Abstract
The nucleolus is a stress sensor and compromised nucleolar activity may be considered as an attractive anticancer strategy. In the present study, the effects of three plant-derived natural compounds, i.e., sulforaphane (SFN), ursolic acid (UA) and betulinic acid (BA) on nucleolar state were investigated in breast cancer cell lines of different receptor status, namely MCF-7, MDA-MB-231 and SK-BR-3 cells. Cytostatic action of phytochemicals against breast cancer cells was observed at low micromolar concentration window (5–20 µM) and mediated by elevated p21 levels, and cell proliferation of SFN-, UA- and BA-treated normal human mammary epithelial cells (HMEC) was unaffected. Phytochemical-mediated inhibition of cell proliferation was accompanied by increased levels of superoxide and protein carbonylation that lead to disorganization of A- and B-type lamin networks and alterations in the nuclear architecture. Phytochemicals promoted nucleolar stress as judged by the nucleoplasmic translocation of RNA polymerase I-specific transcription initiation factor RRN3/TIF-IA, inhibition of new rRNA synthesis and decrease in number of nucleoli. Phytochemicals also decreased the levels of NOP2, proliferation-associated nucleolar protein p120, and WDR12 required for maturation of 28S and 5.8S ribosomal RNAs and formation of the 60S ribosome, and phosphorylation of S6 ribosomal protein that may result in diminished translation and inhibition of cell proliferation. In summary, three novel ribotoxic stress stimuli were revealed with a potential to be used in nucleolus-focused anticancer therapy.
Keywords: Pentacyclic triterpenotids, Sulforaphane, Breast cancer cells, Nucleolus, Protein carbonylation, Cell proliferation
Highlights
-
•
SFN, UA and BA inhibit breast cancer cell proliferation.
-
•
SFN, UA and BA induce oxidative stress and protein carbonylation.
-
•
SFN, UA and BA cause disorganization of A- and B-type lamin networks.
-
•
SFN, UA and BA inhibit rRNA synthesis by nucleoplasmic translocation of RRN3.
-
•
SFN, UA and BA promote nucleolar stress that limit proliferative activity.
1. Introduction
More recently, non-ribosomal functions for the nucleolus have been established [1], [2]. Beyond its primary role in ribosome biosynthesis, the nucleolus is also involved in the regulation of cell cycle progression and stress signaling [3], [4], [5]. Oxidative and ribotoxic stress stimuli have been reported to inhibit RNA polymerase I (Pol I) transcription by inactivation of the Pol I-specific transcription factor RRN3/TIF-IA [6]. The inactivation of TIF-IA is achieved by phosphorylation of TIF-IA by c-Jun N-terminal kinase (JNK) at a single threonine residue (Thr200) that result in both impaired interaction of TIF-IA with Pol I and SL/TIF-IB, thus preventing transcription initiation complex formation at the rDNA promoter, and relocation of TIF-IA from the nucleolus to the nucleoplasm where it is sequestered from Pol I [6]. Several stressors can also promote the nucleoplasmic translocation of nucleolar proteins such as ARF, L5, L11, L23 or B23/nucleophosmin that is considered as a hallmark of nucleolar stress [3], [7], [8], [9]. Relocated proteins bind MDM2 (HDM2 in human, E3 ubiquitin ligase) that block the ubiquitinylation of p53 and induce p53-dependent cell cycle arrest and/or apoptosis [3], [10]. Genetic inactivation of RRN3/TIF-IA may also result in nucleolar disruption, cell cycle arrest and p53-mediated apoptosis [11]. Moreover, p53-independent responses to nucleolar stress have been documented [12], [13], [14], [15], [16]. As more than a half of human cancers lack functional p53 [17], these p53-independent pathways could potentially reveal additional cancer therapies that are based on drugs targeting the rDNA transcription machinery and inducing nucleolar stress.
Dietary phytochemicals are considered as promising candidates for anticancer therapy [18] and the mechanisms of action of plant-derived anticancer drugs are numerous including apoptosis, autophagy, necrosis-like programmed cell death, mitotic catastrophe and cellular senescence [19]. However, the ability of dietary agents to provoke nucleolar stress response that would block the proliferation of cancer cells has not been addressed.
In the present study, we have investigated the mechanism of cytostatic activity of two pentacyclic triterpenoids, namely ursolic acid (UA) and betulinic acid (BA) and sulforaphane (SFN), an isothiocyanate, against phenotypically distinct breast cancer cells MCF-7 (ER+, PR+/-, HER2-, wild type p53), MDA-MB-231 (ER-, PR-, HER2-, mutant p53) and SK-BR-3 (ER-, PR-, HER2+, mutant p53). We found that phytochemicals induced oxidant-based nucleolar stress that resulted in the nucleoplasmic translocation of RRN3/TIF-IA and inhibition of rRNA synthesis, and decreased phospho-S6 ribosomal protein signals leading to diminished translation efficiency and p21-mediated inhibition of cell proliferation.
2. Materials and methods
2.1. Reagents
The reagents used, if not otherwise mentioned, were purchased from Sigma-Aldrich (Poland) and were of analytical grade. Sulforaphane (1-isothiocyanato-4-(methylsulfinyl)-butane, SFN), ursolic acid (3β-hydroxy-12-ursen-28-ic acid, UA) and betulinic acid (3β-hydroxy-20(29)-lupaene-28-oic acid, BA) were dissolved in dimethyl sulfoxide (DMSO). DMSO concentrations did not exceed 0.1% and had no effect on parameters analyzed.
2.2. Cell culture
Human breast cancer cells MCF-7, MDA-MB-231 and SK-BR-3 were obtained from ATCC (Manassas, VA, USA). Cells (10,000 cells/cm2) were cultured at 37 °C in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and antibiotic and antimycotic mix solution (100 U/ml penicillin, 0.1 mg/ml streptomycin and 0.25 μg/ml amphotericin B) in a humidified atmosphere in the presence of 5% CO2 until they reached confluence. Typically, cells were passaged by trypsinization and maintained in DMEM. Normal human mammary epithelial cells (HMEC) were obtained from Lonza (Basel, Switzerland). Cells (10,000 cells/cm2) were cultured in Mammary Epithelial Growth Medium (MEGM) supplemented with BPE, hydrocortisone, hEGF, insulin and gentamicin/ amphotericin B according to manufacturer's instructions.
2.3. Cell proliferation
DNA content was assessed using CyQUANT® Cell Proliferation Assay Kit (Thermo Fisher Scientific). Briefly, the cells were cultured in a 96-well plate (5000 cells per well) and treated with SFN, UA and BA (5, 10, 20 µM) for 24 h and cells were then frozen, thawed and CyQUANT® GR dye/cell lysis buffer was added. After 5-min incubation in the dark, fluorescence (CyQUANT® GR dye bound to nucleic acids) was measured using a fluorescence microplate reader (λex=480 nm, λem=520 nm). DNA content was normalized to control.
2.4. Cell viability
Cell viability was assessed using Muse™ Cell Analyzer and Muse™ Count and Viability Kit according to manufacturer's instructions (Merck Millipore, Poland). Briefly, the cells were cultured in a 6-well plate (10,000 cells/cm2) and treated with SFN, UA and BA (5, 10, 20 µM) for 24 h and viable and non-viable cells were then differentially stained based on their permeability to the two DNA-binding dyes present in the reagent. The calculations were performed automatically and viability profiles (dot plots) were displayed using the Muse™ Count and Viability Software Module.
2.5. Immunostaining
An immunostaining protocol was used as previously described [20]. Briefly, the cells were cultured in a 96-well plate (5000 cells per well) and treated with SFN, UA and BA (5, 10, 20 µM) for 24 h and cells were then fixed and incubated with the primary antibodies anti-Ki67 (1:500), anti-p21 (1:400), anti-lamin A/C (1:100), anti-lamin B1 (1:500), anti-cofilin (1:200), anti-RRN3 (1:200), anti-NOP2 (1:1000), anti-WDR12 (1:200), anti-S6 ribosomal protein (1:50), anti-phospho-S6 ribosomal protein (Ser235/236) (1:100), anti-nucleolar antigen (Thermo Fisher Scientific, Novus Biologicals) and secondary antibodies conjugated to Texas Red or FITC (1:1000) (Thermo Fisher Scientific). Nuclei were visualized using Hoechst 33342 staining, F-actin was assayed using phalloidin staining or β-actin was detected using anti-β-actin antibody and nucleolus was immuno-stained using anti-nucleolar antigen antibody (Thermo Fisher Scientific). Digital cell images were captured using an imaging cytometer In Cell Analyzer 2000 (GE Healthcare, UK) equipped with a high performance CCD camera. Quantitative analysis was conducted using In Cell Analyzer software (GE Healthcare). In general, immuno-fluorescent signals are presented as relative fluorescence units (RFUs). Ki67 signals were scored and normalized to control. Cells with aberrant lamin signals were scored [%]. Compartment-specific immuno-fluorescent signals were also considered and are presented as relative fluorescence units (RFUs) or were scored [%] or were normalized to control.
