Summary
Most broadly neutralizing antibodies (BNAbs) elicited in response to HIV-1 infection are extraordinarily mutated. One goal of HIV-1 vaccine development is to induce antibodies that are similar to the most potent and broad BNAbs isolated from infected subjects. The most effective BNAbs have very high mutation frequencies, indicative of the long periods of continual activation necessary to acquire the BNAb phenotype through affinity maturation. Understanding the mutational patterns that define the maturation pathways in BNAb development is critical to vaccine design efforts to recapitulate through vaccination the successful routes to neutralization breadth and potency that have occurred in natural infection. Studying the mutational changes that occur during affinity maturation, however, requires accurate partitioning of sequence data into B-cell clones and identification of the starting point of a B-cell clonal lineage, the initial V(D)J rearrangement. Here, we describe the statistical framework we have used to perform these tasks. Through the recent advancement of these and similar computational methods, many HIV-1 ancestral antibodies have been inferred, synthesized and their structures determined. This has allowed, for the first time, the investigation of the structural mechanisms underlying the affinity maturation process in HIV-1 antibody development. Here we review what has been learned from this atomic-level structural characterization of affinity maturation in HIV-1 antibodies and the implications for vaccine design.
Keywords: HIV-1, broadly neutralizing antibodies, affinity maturation, computational immunology
Genetic Analysis of Affinity Maturation in HIV-1
Introduction
Antibodies are the defining elements of humoral immunity, serving simultaneously as the primary recognition molecules and key components of effector function. A monomeric antibody molecule comprises four polypeptide chains: two identical heavy chains and two identical light chains, and exhibits symmetry under a 180-degree rotation about one axis. The molecule has three functional domains: two identical antigen binding domains (Fab, “fragment antigen-binding”), and a single domain (Fc, “fragment crystallizable”) that binds to receptors on phagocytes and to other effector molecules of innate immunity. The antigen binding domain can be found in an enormous number of specific forms within any given individual person or mouse, and each has a unique antigenic specificity. The antibody repertoire—the collection of all such binding specificities within an individual organism, collectively provides protection against an essentially unlimited universe of antigens (1).
The diversity of antigen-binding domains is generated through an extraordinary system of intra-individual genetic recombination. Each heavy chain gene is constructed from the recombination of one each of variable (IGHV), diversity (IGHD), and joining (IGHJ) genes. The light chains, kappa and lambda, do not use diversity gene segments. The stochastic choice of these germline genes constitutes antibody combinatorial diversity. In addition to combinatorial diversity, immunoglobulin variable region genes (IgVRG) exhibit junctional diversity. The recombination sites are stochastically chosen and sequences of non-templated nucleotides, of variable length and content, may be added between the gene segments.
Once the nascent B cell has completed the recombination of its immunoglobulin genes, the proteins they encode become the central component of the B-cell receptor (BCR). Following development in the bone marrow, it circulates within the periphery awaiting ligation of its receptor by an as-yet-unknown antigen. Upon activation, it divides several times and its progeny differentiate into several different cell types, including short-lived plasma cells and germinal center cells. The collected progeny of a single B cell is referred to as a clone. Germinal center cells undergo affinity maturation, eventually becoming the memory cells and long-lived plasmacytes that provide protection against subsequent encounters with the pathogen from which the antigen was derived (2).
Affinity Maturation
The germinal center (GC) is a transient anatomical entity formed in the secondary lymphoid tissues (3). It is divided into two distinct zones, populated by distinct cell types. The dark zone is populated by rapidly dividing centroblasts. Dividing centroblasts upregulate activation-induced cytidine deaminase (AID) which induces DNA lesions. These lesions are subsequently repaired, becoming nucleotide substitutions, insertions, and deletions. After several generations of error-prone cell division, centroblasts differentiate into centrocytes, and migrate from the dark zone into the light zone. In the light zone, centrocytes interact with follicular dendritic cells, from which they acquire stored antigen, and T follicular helper cells (TFH), with which they interact via MHC-peptide complexes and co-stimulatory molecules. Those centrocytes that have, by chance, acquired altered B cell receptors that allow them to more effectively receive help from TFH gain reproductive advantage. As a result, the affinity for the eliciting antigen of the antibodies produced by the responding B cell populations increases dramatically over the two weeks of the GC reaction.
In ongoing or repeated exposures, B cells may be driven through the GC multiple times. It is through such sequential rounds of affinity maturation that broadly-neutralizing antibodies against HIV-1 are produced.
Sequencing technology
Recent developments in DNA sequencing technology have greatly accelerated the pace of research into B cell population dynamics. High-throughput sequencing, together with innovations in the preparation of genomic DNA and mRNA libraries, has made it possible to examine IgVRG repertoires—the sequences of millions of B-cell receptor genes simultaneously (4). By sampling over time, we are able to characterize the shifts in clonal complexion and affinity maturation of the humoral response to vaccines and infections. This ability to capture very broad B cell cross-sections is complemented by the ability to isolate paired heavy-chain/light-chain genes from individual B cells and to synthesize the encoded antibody recombinantly (5). This process enables the study of antibodies phenotypically, including the kinetic constants and equilibrium binding constants of their antigen-binding reactions. This technology also allows for relatively inexpensive synthesis of designed antibodies, including those inferred to have been ancestral to the sequences of an isolated clone (6). By judiciously combining the two processes, unprecedented insight into the B cell response can be gained. But as is typical with breakthrough technologies, the full promise of the new methods depends on the development of new statistical tools for the analysis of the resulting data.
Statistical Analysis
The immediate aims of statistical analysis of IgVRG repertoire sequencing are three-fold. 1. To infer the original, unmutated ancestor for a given mature IgVRG or IgVRG clone. 2. To determine whether two IgVRG are clonally related. 3. To partition a set of IgVRG genes into clones. These tasks are related to each other, and each is inherently statistical, in that each involves the careful accounting of uncertainty.
The uncertainty arises because of the combination of the stochasticity of the initial recombination and of the somatic mutations acquired during maturation. Indeed, because somatic hypermutation can change any nucleotide to any other nucleotide and can produce arbitrary insertions and deletions, it is possible for any unmutated ancestor to give rise to any mature antibody. Some pathways are much more likely than others, but none are completely ruled out. By definition, then, the solutions involve estimating the relevant probabilities and drawing conclusions from them as required.
Modeling approaches
Generally, there are two approaches that can be used for the analysis of IgVRG sequences: discriminative modeling and generative modeling. In generative modeling, the goal is to produce a model that is in principle capable of generating data indistinguishable from that to be analyzed. One utilizes all of the information available regarding the processes giving rise to the data and incorporates it into the model. A generative model represents an attempt to represent the biology underlying the data accurately and conduct inferences based on this understanding. A discriminative model, on the other hand, seeks to identify those features of the data that will prove informative in a given analytical task, without reference to the underlying mechanism of their generation. If the generative model is sufficiently accurate, it will provide the more effective inferential tool. But model misspecification can cause bias and lead to erroneous inferences. A discriminative model is less susceptible to misspecification, but requires substantial labeled data to serve as training sets for learning. Such datasets, however, are not available for the analyses of interest in the present context. In what follows, therefore, we focus exclusively on generative models and their applications.
Models for VDJ recombination and affinity maturation
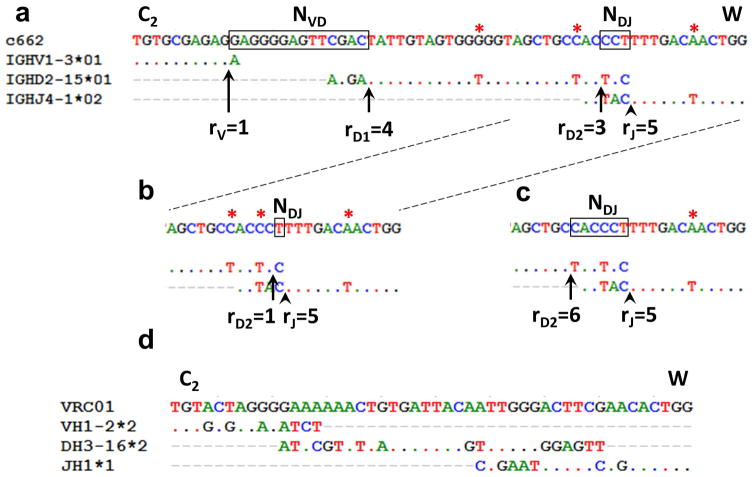
Our statistical model for IgVRG repertoire analysis comprises two sub-models: one for VDJ rearrangements and the other for affinity maturation (7). As described above, the complete rearrangement of a heavy-chain IgVRG is specified completely by the three categorical variables V, D, J naming the gene segments from among a known set of alternatives, the four integer variables RV, RD1, RD2, and RJ (the recombination points), and two short DNA sequences NVD and NDJ (the non-templated nucleotides). Negative recombination points correspond to the use of p-nucleotides. For example, RV = −1 indicates the presence of a single p-nucleotide at the 3′ end of the V gene. Figure 1 illustrates the definitions of recombination points and their relationship to N nucleotide regions, as well as the possible uncertainty inherent in estimating them.
Figure 1. Estimation of recombination parameters.
a) A mature heavy chain VRG (c662) is shown aligned against the maximum likelihood V, D, and J gene segments, with the maximum likelihood recombination points (rV, rD1, rD2, rJ) shown along with the N regions. Dots indicate nucleotide identity. C2 and W indicate the codons for the invariant second cysteine and tryptophan, which mark the outside boundaries of CDR3. b) and c) show alternative classifications for the 3′ end with slightly lower likelihoods. d) Alignment of heavy chain CDR3 of VRC01 against its maximum-likelihood gene segments.
The second component is a model for affinity maturation. We use nucleotide substitution models borrowed from statistical phylogenetics (8), and assume, as is done, that the positions in the gene evolve independently and that selection may be neglected. One shortcoming of the available models is that they do not account for the sequence specificity of AID which is quite pronounced and involves overlapping sequence neighborhoods or motifs. Not only does the mutation rate vary by position in an IgVRG, the mutation rate at a given position changes as the gene evolves. Models respecting this feature of somatic hypermutation are theoretically straightforward to write down, but require wholly impractical computational resources to use. To balance the need for computationally tractable methods of analyzing up to millions of reads with the need for adequate realism, we use a position-dependent mutation frequency and Kimura nucleotide evolution model. We believe that even such a relatively simple model can be used effectively.
