Abstract
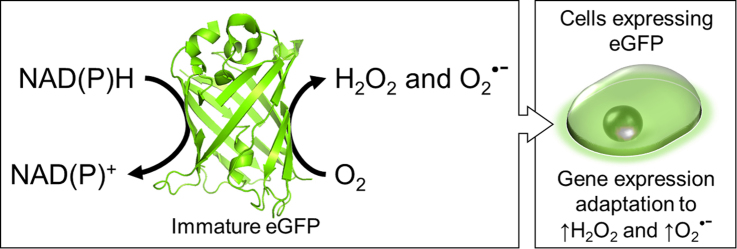
Fluorescent proteins are an important tool that has become omnipresent in life sciences research. They are frequently used for localization of proteins and monitoring of cells [1], [2]. Green fluorescent protein (GFP) was the first and has been the most used fluorescent protein. Enhanced GFP (eGFP) was optimized from wild-type GFP for increased fluorescence yield and improved expression in mammalian systems [3]. Many GFP-like fluorescent proteins have been discovered, optimized or created, such as the red fluorescent protein TagRFP [4]. Fluorescent proteins are expressed colorless and immature and, for eGFP, the conversion to the fluorescent form, mature, is known to produce one equivalent of hydrogen peroxide (H2O2) per molecule of chromophore [5,6]. Even though it has been proposed that this process is non-catalytic and generates nontoxic levels of H2O2 [6], this study investigates the role of fluorescent proteins in generating free radicals and inducing oxidative stress in biological systems. Immature eGFP and TagRFP catalytically generate the free radical superoxide anion (O2•–) and H2O2 in the presence of NADH. Generation of the free radical O2•– and H2O2 by eGFP in the presence of NADH affects the gene expression of cells. Many biological pathways are altered, such as a decrease in HIF1α stabilization and activity. The biological pathways altered by eGFP are known to be implicated in the pathophysiology of many diseases associated with oxidative stress; therefore, it is critical that such experiments using fluorescent proteins are validated with alternative methodologies and the results are carefully interpreted. Since cells inevitably experience oxidative stress when fluorescent proteins are expressed, the use of this tool for cell labeling and in vivo cell tracing also requires validation using alternative methodologies.
Keywords: GFP, H2O2 (hydrogen peroxide), O2•– (superoxide free radical anion), Free radicals, Oxidative stress, Redox biology
Graphical abstract
Highlights
-
•
Immature eGFP and TagRFP catalytically generate O2• – and H2O2 in the presence of NADH.
-
•
Cells expressing eGFP show gene expression associated with oxidative stress.
-
•
Experiments using fluorescent proteins should be alternatively validated.
1. Introduction
Fluorescent proteins are commonly used in biology for protein labeling and cell tracing [1], [2]. Green fluorescent protein (GFP) was the first and has been the most used fluorescent protein. Drs. Shimomura, Chalfie and Tsien were awarded a Nobel Prize in chemistry “for the discovery and development of the green fluorescent protein, GFP,” in 2008 [7]. Enhanced GFP (eGFP) was developed for increasing the fluorescence yield and improving its expression in mammalian systems [3]. More recently, many optimized GFP-like fluorescent proteins have been discovered, developed or created, such as the red fluorescent protein TagRFP [4].
The fluorescent chromophore of fluorescent proteins is formed through an intramolecular reaction between the side chains of certain amino acids localized inside the barrel structure of the protein [5], [8]. Even though it is known that chromophore formation requires cyclization and air-mediated oxidation, proposed mechanisms for this complex reaction remain to be proved. It has been calculated that one molecule of hydrogen peroxide (H2O2) is generated independently of NAD(P)H during the maturation of each chromophore of eGFP. This non-catalytic H2O2 generation is within the cellular baseline level and, consequently, should not be toxic to cells or organisms [6]. Maturation of the fluorescent chromophore of TagRFP has been proposed to generate two molecules of H2O2 independently of NAD(P)H [9], which is believed to be low and nontoxic to cells. However, many reports show that eGFP expression is cytotoxic [10], [11], [12], [13], [14] and, even though it is believed that it does not induce lethality in animals, other GFP-like fluorescent proteins and eGFP are shown to induce abnormalities in skeletal muscle [15] and heart [16]. Since chemical fluorescent probes such as DCFH are known to catalyze the formation of superoxide anion free radical (O2•–) and H2O2 in biological systems by a variety of mechanisms [17], we have searched for the formation of these reactive oxygen species by fluorescent proteins.
2. Material and methods
2.1. Chemicals and buffer
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless stated otherwise. Chelex-100 resin was purchased from Biorad (Hercules, CA). DMPO was purchased from Dojindo (Rockville, MD). Amplex Red was purchased from Invitrogen (Life Technologies, Grand Island, NY). To suppress trace metal contamination, phosphate buffer was Chelex-100-treated and contained 25 μM diethylene triamine pentaacetic acid (DTPA).
2.2. eGFP and TagRFP preparation in E. coli
Gateway® technology (Invitrogen, Thermo Fisher Scientific, Carlsbad, CA) was used to generate the protein-expression vectors. The pDEST vectors with eGFP and TagRFP genes were transformed by heat shock into Rosetta 2 DE3 pLacI competent cells. Ampicillin- and chloramphenicol-resistant colonies were grown in LB with the selecting agents. After cultures of OD 0.05 reached OD 0.6–0.8 (2 h, 220 rpm at 37 °C), protein expression was induced by the addition of 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG); and then cultures were incubated for 4 h at RT (220 rpm). eGFP and TagRFP were purified using an ÄKTA FPLC system (GE Healthcare Life Science, Pittsburgh, PA). Concentrations of the fluorescent proteins were determined based on protein content using the BCA Protein Assay Kit from Pierce® (Rockford, IL) following the manufacturer's instructions. The molecular mass of eGFP was adopted as 27,333.8g/mol and that of TagRFP was adopted as 26,732.5g/mol. No difference was detected when eGFP or TagRFP concentrations were calculated using protein content or absorbance at 280 nm (ε for eGFP=21,890 M−1 cm−1 and ε for TagRFP=27,640 M−1 cm−1). eGFP was expressed and purified 7 independent times.
