Synopsis
Vitiligo is “complex disorder” (also termed polygenic and multifactorial), reflecting simultaneous contributions of multiple genetic risk factors and environmental triggers. Large-scale genome-wide association studies, principally in European-derived whites and in Chinese, have discovered approximately 50 different genetic loci that contribute to vitiligo risk, some of which also contribute to other autoimmune diseases that are epidemiologically associated with vitiligo. At many of these vitiligo susceptibility loci the corresponding relevant genes have now been identified, and for some of these genes the specific DNA sequence variants that contribute to vitiligo risk are also now known. A large fraction of these genes encode proteins involved in immune regulation, a number of others play roles in cellular apoptosis, and still others are involved in regulating functions of melanocytes. For this last group, there appears to be an opposite relationship between susceptibility to vitiligo and susceptibility to melanoma, suggesting that vitiligo may engage a normal mechanism of immune surveillance for melanoma. While many of the specific biologic mechanisms through which these genetic factors operate to cause vitiligo remain to be elucidated, it is now clear that vitiligo is an autoimmune disease involving a complex relationship between programming and function of the immune system, aspects of the melanocyte autoimmune target, and dysregulation of the immune response.
Keywords: Vitiligo, Autoimmunity, Gene, Genomewide association study, Genetic linkage, Genetic epidemiology
Introduction, background, and genetic epidemiology
The disorder now known as vitiligo was first described by Claude Nicolas Le Cat in 1765 [1]. However, the first specific consideration of a genetic component in vitiligo did not come until 1950, when Stűttgen [2] and Teindel [3] simultaneously reported a total of eight families with multiple relatives affected by vitiligo. Stűttgen noted that in his affected family, vitiligo appeared to exhibit dominant inheritance, after intermarriage to a family with apparent recessive thyroid disease, a very early recognition of what would now be considered complex (polygenic, multifactorial) inheritance. Mohr [4], Siemens [5], and Vogel [6] subsequently reported concordant identical twin-pairs affected by vitiligo, pointing to a major role for genetic factors. Early clinical case series reported a frequency of vitiligo in probands’ relatives of 11 to 38 percent [7–10], highlighting the importance of genetic factors even in typical vitiligo cases.
Nevertheless, formal genetic epidemiologic studies of vitiligo came much later. Hafez [11] and Das [12] suggested a polygenic, multifactorial mode of inheritance, and estimated vitiligo heritability at 46% [12] to 72% [11]. Subsequent investigations likewise supported a polygenic, multifactorial model [13–18], with heritability approximately 50% [18]. A twin study of vitiligo in European-derived whites [17] found that the concordance of vitiligo was 23% in monozygotic twins, underscoring the importance of non-genetic factors as well as genetic factors in vitiligo pathogenesis. In this same study, large-scale genetic epidemiologic analyses [17] indicated that in European-derived whites, the overall frequency of vitiligo in probands’ first-degree relatives was 7%, with the risk 7.8% in probands’ parents and 6.1% in siblings, consistent with polygenic, multifactorial inheritance and age-dependency of vitiligo onset. Importantly, among vitiligo probands’ affected relatives, the frequency of vitiligo was equal in males and females, eliminating the female sex bias found in most vitiligo clinical case series. Moreover, a careful study of families with multiple relatives affected by vitiligo [19] showed earlier age-of-onset and greater skin surface involvement than in singleton cases [17], as well as greater frequency of other autoimmune diseases, suggesting that in such “multiplex families” genes likely contribute more to vitiligo risk than in singleton cases.
Relationship to other autoimmune diseases
The genetic basis of vitiligo is deeply intertwined with the genetic basis of other autoimmune diseases with which vitiligo is epidemiologically associated. Indeed, the earliest clue to the autoimmune origin of vitiligo came in the original 1855 report of Addison’s disease [20], which included a patient with idiopathic adrenal insufficiency, generalized vitiligo, and pernicious anemia, a co-occurrence of autoimmune diseases that suggested shared etiologic factors. Subsequently, the co-occurrence of different autoimmune diseases, including vitiligo, was reported by many investigators, particularly Schmidt [21], and key combinations of concomitant autoimmune diseases were later codified by Neufeld and Blizzard [22]. Beginning with the vitiligo case series reported by Steve [23], numerous investigators have since documented prevalent co-occurrence of vitiligo with various other autoimmune diseases, particularly autoimmune thyroid disease (both Hashimoto’s disease and Graves’ disease), pernicious anemia, Addison’s disease, systemic lupus erythematosus [17], rheumatoid arthritis, adult-onset type 1 diabetes mellitus, and perhaps psoriasis [19]. Of particular importance, these same vitiligo-associated autoimmune diseases also occur at increased frequency in first-degree relatives of vitiligo probands who do not themselves have vitiligo, indicating that these autoimmune diseases share at least some of their genetic underpinnings with vitiligo [19].
Early genetic marker studies
The earliest attempts to identify genetic markers associated with vitiligo began in the mid-1960s, assaying polymorphic blood proteins such as the ABO and other blood group antigens [24–30], secretor status [26,27,31], and later serum alpha 1-antitrypsin and haptoglobin phenotypes [32], with no positive results. A decade later, numerous investigators reported association studies of vitiligo with HLA types, which have also been associated with many other autoimmune diseases. Initial association studies of vitiligo and HLA yielded inconsistent and largely spurious findings due to testing different ethnic groups, inadequate statistical power, and inadequate correction for multiple-testing of many different HLA types [33–37]. Nevertheless, Foley and co-workers [38] correctly identified association of the HLA-DR4 class II serotype with vitiligo, borne out by subsequent studies, the first known genetic association for vitiligo. Importantly, HLA-DR4 is also strongly associated with several other autoimmune diseases.
A large number of additional HLA association studies of vitiligo were published subsequently, again with generally inconsistent findings. Nevertheless, Liu and co-workers [39] conducted a careful meta-analysis of eleven previous studies of HLA class I serotypes, and found robust association of vitiligo with HLA-A2 with odds ratio (OR) 2.07, a finding borne out by subsequent studies. Specific associations of vitiligo with the class I and class II gene regions of the Major Histocompatibility Complex (MHC) were subsequently replicated and refined by detailed molecular genetic and genomewide association studies (GWAS), even to the point of identifying apparently causal genetic variation, as will be discussed below.
Non-MHC candidate gene association studies
The development of DNA technology in the late 1970s ushered in an era of testing candidate genes for association with a great many diseases, including vitiligo. Unfortunately, numerous retrospective studies have shown that well over 95% of published case-control genetic association studies represent false-positives, due to inadequate sample size and statistical fluctuation, genotyping errors, occult population stratification, inadequate correction for multiple-testing, and publication bias of positive results [40,41]. As the result, this type of study is no longer considered appropriate for primary “discovery” of genetic association. Accordingly, of the approximately 70 genes for which association with vitiligo is claimed on the basis of such studies, this review will discuss only those two non-MHC candidate gene associations that have received widespread independent confirmation, including by unbiased GWAS.
Kemp et al. [42] reported the first vitiligo non-MHC candidate gene association with CTLA4, which encodes a T-cell co-receptor involved in regulation of T-cell activation and which is associated with several of the other autoimmune diseases that are epidemiologically associated with vitiligo. In fact, CTLA4 association was strongest in vitiligo patients who also had other concomitant autoimmune diseases [43], a finding subsequently replicated by another study and meta-analysis [44]. Association of CTLA4 with vitiligo has been variable among studies of different populations, but at least in European-derived whites has been demonstrated by GWAS.