Insertions and deletions also occur under somatic hypermutation and play key roles in the affinity maturation of some antibodies of interest, such as many of the broadly neutralizing antibodies against HIV-1 (9). Zero-inflated negative-binomial models for insertions and for deletions are consistent with the available data (10) and are widely used in pairwise alignment because of their computational tractability. These models are used in the current implementation of our system.
Inference of the unmutated ancestor
For any hypothesized ancestral sequence, a, and any mature IgVRG, s. The likelihood that a is the ancestral form of s requires that a is a valid unmutated recombination product and that s arose from a. The likelihood is then the product of the probability that a randomly observed recombination event produces a, and that a randomly selected maturation pathway from a gives rise to s.
With this apparatus in hand, we are able to infer the unmutated ancestor UA in several ways using conventional statistical methods. We can find the maximum-likelihood UA as that single sequence that corresponds to the largest likelihood among all sequences. In order to capture information about the uncertainty in our estimate, we may use Bayesian methods to sum over all potential UAs, weighted by their likelihoods under the model. With this procedure we may compute the probability over nucleotides at each position independently (the marginal probabilities) in the UA or compute the probabilities of individual amino acid sequences. The former computation reveals the uncertainty at individual positions directly, but discards information about correlations among positions. If, for example, there are two IGHV alleles differing in two positions that have the same likelihood value, the marginal probabilities at each of the two divergent positions would assign a value of ½ to each of the two alternatives. But they would not indicate that resolving the ambiguity at one position would at the same time resolve the ambiguity at the other. The latter computation preserves the correlations at the expense of a direct measure of nucleotide uncertainty.
The procedure for inferring the unmutated common ancestor for an aligned set of sequences, taken to be clonally related, is directly analogous. The difference is that the affinity maturation component of the model is now based on a complete phylogenetic tree. Additional approximations are invoked to preserve computational tractability in this case. In particular, we compute a single maximum-likelihood tree based on the maximum-likelihood UCA and keep that tree fixed while performing the averages over ancestors (we would ideally sum over trees as well as over ancestors). Work by others suggests that such approximations should have relatively little effect on the inferred UCA under typical circumstances (11).
Kinship analysis
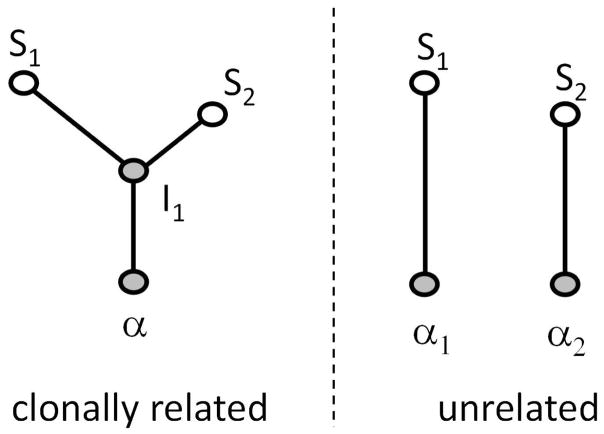
Similar methods are used to assess the kinship of two IgVRGs, s1 and s2. If they are clonally related, then by definition there is some ancestral sequence, a, that was generated by VDJ recombination and gave rise through somatic hypermutation to both s1 and s2. To evaluate the evidence in favor of clonal kinship, we compare two simple phylogenetic models (Fig. 2). In each case there are two unknown genes. In the former case, where a trefoil—a tree with two leaves, a root and an interior node—defines the relationship between the observed genes, both the ancestral sequence (the root) and the most recent common ancestor (the interior node) are unknown. In the latter case, where the simple descent from independently rearranged ancestors is assumed, both ancestral sequences are unknown. In both cases, we perform a Bayesian average over the unknown sequences. The weight of evidence in favor of clonal kinship is the ratio of the averaged likelihood under the kinship model to that under the independence model.
Figure 2. Two simple phylogenetic models for determining clonal kinship of two sequences.
S1 and S2. On the left, the sequences are related through their most recent common ancestor I1, and the founding ancestor a. On the left, each sequence arises by affinity maturation from independent founding ancestors.
As with UCA inference, this procedure may be applied to sets of aligned sequences. This generalization forms the basis for clonal partitioning.
Clonal Partitioning
Given an IgVRG repertoire dataset, it is desirable, and perhaps essential, to partition the sequence set into clones. Such a partitioning is crucial for estimating recombination probabilities (there is one recombination event per clone, not per sequence) and for understanding patterns of clonal initiation, growth, and decline.
Our approach to the problem is based again on our likelihood model. We described above how one computes the likelihood for any given sequence set treated as a clone. The likelihood for a set of clones is the product of the likelihoods for each individual clone. The goal of our approach is to maximize the likelihood over all assignments of sequences to clones. This is conceptually straightforward and statistically appealing, but there are computational challenges.
The number of ways of assigning even a modest number of sequences (for IgVRG repertoire sequencing) into clones is astronomical. Fortunately, the vast majority of such assignments can be ruled out without performing the very expensive computations needed to compute the maximum-likelihood phylogenetic trees. This is because we are usually able to decide without such computations whether two IgVRG could share the same V and J genes. This of course, is a necessary condition for clonal relatedness. It is still prohibitive for typical data sets to evaluate all but a very small number of assignments. This situation is encountered in cluster analysis more generally; we have adopted a method taken from this field of statistics: progressive optimization.
We take each sequence one at a time, keeping track of the clonal sets and assignments making up the partial partition as we proceed. For each sequence, we test its kinship with each of the extant clones for which membership is plausible by V and J gene usage. If the evidence for kinship in one of these clones is positive, we place it into the clonal set for which the evidence is strongest. If there is no evidence for membership in any clonal set, the sequence becomes the first member of a new clonal set.
Like other clustering procedures, such as k-means clustering, and like maximum-likelihood phylogenetic tree inference, this procedure does not guarantee that the final configuration will have maximum likelihood among all configurations. But also like these methods, it seems to do well in practice.
The software suite that implements these procedures is called Cloanalyst (http://www.bu.edu/computationalimmunology/research/software/). We will refer to these procedures using this name as we turn now to the analysis of the protein-structural correlates of affinity maturation.
Structural Analysis of HIV-1 Affinity Maturation
Somatic hypermutation generates mutations in the B-cell receptor (BCR) that are either synonymous (do not result in amino acid substitution) or non-synonymous (result in amino acid substitution). Only non-synonymous mutations can affect affinity maturation as the amino acid substitutions they produce directly lead to structural changes in the BCR. Structural changes that alter the antigen-binding site of the BCR can in turn lead to improvement of BCR binding affinity to the antigen. BCR binding affinity for antigen is the criteria for selection in affinity maturation and the structural alteration of the antigen binding site is the source of variation in this evolutionary process.
Binding affinity for antigen can be improved by individually or in combination increasing the strength of any of the key components of protein-protein recognition. These components can be classified thermodynamically into two groups: enthalpic or entropic. There are several routes in which amino acid substitutions resulting from somatic hypermutation can lead to more favorable enthalpic contributions across the interface of the BCR-antigen complex. These routes to improving binding include increasing shape complementarity, creating more attractive electrostatic interactions, adding hydrogen bonds and increasing the burial of hydrophobic surface across the interface. More favorable binding can also be achieved by decreasing the entropic penalty accrued during BCR-antigen complex formation. Entropy is the tendency of a system towards disorder and according to Gibbs law, the free energy of binding increases when entropy is increased. As two proteins come together to form a complex, the packing of protein-protein interfaces inherently introduces order and results in a loss of entropy. In protein binding, this unfavorable loss of entropy is typically overcome by an increase in enthalpy using the components described above. Proteins that are more flexible prior to complex formation accrue a greater entropic penalty upon binding. Thus, mutations that lead to the rigidification or stabilization of the antigen-binding site prior to binding result in a reduction of the entropic penalty and increase BCR-antigen binding affinity.
Understanding how the structure of BCR antigen-binding sites or their antibody counterpart, paratopes, evolve during affinity maturation from germline rearrangements to mutated, mature forms in the context of HIV-1 is essential for gaining insight into the process that can yield broadly neutralizing antibodies (BNAbs), the target of HIV-1 vaccine development. As we shall discuss, atomic level characterization of affinity maturation of antibodies in HIV-1 infection has profound implications for vaccine design strategies that aim to effectively and efficiently recapitulate the elicitation of BNAbs in the vaccine setting. Until recently very little was known about the effects of affinity maturation on antibody structure in the context of recognition of a complex, evolving antigen such as the HIV-1 envelope protein (Env). The recent development of the statistical framework to accurately determine clonal relatedness and infer the unmutated and intermediate ancestors of B-cell clonal lineages (6, 7) along with the development of software that implements these computational methods has allowed for the synthesis of these ancestral antibodies and their structural characterization (Table 1). We now turn to reviewing what is known about the effects affinity maturation has on the structure of HIV-1 antibodies.
Table 1.
