Abstract
Background
Oxidant stress is arguably a universal feature in toxicology. Research studies on the role of oxidant stress induced by xenobiotic exposures have typically relied on the identification of damaged biomolecules using a variety of conventional biochemical and molecular techniques. However, there is increasing evidence that low-level exposure to a variety of toxicants dysregulates cellular physiology by interfering with redox-dependent processes.
Scope of review
The study of events involved in redox toxicology requires methodology capable of detecting transient modifications at relatively low signal strength. This article reviews the advantages of live-cell imaging for redox toxicology studies.
Major conclusions
Toxicological studies with xenobiotics of supra-physiological reactivity require careful consideration when using fluorogenic sensors in order to avoid potential artifacts and false negatives. Fortunately, experiments conducted for the purpose of validating the use of these sensors in toxicological applications often yield unexpected insights into the mechanisms through which xenobiotic exposure induces oxidant stress.
General significance
Live-cell imaging using a new generation of small molecule and genetically encoded fluorophores with excellent sensitivity and specificity affords unprecedented spatiotemporal resolution that is optimal for redox toxicology studies. This article is part of a Special Issue entitled Air Pollution, edited by Wenjun Ding, Andrew J. Ghio and Weidong Wu.
Keywords: Live-cell imaging, Oxidant stress, Toxicology, Xenobiotic, Reactive oxygen species, Glutathione, Redox potential, Fluorescence
1. Introduction
Oxidant stress is one of the most commonly cited mechanistic features of the adverse action of xenobiotics. Methodological approaches of varying efficiency have shown a wide variety of oxidant events associated with the toxicological effects of environmental contaminants [1, 2], pharmaceutical agents [3,4] and natural toxins [5,6]. In addition, the field of redox biology continues to elucidate physiological redox processes that represent previously unappreciated targets for toxicological disruption.
As with any analytical endpoint, the detectability of xenobiotic oxidant stress is a function of the magnitude, duration and specificity of the signal that the effect of interest produces, as well as the sensitivity, processing speed and resolution of the method used to detect it. Experimentally, it naturally follows that more sensitive methods allow the detection of oxidant effects induced by lower levels of exposure that may more closely approximate real-world exposure scenarios. This review will focus on imaging methods available for toxicological research that employ a new generation of fluorogenic small molecule and genetically encoded sensors that, when combined with light microscopy, enable monitoring of reactive species and markers of intracellular oxidant stress in living cells with high specificity and sensitivity, offering unparalleled spatiotemporal resolution that is optimal for the study of xenobiotic-induced oxidant stress.
2. Redox toxicology
The term “oxidant stress” is often used non-specifically in the toxicological literature to refer to a wide variety of chemical reactions involving electrophilic attack on biomolecules, as well as disparate biological outcomes. “Oxidant stress” is used to refer to the generation of supra-physiological levels of reactive oxygen or nitrogen species (ROS, RNS), free radical-mediated damage to macromolecules, oxidation of glutathione and other intracellular “antioxidants”, and the activation of signaling cascades, most notably the KEAP1/Nrf2 pathway.
In practical terms and for the purposes of this review, oxidant stress is defined as an increase in the concentration of oxidants or oxidized biomolecules in the cell relative to a homeostatic baseline condition. Fundamentally, this stress is understood to be the result of an accumulation of oxidants or oxidant damage at a rate that exceeds the homeostatic capacity of the cell to dissipate or repair it. In toxicology, this damage may be caused directly by the xenobiotic compound itself [7–11], and/or secondarily by oxidant species produced by cellular processes [12–16], or by endogenous oxidant species that are of cellular origin [17–20]. Although the concept is evolving [21], a diminished capacity of defense and repair processes in the cell, as occurs in aging [22,23], may also be considered a form of oxidant stress.
Exposure to a broad range of structurally disparate environmental and therapeutic agents has been reported to induce oxidative modification of cellular biomolecules [3,16,24–36]. A significant fraction of these xenobiotics or their metabolites have been reported to generate reactive oxygen species through redox cycling [37–40], by perturbing energy metabolism [41–45], or by altering redox-dependent processes in the cell [46–48], including the induction of “reductive stress”, a situation in which limited availability of electron acceptors leads to the reduction of oxygen to form ROS [49]. Collectively, the study of the adverse outcomes induced by the reductive or oxidative effects of xenobiotic exposure can thus be referred to as redox toxicology.