2.6. Superoxide levels
After SFN, UA and BA treatments, intracellular total superoxide levels were measured using a fluorogenic probe dihydroethidium and imaging cytometry (In Cell Analyzer 2000 equipped with a high performance CCD camera, GE Healthcare, UK). Briefly, the cells were incubated in DPBS containing 5 µM dihydroethidium for 15 min in the dark, cells were then washed and intracellular fluorescent signals were acquired and quantified using In Cell Analyzer 2000 Software (GE Healthcare). The level of superoxide is presented as relative fluorescence units (RFUs).
2.7. Protein carbonylation
Actin, lamin A/C and total nuclear protein carbonylation was considered. Protein derivatization was conducted according to Lazarus et al. [21]. SFN-, UA- and BA-treated, fixed and derivatized cells were immuno-stained using anti-β-actin antibody (1:500) or anti-lamin A/C antibody (1:100) (Thermo Fisher Scientific) or stained using Hoechst 33342 staining and incubated with anti-DNP antibody (1:200) (Abcam) and the secondary antibodies conjugated to FITC or TR (1:1000) (Thermo Fisher Scientific). Digital cell images were captured using imaging cytometry (In Cell Analyzer 2000 equipped with a high performance CCD camera, GE Healthcare, UK). Co-localization analysis was performed using In Cell Analyzer 2000 Software (GE Healthcare). Protein carbonylation is presented as relative fluorescence units (RFUs).
2.8. Immunodetection of nascent RNA (5-fluorouridine labeling)
After SFN, UA and BA treatments, cells were incubated with a halogenated RNA precursor, 2 mM 5-fluorouridine (5-FU) for 15 min and fixed in 3.7% formaldehyde in PBST (PBS with 0.01% Triton X-100). Indirect immunofluorescence with an anti-BrdU antibody (1:500, BD Biosciences) and the appropriate secondary antibody coupled to FITC (1:1000, BD Biosciences) were used to detect halogenated RNA [22]. 5-FU in the nucleolus is presented as relative fluorescence units (RFUs).
2.9. Western blotting
Whole cell protein extracts were prepared according to Lewinska et al. [23]. Polyvinylidene difluoride (PVDF) membranes were incubated with the primary antibodies anti-cofilin (1:200), anti-RRN3 (1:2000), anti-S6 ribosomal protein (1:100), anti-phospho-S6 ribosomal protein (Ser235/236) (1:1000) or anti-β-actin (1:1000) (Thermo Fisher Scientific, Sigma-Aldrich) and a secondary antibody conjugated to HRP (1:50000, Sigma-Aldrich). The respective proteins were detected using a Clarity™ Western ECL Blotting Substrate (Bio-Rad) and a G: BOX imaging system (Syngene, Cambridge, UK) according to the manufacturer's instructions. Densitometry measurements of the bands were performed using GelQuantNET software (http://biochemlabsolutions.com/GelQuantNET.html). The data represent the relative density normalized to β-actin. Phospho-S6 signals were also normalized to S6 signals.
2.10. Statistical analysis
The results represent the mean±SD from at least three independent experiments. Alternatively, box and whisker plots with median, lowest and highest values were used. Statistical significance was assessed by 1-way ANOVA using GraphPad Prism 5, and with the Dunnett's multiple comparison test.
3. Results
3.1. SFN, UA and BA inhibit cell proliferation in breast cancer cells
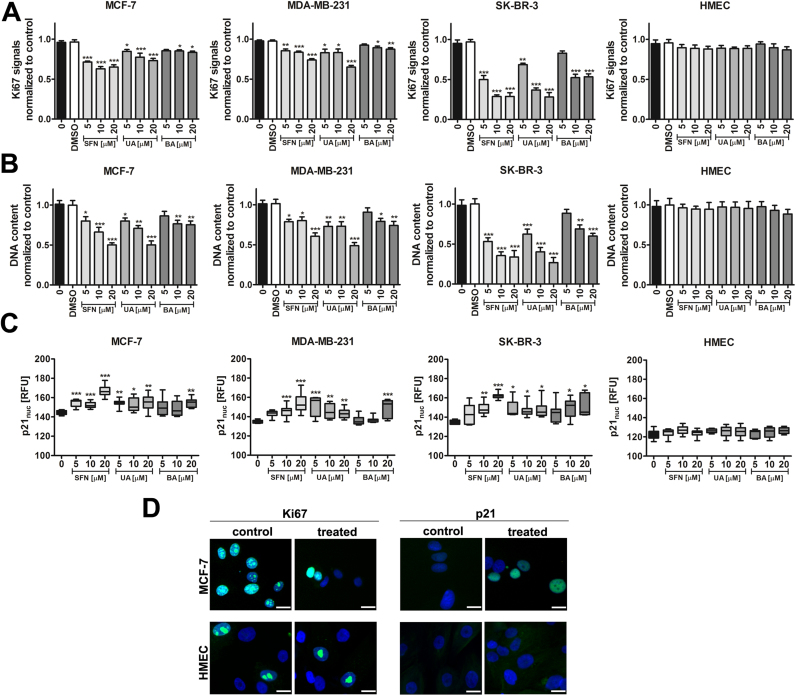
Initially, we have considered thirty plant-derived natural compounds with anticancer properties and investigated their cytostatic activity against phenotypically different breast cancer cell lines, i.e., MCF-7 (ER+, PR+/-, HER2-), MDA-MB-231 (ER-, PR-, HER2-) and SK-BR-3 (ER-, PR-, HER2+) (data not shown). Based on Ki67 immunostaining data, we have selected three phytochemicals with the ability to inhibit cancer cell proliferation when used at low micromolar range (5–20 µM) and 24 h treatment, namely sulforaphane (SFN), an isothiocyanate, and two pentacyclic triterpenotids, ursolic acid (UA) and betulinic acid (BA) (Fig. 1A).
Fig. 1.
SFN-, UA- and BA-mediated inhibition of breast cancer cell proliferation. (A) Ki67 immunostaining. Ki67 signals were normalized to Ki67 signals in the control conditions. The effect of solvent used (0.1% DMSO) is also presented. Bars indicate SD, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (B) CyQUANT® GR dye staining. DNA content was normalized to DNA content in the control conditions. The effect of solvent used (0.1% DMSO) is also presented. Bars indicate SD, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (C) p21 immunostaining. The levels of nuclear p21, a cell cycle inhibitor, are presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (D) Ki67 immunostaining (green, left), p21 immunostaining (green, right). MCF-7 and HMEC cells were treated with 10 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. Nuclei were visualized using Hoechst 33342 staining (blue). SFN, sulforaphane, UA, ursolic acid; BA, betulinic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
After treatment with all three phytochemicals, decreased Ki67 immuno-signals were the most pronounced in SK-BR-3 cells compared to other breast cancer cells used (Fig. 1A). In general, cytostatic activity of UA was found to be more potent than cytostatic activity of BA against three breast cancer cells used (Fig. 1A). In contrast, cytostatic activity of SFN, UA and BA against normal human mammary epithelial cells (HMEC) was limited and no statistically significant differences between control and treated cells were revealed (Fig. 1A). Cell proliferation was also analyzed using CyQUANT® GR dye staining and similar results were obtained (Fig. 1B). SFN-, UA- and BA-mediated inhibition of breast cancer cell proliferation was accompanied by elevated levels of nuclear p21, a cell cycle inhibitor (Fig. 1C). Treatment with SFN resulted in the most accented increase in p21 pools in all three breast cancer cell lines (Fig. 1C). In contrast, an increase in p21 levels was not observed in normal HMEC cells (Fig. 1C and D). Thus, it may be concluded that cytostatic activity of SFN, UA and BA is specific to breast cancer cells and SFN, UA and BA do not affect the proliferation of normal mammary epithelial cells (Fig. 1).