2.3. Quantification of H2O2 in vitro
Samples containing eGFP and NADH were kept under continuous agitation (700 rpm) at room temperature, and H2O2 was quantified using the FOX1 assay [18] adapted to a microplate format. Briefly, 5 μL of samples or H2O2 standards (1–100 μM, ε at 240 nm=43.6 M−1cm−1) were added to 200 μL of FOX assay reagent (100 μM xylenol orange, 250 μM ammonium ironII sulfate, 100 mM sorbitol in 25 mM H2SO4) in wells of 96-well microplates. After 30 min, microplates were read for absorbance at 560 nm. The concentration of H2O2 was calculated after subtracting the background absorbance of wells containing 200 μL FOX1 assay reagent where 5 μL buffer blank was added.
2.4. eGFP preparation in HEK 293 cells
We prepared a pDEST26-eGFP plasmid using Gateway® technology from Invitrogen (Thermo Fisher Scientific, Carlsbad, CA). This plasmid makes an N-terminal 6xHis tagged eGFP under the control of the cytomegalovirus (CMV) promoter. FreestyleTM293-F cells (Thermo Fisher Scientific, Carlsbad, CA) were grown in FreestyleTM293 expression media as a suspension culture using Thomson Optimum Growth™ flasks (Oceanside, CA). For eGFP expression, one liter of 1×106 cells/mL was transfected with 1 mg of pDEST26-eGFP in 2 mL of 1.6 mg/mL PEI MAX (Polysciences, Inc., Warrington, PA). Cultures were allowed to express eGFP for 48 h. Cells were harvested by centrifugation, washed with PBS, flash frozen and stored at −80 °C. For protein extraction, M-Per extraction reagent was added to frozen cell pellets (Thermo Fisher Scientific, Carlsbad, CA) containing 0.2 M NaCl, 5 mM imidazole and EDTA-free protease inhibitors (Roche, Indianapolis, IN). Lysates were prepared with 15 strokes of a Dounce homogenizer and sonication on ice, and insoluble material was removed by centrifugation at 28,000g, for 30 min at 4 °C. Cleared lysate was applied to a NiNTA-agarose column (QIAGEN, Valencia, CA) pre-equilibrated in wash buffer (50 mM Hepes, 100 mM NaCl and 10 mM imidazole, pH 7.5). An ÄKTA FPLC system was used (GE Healthcare Life Science, Pittsburgh, PA). The column was then washed with 10 column volumes of wash buffer containing 20 mM imidazole. Proteins bound to the column were eluted with a step gradient of imidazole in wash buffer (50, 100, and 200 mM). Fractions were analyzed by SDS-PAGE. Selected fractions were pooled and dialyzed into anion exchange buffer (50 mM Hepes and 10 mM NaCl, pH 7.5) before being applied to a HiTrap Q column (GE Healthcare Life Science, Pittsburgh, PA). eGFP was eluted with a linear gradient from 50 to 1000 mM NaCl over 20 column volumes. Pure fractions of eGFP were buffer exchanged into PBS, pH 7.4, and concentrated using 10 kDa cutoff centrifugal concentrators (Millipore, Billerica, MA). The purity of eGFP was examined by looking at the homogeneity of peaks in the FPLC-chromatograms and by SDS-PAGE.
2.5. ESR spectroscopy
ESR spectra were recorded with a Bruker EleXsys E-500 spectrometer (Bruker, Billerica, MA). The instrument operates at 9.80 GHz (X-Band). We used 20 mW microwave power, a modulation frequency of 100 kHz and a modulation amplitude of 1.0 G. The time constant and conversion time were 163.84 ms and the receiver gain was 1×104. For anaerobic measurements, the solutions were bubbled with nitrogen for at least 20 min. All experiments were repeated twice (n=2) on three different days.
2.6. Expression of eGFP and MBP in E. coli
The gene for eGFP was recombined from pDONR 221 into a pDEST 527 expression plasmid (destination vector kindly supplied by Dr. Dominic Esposito, SAIC, MD). The pDEST 527 vector was transformed by heat shock into BL21-AI™ competent cells. The mbp gene was recombined from pDONR 221 into a pDEST 566-expressing plasmid and transformed by heat shock into Origami B DE3™ competent cells (Novagen, Darmstadt, Germany). Bacteria were routinely grown in Luria-Bertani broth with ampicillin (100 μg/mL). Cultures were prepared with Luria-Bertani broth at OD 0.05. After the cultures reached OD 0.6–0.8 (2 h, 220 rpm at 37 °C), IPTG (0.1 mM) was added and protein expression was induced by the addition of 0.002% and 0.02% arabinose (w/v). For the expression of MBP, IPTG alone (0.1 mM) was used as the inducer for high expression in bacteria. Cultures were then incubated for 2 h at 37 °C (220 rpm). Three independent cultures were prepared, and cell pellets were collected and frozen at −80 °C for RNA isolation or Amplex Red assay for the determination of H2O2 production.
2.7. Bacterial RNA isolation and real-time quantitative PCR
Total RNA was isolated from three biological replicate cultures of bacterial cells with the use of the TRIzol Max Bacterial RNA Isolation Kit from Ambion (Life Technologies, Grand Island, NY) and purified with the use of RNeasy Mini Kit Columns from QIAGEN (Valencia, CA) according to the manufacturer's instructions. Purified RNA was reverse transcribed to complementary DNA with the use of random hexamers and Moloney Murine Leukemia Virus Reverse Transcriptase from Applied Biosystems (Foster City, CA). Gene-specific primers (Supplementary Data Table S2) were designed using the NCBI/Primer BLAST Designing Tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The ABsolute SYBR Green ROX Mix from Abgene (Rockford, IL) was used for amplifications. Conditions for amplification were as follows: 15 min at 95 °C, followed by 40 cycles of 95 °C for 1 min and 60 °C for 1 min. Cycle time (Ct) values for the selected genes were normalized to values for 16 S rRNA (n=3 in pseudo triplicates). Ct values for all controls were set at 100%.