3. Methodological approaches to redox toxicology
Given the widespread relevance of oxidant stress endpoints to the toxicology of xenobiotics, there is a constant need for improved methodologies for the detection of oxidant species and stress markers. Analytically, the study of oxidative stress in vitro has most commonly been based on the quantification of concentrations of reactive species [50], such as superoxide [51], hydrogen peroxide [52], hypochlorite [53], free radicals [54], and peroxynitrite [55].
Alternatively, in vitro evidence of oxidant stress has also been based on the measurement of levels of oxidized biomolecules, principally oxidized and adducted proteins [56–65], lipids [66–68], and nucleic acids [69–72]. Another commonly employed approach relies on the measurement of the concentrations and form of intracellular antioxidants [73] such as glutathione (GSH) [74–76], ascorbate [77,78], tocopherols [79,80], carotenoids [79,81,82], and the energy intermediates NADPH and NADH [83,84].
More recently, functional measurements based on the effects of oxidant stress on cells have been developed. For instance, oxidant stress has been shown to activate intracellular signaling through multiple mechanisms, including the activation of kinases and the loss of phosphatase activity [85–88]. The state of activation of signaling intermediates in oxidant-sensitive pathways (e.g., KEAP1/NrF2) is increasingly used as a marker of oxidant stress [89–91]. Similarly, induction of transcriptional expression of hemoxygenase 1 (HO-1) [92–95] and NADPH quinone oxidoreductase 1 (NQO1) [96,97], both of which are genes regulated by the transcription factor NrF2, is gaining in acceptance as a relatively specific marker of the cellular response to oxidant stress. A genomic approach that builds on this concept monitors profiles of gene expression known to be involved in the sensing, signaling and response to oxidant stress [98–100].
A major limitation inherent in all of the aforementioned biochemical and analytical assays used for the study of oxidant stress is that they consume the sample, requiring the use of extraction procedures to isolate the oxidized biomolecules, proteins or mRNA of interest, which precludes repeated collection of data points from the same cells undergoing a response over time. Moreover, disrupting compartments within the cell inevitably removes physical and kinetic constraints that normally modulate the equilibration of redox pairs and regulate interaction between enzymes and their substrates, thus potentially leading to loss of information and, more pernicious, the introduction of artifacts [101].
4. Advantages of live-cell imaging for redox toxicology studies
The generation of reactive oxygen species and the cellular responses induced by oxidant stress are often short-lived events that can be difficult to capture. This may be especially true in cases where the duration of the oxidant event is insufficient to result in the death of the cell but instead initiates adverse cellular responses, a scenario that is often of concern in toxicology. The ability to monitor events by collecting readouts at close time intervals in the same cells is therefore essential to capturing transient oxidative events. Examples include a sharp increase in the concentration of an ROS of interest such as H2O2, or a change in the relative concentration of glutathione (GSH) and its oxidized derivative (GSSG). The transient and localized nature of biochemical redox reactions places unprecedented demands on the spatial and temporal resolution required to study them under physiological and toxicological conditions.
Live-cell imaging is a light microscopy technique for monitoring physiological parameters in living cells in real time [102,103]. Environmental control hardware is used to maintain physiological temperature, pH and humidity levels to support cell viability throughout the period of data collection [103]. By preserving the integrity of the cell throughout the experiment, live-cell imaging avoids many of the limitations of extractive techniques, including loss of spatial information and the introduction of methodological artifacts. Since cellular compartments are not disturbed, live-cell imaging shows the effects of oxidant stress as they unfold in time and space relative to a physiological, baseline or resting state established during a suitable period just prior to the introduction of the exposure agent. This approach obviates the need to consider artifacts or aberrations introduced by sample extraction and analysis as potential alternative explanations for the effects observed.
