep learning using neural networks (0.67) have similar accuracy rate with >1 million patients and > 3 million admission. However, Penalized Logistic Regression had similar accuracy rates as we have shown in our orthogonal validation methods. Compared to existing predictive models for HF readmission rates (AUCs in the range of 0.6–0.7), results from our EMR-wide predictive model (AUC=0.78; Accuracy=83.19%) and phenome-wide feature selection strategies are encouraging and reveal the utility of such data-driven, EMR-wide machine learning.
4. Discussion
Readmission rate is a quality assessment metric routinely used to infer the quality of life index of patient population and the quality of healthcare delivery. Irrespective of the advances in biomedical and healthcare research practices, hospital quality control offices still use traditional readmission risk algorithms and predefined sets of variables to infer the probability patient readmission. However, predictive modeling using big data sourced from different facets of healthcare operations could provide clues to improve the quality of healthcare delivery. Combining predictive analytics with preventive measures would also engage patients, physicians, and payers to participate proactively in improving the health and wellness. Recently we have combined EMR data and genomic data to cluster patients into subtypes with specific genetic variants, disease comorbidities, and medications in a diabetes cohort. Application of deep learning31,32 in healthcare also shows promise for performing EMR-wide analytics using approaches like Deep Patient33. In a recent study, we have created temporal models of disease trajectories that could potentially reveal how the population could cluster into subgroups based on age, gender, self-reported ancestry and comorbidities34. Further, we have shown that cognitive machine learning can be utilized for precise phenotyping of high volume echocardiography datasets35. We have also applied machine learning to understand various features driving patient satisfaction36. Our collective experience in large-scale, automated mining of EMR data suggests that such approaches are useful for both discovery research and the identification of actionable clinical parameters driving diseases or outcomes.
5. Limitations of the current study
In this study, we use all codes without further comprehension; for example, coding systems other than ICD-9 provide an easy way to combine disease. Such an approach could also lead to compiling of similar conditions and hence may not reveal true predictors. For example, we have identified enteritis as a potential diagnosis with readmission. This term would be summarized under gastroenterological conditions. Grouping medication by class or category may also reduce the feature space at the cost of feature resolution. We attempt to capture the best characteristic elements from the real-world data set and hence no data imputation or normalization has been used in our study. The feature selection method may also influence the composition of the models; a systematic assessment of various feature selection algorithms could further enhance the robustness of the model. Healthcare datasets are highly sparse, for example, all patients are not being tested using same laboratory tests except for a few generic tests. Hence, several features may have sparse representations. Even though we had access to EMR-linked genomic data (See BioMe: http://icahn.mssm.edu/research/ipm/programs/biome-biobank), genomic data was not used in this study. Due to a small number of cases; a dramatic increase in feature space would lead to overfitting and high error rates during predictive modeling. We hope to utilize genomic information in a revised version of the model with a larger case dataset. In the current study, we used data from one year of healthcare operations from a single tertiary care healthcare institution. The model should be tested using data from multiple sites and several data-years. Designing of harmonized phenotyping algorithms and data dictionaries leveraging various health information exchanges could help to gather a large number of samples and scale the study using large cohort.
6. Conclusions and Future Directions
A data-driven predictive model is developed to predict readmission rates in heart failure patients. Cases and controls were compiled based on 30-day readmission evidence to the same location. Compared to the existing repertoire of predictive models to assess readmission, our model shows better accuracy using one year of readmission data from a single site. However, the model needs to be updated and calibrated using multiple years of datasets from different sites across the nation. Feature selection provides insights into several novel factors that could help to delineate readmission rates associated with HF. Implementing data-driven methods that EMR-wide variables in a hypothesis-free approach could help us to find new features underlying clinical outcomes. Designing predictive and prescriptive models would help to accelerate stratification of patients at risk for improved care. Such findings and predictive assessments have significant implications for the quality of healthcare delivery and impact on patient outcomes. We envisage that our finding will improve the attempts to develop EMR-wide and scalable phenomics based predictive modeling to find critical events relevant to healthcare delivery and patient outcomes.