3.2. SFN, UA and BA are cytotoxic to breast cancer cells when used at the concentration of 20 µM
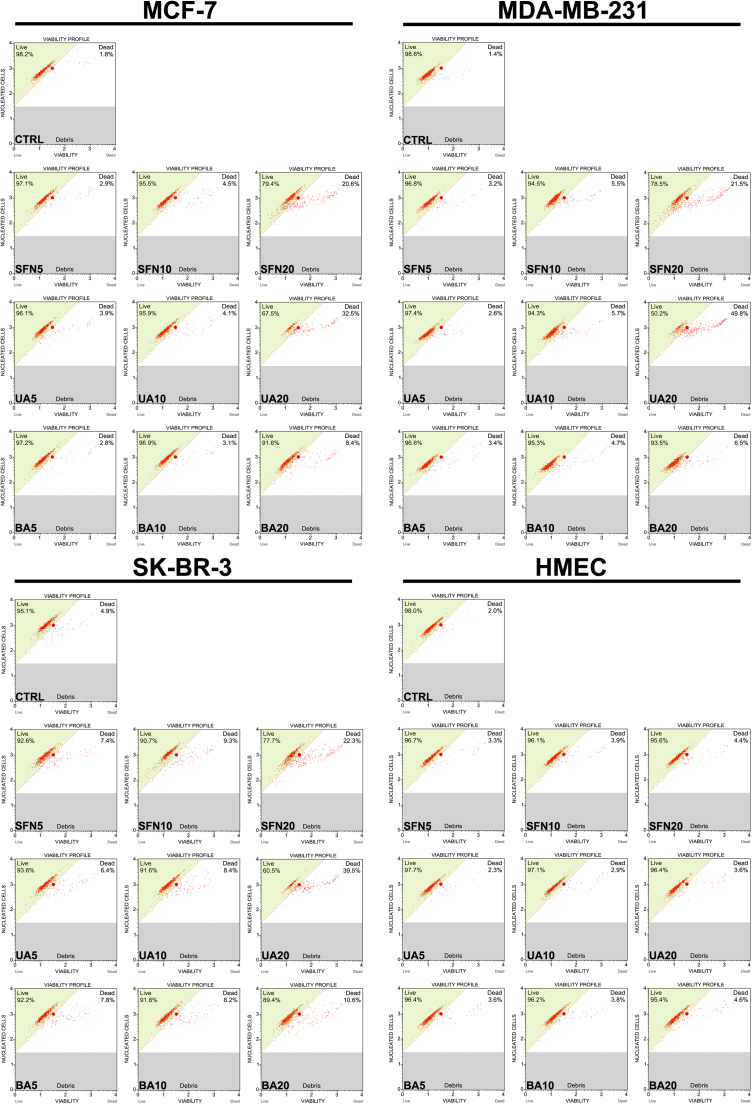
A simple cell membrane permeability test was then used to assess cytotoxic action of SFN, UA and BA (Fig. 2).
Fig. 2.
SFN-, UA- and BA-mediated cytotoxicity in breast cancer cells. Cell viability was assessed using Muse™ Count and Viability Kit. Representative viability profiles (dot plots) are shown. SFN, sulforaphane, UA, ursolic acid; BA, betulinic acid.
SFN, UA and BA were found to be cytotoxic to breast cancer cells when used at the concentration of 20 µM (Fig. 2). Cytotoxicity of 20 µM UA was the most evident and MDA-MB-231 cells were the most sensitive to 20 µM UA treatment (Fig. 2). In contrast, the cytotoxic effects of SFN, UA and BA on normal human mammary epithelial cells (HMEC) were minimal (Fig. 2).
3.3. SFN, UA and BA induce oxidative stress and protein carbonylation
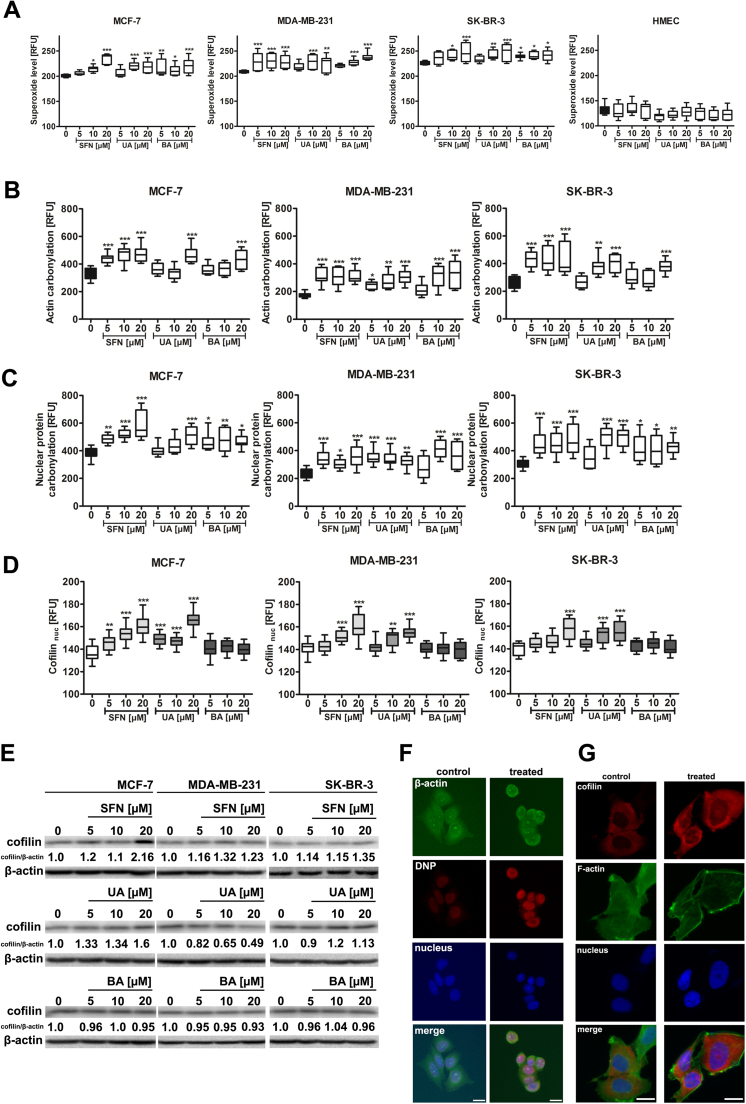
As phytochemicals may be considered as redox active compounds, we have then evaluated if SFN, UA and BA may induce oxidative stress in breast cancer cells (Fig. 3). Indeed, treatment with SFN, UA and BA resulted in elevated superoxide levels in breast cancer cells (Fig. 3A). In contrast, no increase in the levels of superoxide was observed in normal human mammary epithelial cells (HMEC) (Fig. 3A). SFN-, UA- and BA-mediated oxidative stress resulted in protein carbonylation (oxidative protein damage) in breast cancer cells (Fig. 3B, C and F).
Fig. 3.
SFN-, UA- and BA-induced oxidative stress results in oxidative protein damage and cofilin nuclear translocation in breast cancer cells. (A) Superoxide levels were assessed using fluorogenic probe dihydroethidium and are presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (B) Actin carbonylation was assessed using actin and DNP co-immunostaining and is presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (C) Nuclear protein carbonylation was assessed using Hoechst 33342 staining and anti-DNP antibody and is presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (D) Cofilin nuclear levels are presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01 compared to the control (ANOVA and Dunnett's a posteriori test). (E) Western blot analysis of cofilin levels. Anti-β-actin antibody was used as a loading control. The data represent the relative density normalized to β-actin. (F) Actin immunostaining (green), DNP immunostaining (red) and nucleus staining (blue). MCF-7 cells were treated with 10 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. (G) Cofilin nuclear translocation (red) in MCF-7 cells upon stimulation with 10 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. F-actin staining (green) and nucleus staining (blue). SFN, sulforaphane, UA, ursolic acid; BA, betulinic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
SFN, UA and BA promoted both the carbonylation of cytoplasmic proteins, i.e., actin (Fig. 3B and F) as well as the carbonylation of total nuclear protein pools (Fig. 3C and F). SFN- and UA-induced oxidative stress also promoted an increase in the levels of cofilin, an actin-binding protein, and cofilin nuclear translocation as an adaptive stress response (Fig. 3D, E and G).
3.4. Phytochemical-mediated disorganization of A- and B-type lamin networks
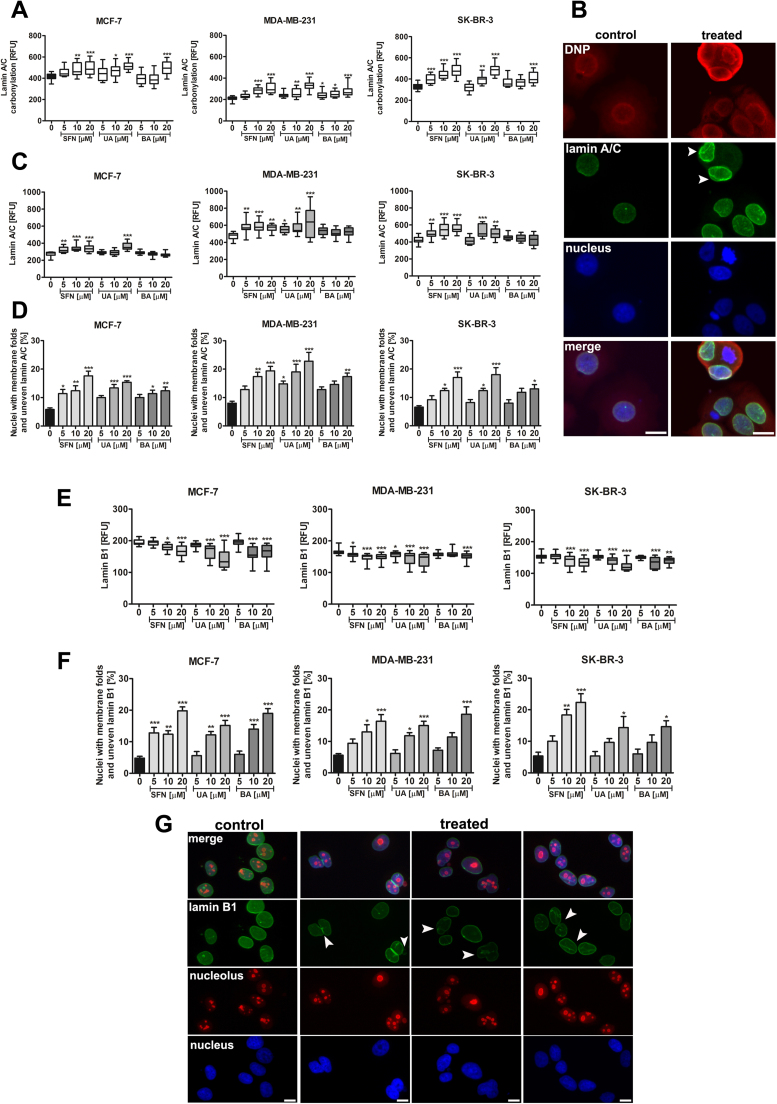
Increased carbonylation of lamin A/C was accompanied by changes in the levels of lamin A/C and its organization (Fig. 4A–D).