A second important non-MHC candidate gene association, also reported by Kemp [45], was with PTPN22, encoding LYP protein tyrosine phosphatase, which likewise has been genetically associated with many different autoimmune diseases. Again, this association was replicated in most other studies of European-derived whites [46,47], and by GWAS, but not in most other populations. Thus, along with HLA class II, CTLA4 and PTPN22 likely are two of the genes that underlie epidemiologic association of vitiligo with other autoimmune diseases, at least in European-derived whites.
Genomewide studies
Candidate gene analyses carry an intrinsic a priori bias by selection of genes for study. In contrast, genomewide analyses of polygenic, multifactorial diseases are in principle unbiased beyond the assumption that genetic factors play some role. There are three approaches to genomewide genetic analysis. Genomewide linkage analysis tests for co-segregation of polymorphic markers with disease within families with multiple affected relatives and across such families. Such families are uncommon, the genetic resolution of linkage is low, and the genetic analyses require several important assumptions that may not be correct. Genomewide association studies, the current “gold standard”, require large numbers of cases and controls, but are reasonably powerful, can detect many genotyping errors, can provide fine-mapping, can detect population outliers and correct for population stratification, can appropriately account for multiple-testing, and require independent replication and a stringent “genomewide significance” criterion (P < 5 × 10−8) to declare “discovery”. For reasons that are not clear, linkage and GWAS often do not detect the same genetic signals. Genomewide or exome DNA sequencing studies can be configured similarly to linkage or GWAS, but are far more expensive and have not yet been applied to vitiligo.
Genetic linkage studies
Initial linkage studies of vitiligo were not genomewide, focusing on the MHC and other specific candidate regions of the genome, and will not be discussed here. The first genomewide linkage study of vitiligo was indirect; Nath and co-workers mapped [48] a locus on chromosome 17p13 called they called SLEV1 in a subset of lupus families that also had relatives with vitiligo. Spritz and co-workers [49] subsequently confirmed the SLEV1 linkage signal by genomewide linkage analysis of vitiligo families in which various other autoimmune diseases also occurred, and that group eventually fine-mapped and identified the corresponding gene as NLRP1 [50], which encodes an inflammasome regulatory protein.
In a unique large European-derived white kindred with near autosomal dominant vitiligo, Spritz and co-workers [51,52] used genomewide linkage to map a locus they termed “Autoimmune Susceptibility 1” (AIS1) at chromosome 1p31.3–p32.2. That group subsequently identified the corresponding gene as FOXD3 [53], encoding a key regulator of melanoblast differentiation. This vitiligo kindred was found to segregate a private sequence variant in the FOXD3 promoter that up-regulated transcription in vitro, which would be expected to reduce melanoblast differentiation. Recently, Schunter and co-workers [54] identified another FOXD3 promoter variant associated with vitiligo that also increases transcriptional activity. Spritz and co-workers also mapped two additional vitiligo linkage signals in European-derived white vitiligo families, AIS2 on chromosome 7 and AIS3 on chromosomes 8 [49,52,55]. Specific genes corresponding to AIS2 and AIS3 have not yet been identified.
In parallel linkage studies of Han Chinese vitiligo families, Zhang and co-workers identified three loci, AIS4 on chromosome 4q12–q21 [56] and two unnamed loci on 6p21–p22 and 22q12 [57]. These investigators suggested that AIS4 might be PDGFRA [58], though this seems much less likely than KIT. The chromosome 6 locus may correspond to the MHC. And Ren et al. [59] found that the chromosome 22 locus may correspond to XBP1.
Genomewide association studies
The first GWAS of vitiligo was of a unique population isolate in Romania in which there is a very high prevalence of vitiligo and other autoimmune diseases [60]. This study identified association with a SNP within SMOC2 on chromosome 6q27, in close vicinity to IDDM8, a linkage and association signal for type 1 diabetes mellitus and rheumatoid arthritis [61].
The Spritz group has also carried out three successive GWAS of vitiligo of USA and European-derived whites [62–65]. As shown in Table 1, these analyses have identified confirmed associations of vitiligo with 48 distinct loci, altogether accounting for 22.5% of vitiligo heritability in European-derived whites, as well as several additional loci with suggestive significance [65]. About half of the confirmed vitiligo loci encode immunoregulatory proteins, consistent with the autoimmune nature of vitiligo, a number of others encode regulators of apoptosis, and at least six encode either melanocyte components or regulators of melanocyte function. Of this last group, all have also been implicated in both normal pigmentary variation and risk of melanoma, and all show a remarkable inverse genetic relationship between vitiligo risk and melanoma risk [62,63,65], suggesting that vitiligo may result from dysregulation of a normal mechanism of immune surveillance for melanoma [66,67]. Many of these proteins encoded by the confirmed vitiligo genes interact directly in functional biological pathways that are key to vitiligo pathogenesis (Figure 1), suggesting that a majority of the pathways involved in vitiligo susceptibility may have already been discovered.
Table 1.
Vitiligo susceptibility genes identified by GWAS
| Chr. | Locus | Protein | Function |
|---|---|---|---|
| 1 | RERE | Arginine-Glutamic acid dipeptide repeats | regulator of apoptosis |
| 1 | PTPN22 | protein tyrosine phosphatase, non-receptor type | alters responsiveness of T cell receptors |
| 1 | FASLG | FAS ligand | regulator of immune apoptosis |