Figure 3.
ROC curves a) logistic regression models and b) composite model with 105 features
Acknowledgments
The authors would like to thank the members of the Mount Sinai Health System—Hospital Big Data initiative. This work was supported by a grant from the National Institutes of Health, National Center for Advancing Translational Sciences (NCATS), Clinical and Translational Science Awards (UL1TR001433-01) to KS and JTD.
References
- 1.Shameer K, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2014;133:95–109. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glicksberg BS, et al. An integrative pipeline for multi-modal discovery of disease relationships. Pac Symp Biocomput. 2015:407–418. [PMC free article] [PubMed] [Google Scholar]
- 3.Badgeley MA, et al. EHDViz: clinical dashboard development using open-source technologies. BMJ Open. 2016;6:e010579. doi: 10.1136/bmjopen-2015-010579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shameer K, et al. Translational bioinformatics in the era of real-time biomedical, health care and wellness data streams. Brief Bioinform. 2016 doi: 10.1093/bib/bbv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamad R, Modrek S, Kubo J, Goldstein BA, Cullen MR. Using “big data” to capture overall health status: properties and predictive value of a claims-based health risk score. PloS one. 2015;10:e0126054. doi: 10.1371/journal.pone.0126054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roski J, Bo-Linn GW, Andrews TA. Creating value in health care through big data: opportunities and policy implications. Health affairs. 2014;33:1115–1122. doi: 10.1377/hlthaff.2014.0147. [DOI] [PubMed] [Google Scholar]
- 7.Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 8.Karasik D. How pleiotropic genetics of the musculoskeletal system can inform genomics and phenomics of aging. Age (Dordr) 2011;33:49–62. doi: 10.1007/s11357-010-9159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thavendiranathan P, et al. Prediction of 30-day heart failure-specific readmission risk by echocardiographic parameters. Am J Cardiol. 2014;113:335–341. doi: 10.1016/j.amjcard.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 10.Padhukasahasram B, Reddy CK, Li Y, Lanfear DE. Joint impact of clinical and behavioral variables on the risk of unplanned readmission and death after a heart failure hospitalization. PloS one. 2015;10:e0129553. doi: 10.1371/journal.pone.0129553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kansagara D, et al. Risk prediction models for hospital readmission: a systematic review. JAMA : the journal of the American Medical Association. 2011;306:1688–1698. doi: 10.1001/jama.2011.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inouye S, et al. Predicting readmission of heart failure patients using automated follow-up calls. BMC medical informatics and decision making. 2015;15:22. doi: 10.1186/s12911-015-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adib-Hajbaghery M, Maghaminejad F, Abbasi A. The role of continuous care in reducing readmission for patients with heart failure. J Caring Sci. 2013;2:255–267. doi: 10.5681/jcs.2013.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourge RC, et al. Randomized controlled trial of an implantable continuous hemodynamic monitor in patients with advanced heart failure: the COMPASS-HF study. J Am Coll Cardiol. 2008;51:1073–1079. doi: 10.1016/j.jacc.2007.10.061. [DOI] [PubMed] [Google Scholar]
- 15.Whellan DJ, et al. Development of a method to risk stratify patients with heart failure for 30-day readmission using implantable device diagnostics. Am J Cardiol. 2013;111:79–84. doi: 10.1016/j.amjcard.2012.08.050. [DOI] [PubMed] [Google Scholar]
- 16.Hosseinzadeh A, Izadi M, Verma A, Precup D, Buckeridge D. Proceedings of the Twenty-Seventh AAAI Conference on Artificial Intelligence; Bellevue, Washington: AAAI Press; 2013. pp. 1532–1538. [Google Scholar]