2.8. Mammalian cell culture
The HeLa cell line stably expressing eGFP (HeLa/eGFP) was purchased from Cell Biolabs, Inc. (San Diego, CA) and grown in parallel with wild-type HeLa cells. Those cell lines were certified to be free of mycoplasma. HeLa.tet and HeLa.tet.eGFP were purchased from GenTarget Inc. (San Diego, CA) and were also certified to be free of mycoplasma. Cells were cultured in high glucose Dulbecco's Modified Eagle's Medium, supplemented with 10% heat-inactivated fetal bovine serum (v/v), 50 U/mL penicillin, 50 μg/mL streptomycin, 2 mM L-glutamine and 0.1 mM MEM nonessential amino acids (NEAA). Cells were maintained in a humidified atmosphere of 5% CO2 at 37 °C and cultured in T-75 Falcon tissue culture flasks at an initial density of 1×106 cells/flask. The culture medium was replaced every 2 days and passaged when confluence was reached. Three independent cell cultures were prepared. Cultures of HeLa.tet and HeLa.tet.eGFP at 70% confluency were exposed to 1 μg/mL doxycycline for 48 h before collection. Confluent cells were collected for RNA isolation by trypsinization, washed with PBS and stored at −80 °C.
2.9. Amplex Red assay for determination of H2O2 production by HeLa cells
Mammalian cells were trypsinized and 1×105 cells were seeded to a black 96-well plate, incubated overnight and washed with PBS. Wells with cells (n=8 for each cell line) were filled with 100 μL of 50 μM Amplex Red, 0.2 U/mL HRP in PBS and 0.5% DMSO (v/v) from the Amplex Red stock solution. Catalase was added to control wells (1 μL/well, 1 kU/mL). Exposure to light was avoided by preparing the assay in a dark room [19]. A microplate plate reader was prewarmed and kept at 37 °C, and fluorescence intensities (filter: λex=530 nm, λem=590 nm) were measured from the top of each well every 10 min for 120 min. E. coli was also used for this assay, but Luria-Brentani broth medium was used at 1.0 of OD at 600 nm/mL of bacterial cells. Experiments were repeated three independent times with mammalian cells and two independent times with bacterial cells.
2.10. RNA isolation from HeLa cells, microarray and GSEA analyses
Total RNA was isolated from three independent triplicates using the QIAshredder and RNeasy Midi® Kit (QIAGEN, Valencia, CA) with in-column DNAse treatment (QIAGEN, Valencia, CA). Gene expression analysis was conducted using Agilent Whole Human Genome 4×44 Multiplex Arrays (014850, Agilent Technologies, CA). Total RNA samples were labeled with Cy3 according to the manufacturer's protocol. Data were obtained using the Agilent Feature Extraction Software (v. 12). Data was further processed using R (version 3.1.3) and RStudio (version 0.98.1103). Raw microarray data was normalized and the background subtracted using the limma package (version 3.22.6). Annotation was made using the hgug4112a database. Heat maps were prepared in R using the gplot package (version 2.16.0).
Two different 2-contrast GSEA analyses [20] were conducted: 1. HeLa/eGFP versus HeLa and 2. the differences in the log2-transformed intensities between HeLa.tet.eGFP incubated with and without doxycycline versus the differences of HeLa.tet incubated with and without doxycycline. Both analyses were performed with the full table using the hallmark database, h.all.v5.0.symbols, 1000 permutations, and defaults for other parameters. The microarray raw data can be accessed on the Gene Expression Omnibus with the accession number GSE96671.
2.11. Statistical analyses
Experiments were routinely prepared in three independent replicates (n=3) and repeated on two different days. Mean and error bars as standard deviation are shown in graphs. ANOVA statistics and the post-hoc two-sided t-test were used for comparisons between groups. Comparisons with p-value lower than 0.05 were considered statistically significant.