In addition to capturing fleeting events, the high temporal resolution of live-cell microscopy systems can provide valuable insights on kinetic indices of interest, which can produce evidence of cause and effect by establishing the existence of a time lag or temporal difference between events that may appear to occur simultaneously when observed by other techniques. Properly equipped live-cell imaging systems are capable of temporal resolution that is impractical to replicate using non-imaging analytical approaches, making this approach in many cases the gold standard for the study of oxidative events associated with xenobiotic exposure. Similarly, given the intrinsic spatial resolution of microscopy techniques for examining subcellular sites and structures, live-cell imaging is an optimal technique for identifying cellular organelles that are targeted by oxidant stress, or compartments that are involved in the production of reactive species. Spatiotemporal information afforded by live-cell microscopy can contribute comprehensively towards a mechanistic understanding of the role of oxidant events in cellular responses to a physiological or xenobiotic stimulus.
5. Fluorogenic sensors for live-cell imaging of redox toxicology
While the sensitivity and dynamic range of a live-cell assay are characteristics determined by the performance specifications of the components of the imaging system (e.g., light source, quality of the optics, detector sensitivity), specificity, selectivity and dynamicity are properties imparted entirely by the sensor molecule that reports the event of interest. Thus, the potential of live-cell microscopy is largely dependent on the availability and performance of sensor molecules that can report the endpoint of interest with suitable efficiency.
Most of the sensors used in live-cell microscopy are fluorogenic. These sensors are designed to respond to a specific physicochemical characteristic in their environment with a proportional change in the intensity of the fluorescence light that they emit when excited with an appropriate electromagnetic energy source (e.g., laser light). Some sensors have two excitation maxima, meaning that their fluorescence emission can be induced by excitation light of two different wavelengths, permitting their fluorescence intensity to be expressed as a ratio. As detailed below, such “ratiometric” sensors are highly desirable as they offer important advantages. Specifically, ratiometry allows for correction of a number of problematic artifacts and aberrations in fluorescence microscopy. For instance, expressing fluorescence intensity data ratiometrically corrects for photodamage (also known as “photobleaching”) to the fluorophore that can be caused by repeated cycles of excitation with high energy (short) wavelengths. Similarly, ratiometric data are resistant to variations in fluorescence intensity caused by differences in the concentration of the probe within the cells secondary to alterations in cell morphology, rearrangement of cytoskeletal proteins, or a compartmentalization of probe molecules, the premise being that the intensity of fluorescence excited by each wavelength is affected equally by these factors. Experimentally, in a live-cell imaging study the ratiometric sensor is excited sequentially with two wavelengths corresponding to its excitation maxima (or close to them). This allows the ratio of the intensity of the fluorescence emitted upon excitation with each wavelength to be monitored. When normalizing to the maximal signals obtained by positive controls (e.g., full oxidation with H2O2 and full reduction with DTT) applied to the cells at the end of the experiment, the following equation may be used to obtain a measure of the sensor response
where I is the fluorescence intensity obtained with excitation at each wavelength (λ).
Depending on the purpose of the experiment, it may be more informative to express changes induced by exposure relative to “at rest” or baseline readings. Alternatively, it may be preferable to show the magnitude of the change induced by an exposure relative to the maximal signal output possible. Thus, both ways are useful and consideration must be given to each in selecting the most appropriated way of expressing the data to suit the objective of the study.
6. Small molecule sensors used in oxidant stress research
At the time of this publication, a general search of the term “oxidative stress” on the Life Technologies (Molecular Probes, ThermoFisher Scientific) on-line catalog returns over 90 small molecule sensors that are marketed for the assessment of intracellular “ROS” or “oxidant stress”. Since the development of the earliest fluorescent oxidant sensors, the use of some of these probes has become commonplace in the study of xenobiotic-induced oxidant stress. The general design involves the use of a parent compound that acts as a membrane permeable precursor that is typically non-fluorescent until it becomes oxidized, ostensibly by a specific ROS, to yield a fluorescent product [104]. The fluorescence intensity is thus proportional to the concentration of the ROS. Among sensors or this type are variants of dichlorodihydrofluorescein (DCF), N-Acetyl-3.7-dihydroxyphenoxazine (Amplex Red), and CellROX. Each of these fluorophores was independently developed for the assessment of “oxidative stress” endpoints, claiming specificity (at least initially) for a single ROS species.