| 1 | PTPRC | protein tyrosine phosphatase, receptor type C | regulator of T- and B-cell antigen receptor signaling |
| 2 | PPP4R3B | protein phosphatase 4 regulatory subunit 3B | Unknown |
| 2 | BCL2L11? | BCL2 like 11 | regulator of apoptosis in thymocyte negative selection |
| 2 | IFIH1 | interferon induced with helicase C domain 1 | innate immune receptor |
| 2 | CTLA4 | cytotoxic T-lymphocyte-associated protein 4 | T lymphocyte checkpoint regulator |
| 2 | FARP2-STK25 | ? | ? |
| 3 | UBE2E2 | ubiquitin-conjugating enzyme E2 E2 | Protein ubiquitination pathway; damage response |
| 3 | FOXP1 | forkhead box protein P1 | transcriptional regulator of B-cell development |
| 3 | CD80 | T-lymphocyte activation antigen CD80 | T-cell co-stimulatory signal |
| 3 | LPP | lipoma-preferred partner | Unknown |
| 3 | FBXO45-NRROS | ? | ? |
| 4 | PPP3CA | Serine/threonine-protein phosphatase 2B catalytic subunit alpha isoform |
T lymphocyte calcium-dependent, calmodulin-stimulated protein phosphatase |
| 6 | IRF4 | interferon regulatory factor 4 | transcriptional activator in immune cells and melanocytes |
| 6 | SERPINB9 | serpin B9 | endogenous inhibitor of granzyme B |
| 6 | HLA-A | HLA class I histocompatibility antigen, A | presents peptide antigens to the immune system |
| 6 | HLA-DRB1/DQA1 | HLA class II histocompatibility antigens, DRB1 and DQA1 |
present peptide antigens to the immune system |
| 6 | BACH2 | BTB domain and CNC homolog 2 | transcriptional activator, regulator of apoptosis |
| 6 | RNASET2-FGFR1OP-CCR6 | ? | ? |
| 7 | CPVL | probable serine carboxypeptidase CPVL | inflammatory protease; trims antigens for presentation |
| 8 | SLA | Src-like-adapter | regulator of T cell antigen receptor signaling |
| 9 | NEK6 | NIMA-related serine/threonine-protein kinase Nek6 | regulator of apoptosis |
| 10 | IL2RA | interleukin-2 receptor subunit alpha | IL2 receptor; regulates regulator T lymphocytes |
| 10 | ARID5B | AT-rich interactive domain-containing protein 5B | transcriptional co-activator |
| 10 | ZMIZ1 | zinc finger MIZ domain-containing protein 1 | possible PIAS-family transcriptional or sumoylation regulator |
| 10 | CASP7 | caspase-7 | apoptosis executor protein |
| 11 | CD44 | CD44 antigen | regulator of FOXP3 expression |
| 11 |
PPP1R14B-PLCB3-BAD-GPR137-KCNK4- TEX40-ESRRA-TRMT112-PRDX5 |
? | ? |
| 11 | TYR | Tyrosinase | melanocyte melanogenic enzyme; vitiligo autoantigen |
| 11 | Gene desert | ? | ? |
| 12 | PMEL | pre-melanosome protein PMEL | melanocyte melanosomal type I transmembrane glycoprotein |
| 12 | IKZF4 | zinc finger protein Eos | transcriptional repressor; regulates FOXP3 transcription in regulatory T lymphocytes |
| 12 | SH2B3 | SH2B adapter protein 3 | links T lymphocyte receptor activation signal to phospholipase C-gamma- 1, GRB2 and phosphatidylinositol 3-kinase |
| 13 | TNFSF11 | tumor necrosis factor ligand superfamily member 11 | T lymphocyte cytokine that binds to TNFRSF11A and TNFRSF11B |
| 14 | GZMB | granzyme B | apoptosis executioner protein of cytotoxic T lymphocytes |
| 15 | OCA2-HERC2 | oculocutaneous albinism 2 | melanocyte melanogenic protein; vitiligo autoantigen |
| 16 | MC1R | melanocortin 1 receptor | melanocyte melanogenic protein; vitiligo autoantigen |
| 17 | KAT2A-HSPB9-RAB5C | ? | ? |
| 18 | TNFRSF11A | tumor necrosis factor receptor superfamily member 11A |
regulates interactions between T lymphocytes and dendritic cells |
| 19 | TICAM1 | TIR domain-containing adapter molecule 1 | TLR3/TLR4 adapter; mediates NF-kappa-B and interferon-regulatory factor (IRF) activation; induces apoptosis |
| 19 | SCAF1-IRF3-BCL2L12 | ? | ? |
| 20 | RALY- ASIP | agouti signaling protein | regulator of melanocytes via MC1R |
| 20 | PTPN1 | tyrosine-protein phosphatase non-receptor type 1 | dephosphorylates JAK2 and TYK2 kinases; cellular response to interferon? |
| 21 | UBASH3A | ubiquitin-associated and SH3 domain-containing protein A |
promotes accumulation of activated T cell receptors on surface |
| 22 | C1QTNF6 | complement C1q tumor necrosis factor-related protein 6 |
Unknown |
| 22 | ZC3H7B-TEF | ? | ? |
| X | IL1RAPL1 | Interleukin-1 receptor accessory protein-like 1 | Unknown |
| X | CCDC22-FOXP3-GAGE | ? | FOXP3 regulates development and inhibitory function of regulatory T- lymphocytes |
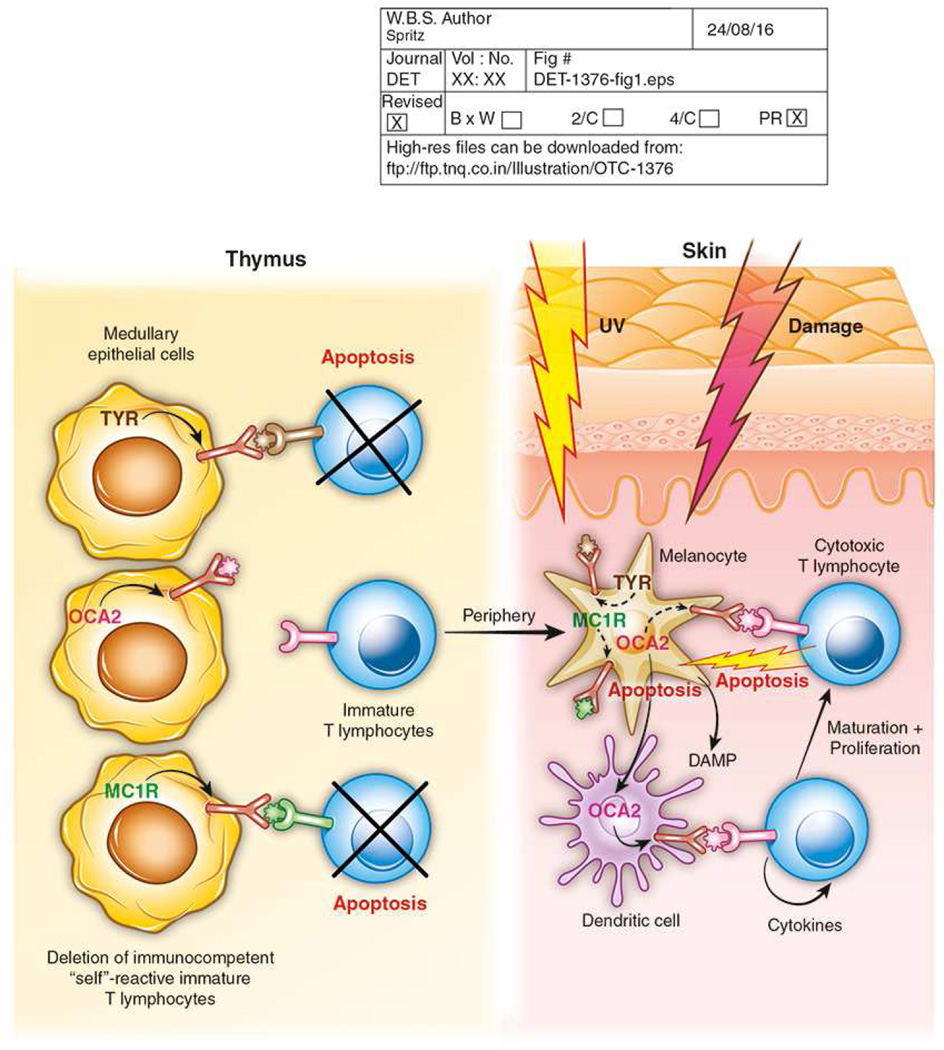
Figure 1.
General framework of vitiligo pathogenesis. During early development, a T lymphocyte repertoire is selected by positive selection of immunocompetent immature T lymphocytes in the thymic cortex. Immunocompetent T lymphocytes that recognize “self” antigens expressed by medullary epithelial cells undergo negative selection and undergo apoptosis. Immunocompetent immature T lymphocytes that do not encounter a cognate “self” antigen then exit the thymus to the peripheral circulation. In the skin, many or most cases of vitiligo initiate with skin damage, often UV exposure or trauma, a process termed “Koebnerization.” Damaged melanocytes apoptose and release molecules that act as “damage-associated molecular patterns” (DAMPs), which stimulate activation of local dendritic cells. Dendritic cells engulf melanosomal proteins, which are degraded in the proteasome, and fragments that act as peptide antigens are presented by HLA class II molecules on the dendritic cell surface. Immature T lymphocytes that express cognate T cell receptors bind these “self” antigens and are activated to express co-stimulatory molecules that result in cell proliferation and differentiation into CD8+ effector cytotoxic T lymphocytes, with the assistance of CD4+ T helper cells. The resultant activated cytotoxic T lymphocytes recognize and bind the cognate “self” antigen presented by HLA class I molecules on the melanocyte surface, assisted by interaction of FAS ligand on the T cell and FAS on the target melanocyte. The cytotoxic T lymphocyte then elaborates granzyme B and perforin, which induce apoptosis of the target melanocyte. Almost all of these processes involve proteins that are encoded by one or more genes associated with genetic susceptibility to vitiligo.