Inferred Ancestral Antibodies with Structures Determined by X-ray Crystallography
| Antibody | Antigen | Ancestor Typea | PDB Code | Ref |
|---|---|---|---|---|
| PGT121.GL | Unliganded | GL | 4FQQ | (60) |
| NIH45–46.GL | 93TH057 gp120 | GL | 4JDT | (98) |
| NIH45–46.GL | Unliganded | GL | 4JDV | (98) |
| VRC01 GL | gp120 eOD-GT6 | GL | 4JPK | (99) |
| VRC01 GL | Unliganded | GL | 4JPI | (99) |
| PGT121 | Unliganded | GL | 4NPY | (85) |
| VRC26.UCA | Unliganded | GL | 4ODH | (21) |
| CH103 UCA | Unliganded | UCA | 4QHK | (31) |
| CH103 I3.1 | Unliganded | INT | 4QHM | (31) |
| CH103 I2 | Unliganded | INT | 4QHN | (31) |
| CH103 I3.2 | Unliganded | INT | 4QHL | (31) |
| CH59.UA | Unliganded | UCA | 4QF1 | (47) |
| CH58.UA | Unliganded | UCA | 4RIR | (35) |
| CH58.UA | V2 peptide | UCA | 4RIS | (35) |
| PGT121 3H+109L | BG505 SOSIP Trimer | INT | 5CEZ | (58) |
| PGT121 9H+3L | Unliganded | INT | 5CEY | (58) |
| PGT121 32H+109L | Unliganded | INT | 5CEX | (58) |
| NIH45–46.GL | gp120 426c.TM4dV1-3 | GL | 5IGX | (46) |
| 3BNC60.GL | Unliganded | GL | 5F7E | (46) |
| 3BNC60.GL | gp120 426c.TM4dV1-3 | GL | 5FEC | (46) |
| 10E8 gHV-gLV | Unliganded | GL | 5JO5 | (100) |
| VRC03 gHVgLV | Unliganded | GL | 5JOF | (48) |
Unmutated Common Ancestor (UCA) defined as containing an inferred unmutated CDR3; Germline Reverted (GL) defined as containing a mature (mutated) CDR3; Ancestral Intermediate (INT)
Mapping affinity maturation mutations onto BNAb complexes
A primary goal in HIV-1 vaccine development is the elicitation of antibodies that can neutralize diverse strains of the virus, so-called BNAbs. The kind of antibodies necessary for providing lasting sterilizing HIV-1 immunity, BNAbs with high potency and exquisite breadth (>90% breadth against Group M viral isolates) are rarely elicited in chronically infected individuals (12). The isolation and structural characterization of such BNAbs over the past decade(13–17) therefore represented a breakthrough and provided the HIV-1 vaccine development field with target cases to attempt to recapitulate by vaccination. HIV-1 BNAbs typically possess rare characteristics that contribute to their infrequency(14, 18) including high mutation frequency in their variable domains. High mutation frequency indicates a sustained, and likely long, affinity maturation process which is consistent with the length of time it takes after infection for BNAbs to typically be elicited(19–23). The accumulation of this tremendous number of mutations can have a profound effect on the structure of the antibody paratope and often is indicative of the great lengths clonal lineages must go to recognize a diverse collection of Env proteins presented by a constantly evolving virus.
One of the most well-characterized BNAbs, VRC01, has a remarkably high mutation frequency of about 30% in its heavy chain variable domain(24). Structural characterization of VRC01 in complex with gp120 showed that it recognizes Env at the CD4-binding site by mimicking CD4(24). Strikingly, two additional BNAbs, deriving from the same germline heavy chain gene segment, VH1-2*02, were then discovered with nearly an identical mode of recognition from separate subjects(25). This remarkable observation of functional convergence of antibody recognition across multiple individuals suggested extraordinary selection pressure for this mode of binding. Wu et al. used maximum likelihood phylogenetic methods to infer the ancestral antibody intermediates of two of these VRC01-like antibodies and mapped the accumulation of mutations from the inferred lineage onto a structural model of the germline-reverted (GL) antibody of VRC01(25). They demonstrated that the two lineages had a similar pattern of the types of mutations acquired during affinity maturation.
There are two distinct methods employed in the HIV-1 vaccine field to infer and generate ancestral antibodies. Here we will refer to antibodies that have had their V and J gene segment regions reverted to the associated germline sequence as “germline-reverted (GL)”. The difference between a GL or UCA is that a GL contains the mature (mutated) CDR3 whereas a UCA contains an inferred unmutated CDR3.
We applied a similar approach to Wu et al. in mapping affinity maturation mutations onto the structure of a BNAb complex in the first-ever study investigating virus-antibody co-evolution in an HIV-1 subject from the time of infection to the development of BNAbs(20). We isolated a CD4 binding site targeting BNAb clonal lineage and determined the structure of clonal member, CH103, in complex with the gp120 outer domain. We then used Cloanalyst to confirm clonal relatedness of multiple observed members of the CH103 clone, reconstruct the clonal history and infer a UCA and ancestral intermediates. Mutations from each node of the clonal lineage tree were then mapped onto structural models of antibody ancestral intermediates. Early mutations in the CH103 accumulated in or near gp120 contact positions with later mutations occurring towards the periphery of the paratope and with some occurring outside of it. Doria-Rose et al. similarly followed the development of a V1V2-glycan binding BNAb clonal lineage, CAP256-VRC26, in a super-infected HIV-1 subject and determined the structure of the GL and 6 mutated clonal members isolated at early and late timepoints(21). Two critical cysteine mutations in the CDR H3 were acquired in clonal members that arose late in maturation and the antibody structures revealed that these residues formed a disulfide bond which acted to stabilize a long, protruding CDR H3. Reversion of this disulfide bond resulted in a loss of neutralization breadth and demonstrated how maturation of the clone led to acquisition of heterologous breadth by gaining a critical structural property via affinity maturation (21).
Conformational Change in anti-HIV-1 Antibodies During Affinity Maturation
Activation-induced cytidine deaminase (AID) is the enzyme responsible for introducing mutations in B-cell receptor sequences during affinity maturation(26). AID preferentially targets sequence motifs (called “hot spots”) that are encoded predominantly in the CDRs (27). Because antigen recognition is typically mediated by the CDR loops, it is thought that enrichment for hot spots located in the CDRs allows the recognition component of the antibody to vary(28) while reducing the potential for deleterious mutations to be acquired in the framework of the antibody which acts as a scaffold holding the CDR loops in place(29). Framework mutations are more likely to destabilize the overall structure of the antibody and thus reduce clonal fitness(30). Nevertheless, mutations can occur outside of the CDRs and their role in affinity maturation has been explored in HIV-1 BNAb development.
In our structural analysis of the CH103-gp120 complex we found that mutations occurred not only in points of contact with gp120 but also outside the paratope as well (20). To address the potential that other factors might have driven affinity maturation that were not apparent from the structure of the complex, our collaborators determined the crystal structures of the unbound Fabs of the CH103 UCA and two ancestral intermediates that were inferred using Cloanalyst (31). Superposition of the UCA Fab structure onto the CH103-gp120 complex revealed a modification of the orientation of the heavy-chain and light-chain variable domains (VH and VL, respectively). This conformational change appeared to alleviate steric clashes that could occur in viral Envs with longer V5 loops. Viral sequencing of the infected subject in which CH103 was isolated showed that the V5 loop, a critical component of the CH103 epitope, indeed grew progressively longer by insertions that started during the first year of infection(20, 32). We altered the V5 length in several gp120 constructs and demonstrated that the UCA binds preferentially to shorter V5 loops, which was consistent with the hypothesis that the VH:VL orientation shifted during affinity maturation to accommodate V5 loop insertions(31). We further showed that the conformational change was correlated with mutations at the VH:VL interface that were in the framework region of the antibody. Thus, mutations outside of the CDRs were associated with a substantial conformational change during affinity maturation that was critical to the developmental pathway of the HIV-1 BNAb. Indeed, the structural insights on framework mutations yielded by the CH103 structural analysis underscore the findings that framework mutations in many BNAbs can play an essential role in antigen binding either through allosteric effects or by direct contacts (33, 34). Thus, framework mutations are not just coincidentally acquired along with CDR mutations and merely a product of the length of time required to induce BNAbs. Rather, mutations in the framework regions are often critical waypoints along antibody maturation pathways.
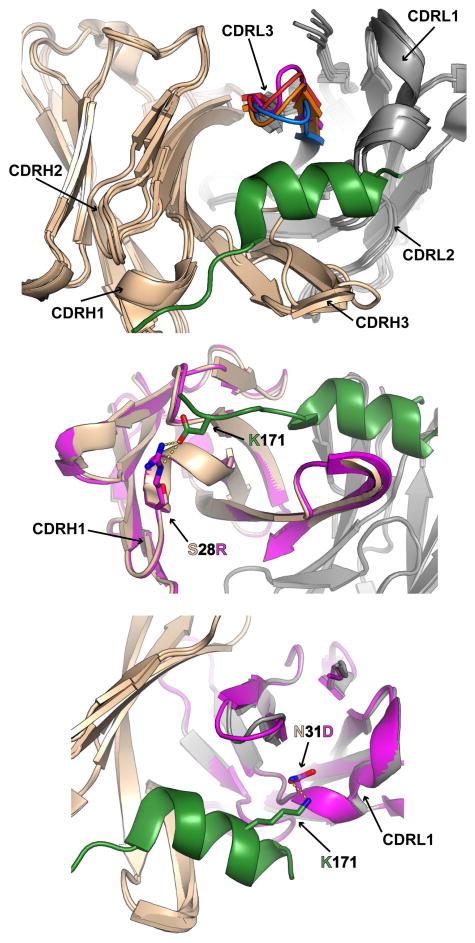
Structural changes that occur during the antibody maturation process can lead to improved binding by providing more favorable enthalpic or entropic contributions as we described above. By performing a comprehensive structural analysis, we investigated how enthalpy and entropy changes during antibody maturation can work in concert to increase affinity in an HIV-1 antibody, CH58 (35). CH58 is a strain-specific neutralizing antibody isolated from the RV144 HIV-1 vaccine trial (36), a vaccine that was shown to have moderate efficacy (37). Analysis of the vaccine trial showed that V2 antibodies were an inverse correlate of transmission risk (38) and that a specific residue in the V2, lysine at position K169, was a site of immune pressure(39). CH58 is a V2-binding antibody that targets K169 and thus has been an important antibody for studying the RV144 vaccine-induced immune response (36). We determined the crystal structures of the unbound CH58 UA as well as the CH58 UA in complex with a V2 peptide (35). We then compared these to previously determined structures of the unbound, mature CH58 and mature CH58 in complex with a V2 peptide (36). CH58 possessed a low frequency of mutations (~2%). Despite the lack of accumulated mutations, we observed a large change in binding affinity (~2000 fold) from UA to mature CH58. Our structural comparison of the various states of CH58 revealed that two primary mechanisms were responsible for the substantial increase in binding strength during affinity maturation of CH58 (Fig 3). First, mutations led to more favorable ethalpic contributions. CH58 acquired two mutations that led to the formation of two additional salt bridges with the V2 peptide. Salt bridges are the strongest non-covalent paired amino acid interactions possible across a protein-protein interface(40) and thus salt bridge formation provides the largest enthalpic cost-effectiveness of any type of affinity maturation mutation. Second, mutations led to more favorable entropic contributions. We observed that the CDR L3 conformation changed between the unbound and bound state for CH58 UA suggesting flexibility of the CDR L3 in the unmutated antibody. There was no difference in the CDR L3 conformation observed between the mature CH58 in the unbound and bound state. Thus, accumulation of mutations in the light chain during affinity maturation resulted in the rigidification of the CDR L3, a critical component of the CH58 paratope. As discussed previously, such rigidification of loops in the unbound conformation reduces the entropic penalty incurred upon complex formation and increases binding affinity. Early work on affinity maturation led to a basic structural model in which unmutated and early ancestors utilize an induced-fit mode of binding in order to retain polyspecificity and bind multiple antigens with low affinity (41). Later as the clone matures, specificity is gained by the rigidification of the paratope that adopts a fixed shape complementary to the antigen and binding can then occur with high specificity through a lock-and-key mode of recognition (42–44). Indeed, we have observed such rigidification of the paratope, particuraly in the CDR H3, during affinity maturation in influenza BNAbs (45). However, HIV-1 antibodies do not appear to consistently follow this maturation route. Sharf et al. (46) found that the binding sites of VRC01-class of antibodies are largely pre-formed, an observation that was in agreement with our conclusion that an essential recognition motif in the strain-specific RV144 vaccine-induced antibody was pre-configured in the structure of the unmutated ancestor (47). Recent work characterizing HIV-1 BNAb structural dynamics during maturation revealed that rigidification mainly occurred in the periphery but not the core of the paratope (48). Together these structural data support a BNAb maturation model in which the initial rearrangement event sets an anchor point within the core of the paratope that makes contacts with conserved Env residues. The size of this core epitope however is likely to be too small for even optimal mutations within it to improve affinity enough to outcompete other strain-specific antibody responses. Thus, mutations on the periphery of the paratope are needed to accommodate the Env variation in the corresponding region of the epitope. This then increases the size of the interface which may be a necessity to improve binding affinity. Nevertheless, given the tremendous ability of HIV-1 to escape the grasp of the antibody response as evidenced by the unusual features shared among BNAbs, it is likely that following conventional and common routes of affinity maturation is not sufficient for the development of BNAbs. Maturation routes that instead employ unconventional mechanisms and follow pathways that utilize large jumps in evolutionary distance, such as we will see with the acquisition of insertions and deletions, may be required for clones to match the pace of the rapidly evolving virus.