3. Results
3.1. Generation of O2•– and H2O2 by eGFP and TagRFP in the presence of NAD(P)H
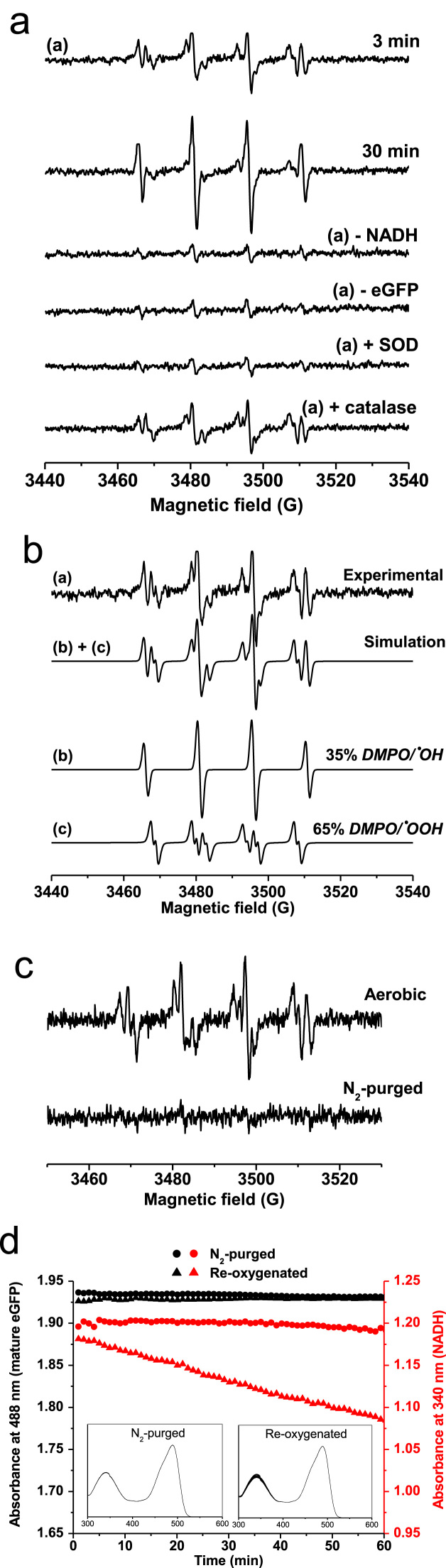
Initially we used purified recombinant eGFP expressed in E. coli. As shown in Fig. 1a, we detected for the first time the generation of the free radical O2•– in samples of eGFP and NADH using ESR spin trapping with 5,5-dimethyl-1-pyrroline N-oxide (DMPO). The signal was a superimposition of the adducts DMPO/•OH and DMPO/•OOH (Fig. 1b; analysis using spectral simulation [21]), was present for over 30 min and was totally dependent on eGFP and NADH. DMPO/•OOH is known to decay to DMPO/•OH by a number of pathways. The signal was sensitive to superoxide dismutase (SOD), but not to catalase, as expected. We demonstrated that molecular oxygen is required for O2•– formation (Fig. 1c) since the consumption of NADH did not occur in the absence of oxygen (Fig. 1d). No change in the light absorption for the chromophore of mature eGFP was detectable in samples either aerobically or anaerobically prepared (Fig. 1d). The reaction restarted as soon as the sample was oxygenated.
Fig. 1.
Reaction of NADH with eGFP leads to the formation of superoxide free radical anion. (a) Spin trapping of superoxide free radical anion (O2•–) generated by eGFP in the presence of NADH. Samples containing 50 μM GFP, 500 μM NADH and 100 mM DMPO showed a multiline-signal, trace (1). The effect of incubation time and eGFP or NADH was also studied. SOD was added to samples at 500 U/mL and catalase at 1 kU/mL. (b) Spectral simulation of trace (1) from panel A. The signal (1) is a superposition of (a) 65% DMPO/•OOH and (b) 35% DMPO/•OH. (c) ESR spin trapping for O2•– was prepared with samples containing 50 μM eGFP, 500 μM NADH and 100 mM DMPO using air-equilibrated solutions (aerobic) or anaerobic solutions (nitrogen-purged solutions). (d) A sample containing 25 μM eGFP was prepared without oxygen and tested for the ability to consume NADH at 250 μM (sample purged with nitrogen, •). This sample was then aerated with atmospheric air, and NADH consumption was followed (re-oxygenated sample, ▲). NADH was measured by its absorbance at 340 nm (ε340 nm=6220 M−1cm−1[33]). Black filled symbols were used to show the absorbance of the fluorescence-active, mature eGFP (• for the nitrogen-purged sample, and ■ for the sample after re-oxygenation, ε489 nm=55,000 M−1 cm−1).
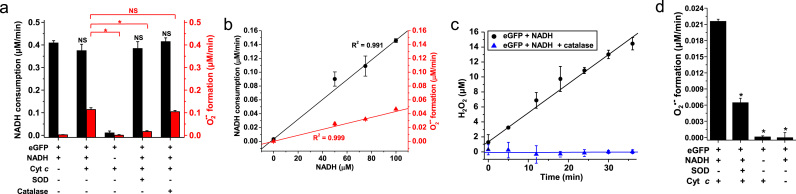
Generation of O2•– by eGFP in the presence of NADH was also determined independently by the cytochrome c reduction assay (Fig. 2a, O2•–-mediated reduction of cytochrome c was followed optically). As expected, cytochrome c reduction was inhibited by SOD but was insensitive to catalase (Fig. 2a, red bars). Cytochrome c did not interfere with the rate of NADH consumption by eGFP (Fig. 2a, black bars). Formation of O2•– was calculated over a range of NADH concentrations and was formed at approximately one third of the rate of NADH consumed (Fig. 2b). By using the same eGFP preparation, we also showed that the consumption of NADH was nearly equimolar to the formation of H2O2 (412 nM/min of NADH consumption and 388 nM/min of H2O2 production, Fig. 2c). The catalytical activity of eGFP in this reaction was further tested in samples containing 25 μM eGFP and 1 mM NADH (Supplemental Data Fig. S1a). The concentration of H2O2 was much greater than stoichiometric at the end of this experiment (478 μM H2O2 or 19 equivalents to total eGFP). In conclusion, eGFP catalytically consumes NADH to produce O2•– and H2O2 with an overall stoichiometry of one NADH oxidized to NAD+ and one O2 reduced to H2O2. It is noteworthy that in all experiments, NADH could be substituted for NADPH with no changes in the chemical reaction (data not shown).
Fig. 2.
Generation of superoxide free radical anion (O2•–) and H2O2 by eGFP in the presence of NADH. (a) Using eGFP expressed by E. coli, rates of NADH consumption and O2•– formation were determined in samples containing 15 μM eGFP, 150 μM NADH and 20 μM cytochrome c. The effects of catalase (1 kU/mL) and SOD (250 U/mL) were also studied. O2•– formation was determined by the cytochrome c reduction measured at 550 nm (ε550 nm=21,000 M−1 cm−1[34]). (b) Rates of NADH consumption and O2•– formation were studied in samples containing 15 μM GFP, 20 μM cytochrome c and different concentrations of NADH. (c) H2O2 production was determined in samples containing 25 μM GFP and 250 μM NADH. Every 5–6 min, an aliquot was withdrawn and H2O2 measured. Samples to which catalase (1 kU/mL) had been added were assayed in parallel. (d) Using eGFP expressed by the mammalian cell line HEK 293, the rate of O2•– formation was determined in samples containing 15 μM eGFP, 150 μM NADH and 20 μM cytochrome c, such as described in (a). The effect of SOD (250 U/mL) was also studied. * is used to show statistically significant differences between samples and controls.