Fig. 4.
SFN-, UA- and BA-induced lamin A/C carbonylation and changes in lamin A/C and B1 levels and organization of lamin network and nucleus morphology. (A) Lamin A/C carbonylation was assessed using lamin A/C and DNP co-immunostaining and is presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (B) Lamin A/C immunostaining (green), DNP immunostaining (red) and nucleus staining (blue). MCF-7 cells were treated with 10 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. Aberrant organization of lamin A/C is presented (arrowheads). (C) Lamin A/C levels are presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (D) Quantitative analysis of abnormal lamin A/C signals [%]. Bars indicate SD, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (E) Lamin B1 levels are presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (F) Quantitative analysis of abnormal lamin B1 signals [%]. Bars indicate SD, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (G) Lamin B1 immunostaining (green), nucleolus immunostaining (red) and nucleus staining (blue). MCF-7 cells were treated with 10 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. Aberrant organization of lamin B1 is presented (arrowheads). SFN, sulforaphane, UA, ursolic acid; BA, betulinic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
SFN and UA caused an increase in the levels of lamin A/C (Fig. 4C) and SFN, UA and BA induced aberrant nuclear morphology, i.e., increased fraction of nuclei with membrane folds and uneven lamin A/C (Fig. 4D). Moreover, the levels of lamin B1 were decreased after SFN, UA and BA treatments and aberrant organization of lamin B1 was also observed (Fig. 4E–G).
3.5. Phytochemical-induced nucleolar stress
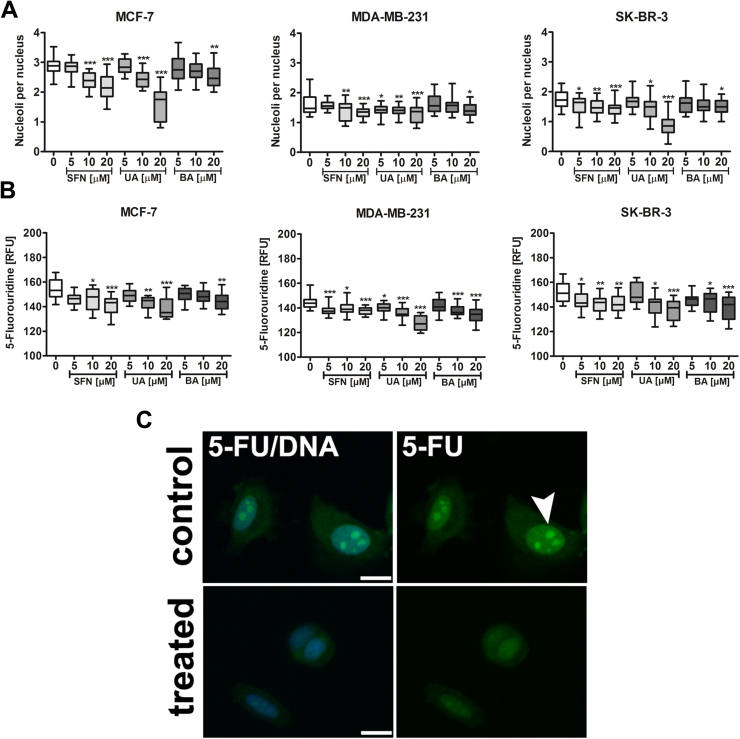
Changes in nuclear morphology was also accompanied by changes in the nucleolus that may be considered as a nucleolar stress. After phytochemical treatment, the number of nucleoli was decreased (Fig. 5A).
Fig. 5.
SFN-, UA- and BA-mediated decrease in the nucleolus number (A) and nucleolar activity (B, C). (A) The nucleolus was visualized using anti-nucleolar antigen. Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (B, C) Transcription of nascent RNA (synthesis of new rRNA) was evaluated using 5-fluorouridine (5-FU) labeling. 5-FU incorporation into nascent RNA was visualized using immunostaining with an anti-bromo-deoxyuridine (BrdU) antibody. (B) Nucleolar 5-FU signals are presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (C) MCF-7 cells were treated with 10 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. 5-FU was detected in nucleoli and throughout the nuclei (green, arrowhead). Lack of nucleolar 5-FU incorporation indicates inhibition of new rRNA synthesis. Nucleus staining (blue). SFN, sulforaphane, UA, ursolic acid; BA, betulinic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Moreover, phytochemicals promoted the inhibition of new rRNA synthesis in breast cancer cells as judged by decreased 5-FU labeling in the nucleolus (Fig. 5B and C). As loss of nucleolar proteins from the nucleolus is a marker of nucleolar stress following inhibition of RNA-Pol-I-driven transcription, we have then analyzed the levels and localization of RNA polymerase I-specific transcription initiation factor RRN3/TIF-IA (Fig. 6).
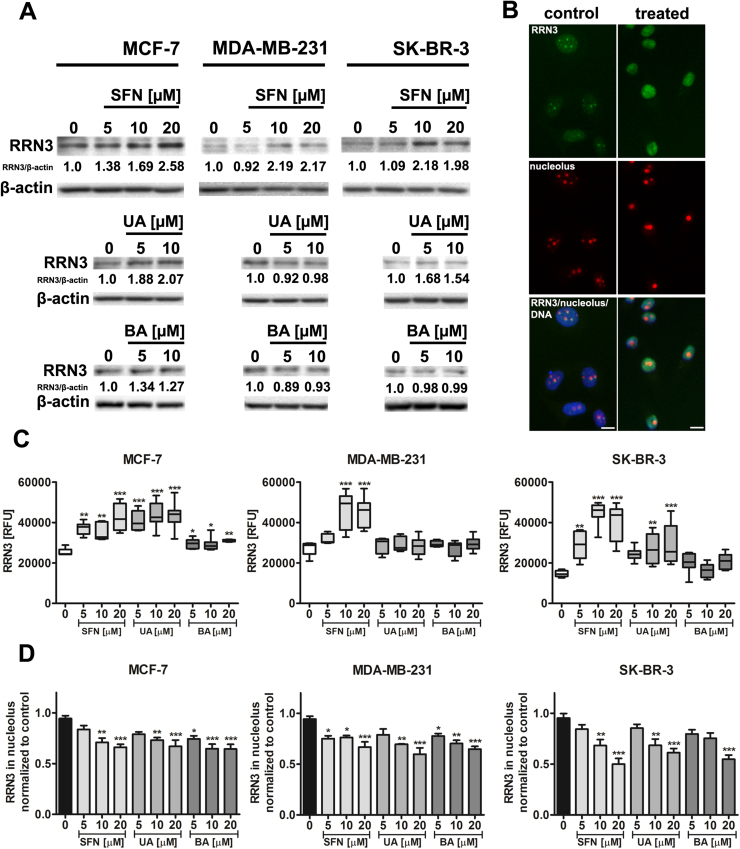
Fig. 6.
SFN-, UA- and BA-induced changes in RRN3 levels (A, C) and nucleoplasmic translocation of RRN3 (B, D). (A) Western blot analysis of RRN3 levels. Anti-β-actin antibody was used as a loading control. The data represent the relative density normalized to β-actin. (B) MCF-7 cells were treated with 10 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. RRN3 immunostaining (green), nucleolus immunostaining (red), nucleus staining (blue). (C) RRN3 signals are presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (D) Quantitative analysis of nucleoplasmic translocation of RRN3. RRN3 signals in nucleoli were normalized to the control conditions. Bars indicate SD, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). SFN, sulforaphane, UA, ursolic acid; BA, betulinic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
In general, total levels of RRN3 (Western blotting) (Fig. 6A) as well as nuclear pools of RRN3 were increased after SFN and UA treatments (imaging cytometry) (Fig. 6C). As increased nuclear levels of RRN3 were accompanied by decreased nucleolar levels of RRN3, one can conclude that RRN3 is relocated from the nucleolus to the nucleoplasm upon phytochemical stimulation (Fig. 6B and D).