Figure 3. Dual Mechanisms for Improving Affinity During Maturation of an HIV-1 Antibody.
Antibodies mature by improving their affinity either by increasing enthalpy by adding favorable interactions at the antibody-antigen interface or by decreasing the entropic penalty paid upon complexation. The HIV-1 antibody CH58 (heavy chains colored beige and light chains colored gray unless otherwise noted) utilized both mechanisms in order to increase affinity to a V2 peptide (green). A) The CDR L3 in the unliganded CH58 UA structure (blue) adopts a different conformation (magenta) in the CH58 UA/V2 peptide complex suggesting that the paratope is flexible prior to maturation. The CDR L3 in the unliganded matured CH58 structure (orange) adopts a highly similar conformation as the CDR L3 in the matured CH58/V2 peptide complex structure (red) demonstrating that after maturation the CDR L3 is no longer flexible. Thus, the rigidification of the CDR L3 loop results in a decrease in the entropy penalty and, in turn, an increase in affinity. B) The maturation mutation S28R (shown in stick representation) in the CDR H1 of the matured CH58 heavy chain (magenta) resulted in the gain of a salt bridge to D180 in the V2 peptide. C) An additional salt bridge to K171 (sticks) in the V2 peptide was formed during maturation upon the acquisition of the mutation N31D (sticks) in the CDR L1 of the matured CH58 light chain (magenta). These two additional salt bridges in the matured CH58 antibody greatly contribute to favorable enthalpic interactions and thus increase affinity. Salt bridges are shown as yellow dashed lines. PDB codes: unliganded CH58 UA (4RIR), CH58 UA/V2 peptide complex (4RIS), unliganded matured CH58 (4HQQ); matured CH58/V2 peptide complex (4HPO).
Angle of recognition of HIV-1 BNAbs remains largely unchanged during affinity maturation
The epitopes of all HIV-1 BNAbs isolated to date can be clustered into five groups (49, 50). These five regions of vulnerability on the surface of the Env are the V1V2 loop and associated glycans at the apex of the Env trimer (V1V2-glycan), the high-mannose glycan patch near the base of the V3 loop (glycan-V3), the CD4 binding site (CD4bs), the gp120-gp41 interface including the fusion peptide site, and the membrane proximal external region in gp41 at the foot of the Env trimer near the viral membrane (MPER). These sites of vulnerability are distinguished by their high sequence conservation, however due to several immune evasion mechanisms, mainly N-linked glycosylation and insertions and deletions in the variable loops, they are highly concealed (51, 52). Structural characterization of HIV-1 antibodies has shown that there are very limited sets of angles of approach in which the Fab portion of antibodies can optimally interact with these epitopes without coming into direct contact with glycans or clashing with variable loops (53–55). One of the key structural components for breadth of neutralization is the utilization of recognition angles that limit antibody contact with the highly variable terminal portions of glycans and/or allow for accommodation of variable loop lengthening (49).
A fundamental question in HIV-1 antibody maturation is whether clonal members can sample multiple recognition angles, thereby allowing the clone to “learn” to approach the epitope in an optimal orientation. We addressed this question in our study of the maturation of a CD4bs-targeting BNAb lineage, CH235 from the same infected subject from which CH103 was isolated (56). Unlike CH103, the CH235 clone interacts with the CD4bs by mimicking the CD4 interaction with gp120. However, because Fab portions of antibodies are larger than the CD4 molecule, there is more potential for antibodies to sterically clash with glycans surrounding the CD4bs and the V5 loop, as well as with the neighboring subunits in the Env trimer. Thus even when antibodies go the extraordinary length of acquiring characteristics to mimic CD4, adopting an optimal angle of recognition is still critical for breadth of neutralization (53). Our collaborators determined the structures of CH235, and two clonal members CH235.9 and CH235.12, isolated 2 and 5 years after CH235 arose, respectively (56). While these members more than doubled (CH235.9) or tripled (CH235.12) the accumulated number of mutations as they matured, their recognition angles were virtually identical. These results suggested that orientation of the antibody was set early in the lineage development, perhaps as early as the initial rearrangement, and that it remained largely unchanged during affinity maturation. The conservation of the angle of approach in CH235 was consistent with the remarkable observation that epitope recognition of another CD4-mimicking BNAb, VRC01, was virtually unchanged after 13 years of maturation of the clone (57).
A structural study of the glycan-V3 epitope targeting PGT121 BNAb clone demonstrated that although the overall angle of approach was not altered substantially during maturation of the lineage, members of one branch did rotate approximately 10 degrees to avoid a critical glycan (58). The glycan-V3 supersite is thought to be less sterically restricted than the CD4 binding site (59) and glycan-V3 BNAbs use a diverse set of VH/VL gene segments (60–63) suggesting multiple structural solutions are available in the repertoire for recognizing this epitope. In contrast, the CD4 binding site mimicking BNAbs such as CH235, which have been isolated from multiple subjects, derive from just two VH gene segments and employ virtually identical angles of approach. While it is difficult to generalize based on the characterization of just two epitopes, it is likely that immunogenetic restriction is related to the angle of approach restriction for HIV-1 BNAb epitope recognition. For CD4 mimicking BNAbs, they must approach at a highly restricted angle in order to faithfully recapitulate the CD4 interaction with gp120. Therefore, this angle must be set initially by the UCA and strictly maintained throughout maturation. For the glycan-V3 supersite, and perhaps other epitopes where multiple angles of recognition are possible, a multitude of structural solutions exist and therefore genetic restriction is lower. Indeed, in a recent study looking at a large longitudinal HIV-1 infection cohort, the glycan-V3 site was shown to be the most commonly targeted BNAb epitope (64). By contrast, the CD4bs epitope was only targeted in a few individuals (64). The lower genetic restriction and prevalence of the glycan-V3 response in favor over the CD4bs targeting response suggests there is more room for antibodies to maneuver within the glycan-V3 site. Correspondingly, there may be more of an allowance for the angle of approach to be fine-tuned during affinity maturation. However, it is likely that the angle of approach cannot change without significant selection pressure being exerted by the virus. Even small moves in angular space will result in large structural movement within the initial epitope. The potential consequence of which would be the disruption of many of the interface contacts that were selected for during the initial engagement by the unmutated ancestor and optimized during affinity maturation.
Structural ramifications of immune tolerance
HIV-1 BNAbs to the gp41 MPER site of vulnerability have been shown to be autoreactive (65–67). These BNAbs, including 2F5 and 4E10, have a highly hydrophobic CDR H3 that is used to tether the antibody to the nearby viral membrane prior to docking to the MPER (68, 69). Knock-in mice studies have shown that the genes for 2F5 and 4E10 can be expressed, but B cell development is limited by immune tolerance mechanisms (70, 71). To determine whether rhesus macaques experienced similar blockades or whether they could overcome these tolerance controls by repetitive boosting, we immunized rhesus macaques repeatedly with an MPER-liposome immunogen designed to engage the 2F5 UA with high affinity (72). We found that this regimen could break tolerance to an autoantigen that shares linear sequence with the 2F5 epitope in the MPER. We characterized one clonal lineage, DH570, isolated from a monkey that included some members that demonstrated breadth of neutralization in an assay that is highly sensitive for MPER antibodies as it provides a kinetic advantage to access the transiently exposed MPER epitope. To shed further light on the affinity maturation of the DH570 clone, the we determined the crystal structures of three mature members, with one member solved in both the unbound state and in complex with an MPER peptide. The complex structure revealed that the MPER peptide bound at the base of a protruding, hydrophobic CDR H3. This mode of recognition was shared by 2F5 suggesting that the hydrophobic apex of the DH570 CDR H3 could play a similar role in lipid binding as 2F5 (69). The three members shared identical CDR H3 sequences and virtually superimposable CDR H3 conformations, but one member lacked neutralization capacity suggesting that mutations outside the CDR H3 played a role in neutralization (72). We computed lipid insertion propensity scores based on the hydrophobicity of the linear amino acid sequences of the clonal members. Hydrophobicity of CDRs was shown in the past to be correlated with neutralization capacity in 2F5 (73). We found that the lipid insertion propensity scores of the CDR H3 and CDR L3 were significantly more favorable for neutralizing members of the DH570 clone (72). We then mapped the kinetics of the clone and observed that the best neutralizing branch of the clone followed a pattern of increased association rate upon descent from the UA but peaked at an ancestral intermediate node and reverted upon further maturation. This abrupt change in affinity was correlated with a similar decrease in the lipid propensity score of these antibodies. We hypothesized that there was a kinetic rate ceiling on the affinity maturation likely mediated by tolerance mechanisms working against further increases in antibody hydrophobicity, a known characteristic of auto-antibodies.