Fine-mapping and functional analyses of these vitiligo loci identified in European-derived whites indicates that, for vitiligo as for other complex diseases, about half of causal variants appear to affect gene regulatory regions, while only about 15% are located within exons, many resulting in missense substitutions. The Spritz group has shown that for both MHC class I (HLA-A) [68,69] and MHC class II (HLA-DRB1/-DQA1) [70,71], the vitiligo-associated causal SNPs are located in transcriptional enhancer elements that up-regulate expression of the corresponding MHC genes, resulting in gain of function. Interestingly, the MHC class II association signal also constitutes a quantitative trait locus (QTL) for vitiligo age-of-onset [72]. For NLRP1, which encodes an inflammasome regulatory component, the vitiligo-associated causal SNPs constitute haplotypes of missense variants in almost complete linkage disequilibrium, which together synergize to result in constitutive gain of NLRP1 function and thus activation of interleukin-1 beta [73]. For GZMB, encoding granzyme B, an apoptotic effector protein used by cytotoxic T-cells to kill their targets, the causal vitiligo-associated SNP is a common missense variant R55Q [74] that alters GZMB function. For TYR, encoding tyrosinase, a key melanogenic enzyme and the major vitiligo autoimmune antigen, the vitiligo-associated SNPs are protective, and represent the missense variants S192Y and R402Q that are common in European-derived whites but not in other populations and which reduce thermal stability and catalytic function [75].
In addition to studies in European-derived whites, there have been several GWAS of vitiligo in Asian populations. Zhang and co-workers carried out a large GWAS of vitiligo in the Han and Uygur populations of China, detecting complex association signals in the class I and class II regions of the MHC and with the RNASET2-FGFR1OP-CCR6 region of chromosome 6q24 [76]. Deeper analysis of this GWAS [77] detected additional association signals in the region of PMEL, 10q22.1 and a nearby locus suggested to be ZMIZ1 [78], and 11q23.3. Of these associations in Chinese, only that in the RNASET2-FGFR1OP-CCR6 region corresponds to an association detected in Caucasian patients [63]. Indeed, while MHC class I and class II region associations were detected in Caucasians [62,65,68,69,71] and Chinese [76], the specific underlying associations appear to be somewhat different. This is surprising, as a GWAS of vitiligo in Japanese [79] detected an MHC class I association with HLA-A*02:01 that appears identical to that in European-derived whites, while an immunocentric GWAS of vitiligo in Asian Indian and Pakistani patients detected an MHC class II association that similar to that European-derived whites [80], though more detailed MHC analysis in Indians [81] found class II association that was the same as in Chinese [76]. A very small GWAS of vitiligo in Koreans [82] was severely underpowered, and detected no association signals that met the genomewide significance threshold.
Where are we now?
The main purpose of identifying genes associated with disease risk is that such genes are causal, providing solid starting points for defining pathobiological mechanisms and approaches to treatment. To date, approximately 50 different genetic loci have been discovered that contribute to risk of vitiligo, most in European-derived whites (Table 1). For most of these loci specific genes have been identified, which have moved us far along in our understanding of the biological causation of vitiligo. Almost all of the identified genes encode proteins involved in immunoregulation, apoptosis, and melanocyte function, underscoring the autoimmune basis of vitiligo, with dysregulated immune programming, cellular activation, and melanocyte target cell recognition and killing (Figure 1). Thus far, the vitiligo susceptibility genes that have been identified provide little or no support for various alternative non-autoimmunity theories that have been proposed for vitiligo causation, such as oxidative stress, neural mechanisms, melanocytorrhagy, and others.
As anticipated, many of the vitiligo susceptibility genes that encode proteins with immunoregulatory and apoptotic functions have also been associated with other autoimmune diseases with which vitiligo is epidemiologically associated (Figure 2). These shared genetic associations thus account for these long recognized clinical epidemiologic associations. Unexpectedly, however, most of vitiligo susceptibility genes that encode proteins involved in melanocyte function have also been associated with melanoma, and in some cases nonmelanoma skin cancers, in each case involving the same associated SNPs, but with opposite effects. While the precise meaning of this observation is not yet clear, it suggests that vitiligo might involve a dysregulated mechanism that has evolved for immune surveillance for melanoma [66,67] and other skin cancers, consistent with the approximately threefold reduced incidence of melanoma and nonmelanoma skin cancers observed in patients with pre-existing vitiligo [83,84]. Alternatively, it might be that gene variants that reduce skin pigmentation elevate risk for all forms of skin cancer, melanoma and nonmelanoma.
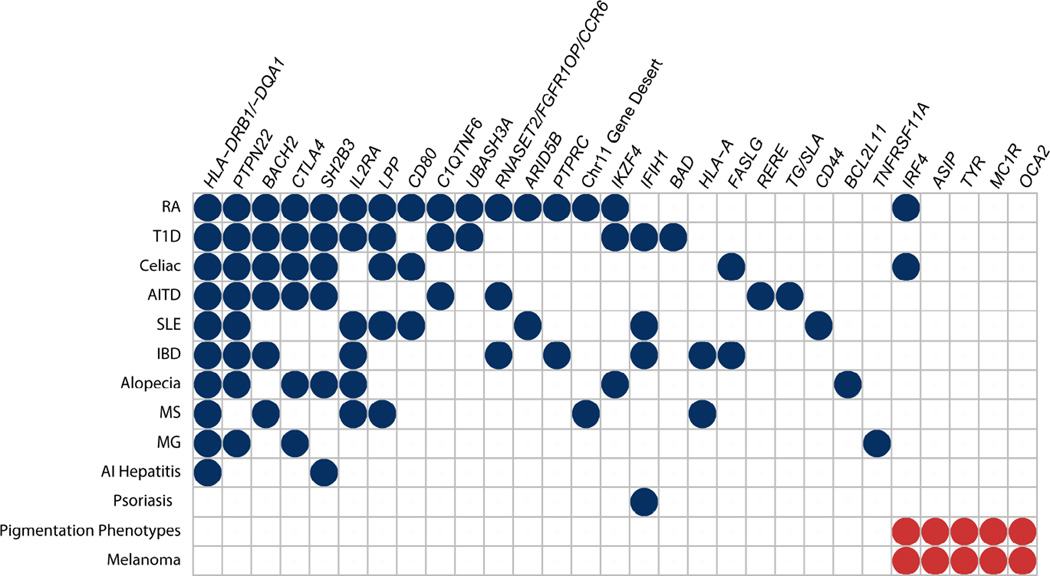
Figure 2.
Shared genetic associations of vitiligo with other autoimmune diseases and with pigmentation and melanoma phenotypes. Blue circles indicate shared genetic associations between vitiligo and other autoimmune diseases. Red circles indicate shared genetic associations between vitiligo and normal pigmentary variation phenotypes and melanoma. Only associations identified by GWAS and meeting the genomewide significance criterion (P < 5 × 10−8) are shown; associations claimed on the basis of candidate gene case-control studies are not included. RA, rheumatoid arthritis; T1D, type 1 diabetes mellitus; AITD, autoimmune thyroid disease; SLE, systemic lupus erythematosus; IBD, inflammatory bowel disease; MS, multiple sclerosis; MG, myasthenia gravis; AI hepatitis, autoimmune hepatitis.