3.6. Phytochemical-mediated changes in ribosome-associated proteins
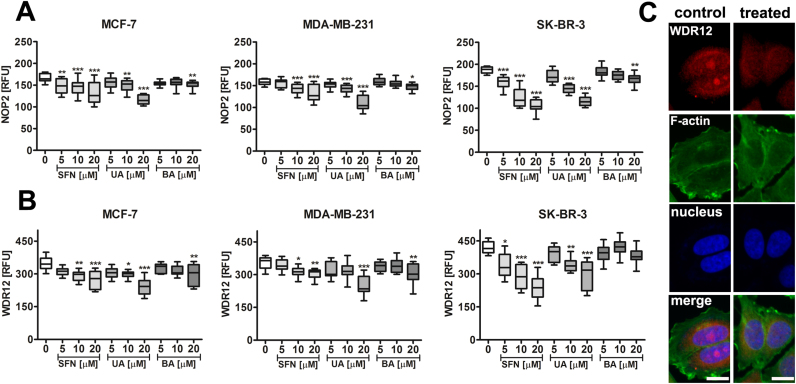
We have then analyzed the levels of selected nucleolar proteins involved in ribosome biogenesis, translation and the regulation of cell proliferation. We found that phytochemicals caused a decrease in the levels of NOP2, proliferation-associated nucleolar protein p120 [24] (Fig. 7A) and WDR12, a component of the PeBoW complex, which is required for maturation of 28S and 5.8S ribosomal RNAs and formation of the 60S ribosome and cell proliferation [25], [26] (Fig. 7B and C).
Fig. 7.
SFN-, UA- and BA-mediated decrease in the levels of nucleolar proteins NOP2 (A) and WDR12 (B, C). NOP2 signals (A) and WDR12 signals (B) are presented as relative fluorescence units (RFUs). Box and whisker plots are shown, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). (C) WDR12 immunostaining (red). MCF-7 cells were treated with 10 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. F-actin staining (green), nucleus staining (blue). SFN, sulforaphane, UA, ursolic acid; BA, betulinic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
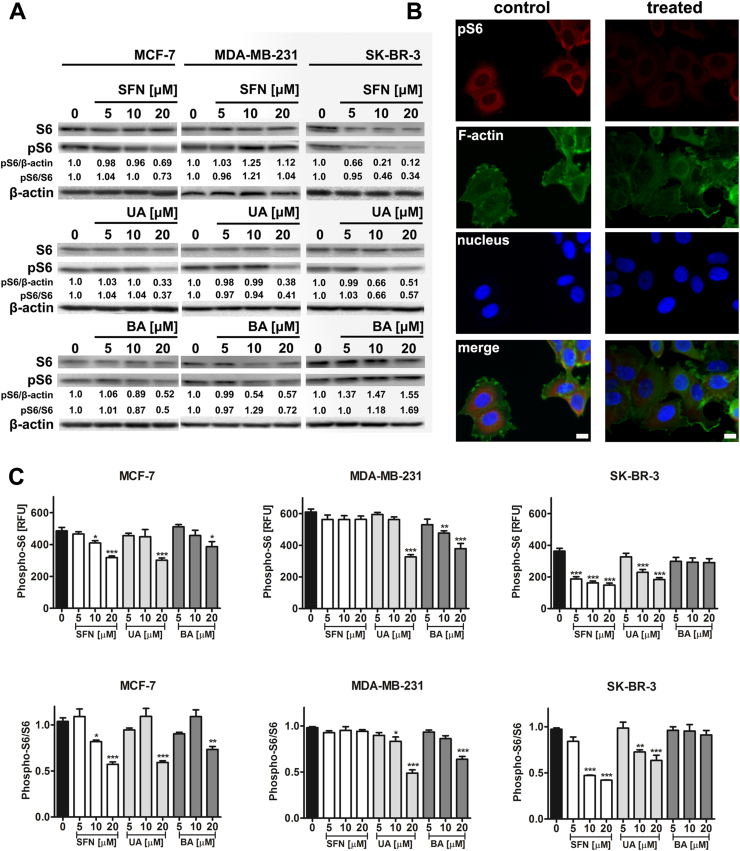
As phosphorylation of S6 ribosomal protein correlates with an increase in translation of mRNA transcripts, we have then investigated the phosphorylation status of S6 ribosomal protein (Ser235/236) upon phytochemical stimulation (Fig. 8).
Fig. 8.
SFN-, UA- and BA-mediated decrease in phospho-S6 signals. (A) Western blot analysis of S6 and phospho-S6 ribosomal protein levels. Anti-β-actin antibody was used as a loading control. The data represent the relative density normalized to β-actin and S6 signals. (B) Phospho-S6 immunostaining (red). MCF-7 cells were treated with 20 µM SFN. Representative micrographs are shown, objective 10×, scale bars 10 µm. F-actin staining (green), nucleus staining (blue). (C) Quantitative analysis of phospho-S6 immunostaining. Phospho-S6 signals are presented as relative fluorescence units (RFUs) (upper panel) and normalized to S6 signals (lower panel). Bars indicate SD, n=100 per one replicate, three independent experiments were considered, ***p<0.001, **p<0.01, *p<0.05 compared to the control (ANOVA and Dunnett's a posteriori test). SFN, sulforaphane, UA, ursolic acid; BA, betulinic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Except of SFN-treated MDA-MB-231 cells and BA-treated SK-BR-3 cells, decreased phospho-S6 signals were observed after treatment with SFN, UA and BA (Fig. 8A). Western blot data were also confirmed using imaging cytometry and S6 and phospho-S6 immunostaining (Fig. 8B and C).
4. Discussion
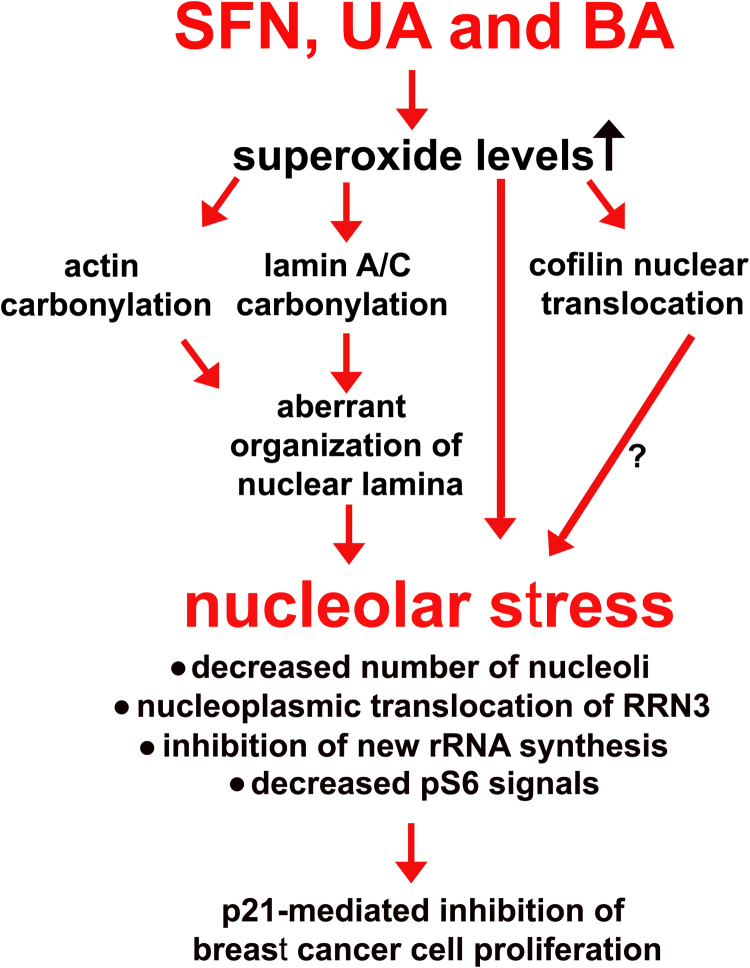
The nucleolus-centered adaptive response was revealed in three breast cancer cell lines, namely MCF-7 (ER+, PR+/-, HER2-, wild type p53), MDA-MB-231 (ER-, PR-, HER2-, mutant p53) and SK-BR-3 (ER-, PR-, HER2+, mutant p53) upon stimulation with low concentrations of an isothiocyanate sulforaphane (SFN) and two pentacyclic triterpenoids, ursolic acid (UA) and betulinic acid (BA) (Fig. 9). Phytochemicals promoted oxidant-based nucleolar stress that limit transcription and translation efficiency and cell proliferation (Fig. 9).
Fig. 9.