We sought to prove that outside the context of tolerance constraints, further increasing hydrophobicity would improve the neutralization capacity of the clone. We used two natural mutations observed in the CDR H3 of the ancestral intermediate that peaked in association rate and made these substitutions in a member with a poor association rate but a more favorable dissociation rate. This mutant antibody was able to neutralize a heterologous virus in the less sensitive assay. Therefore, with just two amino acid substitutions that were observed as part of a natural maturation pathway elsewhere in the clone, neutralization without kinetic assistance could be achieved. This was consistent with the hypothesis that such hydrophobic mutations are selected against during affinity maturation due to immune tolerance control. The phenomenon of the reduction in affinity during descent in the clonal lineage has been observed before and has been referred to as “affinity reversion” (74) or “antibody redemption” (75). We concluded from our work that one structural characteristic with a central role in the affinity reversion phenomenon is hydrophobicity (72).
Affinity maturation in glycan-binding BNAbs
The surface of the HIV-1 Env is highly glycosylated and each glycan site can present a heterogeneous mix of carbohydrates (76). In addition, glycans are poorly immunogenic because they are often presented on the surface of host proteins, thus antibodies that bind glycans can be subject to immune tolerance mechanisms (77, 78). The resulting “glycan shield” (52) protecting the Env protein is one of the principle mechanisms HIV-1 uses to evade the immune system (79). As the virus mutates, glycan sites can be added, subtracted or shifted to different positions and neighboring glycans can influence the type of glycans that are presented at each site (76, 80). This shape-shifting cloak of sugars allows the virus to escape glycan-binding antibodies and often keeps the virus at least one step ahead of the immune system. The Env is predominantly N-linked glycosylated, meaning that glycans are covalently attached to asparagine residues and this attachment occurs at sequence motifs referred to as “sequons”. The sequon sequence motif is N(X)T/S, where the first position is aspargine, the second position is any amino acid residue except proline, and the third position is a threonine or serine residue. Thus, a single mutation to or from N, P, T or S can result in the gain or loss of a glycan site. The implication of the three amino acid motif is that a small step in nucleotide mutational space (via a single non-synonymous nucleotide mutation) can lead to a large change in three-dimensional structural space through increasing or decreasing the steric volume of the glycan shield. This steric volume change is much larger than would otherwise occur with conventional amino acid-substituting mutations because glycans are structurally much larger than amino acids. The payoff of mutating a glycan site and the lack of an equivalently effective, readily-inducible mutational mechanism by B cells to counter it provides a structural rationale for why this route of viral escape is so common in HIV-1 infection (22, 81, 82). Nevertheless, in rare cases, B cell lineages are indeed able to prevent viral escape via glycosylation. We will see how through the affinity maturation process these lineages alter their structure in order to either avoid key glycans or recognize their conserved portions and by doing so gain the capacity to broadly neutralize.
One of the most structurally well-characterized HIV-1 BNAb lineages is the PGT121 family of antibodies (83). The lineage targets the glycan-V3 site of vulnerability, an epitope comprised of the base of the V3 loop and multiple nearby glycans typically including the N332 glycan (51). The PGT121 family is notable for its extremely high potency, a characteristic leveraged in work that showed passive administration of PGT121 protected against HIV-1 infection in macaques (84). Thus, understanding the mutational events and structural changes that occur during the maturation of the PGT121 lineage has critical implications for vaccine design strategies attempting to recapitulate a glycan-V3 BNAb-type response.
A structural comparison between the unbound conformations of inferred GL PGT121 and mature PGT121 revealed that conformational shifts occurred in three CDRs and FR3 upon maturation (85). When superimposed onto the structure of closely-related clonal member PGT122 in complex with a SOSIP trimer (a near-native Env trimer construct) (18), it is evident that these conformational changes would allow for better accommodation of the N332 glycan as well as the V1 loop. By pre-configuring the CDRs in structurally more compatible conformations prior to binding the glycan-V3 epitope, the mature PGT122 would incur less of an entropic penalty upon recognition. This is the same entropy-based mode of improving affinity during maturation as was mentioned with the CDR L3 of CH58 and demonstrated that this type of structural maturation in HIV antibodies is probably common and can also occur with glycan-based epitopes.
Garces and coworkers (86) showed that many of the contacts in the PGT121 family to the conserved GDIR motif at the base of the V3 along with contacts to the conserved N332 glycan are shared among the PGT121 members. Based on reconstruction of the clonal history, most of these contacting residues were either encoded in germline or, in the case of CDR H3 residues, predicted to occur very early in maturation. It is important to note that no CDR H3 inference was attempted for the PGT121 family, therefore the authors were unable to determine whether residues were germline encoded in that region. The authors hypothesized that recognition of GDIR and the N332 glycan was thus an essential event in initiating this lineage. Further structural work has focused on how maturation in the two main branches of the PGT121 lineage diverged leading to the adoption of two different strategies for dealing with glycan heterogeneity in the glycan-V3 site.
The PGT121 lineage diverges into two main branches, one terminating with PGT122 and the other with PGT124 (85). Comparisons of the structures of the complexes of PGT122/SOSIP Env trimer (51), PGT124/gp120 construct (86) and an inferred intermediate PGT121 clonal member bound to a SOSIP lacking the N137 glycan site in V1 (58) revealed two distinct recognition faces in the paratope of the clone. One face is comprised of the residues that contact the GDIR and N332 glycan that are conserved throughout the lineage. Not surprisingly this face adopts a highly similar structure among members across both branches in the lineage. The second face differs between the two branches and is the primary determinant for how PGT121 clonal members interact with the N137 glycan in V1. Mutations early in the line of descent along the PGT122 branch resulted in corresponding residues becoming smaller, likely to accommodate the N137 glycan. Later, mutations in more mutated intermediates then resulted in productive contacts with the N137 glycan. In the PGT124 branch these maturation mutations do not occur and a different strategy is employed to deal with N137 glycan. Instead, members along the PGT124 branch undergo a small rotation of ~10° to avoid the N137 glycan. The consequence of these two recognition strategies is that the PGT122 branch is heavily reliant upon the N137 glycan and thus is highly sensitive to viruses that lack the N137 glycan site but more resistant to viruses that lack the N332 glycan site. Conversely, the PGT124 branch is resistant to escape via N137 glycan site, however its reliance on the N332 glycan makes it sensitive to removal of the N332 glycan site (58).
Of the five sites of vulnerability on the HIV-1 Env, only one is targeted by BNAbs that do not engage glycans (49, 50). And that site, the MPER, unfortunately tends to be recognized by BNAbs that are auto-reactive likely explaining their exceedingly rare elicitation. Therefore, the maturation of paratopes to effectively interact with Env glycan is critical for the development of the most readily-inducible BNAbs. In the next section, we will discuss an extraordinary structural mechanism that allows BNAb lineages to outmaneuver the ever changing Env glycan shield.
The structural role of insertions and deletions in BNAb development
One of the most unusual features shared among many HIV-1 BNAbs is the possession of multibase in-frame insertions or deletions (indels). Using the statistical inference methods outlined in this review, we analyzed BNAb gene sequences and found that indels occur in ~40% of BNAb lineages (87). We observed that indel frequency was associated with antibody mutation frequency which was consistent with indel and substitution mutations sharing the same underlying molecular mechanism. We hypothesized that increased accumulation of mutation can increase the instability of the antibody gene and thus increase the susceptibility to indel events occurring. We also observed that indels tend to occur near the antigen binding site in BNAbs. In one detailed example, we showed that the antibody VRC-CH31 underwent a deletion and tandem insertion with the net effect of adding 9 amino acids to the CDR H1. Reconstruction of the clonal history, aided by high-throughput sequencing, allowed us to isolate lineage members from before and after the indel event. We found that neutralization breadth for the VRC-CH31 clone was acquired after the indel formation. Structural determination of VRC-CH31 in complex with gp120 showed that CDR H1 was in the paratope. However, portions of the CDR H1 that included the insertion were not completely resolved in the crystal structure, most likely due to the length and flexibility of the inserted loop. In addition, the complex was determined with monomeric gp120 core which did not provide context for the structural role of the indel in VRC-CH31 in binding the native trimer.
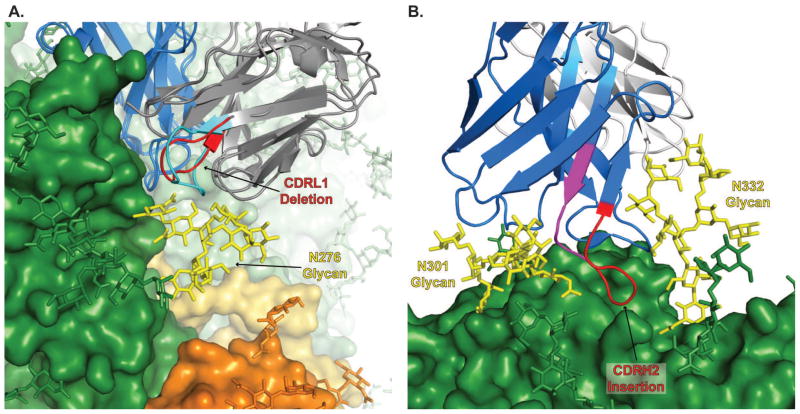
Given the preponderance of indels in HIV-1 BNAbs and the tendency of indels to be included in BNAb paratopes, it is not surprising that there are many additional examples of structural characterizations of BNAbs that acquired indels during their maturation. We focus here on BNAbs with indels of at least 2 amino acids as their structural effect on antibody maturation is more easily interpretable. One of the most prevalent features of the VRC01-like class of CD4bs-targeting BNAbs is a 2 to 3 amino acid deletion in the CDR L1 (88). The role of this deletion, which is observed in the complex structures of VRC01 (24), VRC03 (25), NIH45–46 (89), PG04 (25) and 3BNC117 (88), has been demonstrated to be related to accommodation of the N276 glycan within loop D(46, 88, 90) and reversion of the deletion in VRC01 was associated with a large reduction in binding (24). The conservation of this deletion across multiple lineages from multiple different donors suggests it is an essential component in one of the extremely limited structural solutions to fitting an antibody Fab arm into the tight quarters normally reserved only for the much smaller CD4 molecule that these lineages attempt to emulate.