One of the most commonly used small molecule fluorophores for imaging of xenobiotic-induced oxidant stress is the fluorescein variant dichlorodihydrofluorescein diacetate (DCFH2-DA). DCFH2-DA was originally developed for acellular detection and measurement of hydrogen peroxide (H2O2), but has been utilized in numerous physiological and toxicological studies for the non-specific detection of “ROS” in cells for nearly 50 years [105,106]. Unfortunately, despite their popularity, DCF-family sensors have been demonstrated repeatedly to have characteristics that impact their utility significantly. First, DCF and its congeners have been shown to react with multiple ROS species either directly or indirectly, including H2O2, superoxide anion ( ), hydroxyl radical (HO•), peroxynitrite (ONOO−) and/or RNS [104,105,107]. Moreover, older “ROS sensors” in the DCF family show relatively high susceptibility to photobleaching and photo-oxidation, redox cycling, and a short dynamic range. These deficiencies not only present challenges in experimental settings but, more troubling, can also confound the interpretation of results. In recognition to their demonstrated lack of specificity, the DCF-family probes are sometimes used as “general oxidative stress” sensors which, while adequate for some purposes, are far less informative than the specificity offered by modern sensors. For an in-depth discussion of the benefits and shortcomings of these probes, the reader is referred elsewhere [50,104,105,107–109]. Despite the widely acknowledged limitations of DCF-based sensors, they offer ready commercial availability and relative ease of use. It is not surprising, therefore, that these fluorophores continue to find utility as a tool for the investigation of oxidant stress, including in toxicological studies. However, as explained below, when it comes to small molecule sensors for the detection of ROS species, better choices exist.
7. Advanced fluorogenic sensors for oxidant stress measurements
The fact that DCF-family probes continue to be used widely is also evidence of the persistent demand for fluorogenic small molecule sensors to detect intracellular oxidative events. The properties of an ideal small molecule fluorophore include high sensitivity and relative specificity for one reactive species (oxidant or redox pair of interest), membrane permeability and trapping, efficient localization to a defined cellular compartment, wide dynamic range, low toxicity, photostability with repeated cycles of excitation, dynamic response to changing concentrations of the analyte, water solubility, and insensitivity to pH, ionic strength and temperature (Table 1). While no fluorogenic sensor exists that possesses all these qualities, recent developments in redox chemistry have led to a new generation of advanced fluorogenic sensors specifically designed for oxidant stress measurements with improved performance characteristics. This section highlights a few of these small-molecule fluorophores, grouped by the reactive species to which they respond. (See Table 2.)
Table 1.
Essential and Desirable Characteristics of an Oxidant Sensor.
| Essential characteristics | Desirable characteristics |
|---|---|
| Specific | Cell permeable/trappable |
| (Reacts with only one species) | |
| Biologically inert | Photostable |
| (Does not redox cycle) | (Resistant to auto- or photooxidation) |
| (Low cytotoxicity) | Dynamic |
| (Reversible) | |
| High signal-to-noise ratio | |
| Water solubility | |
| Dual excitation maxima | |
| (Ratiometric) | |
| Targetable to organelles |
Table 2.
Characteristics of notable small molecule and genetically-encoded sensors used in live-cell imaging of oxidant species.
| Sensor | Specificity | Excitation maxima | Emission maxima | Example applications | Quantum yield |
|---|---|---|---|---|---|
| PF6-AM | Hydrogen peroxide | 460 nm | 520 nm | Cytosolic detection of H2O2 Hela cells | 0.94 |
| Hydroethidine | Superoxide | 518 nm | 605 nm | Superoxide detection in neutrophils and endothelial cells | 0.4 (ethidium monoazide) |
| DAF-FM DA | Nitric Oxide | 495 nm | 515 nm | NO detection in cortical neurons and in vivo in zebrafish | 0.81 |
| roGFP2 | EGSH | 390 nm 490 nm |
510 nm | Monitoring GSH/GSSG changes in a variety of compartment in mammalian cells | 0.9 |
| HyPer | Hydrogen peroxide | 420 nm 500 nm |
516 nm | H2O2 detection in cytoplasm and mitochondria in HeLa cells | 0.29 |
7.1. Hydrogen peroxide sensors
While it is typically thought of as an injurious ROS in toxicology, H2O2 is also increasingly recognized as a signaling molecule and effector of protein modification (i.e., cysteinyl sulfenylation) with pivotal roles in normal physiology [110,111]. Thus, monitoring H2O2 is important in elucidating mechanisms of toxicity involving damage to macromolecules as well as the perturbation of normal signaling processes.