Mechanism of SFN-, UA- and BA-mediated antiproliferative action against breast cancer cells. Treatment with SFN, UA and BA promoted an increase in superoxide levels that resulted in oxidative protein damage (protein carbonylation, e.g., actin carbonylation). Carbonylation of lamin A/C and changes in the levels of lamin A/C and lamin B1 (unbalanced lamin B1/lamin A/C ratio) affected the organization of nuclear lamina and nucleus morphology. SFN, UA and BA induced nucleolar stress as judged by decreased number of nucleoli, nucleoplasmic translocation of RRN3, inhibition of rRNA synthesis and decreased phospho-S6 ribosomal protein levels that lead to p21-mediated inhibition of cell proliferation. Moreover, SFN, UA and BA promoted cofilin nuclear translocation as a stress response that may also affect the nucleolus state.
To study phytochemical-induced nucleolar stress response, we have used imaging cytometry and single-cell analysis because cancer cell populations are heterogeneous and some discrete individual cellular changes might be masked using population-based approaches.
Phytochemical-induced inhibition of cell proliferation was mediated by elevated levels of nuclear p21. In general, p21 may be activated by both p53-dependent or p53-independent mechanisms [27]. As we have used breast cancer cells with wild type p53 (MCF-7) as well as mutant p53 (MDA-MB-231, SK-BR-3) [28], one can conclude that phytochemicals induced upregulation of p21 irrespective of p53 status. Antiproliferative activity of SFN, UA and BA has already been documented [29], [30], [31]. SFN (15 µM) has been shown to inhibit cell proliferation of MCF-7 cells [29]. SFN promoted G2/M cell cycle arrest and increase in cyclin B1 protein levels as well as phosphorylation of histone H1, blocked cells in early mitosis and disrupted polymerization of mitotic microtubules in vivo [29]. UA (17.5 µM) caused p21-dependent G0/G1 cell cycle arrest in MCF-7 cells as the effect was nearly abolished after p21 silencing [30]. BA (5–10 µM) induced cell cycle arrest in the G2/M phase in MDA-MB-231 cells that was based on interactions with the microRNA-27a-ZBTB10-Sp-axis [31]. More recently, we have also shown that UA and BA (5–10 µM) caused G0/G1 cell cycle arrest in MCF-7, MDA-MB-231 and SK-BR-3 cells [32]. Cytotoxic action of SFN, UA and BA was limited to the concentration of 20 µM and UA was found to be the most potent cytotoxic agent in MCF-7, MDA-MB-231 and SK-BR-3 cells. This is in agreement with our previous results on UA-induced apoptosis in MCF-7, MDA-MB-231 and SK-BR-3 cells when UA was used at the concentration of 20 µM [32].
Treatment with SFN, UA and BA resulted in elevated superoxide levels that in turn lead to protein carbonylation and oxidant-based nucleolar stress response (this study). In general, SFN, UA and BA are considered as antioxidants [33], [34], [35], [36], [37] and their pro-oxidative action is limited to cancer cells [38], [39], [40]. The effects of reactive oxygen species (ROS) on cancer cell biology depend on their levels [41]. Low to moderate oxidative stress may promote cancer growth and proliferation, whereas excessive oxidative stress induce oxidative damage of biomolecules and cell death [41]. Protein carbonyl groups are biomarkers of oxidative stress [42] and actin is a major oxidation-prone cytoskeletal protein [43], [44], [45]. Actin is involved in numerous cellular processes such as motility, adherence, gene expression, cell secretion and cell division [46], [47], [48], [49], [50], and actin carbonylation may be implicated in cytoskeleton disturbance, cell dysfunction and age-related diseases [45]. Hydrogen peroxide-induced actin carbonylation resulted in actin aggregate formation, decreased proteasome activity and affected T cell proliferation that lead to T cell functional impairment [44]. Oxidatively stressed human T cells were also impaired in chemotaxis- and co-stimulation-induced F-actin modulation through oxidation of the actin-remodeling protein cofilin [51]. Phytochemicals also promoted actin carbonylation in breast cancer cells, but we did not observe a tendency of carbonylated actin to form aggregates. This may be due to the exposure time and the magnitude of phytochemical-induced oxidative stress. However, changes in cofilin levels and cofilin nuclear translocation were documented that may be considered as a adaptive stress response to phytochemical-mediated actin modification. The actin depolymerizing factor (ADF)/cofilin protein family is essential for cytokinesis, phagocytosis, fluid phase endocytosis, and other cellular processes dependent upon actin dynamics [52], [53], [54]. Cofilin possesses nuclear localization signals (NLS) in its protein sequence that allows for the transport of depolymerized actin to the nucleus [55], [56]. Stress conditions such as heat shock, ATP depletion, DMSO treatment, cytochalasin D or high cytosolic G-actin concentration may promote cofilin-associated actin transition from the cytoplasm into the nucleus and the formation of cofilin-actin rods [55], [57]. However, the nuclear functions of cofilin remain elusive. Cofilin may regulate transcription and chromatin structure as cofilin has been reported to be required for RNA polymerase II transcription elongation [58]. The role of mitochondrial translocation of cofilin during cell death signaling is also ambiguous [59], [60], [61], [62]. Cell death induced upon oxidant stimulation and mitochondrial translocation of oxidized cofilin has been assumed to be due to apoptosis [60] or caspase-independent necrosis [61]. Moreover, it has been confirmed that ADF/cofilin proteins are translocated to mitochondria during apoptosis but this is not required for cell death signaling [62].
Phytochemical-induced oxidative stress also promoted the carbonylation of nuclear proteins, e.g., lamin A/C that in turn resulted in aberrant nuclear morphology such as formation of membrane folds and blebs and uneven lamin A/C and lamin B1 signals. Disorganization of lamin networks may be also mediated by oxidatively modified actin-associated cytoskeletal dysfunction and changes in the levels of lamin A/C and lamin B1, namely an increase in lamin A/C and a decrease in lamin B1 (this study). Oxidative stress can affect lamin functions by oxidation of lamin A, accumulation of pre-lamin A or lamin B1 leading to nuclear shape alterations and loss of redox control, which amplify the oxidative stress [63]. It is widely accepted that lamin levels must be tightly controlled for the maintenance of nuclear architecture and for protection against senescence [63]. The associations between changes in lamin levels, oxidative stress and cellular senescence are rather complex [64]. Lamin B1 loss has been considered as a senescence-associated biomarker [65] and lamin B1 silencing resulted in the inhibition of proliferation through a ROS signaling pathway in human fibroblast WI-38 cells [66]. However, both elevated and diminished levels of lamin B1 may be associated with senescence in response to disturbed ROS homeostasis [66], [67], but, in general, healthy early passage cells are characterized by balanced lamin B1/lamin A ratio and stressed/senescent cells are characterized by unbalanced lamin B1/lamin A ratio [64]. Phytochemical-mediated changes in the levels of lamin A/C and lamin B1 and disorganization of lamin networks (this study) may also promote cellular senescence in breast cancer cells. Indeed, we have recently shown that UA and BA, when used at the concentrations of 5 µM and 10 µM, caused an increase in senescence-associated beta-galactosidase staining in breast cancer cells [32].
Finally, elevated levels of superoxide and oxidative stress-mediated alterations in the nuclear architecture promoted nucleolar stress in breast cancer cells upon phytochemical stimulation. Classical biomarkers of nucleolar stress were observed, namely decreased number of nucleoli, nucleoplasmic translocation of RRN3 transcription factor and inhibition of rRNA synthesis [3], [6], [11] that lead to inhibition of breast cancer cell proliferation. Moreover, phytochemicals acted as ribotoxic stress stimuli by decreasing the levels of ribosomal proteins NOP2, a cell proliferation marker [24] and WDR12, required for ribosome biogenesis and cell proliferation [25], [26], and phosphorylated S6 ribosomal protein that may affect translation efficiency and cell growth. Indeed, SFN (10–40 µM) has already been shown to inhibit protein synthesis (diminished [3H]-leucine incorporation) in MCF-7 and MDA-MB-231 cells [68]. More recently, the nucleolus as a new target for anticancer therapy has been proposed [69], [70]. Some traditional chemotherapeutic drugs as well as newer compounds have been demonstrated to target the nucleolar surveillance pathway, either at the level of Pol I transcription of the rDNA genes, or processing of the pre-rRNA, e.g., doxorubicin, actinomycin D, cisplatin, CX-5461 and 5-fluorouracil [70]. CX-5461 has been shown to inhibit Pol I-driven transcription relative to Pol II-driven transcription, DNA replication, and protein translation that diminish rRNA synthesis and solid tumor growth [71]. CX-5461 induced senescence and autophagy, but not apoptosis, through a p53-independent process in solid tumor cell lines [71].
In summary, we have shown for the first that selected phytochemicals (SFN, UA and BA) can be considered as nucleolar stress stimuli in breast cancer cells (Fig. 9). SFN, UA and BA induced oxidative stress and protein carbonylation that resulted in unbalanced lamin B1/lamin A/C ratio, altered organization of nuclear lamina and abnormal nuclear morphology. Phytochemical-mediated nucleolar stress response involved nucleoplasmic translocation of RRN3 and inhibition of rRNA synthesis. Phytochemicals may be also considered as ribotoxic stress stimuli because they caused a decrease in phospho-S6 signals and nucleolar protein levels NOP2 and WDR12 that diminished translation efficiency and in turn lead to the inhibition of breast cancer cell proliferation.