Interestingly, different preparations of our extensively purified eGFP at the same total protein concentration showed different rates of NADH consumption (Supplemental Data Fig. S1b). These preparations had similar quantities of mature eGFP per total protein content (Supplemental Data Fig. S1c) and showed NADH consumption dependent on both NADH and eGFP (Supplemental Data Fig. S1d). By calculating the amount of mature eGFP chromophore per protein, we found that approximately 75% of the total eGFP is in its chromophore-mature state. Apparently, the mature eGFP is not the generator of O2•–.
We next used extensively purified eGFP expressed in the mammalian cell line HEK 293 (Fig. 2d). This protein was found approximately 85% in its chromophore-mature form. eGFP expressed in HEK 293 also generated O2•– in the presence NADH, as shown by the inhibition of cytochrome c reduction when SOD was present (first and second columns, Fig. 2d). Formation of O2•– was dependent on NADH (third column, Fig. 2d).
We also tested the formation of O2•– and consumption of NADH by a different fluorescent protein, the red fluorescent protein TagRFP. Highly purified TagRFP produced in E. coli consumed NADH and generated O2•– (Supplemental Data Fig. S1e). Purified TagRFP was approximately 90% in its chromophore-mature form (ε at 555 nm=100,000 M−1cm−1 4). The yield of O2•– generation by TagRFP lies between the higher yields found for eGFP produced in E. coli and the lower yields for the eGFP produced in HEK 293.
3.2. Cells expressing eGFP show oxidative stress and changes in many biological processes
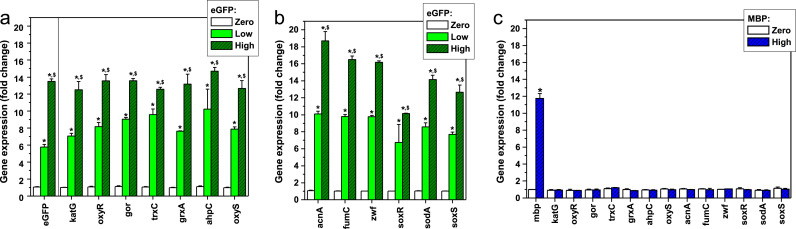
In order to investigate how eGFP can alter gene expression associated with oxidative stress in living systems, we used first the bacterium E. coli to monitor the expression of genes under the control of the transcription factors SoxR and OxyR. SoxR is activated by direct reaction with superoxide anion free radical [22], whereas OxyR induces gene expression when key cysteines are oxidized as a result of an oxidizing intracellular redox status [23]. Experiments were performed with E. coli expressing eGFP under low and high levels. All genes expressed upon activation of SoxR (Fig. 3a) and OxyR (Fig. 3b) were highly upregulated in these cells, proportionally to eGFP expression. A bacterium expressing maltose binding protein (MBP) was used as a control for the possible oxidative stress involved in overexpressing a protein. Notably, the high level of expression of MBP (Fig. 3c, first two columns) did not lead to changes in expression for any of the genes activated by SoxR or OxyR (Fig. 3c, left two panels). Even though the differences in gene expression among the bacteria expressing eGFP were striking, we did not detect differences in extracellular H2O2 production (Supplementary Data Fig. S2). This negative result can be explained by the fact that E. coli is very resistant to H2O2. Even at 1 mM H2O2, E. coli does not lose viability, presumably because this bacterium is equipped with very efficient systems for H2O2 decomposition [24].
Fig. 3.
Oxidative stress response in E. coli follows the expression of eGFP. Oxidative stress response was determined by the quantification of mRNAs by real-time PCR of genes activated by the SoxR/S and OxyR regulons in E. coli. (a) eGFP expression (first three bars on the left) and quantification of genes that are targets for the activation of the SoxR/S regulon are shown. (b) Quantification of gene expression for different genes activated by OxyR. (c) Quantification of mRNAs for MBP (maltose binding protein) and targets of SoxR/S and OxyR were determined in an E. coli strain engineered for the controlled expression of high levels of MBP. * is used to show statistically significant differences between samples and controls, and $ shows statistically significant differences between samples of high and low expression.
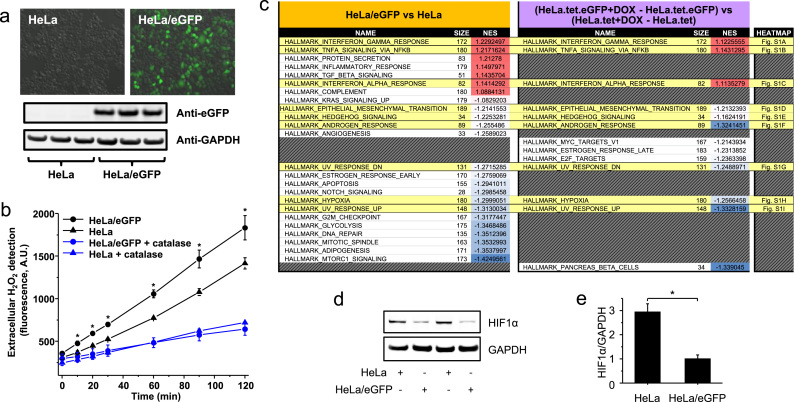
Next, we studied the effect of eGFP expression in mammalian cells using the HeLa cell lines. We discovered that HeLa cells stably expressing eGFP (HeLa/eGFP, Fig. 4a) show significantly higher rates of H2O2 efflux compared to regular HeLa cells (7.3 and 5.15 nM H2O2/min, respectively), indicating a higher generation of intracellular H2O2 (Fig. 4b). The H2O2-decomposing enzyme catalase inhibits the Amplex Red oxidation to levels that do not differ between cell lines (1.8 and 2.3 nM H2O2/min, respectively; black traces on Fig. 4b).