- 17.Duggal R, Shukla S, Chandra S, Shukla B, Khatri SK. Predictive risk modelling for early hospital readmission of patients with diabetes in India. International Journal of Diabetes in Developing Countries. 2016:1–10. doi: 10.1007/s13410-016-0511-8. [DOI] [Google Scholar]
- 18.Gewehr JE, Szugat M, Zimmer R. BioWeka--extending the Weka framework for bioinformatics. Bioinformatics. 2007;23:651–653. doi: 10.1093/bioinformatics/btl671. [DOI] [PubMed] [Google Scholar]
- 19.Hall M, et al. The WEKA Data Mining Software: An Update. SIGKDD Explor Newsl. 2009;11:10–18. [Google Scholar]
- 20.Pyka M, Balz A, Jansen A, Krug A, Hullermeier E. A WEKA interface for fMRI data. Neuroinformatics. 2012;10:409–413. doi: 10.1007/s12021-012-9144-3. [DOI] [PubMed] [Google Scholar]
- 21.Smith TC, Frank E. Introducing Machine Learning Concepts with WEKA. Methods Mol Biol. 2016;1418:353–378. doi: 10.1007/978-1-4939-3578-9_17. [DOI] [PubMed] [Google Scholar]
- 22.Guyon I, Elisseeff André. An introduction to variable and feature selection. J Mach Learn Res. 2003;3:1157–1182. [Google Scholar]
- 23.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. Springer New York Inc; 2001. [Google Scholar]
- 24.Doughty RN, White HD. Carvedilol: use in chronic heart failure. Expert Rev Cardiovasc Ther. 2007;5:21–31. doi: 10.1586/14779072.5.1.21. [DOI] [PubMed] [Google Scholar]
- 25.Richardson LG. Psychosocial issues in patients with congestive heart failure. Prog Cardiovasc Nurs. 2003;18:19–27. doi: 10.1111/j.0889-7204.2003.00835.x. [DOI] [PubMed] [Google Scholar]
- 26.MacMahon KM, Lip GY. Psychological factors in heart failure: a review of the literature. Arch Intern Med. 2002;162:509–516. doi: 10.1001/archinte.162.5.509. [DOI] [PubMed] [Google Scholar]
- 27.Schweitzer RD, Head K, Dwyer JW. Psychological factors and treatment adherence behavior in patients with chronic heart failure. J Cardiovasc Nurs. 2007;22:76–83. doi: 10.1097/00005082-200701000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Ramasamy R, et al. Psychological and social factors that correlate with dyspnea in heart failure. Psychosomatics. 2006;47:430–434. doi: 10.1176/appi.psy.47.5.430. [DOI] [PubMed] [Google Scholar]
- 29.Iribarren C, et al. Glycemic control and heart failure among adult patients with diabetes. Circulation. 2001;103:2668–2673. doi: 10.1161/01.cir.103.22.2668. [DOI] [PubMed] [Google Scholar]
- 30.Futoma J, Morris J, Lucas J. A comparison of models for predicting early hospital readmissions. J Biomed Inform. 2015;56:229–238. doi: 10.1016/j.jbi.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 31.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 32.Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Miotto R, Li L, Kidd BA, Dudley JT. Deep Patient: An Unsupervised Representation to Predict the Future of Patients from the Electronic Health Records. Sci Rep. 2016;6:26094. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glicksberg Benjamin S, LL, Badgeley Marcus A, Shameer Khader, Kosoy Roman, Beckmann Noam D, Pho Nam, Hakenberg Jörg, Ma Meng, Ayers Kristin L, Hoffman Gabriel E, Dan Li Shuyu, Schadt Eric E, Patel Chirag J, Chen Rong, Dudley Joel T. Comparative Analyses of Population-scale Phenomic Data in Electronic Medical Records Reveal Race-specific Disease Networks. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw282. ISCB Special Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sengupta PP, et al. Cognitive Machine Learning Algorithm for Cardiac Imaging: A Pilot Study for Differentiating Constrictive Pericarditis From Restrictive Cardiomyopathy. Circ Cardiovasc Imaging. 2016;9 doi: 10.1161/CIRCIMAGING.115.004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Lee NJ, Glicksberg BS, Radbill BD, Dudley JT. Data-Driven Identification of Risk Factors of Patient Satisfaction at a Large Urban Academic Medical Center. PLoS One. 2016;11:e0156076. doi: 10.1371/journal.pone.0156076. [DOI] [PMC free article] [PubMed] [Google Scholar]