Acknowledgments
This study was supported by Grant from the National Science Center, 2013/11/D/NZ7/00939. Diana Bednarz is a student of Biotechnology at University of Rzeszow, Poland.
References
- 1.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boisvert F.M., van Koningsbruggen S., Navascues J., Lamond A.I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 3.Olson M.O. Sensing cellular stress: another new function for the nucleolus? Sci. STKE. 2004;2004:pe 10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 4.Grummt I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122:487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- 5.Boulon S., Westman B.J., Hutten S., Boisvert F.M., Lamond A.I. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer C., Bierhoff H., Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005;19:933–941. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Wolf G.W., Bhat K., Jin A., Allio T., Burkhart W.A., Xiong Y. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell Biol. 2003;23:8902–8912. doi: 10.1128/MCB.23.23.8902-8912.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurki S., Peltonen K., Latonen L., Kiviharju T.M., Ojala P.M., Meek D., Laiho M. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell. 2004;5:465–475. doi: 10.1016/s1535-6108(04)00110-2. [DOI] [PubMed] [Google Scholar]
- 9.Sherr C.J., Weber J.D. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- 10.Rubbi C.P., Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan X., Zhou Y., Casanova E., Chai M., Kiss E., Grone H.J., Schutz G., Grummt I. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 12.James A., Wang Y., Raje H., Rosby R., DiMario P. Nucleolar stress with and without p53. Nucleus. 2014;5:402–426. doi: 10.4161/nucl.32235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donati G., Brighenti E., Vici M., Mazzini G., Trere D., Montanaro L., Derenzini M. Selective inhibition of rRNA transcription downregulates E2F-1: a new p53-independent mechanism linking cell growth to cell proliferation. J. Cell Sci. 2011;124:3017–3028. doi: 10.1242/jcs.086074. [DOI] [PubMed] [Google Scholar]
- 14.Iadevaia V., Caldarola S., Biondini L., Gismondi A., Karlsson S., Dianzani I., Loreni F. PIM1 kinase is destabilized by ribosomal stress causing inhibition of cell cycle progression. Oncogene. 2010;29:5490–5499. doi: 10.1038/onc.2010.279. [DOI] [PubMed] [Google Scholar]
- 15.Russo A., Esposito D., Catillo M., Pietropaolo C., Crescenzi E., Russo G. Human rpL3 induces G(1)/S arrest or apoptosis by modulating p21 (waf1/cip1) levels in a p53-independent manner. Cell Cycle. 2013;12:76–87. doi: 10.4161/cc.22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cirstea D., Hideshima T., Santo L., Eda H., Mishima Y., Nemani N., Hu Y., Mimura N., Cottini F., Gorgun G., Ohguchi H., Suzuki R., Loferer H., Munshi N.C., Anderson K.C., Raje N. Small-molecule multi-targeted kinase inhibitor RGB-286638 triggers P53-dependent and -independent anti-multiple myeloma activity through inhibition of transcriptional CDKs. Leukemia. 2013;27:2366–2375. doi: 10.1038/leu.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soussi T., Dehouche K., Beroud C. p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum. Mutat. 2000;15:105–113. doi: 10.1002/(SICI)1098-1004(200001)15:1<105::AID-HUMU19>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 18.Aggarwal B.B., Shishodia S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem. Pharmacol. 2006;71:1397–1421. doi: 10.1016/j.bcp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Gali-Muhtasib H., Hmadi R., Kareh M., Tohme R., Darwiche N. Cell death mechanisms of plant-derived anticancer drugs: beyond apoptosis. Apoptosis. 2015;20:1531–1562. doi: 10.1007/s10495-015-1169-2. [DOI] [PubMed] [Google Scholar]
- 20.Lewinska A., Jarosz P., Czech J., Rzeszutek I., Bielak-Zmijewska A., Grabowska W., Wnuk M. Capsaicin-induced genotoxic stress does not promote apoptosis in A549 human lung and DU145 prostate cancer cells. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2015;779:23–34. doi: 10.1016/j.mrgentox.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Lazarus R.C., Buonora J.E., Jacobowitz D.M., Mueller G.P. Protein carbonylation after traumatic brain injury: cell specificity, regional susceptibility, and gender differences. Free Radic. Biol. Med. 2015;78:89–100. doi: 10.1016/j.freeradbiomed.2014.10.507. [DOI] [PubMed] [Google Scholar]
- 22.Boisvert F.M., Hendzel M.J., Bazett-Jones D.P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J. Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewinska A., Adamczyk-Grochala J., Kwasniewicz E., Deregowska A., Wnuk M. Diosmin-induced senescence, apoptosis and autophagy in breast cancer cells of different p53 status and ERK activity. Toxicol. Lett. 2017;265:117–130. doi: 10.1016/j.toxlet.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 24.Fonagy A., Swiderski C., Ostrovsky A.M., Bolton W.E., Freeman J.W. Effect of nucleolar P120 expression level on the proliferation capacity of breast cancer cells. Cancer Res. 1994;54:1859–1864. [PubMed] [Google Scholar]
- 25.Holzel M., Rohrmoser M., Schlee M., Grimm T., Harasim T., Malamoussi A., Gruber-Eber A., Kremmer E., Hiddemann W., Bornkamm G.W., Eick D. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 2005;170:367–378. doi: 10.1083/jcb.200501141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohrmoser M., Holzel M., Grimm T., Malamoussi A., Harasim T., Orban M., Pfisterer I., Gruber-Eber A., Kremmer E., Eick D. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol. Cell Biol. 2007;27:3682–3694. doi: 10.1128/MCB.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gartel A.L., Tyner A.L. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol. Cancer Ther. 2002;1:639–649. [PubMed] [Google Scholar]
- 28.Lacroix M., Toillon R.A., Leclercq G. p53 and breast cancer, an update. Endocr. Relat. Cancer. 2006;13:293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 29.Jackson S.J., Singletary K.W. Sulforaphane inhibits human MCF-7 mammary cancer cell mitotic progression and tubulin polymerization. J. Nutr. 2004;134:2229–2236. doi: 10.1093/jn/134.9.2229. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X., Song X., Yin S., Zhao C., Fan L., Hu H. p21 induction plays a dual role in anti-cancer activity of ursolic acid. Exp. Biol. Med. 2016;241:501–508. doi: 10.1177/1535370215616195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mertens-Talcott S.U., Noratto G.D., Li X., Angel-Morales G., Bertoldi M.C., Safe S. Betulinic acid decreases ER-negative breast cancer cell growth in vitro and in vivo: role of Sp transcription factors and microRNA-27a:ZBTB10. Mol. Carcinog. 2013;52:591–602. doi: 10.1002/mc.21893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lewinska A., Adamczyk-Grochala J., Kwasniewicz E., Deregowska A., Wnuk M. Ursolic acid-mediated changes in glycolytic pathway promote cytotoxic autophagy and apoptosis in phenotypically different breast cancer cells. Apoptosis. 2017 doi: 10.1007/s10495-017-1353-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J., Zheng Y.L., Wu D.M., Luo L., Sun D.X., Shan Q. Ursolic acid ameliorates cognition deficits and attenuates oxidative damage in the brain of senescent mice induced by D-galactose. Biochem. Pharmacol. 2007;74:1078–1090. doi: 10.1016/j.bcp.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Tsai S.J., Yin M.C. Antioxidative and anti-inflammatory protection of oleanolic acid and ursolic acid in PC12 cells. J. Food Sci. 2008;73:H174–H178. doi: 10.1111/j.1750-3841.2008.00864.x. [DOI] [PubMed] [Google Scholar]
- 35.Liobikas J., Majiene D., Trumbeckaite S., Kursvietiene L., Masteikova R., Kopustinskiene D.M., Savickas A., Bernatoniene J. Uncoupling and antioxidant effects of ursolic acid in isolated rat heart mitochondria. J. Nat. Prod. 2011;74:1640–1644. doi: 10.1021/np200060p. [DOI] [PubMed] [Google Scholar]
- 36.Lu Q., Xia N., Xu H., Guo L., Wenzel P., Daiber A., Munzel T., Forstermann U., Li H. Betulinic acid protects against cerebral ischemia-reperfusion injury in mice by reducing oxidative and nitrosative stress. Nitric Oxide. 2011;24:132–138. doi: 10.1016/j.niox.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 37.Angeloni C., Teti G., Barbalace M.C., Malaguti M., Falconi M., Hrelia S. 17beta-Estradiol enhances sulforaphane cardioprotection against oxidative stress. J. Nutr. Biochem. 2017;42:26–36. doi: 10.1016/j.jnutbio.2016.12.017. [DOI] [PubMed] [Google Scholar]