Fig. 4.
eGFP in cells induces gene expression associated with oxidative stress and other biological processes. Images of HeLa cells and stable eGFP-expressing HeLa cells (HeLa/eGFP) under a fluorescent microscope with a green filter can be seen in (a). eGFP was also detected by Western blotting, with GAPDH staining as the loading control. In (b) extracellular H2O2 was determined in HeLa and HeLa/eGFP. Catalase was added to cultures at 500 U/mL. (c) Microarray-based experiments were designed to show the effect of GFP expression in the gene expression of two independent cell models. The two GSEA (Gene Set Enrichment Analyses [20]) were performed using the hallmark dataset (h.all.v5.0.symbols) and 1000 permutations. The contrasts were HeLa/eGFP versus HeLa, and Hela.tet.eGFP + doxycycline subtracted from HeLa.tet.eGFP versus HeLa.tet + doxycycline subtracted from HeLa.tet. The enriched gene sets with p-values lower than 0.001 were ranked, and matches between the two independent analyses are shown highlighted in yellow. The top genes contributing to the enrichment for the commonly enriched gene sets are depicted as heat maps in Supplementary Data Fig. S3. (d) Intracellular levels of HIF1α were determined by Western blotting using whole cell lysates of HeLa cells and HeLa cells stably expressing eGFP. In (e), GAPDH densitometry was used as the loading control to normalize the densitometry of HIF1α. * is used to show statistically significant differences between HeLa and HeLa/eGFP.
Production of H2O2 was further investigated using a HeLa cell line engineered to express eGFP under the control of a tet-mediated promoter (HeLa.tet.eGFP). In this cell model, eGFP is expressed only when doxycycline is added to the cell culture medium (Supplemental Data Fig. S3a). Possible non-specific effects of tet and/or doxycycline were taken into account by analyzing the response of its isogenic parental cell line (HeLa.tet) exposed to doxycycline. We compared HeLa.tet.eGFP cells with induced expression of eGFP to the isogenic parental cell line (HeLa.tet) exposed to doxycycline and found no detectable difference in the low rate of H2O2 generation in both cultures. The level of eGFP in the Hela.tet.eGFP exposed to doxycycline was approximately 13 times lower than the level in the stable eGFP-expressing cell line, HeLa/eGFP. However, when we compared the gene expression and GSEA (Gene Set Enrichment Analyses [20]) of the HeLa/eGFP versus the control HeLa cells, and the specific effects of eGFP expression in HeLa.tet.eGFP versus Hela.tet, we saw that both eGFP-expressing cell models showed significant gene set enrichment for nine biological processes (gene sets with p-values lower than 0.001, Fig. 4c). Three gene sets associated with pathogen response and inflammation are upregulated and are related to interferon-α response, interferon-γ response, and TNF-α signalling via NFkβ (Fig. 4c and Supplemental Data Fig. 3b-d), which are classically known to be activated when cells are producing intracellular H2O2 and suffering from oxidative stress [25]. Another six gene sets are downregulated in HeLa cells that express eGFP (Fig. 4c and Supplemental Data Fig. 3e–j), including hypoxia. In agreement with our analyses of global gene expression, we found three times lower levels of HIF-1α, the master regulator for hypoxia, in HeLa/eGFP compared to HeLa (Fig. 4d and e). It has been reported that under normoxia, the intracellular level of HIF-1α is decreased in response to the toxic effects of higher intracellular H2O2 and O2•– formation [26], [27], such as observed in HeLa/eGFP. This cell line secreted nearly 500 nM of H2O2 into the medium in the course of one hour (Fig. 4b).
4. Conclusions
Our in vitro experiments show that eGFP in the presence of NAD(P)H generates O2•– and H2O2. Collectively, the absence of changes in the visible spectrum of the eGFP-chromophore, the lack of correlation between the concentration of mature chromophore and rates for the reaction with NADH in different batches of eGFP lead us to conclude that an inevitable small portion of non-fluorescent or immature chromophore present in eGFP is responsible for the catalytical consumption of NADH with O2•– and H2O2 formations. This is consistent with the finding that the mimic probe for the mature eGFP chromophore, 3,5′-difluoro-4-hydroxybenzyldene imidazolinone (DFHBI, Lucerna, Inc., New York, NY), fails to consume NAD(P)H (data not shown).