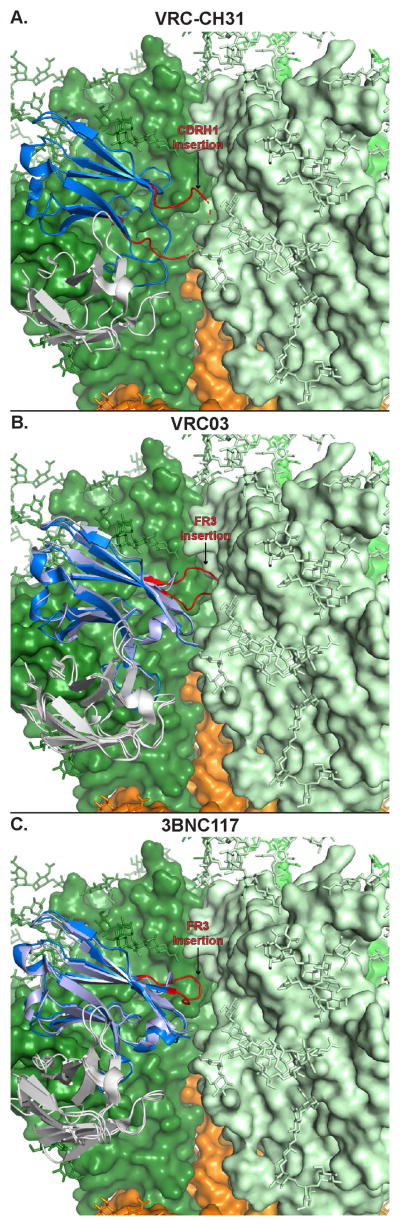
Several of the VRC01-class BNAbs also underwent insertion events during their maturation. Both 3BNC117 and VRC03, a member of the VRC01 clonal lineage, had insertions in their heavy chain framework 3 region (HFR3) of 3 and 7 amino acids, respectively. As mentioned, VRC-CH31 had a large net 9 amino acid insertion in CDR H1 that was important for neutralization capacity of the antibody, however the structural role of the insertion in the context of a native-like trimer was not clear at the time of the initial study (87). Since then, several high resolution structures of the Env trimer have been determined (51, 52) making it possible to superimpose the VRC-CH31/gp120 complex structure onto the Env trimer to gain insight into how the VRC-CH31 could interact with the trimer in a more native structural environment (Fig 4A). Modeling the VRC-CH31 into the Env trimer suggested that the large insertion reaches into the cleft between the gp120 in the complex and a gp120 in the neighboring subunit of the trimer. This quaternary interaction of the inserted CDR H1 would likely include portions of the V3 loop region in the neighboring gp120. Indeed, additional structural modeling with the SOSIP trimer showed that the inter-gp120 cleft including the neighboring subunit V3 is the same secondary epitope potentially contacted by HFR3 insertions of 3BNC117 and VRC03 (Fig 4B–C). The importance of this secondary epitope is underscored by the observation that reverting the HFR3 insertion resulted in loss of neutralization of 3BNC60, a member of the 3BNC117 clone (33). It is therefore striking that three different subjects employed the same strategy to supplement the interaction with a secondary epitope and suggests that increase in interface size through secondary site engagement is a necessary component of the antibody maturation process in many CD4 mimicking BNAb lineages.
Figure 4. Large Insertions in CD4 Binding Site Targeting Antibodies Contact a Secondary Epitope Between gp120 Subunits.
Structural modeling of three matured VRC01-like class HIV-1 antibodies (matured heavy chain colored in blue; GL heavy chain in light blue; all light chains in white) in complex with monomeric gp120 by superposition onto the SOSIP Env trimer structure (surface representation; gp120 in green shades; gp41 in orange shades; glycans shown in stick representation) suggested that large insertions may be contacting a secondary epitope in the cleft between two gp120 subunits. A) An insertion in CDR H1 (red) in VRC-CH31 is directed towards the inter-protomer cleft. Dashed lines indicate the CDR H1 was not completely resolved in the crystal structure. B) An insertion in the framework region 3 (FWR3; red) of VRC03 may allow extension into the inter-subunit cleft. C) An insertion in the FWR3 of 3BNC117 may play a similar role. The addition of a secondary epitope involving a neighboring gp120 in the Env trimer structure suggests affinity maturation in the VRC01-like class of HIV-1 BNAbs may utilize insertions to increase the size of the paratope in order to strengthen binding to Env. The N276 and N332 glycan representations have been removed from view for clarity.
Acquisition of indels has also been demonstrated to be essential in the maturation of BNAbs that target the glycan-V3 supersite (61, 85, 91). Thanks to the recent determination of several antibody structures to this epitope in complex with both monomeric gp120 and Env trimers, the structural roles for many of these indels have now been characterized at the atomic level. PGT128 uses a 6 aa insertion in its CDR H2 to penetrate the glycan shield by reaching between glycans at N301 and N332 (61). Reversion by deletion of this insertion showed that the CDR H2 insertion was critical for the neutralization potency and breadth (91). An insertion in the CDR H1 of 5 aas in PGT135 was observed to make a similar interaction between the N332 and N392 glycans (59). All observed members of the PGT121 family had a 3 aa insertion in the light chain framework 3 region (LFR3) that was a key event in the maturation of the lineage allowing it to interact with the conserved GDIR motif at the base of the V3 loop (86). The PGT121 family insertion extends the LFR3 loop down past the N301 glycan that typically occludes the GDIR motif (58). The PGT121 family also exhibited a deletion event at the N-terminus of the light chain thought to accommodate the N156 glycan(58). A deletion was also observed in PGT128 and occurred in the CDR L1. However, the CDR L1 of PGT128 falls well outside of its paratope, the notable lone exception in which an indel of more than 1 aa did not occur in the antigen binding site of BNAb complex structures (61). Reversion of the PGT128 CDR L1 deletion had no effect on neutralization (91). Thus, the role of this deletion in the PGT128 maturation is unclear and may merely be a stochastic event maintained due to a neutral effect on B-cell fitness.
8ANC195, a BNAb that contacts the gp41-gp120 interface, a rarely observed BNAb site of vulnerability, had a 5 aa insertion in HFR3 (87). Structural characterization of 8ANC195 in complex with the Env trimer revealed that the insertion extends between the glycans at N276 and N234 (92). Another gp41-gp120 interface targeting BNAb, 35O22, had an 8 aa insertion in HFR3 that increased the size of the paratope by extending between gp41 and gp120 molecules, and this insertion was critical for neutralization breadth (93). It is interesting to note that insertions at the HFR3 are used in the recognition modes of multiple distinct BNAb epitopes. In fact, the loop in this region is sometimes referred to as CDR H4 because it has been shown to make contact with antigen from a wide range of pathogens and insertions within HFR3 have been observed to be important for influenza antibodies (94).
Based on the commonalities observed in the plethora of BNAb structures that include indels, we can draw conclusions on how insertions and deletions are employed during BNAb maturation to improve affinity and acquire breadth of neutralization. All but one large (>1 aa) indel characterized occurred within the antibody paratope and most large indels were shown to exhibit an effect on binding and/or neutralization when restored to their germline versions. Taken together these data are consistent with the notion that these typically rarely observed events occur as the result of strong selection pressure during the maturation process of BNAb lineages. The strategies by which HIV-1 BNAbs employ large indels to recognize Env can essentially be divided into three groups. The first group is defined by large insertions that are used to penetrate the glycan shield extending through it to contact conserved portions of the polypeptide surface of the Env as well as the conserved portion of glycans that resides at their base (8ANC195 HFR3; PGT135 CDR H1; PGT121 LFR3; and PGT128 CDR H2). The second group uses insertions to increase the size of the paratope by binding to clefts formed at the interface of two of the Env subunits. In this group, the insertion reaches into either a secondary epitope at the inter-gp120 cleft of the Env trimer (VRC03 and 3BNC117 HFR3; and VRC-CH31 CDR H1) or at the gp41-gp120 interface (35O22 HFR3). The third group uses deletions to avoid or accommodate glycan heterogeneity (VRC01, 3BNC117, PG04 CDR L1; PGT121 N-terminus). Figures 4 and 5 show representative antibody-antigen complex structures that exemplify these three classes of indel-mediated recognition.
Figure 5. Large Insertions and Deletions in HIV-1 BNAbs Mediate Glycan Interactions.
Most large insertions and deletions observed in the structures of HIV-1 BNAbs are involved in mediating glycan interactions. Many of the large deletions that have been observed in crystal structures contribute to maturation by providing steric clearance to nearby glycans such as with A) VRC01 in which a 2 aa deletion in CDR L1 from GL (cyan) to matured VRC01 (red) allows for accommodation of the N276 glycan (yellow sticks). B) Large insertions tend to be employed for penetrating between glycans such as in the maturation of PGT128 where a 6 aa insertion (magenta) in the CDR H2 (red) allowed for the loop to extend between the N301 and N332 glycans (yellow sticks). In both panels, the Env trimer is shown in surface representation with gp120 in green shades and gp41 in orange shades. Glycans are shown as sticks. Antibody heavy chains are colored blue and light chains colored gray.
It is notable that all large deletions in BNAbs with an observed effect on binding and/or neutralization were associated with glycan accommodation. As we previously mentioned, addition, subtraction and shifting of glycan sites is a highly efficient technique for escape by the virus because a small step in mutational space results in a large change in structural space. Therefore, glycans must be properly accommodated in recognition during the maturation of the antibody lineage or else total viral escape will occur and the clone will not survive. The result of this uniquely effective escape artistry by HIV-1 is an immense selective pressure on the antibody to make concomitant changes to adapt its paratope to keep up with the altering structure of the viral Env surface. Indels are the primary mechanism by which antibodies can make large structural changes on par with the change in steric volume that accompanies the addition or removal of Env glycans. Thus we observe from reviewing the structures of BNAbs that contain indels that most occur as a recognition mechanism to either penetrate through the glycan shield or accommodate glycans. In some clonal lineages both strategies are utilized, most likely reflecting the need to balance and maintain an interface size sufficiently large enough for strong antibody affinity. The maintenance of a minimum interface size could also be responsible for the third group of recognition modes associated with indels which was insertion into the inter-gp120 cleft by CD4 mimicking BNAbs. The tight fit required for Fab arms to replicate the smaller CD4 molecules interaction with the receptor binding site on gp120 could in a smaller than usual paratope. Thus, these antibodies may compensate by increasing the size of the paratope via acquisition of contacts to a secondary epitope that includes the neighboring Env subunit. More structural studies involving native-like Envs trimers will be needed to confirm the role of this secondary epitope. Insertions and deletions, while rare in the context of normal antibody response to infection, are quite frequent in HIV-1 BNAb development, and as we shall see in the next section this has important implications for vaccine design.