- 38.Wu C.C., Cheng C.H., Lee Y.H., Chang I.L., Chen H.Y., Hsieh C.P., Chueh P.J. Ursolic acid triggers apoptosis in human osteosarcoma cells via caspase activation and the ERK1/2 MAPK pathway. J. Agric. Food Chem. 2016;64:4220–4226. doi: 10.1021/acs.jafc.6b00542. [DOI] [PubMed] [Google Scholar]
- 39.Shen S., Zhang Y., Zhang R., Tu X., Gong X. Ursolic acid induces autophagy in U87MG cells via ROS-dependent endoplasmic reticulum stress. Chem. Biol. Interact. 2014;218:28–41. doi: 10.1016/j.cbi.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 40.Singh S.V., Srivastava S.K., Choi S., Lew K.L., Antosiewicz J., Xiao D., Zeng Y., Watkins S.C., Johnson C.S., Trump D.L., Lee Y.J., Xiao H., Herman-Antosiewicz A. Sulforaphane-induced cell death in human prostate cancer cells is initiated by reactive oxygen species. J. Biol. Chem. 2005;280:19911–19924. doi: 10.1074/jbc.M412443200. [DOI] [PubMed] [Google Scholar]
- 41.Gupta S.C., Hevia D., Patchva S., Park B., Koh W., Aggarwal B.B. Upsides and downsides of reactive oxygen species for cancer: the roles of reactive oxygen species in tumorigenesis, prevention, and therapy. Antioxid. Redox Signal. 2012;16:1295–1322. doi: 10.1089/ars.2011.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dalle-Donne I., Rossi R., Giustarini D., Milzani A., Colombo R. Protein carbonyl groups as biomarkers of oxidative stress. Clin. Chim. Acta. 2003;329:23–38. doi: 10.1016/s0009-8981(03)00003-2. [DOI] [PubMed] [Google Scholar]
- 43.Dalle-Donne I., Rossi R., Giustarini D., Gagliano N., Lusini L., Milzani A., Di Simplicio P., Colombo R. Actin carbonylation: from a simple marker of protein oxidation to relevant signs of severe functional impairment. Free Radic. Biol. Med. 2001;31:1075–1083. doi: 10.1016/s0891-5849(01)00690-6. [DOI] [PubMed] [Google Scholar]
- 44.Castro J.P., Ott C., Jung T., Grune T., Almeida H. Carbonylation of the cytoskeletal protein actin leads to aggregate formation. Free Radic. Biol. Med. 2012;53:916–925. doi: 10.1016/j.freeradbiomed.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 45.Castro J.P., Jung T., Grune T., Almeida H. Actin carbonylation: from cell dysfunction to organism disorder. J. Proteom. 2013;92:171–180. doi: 10.1016/j.jprot.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 46.Vicente-Manzanares M., Sanchez-Madrid F. Role of the cytoskeleton during leukocyte responses. Nat. Rev. Immunol. 2004;4:110–122. doi: 10.1038/nri1268. [DOI] [PubMed] [Google Scholar]
- 47.Zamir E., Geiger B. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 2001;114:3583–3590. doi: 10.1242/jcs.114.20.3583. [DOI] [PubMed] [Google Scholar]
- 48.DeMali K.A., Wennerberg K., Burridge K. Integrin signaling to the actin cytoskeleton. Curr. Opin. Cell Biol. 2003;15:572–582. doi: 10.1016/s0955-0674(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 49.Samstag Y., Nebl G. Ras initiates phosphatidyl-inositol-3-kinase (PI3K)/PKB mediated signalling pathways in untransformed human peripheral blood T lymphocytes. Adv. Enzym. Regul. 2005;45:52–62. doi: 10.1016/j.advenzreg.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Pollard T.D. Mechanics of cytokinesis in eukaryotes. Curr. Opin. Cell Biol. 2010;22:50–56. doi: 10.1016/j.ceb.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klemke M., Wabnitz G.H., Funke F., Funk B., Kirchgessner H., Samstag Y. Oxidation of cofilin mediates T cell hyporesponsiveness under oxidative stress conditions. Immunity. 2008;29:404–413. doi: 10.1016/j.immuni.2008.06.016. [DOI] [PubMed] [Google Scholar]
- 52.Bamburg J.R. Proteins of the ADF/cofilin family: essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 53.Carlier M.F., Ressad F., Pantaloni D. Control of actin dynamics in cell motility. Role of ADF/cofilin. J. Biol. Chem. 1999;274:33827–33830. doi: 10.1074/jbc.274.48.33827. [DOI] [PubMed] [Google Scholar]
- 54.Elam W.A., Kang H., De la Cruz E.M. Biophysics of actin filament severing by cofilin. FEBS Lett. 2013;587:1215–1219. doi: 10.1016/j.febslet.2013.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abe H., Nagaoka R., Obinata T. Cytoplasmic localization and nuclear transport of cofilin in cultured myotubes. Exp. Cell Res. 1993;206:1–10. doi: 10.1006/excr.1993.1113. [DOI] [PubMed] [Google Scholar]
- 56.Pendleton A., Pope B., Weeds A., Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J. Biol. Chem. 2003;278:14394–14400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- 57.Munsie L.N., Desmond C.R., Truant R. Cofilin nuclear-cytoplasmic shuttling affects cofilin-actin rod formation during stress. J. Cell Sci. 2012;125:3977–3988. doi: 10.1242/jcs.097667. [DOI] [PubMed] [Google Scholar]
- 58.Obrdlik A., Percipalle P. The F-actin severing protein cofilin-1 is required for RNA polymerase II transcription elongation. Nucleus. 2011;2:72–79. doi: 10.4161/nucl.2.1.14508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chua B.T., Volbracht C., Tan K.O., Li R., Yu V.C., Li P. Mitochondrial translocation of cofilin is an early step in apoptosis induction. Nat. Cell Biol. 2003;5:1083–1089. doi: 10.1038/ncb1070. [DOI] [PubMed] [Google Scholar]
- 60.Klamt F., Zdanov S., Levine R.L., Pariser A., Zhang Y., Zhang B., Yu L.R., Veenstra T.D., Shacter E. Oxidant-induced apoptosis is mediated by oxidation of the actin-regulatory protein cofilin. Nat. Cell Biol. 2009;11:1241–1246. doi: 10.1038/ncb1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wabnitz G.H., Goursot C., Jahraus B., Kirchgessner H., Hellwig A., Klemke M., Konstandin M.H., Samstag Y. Mitochondrial translocation of oxidized cofilin induces caspase-independent necrotic-like programmed cell death of T cells. Cell Death Dis. 2010;1:e58. doi: 10.1038/cddis.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rehklau K., Gurniak C.B., Conrad M., Friauf E., Ott M., Rust M.B. ADF/cofilin proteins translocate to mitochondria during apoptosis but are not generally required for cell death signaling. Cell Death Differ. 2012;19:958–967. doi: 10.1038/cdd.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barascu A., Le Chalony C., Pennarun G., Genet D., Zaarour N., Bertrand P. Oxydative stress alters nuclear shape through lamins dysregulation: a route to senescence. Nucleus. 2012;3:411–417. doi: 10.4161/nucl.21674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hutchison C.J. B-type lamins and their elusive roles in metazoan cell proliferation and senescence. EMBO J. 2012;31:1058–1059. doi: 10.1038/emboj.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freund A., Laberge R.M., Demaria M., Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012;23:2066–2075. doi: 10.1091/mbc.E11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shimi T., Butin-Israeli V., Adam S.A., Hamanaka R.B., Goldman A.E., Lucas C.A., Shumaker D.K., Kosak S.T., Chandel N.S., Goldman R.D. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25:2579–2593. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barascu A., Le Chalony C., Pennarun G., Genet D., Imam N., Lopez B., Bertrand P. Oxidative stress induces an ATM-independent senescence pathway through p38 MAPK-mediated lamin B1 accumulation. EMBO J. 2012;31:1080–1094. doi: 10.1038/emboj.2011.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pawlik A., Wiczk A., Kaczynska A., Antosiewicz J., Herman-Antosiewicz A. Sulforaphane inhibits growth of phenotypically different breast cancer cells. Eur. J. Nutr. 2013;52:1949–1958. doi: 10.1007/s00394-013-0499-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Quin J.E., Devlin J.R., Cameron D., Hannan K.M., Pearson R.B., Hannan R.D. Targeting the nucleolus for cancer intervention. Biochim. Biophys. Acta. 2014;1842:802–816. doi: 10.1016/j.bbadis.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 70.Woods S.J., Hannan K.M., Pearson R.B., Hannan R.D. The nucleolus as a fundamental regulator of the p53 response and a new target for cancer therapy. Biochim. Biophys. Acta. 2015;1849:821–829. doi: 10.1016/j.bbagrm.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 71.Drygin D., Lin A., Bliesath J., Ho C.B., O'Brien S.E., Proffitt C., Omori M., Haddach M., Schwaebe M.K., Siddiqui-Jain A., Streiner N., Quin J.E., Sanij E., Bywater M.J., Hannan R.D., Ryckman D., Anderes K., Rice W.G. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71:1418–1430. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]