7.1.1. Peroxy Green 1 (PG-1)
Developed by the laboratory of Christopher Chang at UC Berkeley, Peroxy Green 1 (9-(4-methoxy-2-methyl-phenyl)-6-(4,4,5,5-tetramethyl-1,3,2-dioxaborolan-2-yl)xanthen-3-one, PG-1) was the first fluorogenic H2O2 indicator to offer sufficient sensitivity to detect H2O2 production in non-phagocytic cells responding to physiological signals [112,113]. PG-1 consists of fluorescein conjugated to a “chemoselective boronate switch” that responds to H2O2 with high specificity [112,113]. Although PG-1 is not targeted to specific intracellular compartments, it can be used to identify the intracellular source of H2O2 production when used in conjunction with well-characterized metabolic inhibitors [114]. It has to be noted that PG-1 is relatively susceptible to photobleaching, has been reported to respond to two-electron transfer by peroxynitrite [50], and has a slower reaction time compared to more recently developed H2O2-specific protein-based sensors such as HyPer (discussed below). Despite these shortcomings, PG-1 is a worthy small molecule fluorophore that is sensitive and relatively specific for H2O2. Additionally, the Chang laboratory has modified the PG-1 molecule to emit at different colors with the H2O2-specific sensors PY-1 (Peroxy Yellow 1) and PO-1 (Peroxy Orange 1) in order to allow multiplex detection with green-emitting fluorophores [115].
7.1.2. PF6-AM
Building upon the boronate-based chemistry of PG-1, peroxyfluor-6-acetoxymethyl ester (PF6-AM) was developed to be a cytosolically-trappable H2O2 sensor [116]. Like PG-1, PF6-AM consists of a carboxyfluorescein core that is conjugated to a single phenolic boronate group that masks the fluorescent properties of the fluorophore. As an improvement over PG-1, PF6-AM employs the strategy used in the design of other sensors (e.g., DCF-AM) by introducing two acetoxymethyl ester groups to cap phenol and carboxylic acid functionalities that were engineered into the structure of this sensor in order to enhance its intracellular retention [116]. While attached, the lipophilic AM esters make this fluorophore membrane-permeable. Entry into the cell exposes the AM groups to cleavage by nonspecific intracellular esterases yielding a negatively-charged boronated fluorophore, PF6, that remains confined within the cytoplasm.
Just as with PG-1, exposure to H2O2 removes the sole boron group that serves to quench the fluorescence of the reporter in its inactive state, thus permitting the fluorescein core to emit upon excitation with 482 nm light [116,117]. The primary advantage to using this fluorophore over other boronated H2O2 sensors is the added ability to sensitively detect H2O2 localized within cells over sustained periods of time. In addition, Dickinson and colleagues report a nearly two-fold increase in the florescence of PF6 in comparison to PG-1 using the same methodological parameters [116]. A relatively new fluorophore, PF6-AM has been used to examine H2O2 responses under physiological conditions in several studies that have followed the original report by Dickinson and colleagues [116,118–122].
7.1.3. MitoPY1
Mitochondrial PY-1 (MitoPY1) is a H2O2-specific organelle-targeting fluorogenic sensor. This fluorophore uses the same boronate chemistry as described previously to detect H2O2, but it has an additional positively-charged phosphonium moiety, tetraphenylphosphonium (TPP+), that targets the probe to the mitochondria of treated cells [109,123–126]. As the site of oxidative phosphorylation, mitochondria also represent a potential source of partially-reduced oxygen species that can contribute to xenobiotic-induced oxidant stress. Since its introduction, studies have used MitoPY1 in the investigation of mitochondrial H2O2 production across a variety of physiological and toxicological contexts [118,127–131]
7.2. Superoxide sensors
The detection of superoxide presents a significant challenge due to the fact that the sensor must compete with the very high enzymatic activity of superoxide dismutases that exists in the cell [132]. Additionally, superoxide anion can act as a reductant as well as an oxidant, which can make the development of a specific superoxide sensor more complex.