Implications for vaccine design
A fundamental goal of HIV-1 vaccine development is the recapitulation of an infrequently observed response in HIV-1 infection: the induction of exquisitely potent and broadly neutralizing antibodies. The extraordinary number of mutations that are acquired in HIV-1 BNAbs suggests that a detailed understanding of the affinity maturation process when a BNAb is elicited is paramount for designing vaccine strategies that can recapitulate these complex BNAb developmental pathways. Clearly all mutations acquired are not created equal and some mutations are inessential for the acquisition of neutralization breadth. Recent work has shown that effective levels of breadth can be reached by reverting many of these mutations to germline in the VRC01 BNAb lineage (18). Identifying the essential mutations in the maturation of a BNAb lineage can define the minimal set of mutations that a vaccine may be required to induce. More work will need to be done to determine if the minimum number of mutations is generalizable in other BNAb lineages. If other lineages also require just a handful of essential mutations it will be critical to delineate these shorter pathways as they can act as roadmaps to guide immunogen design strategies to multiple sites of vulnerability on the Env. However, careful attention will need to be paid to how any essential mutations affect the auto-reactivity of clonal members as elicitation of essential mutations may require transient modulation of immune tolerance mechanisms in order to break through such roadblocks (72).
The role of large indels in the maturation of BNAbs is primarily to interact with the glycan shield on the Env trimer. Because the glycan shield is one of the most effective defense mechanisms that the virus employs, it is perhaps not surprising that the antibody response must resort to extraordinary affinity maturation measures in order to combat it. Vaccine development efforts will need to take into account that the usage of indels typically observed in BNAbs is a very atypical maturation event in the adaptive immune response. One strategy is to identify BNAb lineages that do not use large indels and focus on recapitulating responses similar to these more readily-inducable lineages. If attempts to elicit BNAbs that require indels is unavoidable, and it may be given most of the very best BNAbs possess them, then vaccine development strategies will need to approach the problem using a detailed understanding of two components of the evolutionary process of affinity maturation. One strategy would be to focus on the first component: diversification by somatic hypermutation. Here strategies would aim to enrich the pool of indels by utilizing strategies that could induce more indel events to occur. Such a strategy would likely need to be highly specific to clones of interest that were already initiated since increasing the incidence of indels across the entire antibody repertoire could potentially have dire immunological consequences. Given we have observed that the indels can occur, however rarely that they do, targeting the second component of affinity maturation, selection, may be the better strategy. Given a very narrow funnel of selection, even extremely rare diversification events can be promoted and expanded. Here the strategy would be to use immunogens that bind specifically to clonal members with insertions or deletions. In fact, the ability of vaccine strategies to induce indels is just now being evaluated. Sequential immunization strategies in BNAb UCA/GL knock-in mice systems that prime with immunogens lacking glycans and then boost with immunogens that progressively add glycans back have recently been reported (95–97). It is notable that large indels were not observed in any of the antibody responses in these studies and that the lack of indels induced in these lineages likely hindered acquisition of fully matured BNAb neutralization activity. Thus, more targeted vaccine strategies appear to be necessary for selecting these improbable, but critical indel events.
The development of the statistical framework and computational methods to infer antibody ancestral sequences has been an important advance in the field of HIV-1 vaccinology. The atomic-level detail of structures of inferred antibodies has led to a greater understanding of the extraordinary measures that clonal lineages utilize to acquire the capacity to broadly neutralize HIV-1. Given the remarkable insights gained so far, it is clear that genetic and structural analysis of affinity maturation in the HIV-1 response will continue to play an integral role in informing HIV-1 vaccine design strategies.
Acknowledgments
This work was supported in part by NIH awards UM1-AI100645-04 for the Duke Center For HIV/AIDS Vaccine Immunology-Immunogen Design (B. Haynes) and AI117892 (TBK and G. Kelsoe) and by the Bill and Melinda Gates Foundation 38643 (B. Haynes). The authors thank Axin Hua, Feng Feng, and other members of the Kepler laboratory, as well as Larry Liao, Garnett Kelsoe, Todd Bradley, and Barton Haynes for their invaluable contributions.
Footnotes
I declare that I have no conflicts of interest to report.
References
- 1.Janeway C. Immunobiology: the immune system in health and disease. 6. New York: Garland Science; 2005. [Google Scholar]
- 2.McHeyzer-Williams M, Okitsu S, Wang N, McHeyzer-Williams L. Molecular programming of B cell memory. Nat Rev Immunol. 2012;12:24–34. doi: 10.1038/nri3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Victora GD, Nussenzweig MC. Germinal Centers. Annual Review of Immunology. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 4.Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR. The promise and challenge of high-throughput sequencing of the antibody repertoire. Nat Biotech. 2014;32:158–168. doi: 10.1038/nbt.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrammert J, Smith K, Miller J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kepler TB, Munshaw S, Wiehe K, et al. Reconstructing a B-Cell Clonal Lineage. II. Mutation, Selection, and Affinity Maturation. Front Immunol. 2014;5:170. doi: 10.3389/fimmu.2014.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kepler TB. Reconstructing a B-cell clonal lineage. I. Statistical inference of unobserved ancestors. F1000Res. 2013;2:103. doi: 10.12688/f1000research.2-103.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z. Molecular Evolution: A Statistical Approach. Oxford University Press; 2014. [Google Scholar]
- 9.Kepler TB, Liao H-X, Alam SM, et al. Immunoglobulin Gene Insertions and Deletions in the Affinity Maturation of HIV-1 Broadly Reactive Neutralizing Antibodies. Host Cell and Microbe. 2014 doi: 10.1016/j.chom.2014.08.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munshaw S, Kepler TB. SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics. 2010;26:867–872. doi: 10.1093/bioinformatics/btq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson-Smith V, Kolaczkowski B, Thornton JW. Robustness of Ancestral Sequence Reconstruction to Phylogenetic Uncertainty. Molecular Biology and Evolution. 2010;27:1988–1999. doi: 10.1093/molbev/msq081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hraber P, Seaman MS, Bailer RT, Mascola JR, Montefiori DC, Korber BT. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28:163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature’s pathways. Immunol Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macchioro G, Zucconi V, Magris D. On the possibilities of preventing remote sequelae of viral heptatitis. Epatologia. 1966;12:281–287. [PubMed] [Google Scholar]
- 17.Wibmer CK, Moore PL, Morris L. HIV broadly neutralizing antibody targets. Curr Opin HIV AIDS. 2015;10:135–143. doi: 10.1097/COH.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jardine JG, Sok D, Julien JP, et al. Minimally Mutated HIV-1 Broadly Neutralizing Antibodies to Guide Reductionist Vaccine Design. PLoS Pathog. 2016;12:e1005815. doi: 10.1371/journal.ppat.1005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walker LM, Simek MD, Priddy F, et al. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao HX, Lynch R, Zhou T, et al. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doria-Rose NA, Schramm CA, Gorman J, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore PL, Gray ES, Wibmer CK, et al. Evolution of an HIV glycan-dependent broadly neutralizing antibody epitope through immune escape. Nat Med. 2012;18:1688–1692. doi: 10.1038/nm.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikell I, Sather DN, Kalams SA, Altfeld M, Alter G, Stamatatos L. Characteristics of the earliest cross-neutralizing antibody response to HIV-1. PLoS Pathog. 2011;7:e1001251. doi: 10.1371/journal.ppat.1001251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou T, Georgiev I, Wu X, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X, Zhou T, Zhu J, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK. Immunity through DNA deamination. Trends Biochem Sci. 2003;28:305–312. doi: 10.1016/S0968-0004(03)00111-7. [DOI] [PubMed] [Google Scholar]
- 27.Betz AG, Rada C, Pannell R, Milstein C, Neuberger MS. Passenger transgenes reveal intrinsic specificity of the antibody hypermutation mechanism: clustering, polarity, and specific hot spots. Proc Natl Acad Sci U S A. 1993;90:2385–2388. doi: 10.1073/pnas.90.6.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reynaud CA, Garcia C, Hein WR, Weill JC. Hypermutation generating the sheep immunoglobulin repertoire is an antigen-independent process. Cell. 1995;80:115–125. doi: 10.1016/0092-8674(95)90456-5. [DOI] [PubMed] [Google Scholar]
- 29.Wagner SD, Milstein C, Neuberger MS. Codon bias targets mutation. Nature. 1995;376:732. doi: 10.1038/376732a0. [DOI] [PubMed] [Google Scholar]
- 30.Shlomchik MJ, Watts P, Weigert MG, Litwin S. Clone: a Monte-Carlo computer simulation of B cell clonal expansion, somatic mutation, and antigen-driven selection. Curr Top Microbiol Immunol. 1998;229:173–197. doi: 10.1007/978-3-642-71984-4_13. [DOI] [PubMed] [Google Scholar]
- 31.Fera D, Schmidt AG, Haynes BF, Gao F, Liao HX, Kepler TB, Harrison SC. Affinity maturation in an HIV broadly neutralizing B-cell lineage through reorientation of variable domains. Proc Natl Acad Sci U S A. 2014;111:10275–10280. doi: 10.1073/pnas.1409954111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao F, Bonsignori M, Liao HX, et al. Cooperation of B cell lineages in induction of HIV-1-broadly neutralizing antibodies. Cell. 2014;158:481–491. doi: 10.1016/j.cell.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein F, Diskin R, Scheid JF, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georgiev IS, Rudicell RS, Saunders KO, et al. Antibodies VRC01 and 10E8 neutralize HIV-1 with high breadth and potency even with Ig-framework regions substantially reverted to germline. J Immunol. 2014;192:1100–1106. doi: 10.4049/jimmunol.1302515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicely NI, Wiehe K, Kepler TB, et al. Structural analysis of the unmutated ancestor of the HIV-1 envelope V2 region antibody CH58 isolated from an RV144 vaccine efficacy trial vaccinee. EBioMedicine. 2015;2:713–722. doi: 10.1016/j.ebiom.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao HX, Bonsignori M, Alam SM, et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity. 2013;38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 38.Haynes BF, Gilbert PB, McElrath MJ, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rolland M, Edlefsen PT, Larsen BB, et al. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490:417–420. doi: 10.1038/nature11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S, Nussinov R. Close-range electrostatic interactions in proteins. Chembiochem. 2002;3:604–617. doi: 10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 41.Foote J, Milstein C. Conformational isomerism and the diversity of antibodies. Proc Natl Acad Sci U S A. 1994;91:10370–10374. doi: 10.1073/pnas.91.22.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manivel V, Sahoo NC, Salunke DM, Rao KV. Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity. 2000;13:611–620. doi: 10.1016/s1074-7613(00)00061-3. [DOI] [PubMed] [Google Scholar]