Most vitiligo susceptibility genes have been detected in European-derived whites. Some of these genes likewise contribute to vitiligo risk in Asian populations, whereas others apparently do not. In European-derived whites, altogether the identified genes and gene variants account for about 25% of total vitiligo genetic risk. It remains unknown whether the remainder of risk is attributable to additional unknown variation in these same genes, to additional unknown genes that exert smaller effects, to genetic interactions that potentiate gene effects, or to other causes. Understanding this will be essential to achieve personalized medicine for vitiligo, enabling reasonably accurate prediction of risks and classification of patients into genetically-based subgroups that may benefit from specialized approaches to vitiligo treatment or even prevention.
Key Points.
Vitiligo is a “complex disorder” (also termed polygenic and multifactorial), reflecting simultaneous contributions of multiple genetic risk factors and environmental triggers.
Large-scale genome-wide association studies, principally in European-derived whites and in Chinese, have discovered approximately 50 different genetic loci that contribute to vitiligo risk, some of which also contribute to other autoimmune diseases that are epidemiologically associated with vitiligo. At many of these vitiligo susceptibility loci the corresponding relevant genes have now been identified, and for some of these genes the specific DNA sequence variants that contribute to vitiligo risk are also now known.
A large fraction of these genes encode proteins involved in immune regulation, a number of others play roles in cellular apoptosis, and still others are involved in regulating functions of melanocytes.
While many of the specific biologic mechanisms through which these genetic factors operate to cause vitiligo remain to be elucidated, it is now clear that vitiligo is an autoimmune disease involving a complex relationship between programming and function of the immune system, aspects of the melanocyte autoimmune target, and dysregulation of the immune response.
Acknowledgments
They received no funding for writing this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no commercial or financial conflict of interest related to this manuscript.
References
- 1.Le Cat M. Traité de la Couleur de la Peau Humaine. Amsterdam: 1765. [Google Scholar]
- 2.Stűttgen G. Die Vitiligo in erbbiologischer Betrachtung. Z Haut Geschlechtskr. 1950;9(11):451–457. [PubMed] [Google Scholar]
- 3.Teindel H. Familiäre Vitiligo. Z Haut Geschlechtskr. 1950;9(11):457–462. [PubMed] [Google Scholar]
- 4.Mohr J. Vitiligo in a pair of monovular twins. Acta Genet Stat Med. 1951;2(3):252–255. doi: 10.1159/000150674. [DOI] [PubMed] [Google Scholar]
- 5.Siemens HW. Het erfelijkheidsvraagstuk bij vitiligo. Ned Tijdschr Geneeskd. 1953;97(38):2449–2451. [PubMed] [Google Scholar]
- 6.Vogel F. [Dermatological observations on uniovular twins: vitiligo, ichthyosis simplex, psoriasis.] Z Haut Geschlechtskr. 1956;20(1):1–4. [PubMed] [Google Scholar]
- 7.Sidi E, Bourgeois-Gavardin J. The treatment of vitiligo with Ammi majus. Presse Méd. 1953;61(21):436–440. [PubMed] [Google Scholar]
- 8.Behl PN. Leucoderma and its treatment with Ammi majus. J Indian Med Assoc. 1955;24(16):615–618. [PubMed] [Google Scholar]
- 9.Levai M. A study of certain contributory factors in the development of vitiligo in South Indian patients. AMA Arch Derm. 1958;78(3):364–371. doi: 10.1001/archderm.1958.01560090080017. [DOI] [PubMed] [Google Scholar]
- 10.Lerner AB. Vitiligo. J Invest Dermatol. 1959;32(2, Part 2):285–310. [PubMed] [Google Scholar]
- 11.Hafez M, Sharaf L, Abd el-Nabi SM. The genetics of vitiligo. Acta Derm Venereol. 1983;63(3):249–251. [PubMed] [Google Scholar]
- 12.Das SK, Majumder PP, Majumdar TK, Haldar B. Studies on vitiligo. II. Familial aggregation and genetics. Genet Epidemiol. 1985;2(3):255–262. doi: 10.1002/gepi.1370020303. [DOI] [PubMed] [Google Scholar]
- 13.Majumder PP, Das SK, Li CC. A genetical model for vitiligo. Am J Hum Genet. 1988;43(2):119–125. [PMC free article] [PubMed] [Google Scholar]
- 14.Bhatia PS, Mohan L, Pandey ON, Singh KK, Arora SK, Mukhija RD. Genetic nature of vitiligo. J Dermatol Sci. 1992;4(3):180–184. doi: 10.1016/0923-1811(92)90017-6. [DOI] [PubMed] [Google Scholar]
- 15.Majumder PP1, Nordlund JJ, Nath SK. Pattern of familial aggregation of vitiligo. Arch Dermatol. 1993;129(8):994–998. [PubMed] [Google Scholar]
- 16.Nath SK, Majumder PP, Nordlund JJ. Genetic epidemiology of vitiligo: multilocus recessivity cross-validated. Am J Hum Genet. 1994;55(5):981–990. [PMC free article] [PubMed] [Google Scholar]
- 17.Alkhateeb A, Fain PR, Thody A, Bennett DC, Spritz RA. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003;16(3):208–214. doi: 10.1034/j.1600-0749.2003.00032.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang XJ, Liu JZB, Gui JP, Li M, Xiong QG, Wu HB, Li JX, Yang S, Wang HY, Gao M, Yang J, Yang Q. Characteristics of genetic epidemiology and genetic models for vitiligo. J Am Acad Dermatol. 2004;51(3):383–390. doi: 10.1016/j.jaad.2003.12.044. [DOI] [PubMed] [Google Scholar]
- 19.Laberge G, Mailloux CM, Gowan K, Holland P, Bennett DC, Fain PR, Spritz RA. Early disease onset and increased risk of other autoimmune diseases in familial generalized vitiligo. Pigment Cell Res. 2005;18(4):300–305. doi: 10.1111/j.1600-0749.2005.00242.x. [DOI] [PubMed] [Google Scholar]
- 20.Addison T. On the Constitutional and Local Effects of Disease of the Supra-Renal Capsules. London: Samuel Highley; 1855. [Google Scholar]
- 21.Schmidt M. Eine biglanduiare Erkrankung (Nebennieren und Schilddruse) bei Morbus Addisonii. Verh Dtsch Ges Pathol. 1926;21:212–221. [Google Scholar]
- 22.Neufeld M, Blizzard RM. Polyglandular autoimmune diseases. In: Pinchera A, Doniach D, Fenzi GF, et al., editors. Symposium on Autoimmune Aspects of Endocrine Disorders. New York: Academic Press; 1980. pp. 357–365. [Google Scholar]