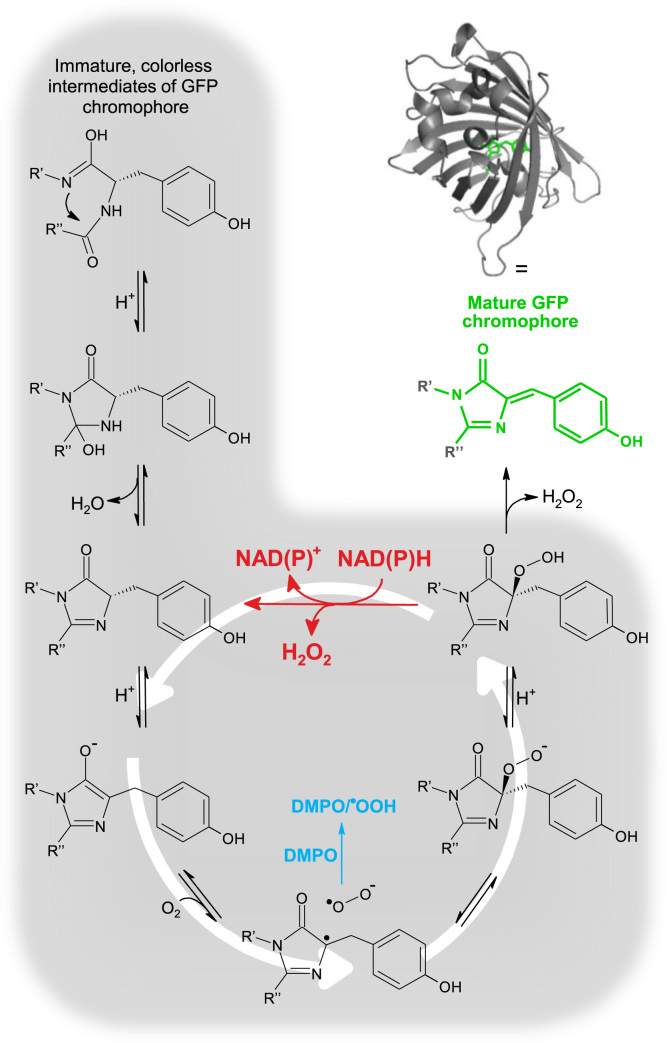
Even though we do not have direct experimental evidence to support a definitive mechanism for this reaction, we speculate that the last immature intermediate in the proposed chromophore maturation process [28] reacts with NAD(P)H. This reaction would result in the regeneration of an earlier immature intermediate, which ultimately leads to the catalytical consumption of NAD(P)H and generation of H2O2, as shown in Scheme 1 for the chromophore of eGFP. It is very likely that the maturation of the last immature intermediate is slow since tertiary hydroperoxides are known to be highly stable, e.g., tert-butyl hydroperoxide.
Scheme 1.
GFP chromophore maturation and proposed mechanism for the catalytical consumption of NAD(P)H and generation of O2•–/H2O2 by GFP immature intermediates. The GFP chromophore maturation process is shown in black (adapted from Barondeau et al. [28]). Shown in red, the proposed reaction of the last immature intermediate of GFP with NAD(P)H leads to the catalytical generation of O2•–/H2O2 and consumption of NAD(P)H. The eGFP structure is shown (PDB# 2y0g).
More importantly, this chemistry likely occurs inside of bacteria and mammalian cells expressing eGFP. Our experiments were carried out with 50, 25 and 10 μM eGFP or 10 μM TagRFP, which is comparable to the concentration found in different cellular systems [29], [30]. NAD(P)H is present at mM concentrations in biological systems; therefore, the chemistry shown here is likely to occur in most biological experiments, which, in turn, leads to oxidative stress and cellular adaptations in gene expression.
The level of extracellular H2O2 detected in HeLa cells stably expressing eGFP appears well in the range of concentration in which H2O2 acts as a global messenger. As a global messenger, H2O2 affects many biological processes beyond oxidative stress response, such as cell metabolism, cellular fate, immunity and homeostasis [31], [32]. The mechanism shown here for the generation of O2•– and H2O2 by fluorescent proteins explains the cytotoxicity and tissue abnormalities seen in cells and animals expressing fluorescent proteins [10], [11], [12], [15], [16]. Given the extensive use of fluorescent proteins, especially eGFP, in studies of different human diseases (Supplemental Data Table S1) and the known role of O2•– and H2O2 in the pathophysiology of those conditions, we call for a careful evaluation of experimental design, validation and interpretation of results in investigations using fluorescent proteins.
Authors’ contributions
D.G. and F.L. designed experiments, performed experiments and wrote the paper. A.K. performed experiments with bacterial cells and wrote the paper. J.J. supervised the ESR experiments. E.T. supervised the experiments with bacterial cells. C.C.M. and R.M.P. expressed and purified eGFP and RFP. R.P.M. provided guidance and wrote the paper.
Competing financial interests
The authors declare no competing financial interests.
Materials and correspondence
Correspondence and materials requests should be addressed to Dr. Ronald P. Mason (mason4@niehs.nih.gov) and Dr. Douglas Ganini da Silva (ganinidasilvad@niehs.nih.gov).
Acknowledgements
We acknowledge Dr. Ann Motten and Ms. Mary Mason for editing the manuscript. We thank Dr. Tom Eling and Dr. Thomas Van'T Erve for their suggestions regarding the manuscript. We are also in debt to Ms. Jean Corbett for her invaluable technical assistance. We declare no conflicts of interest. This research was supported by the Intramural Research Program of the NIEHS, National Institute of Environmental Health Sciences/NIH.
Footnotes
This research was supported by the Intramural Research Program of the NIEHS, National Institute of Environmental Health Sciences/NIH.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2017.03.002.