- 43.Sagawa T, Oda M, Ishimura M, Furukawa K, Azuma T. Thermodynamic and kinetic aspects of antibody evolution during the immune response to hapten. Mol Immunol. 2003;39:801–808. doi: 10.1016/s0161-5890(02)00282-1. [DOI] [PubMed] [Google Scholar]
- 44.Thorpe IF, Brooks CL., 3rd Molecular evolution of affinity and flexibility in the immune system. Proc Natl Acad Sci U S A. 2007;104:8821–8826. doi: 10.1073/pnas.0610064104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmidt AG, Xu H, Khan AR, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci U S A. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scharf L, West AP, Sievers SA, et al. Structural basis for germline antibody recognition of HIV-1 immunogens. Elife. 2016:5. doi: 10.7554/eLife.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiehe K, Easterhoff D, Luo K, et al. Antibody light-chain-restricted recognition of the site of immune pressure in the RV144 HIV-1 vaccine trial is phylogenetically conserved. Immunity. 2014;41:909–918. doi: 10.1016/j.immuni.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davenport TM, Gorman J, Joyce MG, et al. Somatic Hypermutation-Induced Changes in the Structure and Dynamics of HIV-1 Broadly Neutralizing Antibodies. Structure. 2016 doi: 10.1016/j.str.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burton DR, Hangartner L. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol. 2016;34:635–659. doi: 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ward AB, Wilson IA. Insights into the trimeric HIV-1 envelope glycoprotein structure. Trends Biochem Sci. 2015;40:101–107. doi: 10.1016/j.tibs.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pancera M, Zhou T, Druz A, et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stewart-Jones GB, Soto C, Lemmin T, et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell. 2016;165:813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen L, Kwon YD, Zhou T, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou T, Lynch RM, Chen L, et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Julien JP, Lee PS, Wilson IA. Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev. 2012;250:180–198. doi: 10.1111/imr.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonsignori M, Zhou T, Sheng Z, et al. Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell. 2016;165:449–463. doi: 10.1016/j.cell.2016.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu X, Zhang Z, Schramm CA, et al. Maturation and Diversity of the VRC01-Antibody Lineage over 15 Years of Chronic HIV-1 Infection. Cell. 2015;161:470–485. doi: 10.1016/j.cell.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garces F, Lee JH, de Val N, et al. Affinity Maturation of a Potent Family of HIV Antibodies Is Primarily Focused on Accommodating or Avoiding Glycans. Immunity. 2015;43:1053–1063. doi: 10.1016/j.immuni.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong L, Lee JH, Doores KJ, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mouquet H, Scharf L, Euler Z, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109:E3268–3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pejchal R, Doores KJ, Walker LM, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker LM, Huber M, Doores KJ, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.MacLeod DT, Choi NM, Briney B, et al. Early Antibody Lineage Diversification and Independent Limb Maturation Lead to Broad HIV-1 Neutralization Targeting the Env High-Mannose Patch. Immunity. 2016;44:1215–1226. doi: 10.1016/j.immuni.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landais E, Huang X, Havenar-Daughton C, et al. Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLoS Pathog. 2016;12:e1005369. doi: 10.1371/journal.ppat.1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haynes BF, Fleming J, St Clair EW, et al. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 66.Liu M, Yang G, Wiehe K, et al. Polyreactivity and autoreactivity among HIV-1 antibodies. J Virol. 2015;89:784–798. doi: 10.1128/JVI.02378-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang G, Holl TM, Liu Y, et al. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. J Exp Med. 2013;210:241–256. doi: 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alam SM, Morelli M, Dennison SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alam SM, McAdams M, Boren D, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178:4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verkoczy L, Diaz M, Holl TM, et al. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proc Natl Acad Sci U S A. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y, Zhang J, Hwang KK, et al. Common tolerance mechanisms, but distinct cross-reactivities associated with gp41 and lipids, limit production of HIV-1 broad neutralizing antibodies 2F5 and 4E10. J Immunol. 2013;191:1260–1275. doi: 10.4049/jimmunol.1300770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang R, Verkoczy L, Wiehe K, et al. Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages. Sci Transl Med. 2016;8:336ra362. doi: 10.1126/scitranslmed.aaf0618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ofek G, McKee K, Yang Y, et al. Relationship between antibody 2F5 neutralization of HIV-1 and hydrophobicity of its heavy chain third complementarity-determining region. J Virol. 2010;84:2955–2962. doi: 10.1128/JVI.02257-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verkoczy L, Chen Y, Zhang J, et al. Induction of HIV-1 broad neutralizing antibodies in 2F5 knock-in mice: selection against membrane proximal external region-associated autoreactivity limits T-dependent responses. J Immunol. 2013;191:2538–2550. doi: 10.4049/jimmunol.1300971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sabouri Z, Schofield P, Horikawa K, et al. Redemption of autoantibodies on anergic B cells by variable-region glycosylation and mutation away from self-reactivity. Proc Natl Acad Sci U S A. 2014;111:E2567–2575. doi: 10.1073/pnas.1406974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Behrens AJ, Vasiljevic S, Pritchard LK, et al. Composition and Antigenic Effects of Individual Glycan Sites of a Trimeric HIV-1 Envelope Glycoprotein. Cell Rep. 2016;14:2695–2706. doi: 10.1016/j.celrep.2016.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- 78.Crispin M, Doores KJ. Targeting host-derived glycans on enveloped viruses for antibody-based vaccine design. Curr Opin Virol. 2015;11:63–69. doi: 10.1016/j.coviro.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mascola JR, Montefiori DC. HIV-1: nature’s master of disguise. Nat Med. 2003;9:393–394. doi: 10.1038/nm0403-393. [DOI] [PubMed] [Google Scholar]
- 80.Go EP, Hewawasam G, Liao HX, et al. Characterization of glycosylation profiles of HIV-1 transmitted/founder envelopes by mass spectrometry. J Virol. 2011;85:8270–8284. doi: 10.1128/JVI.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klein F, Halper-Stromberg A, Horwitz JA, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krumm SA, Mohammed H, Le KM, et al. Mechanisms of escape from the PGT128 family of anti-HIV broadly neutralizing antibodies. Retrovirology. 2016;13:8. doi: 10.1186/s12977-016-0241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Julien JP, Sok D, Khayat R, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog. 2013;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barouch DH, Whitney JB, Moldt B, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sok D, Laserson U, Laserson J, et al. The effects of somatic hypermutation on neutralization and binding in the PGT121 family of broadly neutralizing HIV antibodies. PLoS Pathog. 2013;9:e1003754. doi: 10.1371/journal.ppat.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Garces F, Sok D, Kong L, et al. Structural evolution of glycan recognition by a family of potent HIV antibodies. Cell. 2014;159:69–79. doi: 10.1016/j.cell.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kepler TB, Liao HX, Alam SM, et al. Immunoglobulin gene insertions and deletions in the affinity maturation of HIV-1 broadly reactive neutralizing antibodies. Cell Host Microbe. 2014;16:304–313. doi: 10.1016/j.chom.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhou T, Zhu J, Wu X, et al. Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies. Immunity. 2013;39:245–258. doi: 10.1016/j.immuni.2013.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Diskin R, Scheid JF, Marcovecchio PM, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McGuire AT, Hoot S, Dreyer AM, et al. Engineering HIV envelope protein to activate germline B cell receptors of broadly neutralizing anti-CD4 binding site antibodies. J Exp Med. 2013;210:655–663. doi: 10.1084/jem.20122824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Doores KJ, Kong L, Krumm SA, et al. Two classes of broadly neutralizing antibodies within a single lineage directed to the high-mannose patch of HIV envelope. J Virol. 2015;89:1105–1118. doi: 10.1128/JVI.02905-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Scharf L, Wang H, Gao H, Chen S, McDowall AW, Bjorkman PJ. Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env. Cell. 2015;162:1379–1390. doi: 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Huang J, Kang BH, Pancera M, et al. Broad and potent HIV-1 neutralization by a human antibody that binds the gp41-gp120 interface. Nature. 2014;515:138–142. doi: 10.1038/nature13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Briney BS, JE Secondary mechanisms of diversification in the human antibody repertoire. Front Immunol. 2013;4:42. doi: 10.3389/fimmu.2013.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Briney B, Sok D, Jardine JG, et al. Tailored Immunogens Direct Affinity Maturation toward HIV Neutralizing Antibodies. Cell. 2016;166:1459–1470 e1411. doi: 10.1016/j.cell.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Escolano A, Steichen JM, Dosenovic P, et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell. 2016;166:1445–1458 e1412. doi: 10.1016/j.cell.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tian M, Cheng C, Chen X, et al. Induction of HIV Neutralizing Antibody Lineages in Mice with Diverse Precursor Repertoires. Cell. 2016;166:1471–1484 e1418. doi: 10.1016/j.cell.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scharf L, West AP, Jr, Gao H, et al. Structural basis for HIV-1 gp120 recognition by a germ-line version of a broadly neutralizing antibody. Proc Natl Acad Sci U S A. 2013;110:6049–6054. doi: 10.1073/pnas.1303682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jardine J, Julien JP, Menis S, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Soto C, Ofek G, Joyce MG, et al. Developmental Pathway of the MPER-Directed HIV-1-Neutralizing Antibody 10E8. PLoS One. 2016;11:e0157409. doi: 10.1371/journal.pone.0157409. [DOI] [PMC free article] [PubMed] [Google Scholar]