- 23.Steve BF. Further investigations in the treatment of vitiligo. Virginia Med Monthly. 1945;71(1):6–17. [Google Scholar]
- 24.Srivistava GN, Shukla RC. ABO blood groups in vitiligo. Indian J Med Res. 1965;53(3):221–225. [PubMed] [Google Scholar]
- 25.Singh G, Shanker P. Vitiligo and blood groups. Preliminary report. Br J Dermatol. 1966;78(2):91–92. doi: 10.1111/j.1365-2133.1966.tb12180.x. [DOI] [PubMed] [Google Scholar]
- 26.Sehgal VN, Dube B. ABO blood groups and vitiligo. J Med Genet. 1968;5:308–309. doi: 10.1136/jmg.5.4.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutta AK, Mondal SB, Dutta SB. ABO blood group and secretory status in vitiligo. J Indian Med Assoc. 1969;53(4):186–189. [PubMed] [Google Scholar]
- 28.Oriente Biondi C, Ruocco V. Vitiligo and blood groups. Rass Int Clin Ter. 1969;49(22):1395–1399. [PubMed] [Google Scholar]
- 29.Kareemullah L, Taneja V, Begum S, Sarma PK, Baig HA. Association of ABO blood groups and vitiligo. J Med Genet. 1977;14(3):211–213. doi: 10.1136/jmg.14.3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wasfi AI, Saha N, El Munshid HA, El Sheikh FS, Ahmed MA. Genetic association in vitiligo: ABO, MNSs, Rhesus, Kell and Duffy blood groups. Clin Genet. 1980;17(6):415–417. doi: 10.1111/j.1399-0004.1980.tb00172.x. [DOI] [PubMed] [Google Scholar]
- 31.Sehgal VN, Dube B. Secretory state in vitiligo. Dermatologica. 1969;138(2):89–92. doi: 10.1159/000253970. [DOI] [PubMed] [Google Scholar]
- 32.Mujahid Ali M, Banu M, Waheed MA, Qadri GS, Habibullah CM. Serum alpha 1-antitrypsin and haptoglobin phenotypes in vitiligo. Arch Dermatol Res. 1990;282(3):206–207. doi: 10.1007/BF00372625. [DOI] [PubMed] [Google Scholar]
- 33.Retornaz G, Betuel H, Ortonne JP, Thivolet J. HL-A antigens and vitiligo. Br J Dermatol. 1976;95(2):173–175. doi: 10.1111/j.1365-2133.1976.tb00822.x. [DOI] [PubMed] [Google Scholar]
- 34.Kachru RB, Telischi M, Mittal KK. HLA antigens and vitiligo in an American black population. Tissue Antigens. 1978;12(5):396–397. doi: 10.1111/j.1399-0039.1978.tb01349.x. [DOI] [PubMed] [Google Scholar]
- 35.Metzker A, Zamir R, Gazit E, David M, Feuerman EJ. Vitiligo and the HLA system. Dermatologica. 1980;160(2):100–105. doi: 10.1159/000250480. [DOI] [PubMed] [Google Scholar]
- 36.Nakagawa H, Otuka F, Kukita A, Mizoguchi M, Ito H, Juji T. Histocompatible antigens in vitiligo vulgaris II. Nihon Hifuka Gakkai Zasshi. 1980;90(10):939–941. [PubMed] [Google Scholar]
- 37.Minev N, Tonkin N, Martinova F. Association of the HLA system with vitiligo. Vestn Dermatol Venerol. 1985 May;(5):41–42. [PubMed] [Google Scholar]
- 38.Foley LM, Lowe NJ, Misheloff E, Tiwari JL. Association of HLA-DR4 with vitiligo. J Am Acad Dermatol. 1983;8(1):39–40. doi: 10.1016/s0190-9622(83)80279-5. [DOI] [PubMed] [Google Scholar]
- 39.Liu J-B, Li M, Chen H, Zhong S-Q, Du W-D, Hao J-H, Zhang T-S, Zhang X-J, Zeegers MP. Association of vitiligo with HLA-A2: a meta-analysis. J Eur Acad Dermatol Venereol. 2007;21(2):205–213. doi: 10.1111/j.1468-3083.2006.01899.x. [DOI] [PubMed] [Google Scholar]
- 40.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4(2):45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 41.Ioannidis JPA, Tarone R, McLaughlin JK. The false-positive to false-negative ratio in epidemiologic studies. Epidemiology. 2011;22(4):450–456. doi: 10.1097/EDE.0b013e31821b506e. [DOI] [PubMed] [Google Scholar]
- 42.Kemp EH, Ajjan RA, Waterman EA, Gawkrodger DJ, Cork MJ, Watson PF, Weetman AP. Analysis of a microsatellite polymorphism of the cytotoxic T-lymphocyte antigen-4 gene in patents with vitiligo. Br J Dermatol. 1999;140(1):73–78. doi: 10.1046/j.1365-2133.1999.02610.x. [DOI] [PubMed] [Google Scholar]
- 43.Blomhoff A, Kemp EH, Gawkrodger DJ, Weetman AP, Husebye ES, Akselsen HE, Lie BA, Undlien DE. CTLA4 polymorphisms are associated with vitiligo, in patients with concomitant autoimmune diseases. Pigment Cell Res. 2004;18(1):55–58. doi: 10.1111/j.1600-0749.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 44.Birlea SA, LaBerge GS, Procopciuc LM, Fain PR, Spritz RA. CTLA4 and generalized vitiligo: two genetic association studies and a meta-analysis of published data. Pigment Cell Melanoma Res. 2009;22(2):230–234. doi: 10.1111/j.1755-148X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cantón I, Akhtar S, Gavalas NG, Gawkrodger DJ, Blomhoff A, Watson PF, Weetman AP, Kemp EH. A single-nucleotide polymorphism in the gene encoding lymphoid protein tyrosine phosphatase (PTPN22) confers susceptibility to generalised vitiligo. Genes Immun. 2005;6:584–587. doi: 10.1038/sj.gene.6364243. [DOI] [PubMed] [Google Scholar]
- 46.LaBerge GS, Bennett DC, Fain PR, Spritz RA. PTPN22 is genetically associated with risk of generalized vitiligo, but CTLA4 is not. J Investig Dermatol. 2008;128:1757–1762. doi: 10.1038/sj.jid.5701233. [DOI] [PubMed] [Google Scholar]
- 47.LaBerge GS, Birlea SA, Fain PR, Spritz RA. The PTPN22-1858C>T (R620W) functional polymorphism is associated with generalized vitiligo in the Romanian population. Pigment Cell Melanoma Res. 2008;21(2):206–208. doi: 10.1111/j.1755-148X.2008.00443.x. [DOI] [PubMed] [Google Scholar]
- 48.Nath SK, Kelly JA, Namjou B, Lam T, Bruner GR, Scofield RH, Aston CE, Harley JB. Evidence for a susceptibility gene, SLEV1, on chromosome 17p13 in families with vitiligo-related systemic lupus erythematosus. Am J Hum Genet. 2001;69(6):1401–1406. doi: 10.1086/324470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spritz RA, Gowan K, Bennett DC, Fain PR. Novel vitiligo susceptibility loci on chromosomes 7 (AIS2) and 8 (AIS3), confirmation of SLEV1 on chromosome 17, and their roles in an autoimmune diathesis. Am J Hum Genet. 2004;74(1):188–191. doi: 10.1086/381134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin Y, Mailloux CM, Gowan K, Riccardi SL, LaBerge G, Bennett DC, Fain PR, Spritz RA. NALP1 in vitiligo-associated multiple autoimmune disease. N Engl J Med. 2007;356(12):1216–1225. doi: 10.1056/NEJMoa061592. [DOI] [PubMed] [Google Scholar]