Appendix A. Supplementary material
Supplementary material
References
- 1.Shaner N.C., Steinbach P.A., Tsien R.Y. A guide to choosing fluorescent proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 2.Zhang J., Campbell R.E., Ting A.Y., Tsien R.Y. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 3.Zhang G., Gurtu V., Kain S.R. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem. Biophys. Res. Commun. 1996;227:707–711. doi: 10.1006/bbrc.1996.1573. [DOI] [PubMed] [Google Scholar]
- 4.Merzlyak E.M. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L., Patel H.N., Lappe J.W., Wachter R.M. Reaction progress of chromophore biogenesis in green fluorescent protein. J. Am. Chem. Soc. 2006;128:4766–4772. doi: 10.1021/ja0580439. [DOI] [PubMed] [Google Scholar]
- 6.Lu C., Albano C.R., Bentley W.E., Rao G. Quantitative and kinetic study of oxidative stress regulons using green fluorescent protein. Biotechnol. Bioeng. 2005;89:574–587. doi: 10.1002/bit.20389. [DOI] [PubMed] [Google Scholar]
- 7.Tsien R.Y. Constructing and Exploiting the Fluorescent Protein Paintbox (Nobel Lecture) Angew. Chem.-Int. Ed. 2009;48:5612–5626. doi: 10.1002/anie.200901916. [DOI] [PubMed] [Google Scholar]
- 8.Ormo M. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 9.Subach O.M. Structural characterization of acylimine-containing blue and red chromophores in mTagBFP and TagRFP fluorescent proteins. Chem. Biol. 2010;17:333–341. doi: 10.1016/j.chembiol.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taghizadeh R.R., Sherley J.L. CFP and YFP, but not GFP, provide stable fluorescent marking of rat hepatic adult stem cells. J. Biomed. Biotechnol. 2008;2008:453590. doi: 10.1155/2008/453590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ansari A.M. Cellular GFP Toxicity and Immunogenicity: Potential Confounders in in Vivo Cell Tracking Experiments. Stem Cell Rev. 2016;12:553–559. doi: 10.1007/s12015-016-9670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koike M., Yutoku Y., Koike A. Ku80 attentuates cytotoxicity induced by green fluorescent protein transduction independently of non-homologous end joining. FEBS Open Bio. 2013;3:46–50. doi: 10.1016/j.fob.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H.S., Jan M.S., Chou C.K., Chen P.H., Ke N.J. Is green fluorescent protein toxic to the living cells? Biochem. Biophys. Res. Commun. 1999;260:712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 14.Goto H. Transduction of green fluorescent protein increased oxidative stress and enhanced sensitivity to cytotoxic drugs in neuroblastoma cell lines. Mol. Cancer Ther. 2003;2:911–917. [PubMed] [Google Scholar]
- 15.Wallace L.M., Moreo A., Clark K.R., Harper S.Q. Dose-dependent toxicity of humanized Renilla reniformis GFP (hrGFP) limits its utility as a reporter gene in mouse muscle. Molecular therapy. Nucleic Acids. 2013;2:e86. doi: 10.1038/mtna.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang W.Y., Aramburu J., Douglas P.S., Izumo S. Transgenic expression of green fluorescence protein can cause dilated cardiomyopathy. Nat. Med. 2000;6:482–483. doi: 10.1038/74914. [DOI] [PubMed] [Google Scholar]
- 17.Wardman P. Fluorescent and luminescent probes for measurement of oxidative and nitrosative species in cells and tissues: progress, pitfalls, and prospects. Free Radic. Biol. Med. 2007;43:995–1022. doi: 10.1016/j.freeradbiomed.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 18.Wolff S.P. Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 1994;233:182–189. [Google Scholar]
- 19.Summers F.A., Zhao B., Ganini D., Mason R.P. Photooxidation of Amplex Red to resorufin: implications of exposing the Amplex Red assay to light. Methods Enzymol. 2013;526:1–17. doi: 10.1016/B978-0-12-405883-5.00001-6. [DOI] [PubMed] [Google Scholar]
- 20.Subramanian A. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duling D.R. Simulation of multiple isotropic spin-trap EPR spectra. J. Magn. Reson. Ser. B. 1994;104:105–110. doi: 10.1006/jmrb.1994.1062. [DOI] [PubMed] [Google Scholar]
- 22.Koo M.S. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 2003;22:2614–2622. doi: 10.1093/emboj/cdg252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michan C., Manchado M., Dorado G., Pueyo C. In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J. Bacteriol. 1999;181:2759–2764. doi: 10.1128/jb.181.9.2759-2764.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dukan S., Touati D. Hypochlorous acid stress in Escherichia coli: resistance, DNA damage, and comparison with hydrogen peroxide stress. J. Bacteriol. 1996;178:6145–6150. doi: 10.1128/jb.178.21.6145-6150.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.I. Dichi, Breganó, J.W., A.A.N. Colado Simão, R. Cecchini Role of oxidative stress in chronic diseases.
- 26.Bunn H.F., Gu J., Huang L.E., Park J.W., Zhu H. Erythropoietin: a model system for studying oxygen-dependent gene regulation. J. Exp. Biol. 1998;201:1197–1201. doi: 10.1242/jeb.201.8.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qutub A.A., Popel A.S. Reactive oxygen species regulate hypoxia-inducible factor 1alpha differentially in cancer and ischemia. Mol. Cell. Biol. 2008;28:5106–5119. doi: 10.1128/MCB.00060-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barondeau D.P., Kassmann C.J., Tainer J.A., Getzoff E.D. The case of the missing ring: radical cleavage of a carbon-carbon bond and implications for GFP chromophore biosynthesis. J. Am. Chem. Soc. 2007;129:3118–3126. doi: 10.1021/ja063983u. [DOI] [PubMed] [Google Scholar]
- 29.Niswender K.D., Blackman S.M., Rohde L., Magnuson M.A., Piston D.W. Quantitative imaging of green fluorescent protein in cultured-cells - Comparison of microscopic techniques, use in fusion proteins and detection limits. J. Microsc.-Oxf. 1995;180:109–116. doi: 10.1111/j.1365-2818.1995.tb03665.x. [DOI] [PubMed] [Google Scholar]
- 30.Tsien R.Y. The green fluorescent protein. Annu Rev. Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 31.Stone J.R., Yang S.P. Hydrogen peroxide: a signaling messenger. Antioxid. Redox Signal. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 32.Murrell G.A., Francis M.J., Bromley L. Modulation of fibroblast proliferation by oxygen free radicals. Biochem J. 1990;265:659–665. doi: 10.1042/bj2650659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.E.J. Wood, Data for biochemical research (third edition) by R M C Dawson, D C Elliott, W H Elliott and K M Jones, pp 580. Oxford Science Publications, OUP, Oxford, 1986. ISBN 0-19-855358-7. Biochemical Education 15, 122, 1987.
- 34.O'Brien P.J. Superoxide production. Methods Enzymol. 1984;105:370–378. doi: 10.1016/s0076-6879(84)05050-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material