7.2.1. Hydroethidine
Hydroethidine (2.7-diamino-10-ethyl-9-phenyl-9.10-dihydrophenanthridine, HE; also known as dihydroethidine, DHE) is a reduced form of the red-fluorescent compound ethidium (ET) that has become a widely-used standard for measuring superoxide generation within cells [50,104,133–136]. The oxidation of HE by yields the red-fluorescing hydroxylated form of ethidium (HO-ET). Unfortunately, HE can also be oxidized enzymatically within the cell to form the ET, which is spectrally very similar to HO-ET. One solution to this problem is the use of HPLC to isolate HO-ET from ET [136]. However, this approach is cumbersome and incompatible with live-cell microscopy. More recently, Robinson and colleagues reported that although both species fluoresce when excited at 514 nm, a second wavelength, 405 nm, can be used to excite HO-ET with considerably greater efficiency relative to ET [133]. Although the emission spectra for HO-ET and ET overlap significantly, their peaks are more than 5 nm apart. In principle, this should make it possible to resolve HO-ET from ET fluorescence in living cells using spectral unmixing [137]. An additional application of HE-based dyes for the detection of involves the use of the cell impermeable sensor hydropropiridine (HPr+). Since the undesirable oxidation of HE occurs largely as a result of intracellular enzymatic reactions, it does not affect the detection of extracellular levels of , which can be monitored using this cell-impermeable HE analog, [50, 138].
7.3. NO sensors
Interest in nitric oxide (NO) has expanded from its adverse effects as an air pollutant to include its role as an important intra- and inter-cellular signaling mediator. Cells maintain a baseline level of NO for normal physiological functions including signal transduction, neurotransmission, and immune responses [139]. High levels of NO induce pathophysiological effects involved in diseases including cancer, diabetes, and stroke [140]. Non-invasive, intracellular NO measurement is therefore critical for understanding the mechanisms of NO-induced adverse effects. The characteristics desirable in an NO sensor are the same as those for an ROS sensor (Table 1).
The backbone of NO sensors utilizes 1.2-diaminobenzene as a functional group to react with NO oxidation products (typically N2O3) for the conversion of diamine to triazole [141], with an emission maximum of 510 nm that can be detected using fluorescein compatible instruments (e.g., flow cytometers, microscopes, microplate readers). Probes based on 2.3-diaminonapthalene (DAN) for NO detection were developed in the 1990s by the Nagano group [142,143]. This probe is converted to the fluorescent compound, 2.3-naphththotriazole, when exposed to NO in an aerobic environment. Under a series of optimization experiments, DAF-FM DA (4-Amino-5-methylamino-2′,7′-difluorofluorescein diacetate) was demonstrated to have high specificity for NO and possess desirable sensor characteristics (cell membrane permeability, water solubility, photostability, and bio-compatibility, with excitation and emission wavelengths in the visible range [142,143]). However, the DAF sensors have a limited functional pH range, between 5.8 and above. The same group reported another series of NO probes based on rhodamine that are usable over a broader pH range (pH 4 and above) [144]. In particular, DAR-4MAM has a low detection limit (7 nM), high specificity, high membrane permeability, and is trappable inside the cell through cleavage of the acetoxymethyl ester by endogenous esterases [144]. Regarding optical properties, DAR-4MAM is excited with longer wavelengths (diamine 543 nm, triazole 554 nm) than DAF-FM DA (diamine 487 nm, triazole 495 nm). Although DAR-4MAM is not as bright as DAF-FM DA, the emission maxima of 574 nm is distant from the emission range of cellular auto-fluorescence and hence exhibits a greater signal-to-noise ratio than DAF-FM DA.
Another small molecule probe for NO detection was designed based on the popular fluorogenic center boron dipyrromethene (BODIPY) [145,146]. Among these, the DAMBO (8-(3.4-diaminophenyl)-2.6-bis(2-carboxyethyl)-4.4-difluoro-1,3,5,7-tetramethyl-4-bora-3a,4a-diaza-s-indacene) sensors exhibits desirable features including high water solubility, high signal to noise ratio, and compatibility with popular instrumentation (excitation for diamine 495 nm, triazole 497 nm) and emission (triazole 510 nm) with a well-established microscopy filter system (fluorescein-4-isothiocyanate (FITC)). DAMBO congeners are also stable over a broad pH range (pH 4–12).