- 51.Alkhateeb A, Stetler GL, Old W, Talbert J, Uhlhorn C, Taylor M, Fox A, Miller C, Dills DG, Ridgway EC, Bennett DC, Fain PR, Spritz RA. Mapping of an autoimmunity susceptibility locus (AIS1) to chromosome 1p31.3p32.2. Hum Mol Genet. 2002;11(6):661–667. doi: 10.1093/hmg/11.6.661. [DOI] [PubMed] [Google Scholar]
- 52.Fain PR, Gowan K, LaBerge GS, Alkhateeb A, Stetler GL, Talbert J, Bennett DC, Spritz RA. A genomewide screen for generalized vitiligo: confirmation of AIS1 on chromosome 1p31 and evidence for additional susceptibility loci. Am J Hum Genet. 2003;72(6):1560–1564. doi: 10.1086/375451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alkhateeb A, Fain P, Spritz RA. Candidate functional promoter variant in the FOXD3 melanoblast developmental regulator gene in autosomal dominant vitiligo. J. Investig. Dermatol. 2005;125:388–391. doi: 10.1111/j.0022-202X.2005.23822.x. [DOI] [PubMed] [Google Scholar]
- 54.Schunter JA, Löffler D, Wiesner T, Kovacs P, Badenhoop K, Aust G, Tönjes A, Müller P, Baber R, Simon JC, Führer D, Pfäffle RW, Thiery J, Stumvoll M, Kiess W1, Kratzsch J, Körner A. A novel FoxD3 variant is associated with vitiligo and elevated thyroid auto-antibodies. J Clin Endocrinol Metab. 2015;100(10):E1335–E1342. doi: 10.1210/jc.2015-2126. [DOI] [PubMed] [Google Scholar]
- 55.Jin Y, Riccardi SL, Gowan K, Fain PR, Spritz RA. Fine-mapping of vitiligo susceptibility loci on chromosomes 7 and 9 and interactions with NLRP1 (NALP1) J Invest Dermatol. 2010;130(3):774–783. doi: 10.1038/jid.2009.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen JJ, Huang W, Gui JP, Yang S, Zhou FS, Xiong QG, Wu HB, Cui Y, Gao M, Li W, Li JX, Yan KL, Yuan WT, Xu SJ, Liu JJ, Zhang XJ. A novel linkage to generalized vitiligo on 4q13–q21 identified in a genomewide linkage analysis of Chinese families. Am J Hum Genet. 2005;76(6):1057–1065. doi: 10.1086/430279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liang Y, Yang S, Zhou Y, Gui J, Ren Y, Chen J, Fan X, Sun L, Xiao F, Gao M, Du W, Fang Q, Xu S, Huang W, Zhang X. Evidence for two susceptibility loci on chromosomes 22q12 and 6p21–p22 in Chinese generalized vitiligo families. J Invest Dermatol. 2007;127(11):2552–2557. doi: 10.1038/sj.jid.5700904. [DOI] [PubMed] [Google Scholar]
- 58.Xu S, Zhou Y, Yang S, Ren Y, Zhang C, Quan C, Gao M, He C, Chen H, Hhan J, Chen J, Liang Y, Yang J, Sun L, Yin X, Liu J, Zhang X. Platelet-derived growth factor receptor alpha gene mutations in vitiligo vulgaris. Acta Derm Venereol. 2010;90(2):131–135. doi: 10.2340/00015555-0820. [DOI] [PubMed] [Google Scholar]
- 59.Ren Y, Yang S, Xu S, Gao M, Huang W, Gao T, Fang Q, Quan C, Zhang C, Sun L, Liang Y, Han J, Wang Z, Zhang F, Zhou Y, Liu J, Zhang X. Genetic variation of promoter sequence modulates XBP1 expression and genetic risk for vitiligo. PLoS Genet. 2009;5(6):e1000523. doi: 10.1371/journal.pgen.1000523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Birlea SA, Fain PR, Spritz RA. A Romanian population isolate with high frequency of vitiligo and associated autoimmune diseases. Arch Dermatol. 2008;144(3):310–316. doi: 10.1001/archderm.144.3.310. [DOI] [PubMed] [Google Scholar]
- 61.Birlea SA, Gowan K, Fain PR, Spritz RA. Genome-wide association study of generalized vitiligo in an isolated European founder population identifies SMOC2, in close proximity to IDDM8. J Invest Dermatol. 2010;130(3):798–803. doi: 10.1038/jid.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, Mailloux CM, Sufit AJ, Hutton SM, Amadi-Myers A, Bennett DC, Wallace MR, McCormack WT, Kemp EH, Gawkrodger DJ, Weetman AP, Picardo M, Leone G, Taïeb A, Jouary T, Ezzedine K, van Geel N, Lambert J, Overbeck A, Spritz RA. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med. 2010;362(18):1686–1697. doi: 10.1056/NEJMoa0908547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin Y, Birlea SA, Fain PR, Mailloux CM, Riccardi SL, Gowan K, Holland PJ, Bennett DC, Wallace MR, McCormack WT, Kemp EH, Gawkrodger DJ, Weetman AP, Picardo M, Leone G, Taïeb A, Jouary T, Ezzedine K, van Geel N, Lambert J, Overbeck A, Spritz RA. Common variants in FOXP1 are associated with generalized vitiligo. Nat Genet. 2010;42(7):576–578. doi: 10.1038/ng.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Riccardi SL, Cole JB, Gowan K, Holland PJ, Bennett DC, Luiten RM, Wolkerstorfer A, van der Veen JP, Hartmann A, Eichner S, Schuler G, van Geel N, Lambert J, Kemp EH, Gawkrodger DJ, Weetman AP, Taïeb A, Jouary T, Ezzedine K, Wallace MR, McCormack WT, Picardo M, Leone G, Overbeck A, Silverberg NB, Spritz RA. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat Genet. 2012;44(6):676–680. doi: 10.1038/ng.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin Y, Birlea SA, Fain PR, Ferrara TM, Ben S, Andersen G, Burgess K, Holland PJ, Siebert J, Hartmann A, Lienert A, Luiten RM, Wolkerstorfer A, van der Veen JPW, van Geel N, Lambert J, Bennett DC, Taïeb A, Ezzedine K, Kemp EH, Gawkrodger DJ, Weetman AP, Koks S, Prans E, Kingo K, Karelson M, Wallace MR, McCormack WT, Overbeck A, Moretti S, Colucci R, Picardo M, Silverg NB, Olssen M, Valle Y, Korobko I, Böhm M, Lim H, Hamzavi I, Zhou L, Mi Q-S, Santorico S, Spritz RA. Enhanced genome-wide association studies of autoimmune vitiligo identify 23 novel loci and highlight key pathobiological pathways and causal regulatory variation. Nat Genet. 2016 doi: 10.1038/ng.3680. (2016) in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spritz RA. The genetics of generalized vitiligo: autoimmune pathways and an inverse relationship with malignant melanoma. Genome Med. 2010;2(10):78. doi: 10.1186/gm199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Das PK, van den Wijngaard RMJGJ, Wankowicz-Kalinska A, Le Poole IC. A symbiotic concept of autoimmunity and tumour immunity: lessons from vitiligo. Trends Immunol. 2001;22:130–136. doi: 10.1016/s1471-4906(00)01844-5. [DOI] [PubMed] [Google Scholar]
- 68.Hayashi M, Jin Y, Yorgov D, Santorico SA, Hagman J, Ferrara TM, Jones KL, Cavalli G, Dinarello CA, Spritz RA. Autoimmune vitiligo is associated with gain-of-function by a transcriptional regulator that elevates expression of HLA-A*02:01 in vivo. Proc Natl Acad Sci USA. 2016;113(5):1357–1362. doi: 10.1073/pnas.1525001113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jin Y, Ferrara T, Gowan K, Holcomb C, Rastrou M, Erlich HA, Fain PR, Spritz RA. Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of generalized vitiligo via antigen presentation. J Invest Dermatol. 2012;132(6):1730–1733. doi: 10.1038/jid.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fain PR, Babu SR, Bennett DC, Spritz RA. HLA class II haplotype DRB1*04-DQB1*0301 contributes to risk of familial generalized vitiligo and early disease onset. Pigment Cell Res. 2005;19:51–57. doi: 10.1111/j.1600-0749.2005.00279.x. [DOI] [PubMed] [Google Scholar]
- 71.Cavalli G, Hayashi M, Jin Y, Yorgov D, Santorico SA, Holcomb C, Rastrou M, Erlich H, Tengesdal IW, Dagna L, Neff CP, Palmer BE, Spritz RA, Dinarello CA. MHC class II super-enhancer increases surface expression of HLA-DR and HLA-DQ and affects cytokine production in autoimmune vitiligo. Proc Natl Acad Sci USA. 2016;113(5):1363–1368. doi: 10.1073/pnas.1523482113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, Bennett DC, Herbstman DM, Wallace MR, McCormack WT, Kemp EH, Gawkrodger DJ, Weetman AP, Picardo M, Leone G, Taïeb A, Jouary T, Ezzedine K, van Geel N, Lambert J, Overbeck A, Spritz RA. Genome-wide analysis identifies a quantitative trait locus in the MHC class II region associated with generalized vitiligo age of onset. J Invest Dermatol. 2011;131(6):1308–1312. doi: 10.1038/jid.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Levandowski CB, Mailloux CM, Ferrara TM, Gowan K, Ben S, Jin Y, McFann KK, Holland PJ, Fain PR, Dinarello CA, Spritz RA. NLRP1 haplotypes associated with vitiligo and autoimmunity increase interleukin-1β processing via the NLRP1 inflammasome. Proc Natl Acad Sci USA. 2013;110(8):2952–2956. doi: 10.1073/pnas.1222808110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferrara TM, Jin Y, Gowan K, Fain PR, Spritz RA. Risk of generalized vitiligo is associated with the common 55R-94A-247H variant haplotype of GZMB (encoding granzyme B) J Invest Dermatol. 2013;133(6):1677–1679. doi: 10.1038/jid.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tripathi RK, Giebel LB, Strunk KM, Spritz RA. A polymorphism of the human tyrosinase gene is associated with temperature-sensitive enzymatic activity. Gene Expr. 1991;1(2):103–110. [PMC free article] [PubMed] [Google Scholar]
- 76.Quan C, Ren YQ, Xiang LH, Sun LD, Xu AE, Gao XH, Chen HD, Pu XM, Wu RN, Liang CZ, Li JB, Gao TW, Zhang JZ, Wang XL, Wang J, Yang RY, Liang L, Yu JB, Zuo XB, Zhang SQ, Zhang SM, Chen G, Zheng XD, Li P, Zhu J, Li YW, Wei XD, Hong WS, Ye Y, Zhang Y, Wu WS, Cheng H, Dong PL, Hu DY, Li Y, Li M, Zhang X, Tang HY, Tang XF, Xu SX, He SM, Lv YM, Shen M, Jiang HQ, Wang Y, Li K, Kang XJ, Liu YQ, Sun L, Liu ZF, Xie SQ, Zhu CY, Xu Q, Gao JP, Hu WL, Ni C, Pan TM, Li Y, Yao S, He CF, Liu YS, Yu ZY, Yin XY, Zhang FY, Yang S, Zhou Y, Zhang XJ. Genome-wide association study for vitiligo identifies susceptibility loci at 6q27 and the MHC. Nat Genet. 2010;42(7):614–618. doi: 10.1038/ng.603. [DOI] [PubMed] [Google Scholar]
- 77.Tang XF, Zhang Z, Hu DY, Xu AE, Zhou HS, Sun LD, Gao M, Gao TW, Gao XH, Chen HD, Xie HF, Tu CX, Hao F, Wu RN, Zhang FR, Liang L, Pu XM, Zhang JZ, Han JW, Pan GP, Wu JQ, Li K, Su MW, Du WD, Zhang WJ, Liu JJ, Xiang LH, Yang S, Zhou YW, Zhang XJ. Association analyses identify three susceptibility Loci for vitiligo in the Chinese Han population. J Invest Dermatol. 2013;133(2):403–410. doi: 10.1038/jid.2012.320. [DOI] [PubMed] [Google Scholar]
- 78.Sun Y, Zuo X, Zheng X, Zhou F, Liang B, Liu H, Chang R, Gao J, Sheng Y, Cui H, Wang W, Andiappan AK, Rotzschke O, Yang S, Sun L, Zhang F, Zhang X, Ren Y, Liu J. A comprehensive association analysis confirms ZMIZ1 to be a susceptibility gene for vitiligo in Chinese population. J Med Genet. 2014;51(5):345–353. doi: 10.1136/jmedgenet-2013-102233. [DOI] [PubMed] [Google Scholar]
- 79.Jin Y, Hayashi M, Fain PR, Suzuki T, Fukai K, Oiso N, Tanemura A, Holcomb CL, Rastrou M, Erlich HA, Spritz RA. Major association of vitiligo with HLA-A*02:01 in Japanese. Pigment Cell Melanoma Res. 2015;28(3):360–362. doi: 10.1111/pcmr.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Birlea SA, Ahmad FJ, Uddin RM, Ahmad S, Pal SS, Begum R, Laddha NC, Dwivedi M, Shoab Mansuri M, Jin Y, Gowan K, Riccardi SL, Holland PJ, Ben S, Fain PR, Spritz RA. Association of generalized vitiligo with MHC class II loci in patients from the Indian subcontinent. J Invest Dermatol. 2013;133(5):1369–1372. doi: 10.1038/jid.2012.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh A, Sharma P, Kar HK, Sharma VK, Tembhre MK, Gupta S, Laddha NC, Dwivedi M, Begum R, Indian Genome Variation Consortium. Gokhale RS, Rani R. HLA alleles and amino-acid signatures of the peptide-binding pockets of HLA molecules in vitiligo. J Invest Dermatol. 2012;132(1):124–134. doi: 10.1038/jid.2011.240. [DOI] [PubMed] [Google Scholar]
- 82.Cheong KA, Kim NH, Noh M, Lee AY. Three new single nucleotide polymorphisms identified by a genome-wide association study in Korean patients with vitiligo. J Korean Med Sci. 2013;28(5):775–779. doi: 10.3346/jkms.2013.28.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Teulings HE, Overkamp M, Ceylan E, Nieuweboer-Krobotova L, Bos JD, Nijsten T, Wolkerstorfer AW, Luiten RM, van der Veen JPW. Decreased risk of melanoma in patients with vitiligo: a survey among 1307 patients and their partners. Br J Dermatol. 2013;168:162–171. doi: 10.1111/bjd.12111. [DOI] [PubMed] [Google Scholar]
- 84.Paradisi A, Tabolli S, Didona B, Sobrino L, Russo N, Abeni D. Markedly reduced incidence of melanoma and nonmelanoma skin cancer in a nonconcurrent cohort of 10,040 patients with vitiligo. J Am Acad Dermatol. 2014;71(6):1110–1116. doi: 10.1016/j.jaad.2014.07.050. [DOI] [PubMed] [Google Scholar]