Many of the NO fluorescent probes employ the conversion of 1.2-diaminobenzene to triazole (with an emission maximum of 515 nm) for NO detection. In the presence of NO inhibitors, no triazole production was observed for the three sensors (DAF-FM DA, DAR-4MAM, DAMBOO-T) which demonstrates the specificity of these sensors. However, it is notable that co-localization with other oxidants such as , ONOO−, and NO− can modulate the detection of NO with these sensors [147–149]. In addition, the probes do not interact with NO directly, but rather the oxidation products, such as and N2O3 [150]. As a result, this limits NO detection based on the concentration of oxygen in the medium. Therefore, it is important to verify findings obtained using this sensor with additional experiments using NO inhibitors in order to modulate functional effects.
7.4. Specialized oxidant sensors
With continued progress in sensor design, the development of new sensors that can be used to investigate a broader range of oxidant species and relevant endpoints can be anticipated. A survey of the current literature reveals several examples of unconventional oxidant sensors that have been applied in physiologically- and toxicologically-relevant studies. For instance, in 2009 the Koide group described a novel fluorophore capable of detecting the ubiquitous tropospheric air pollutant ozone (O3) in biological and atmospheric samples [151]. Of the various iterations of O3 sensor reported, a fluorophore designated as “Compound 8” (C8) was demonstrated to have specificity for this potent oxidizing air pollutant in cultured lung epithelial cells. However, a major disadvantage limiting its use is that this fluorophore is not confined to the intracellular space, which likely contributed to the observed loss of C8 fluorescence intensity immediately following a short O3 exposure period [151]. Despite this drawback, this is a useful sensor that demonstrates the feasibility of using fluorogenic probes to target even the most highly reactive oxidant species.
Several groups have recently described specific sensors to detect peroxynitrite (ONOO−) [152–155]. This is an important development because ONOO− is not only a reactive species produced in response to a broad range of stimuli, but it is also often cited as a confounding reactive species in the detection of H2O2, O2•− and NO by several small-molecule sensors. Thus, fluorescent probes designed to specifically detect ONOO− offer additional utility in that they can be used for validating measurements made by other fluorogenic sensors.
Lastly, another emerging fluorescent probe technology is the packaging of small-molecule sensors into “inert” nanosized particles. A primary advantage in the development of these “nanoprobes” is the ability to combine multiple probes onto a single delivery platform in efforts to monitor various, potentially unrelated, endpoints simultaneously. Similarly, the encapsulation of fluorogenic sensors into nanomaterials may overcome the inherent cytotoxicity issues of certain sensors, while protecting the loaded sensor(s) from nonspecific interactions with the intracellular environment [109]. Although the loading of nanoparticle matrices such as silicate sol-gel is not a particularly new technology, recent applications have generated a line of sol-gel nanoprobes called Photonic Explorer for Bioanalysis with Biologically Localized Embedding (PEBBLEs) for the specific detection of ROS [109,156,157]. The transparent nature of the silicate matrix used to generate PEEBLEs has been demonstrated to have minimal cellular effects, while actually enhancing the photoprotection of the loaded fluorogenic reporter so as to minimize photobleaching [109,157]. Pushing this technology even further, Yang and colleagues report the generation of a new fluorogenic nanosensor capable of reporting fluctuations in mitochondrial H2O2 and pH [158]. Ultimately, the development of more sophisticated scientific questions regarding oxidant stress will continue to drive the evolution of next-generation sensors needed for both physiological and toxicological studies of oxidant processes.
8. Genetically-encoded sensors for redox toxicology
The development, characterization, and application of the growing family of genetically encoded redox sensors based on Aequora victoria-derived green fluorescent protein (GFP) congeners has been extensively reviewed [101,159–162]. Since their introduction, these sensors have spurred a rapid expansion of our understanding of the role of intracellular redox events in physiology [163,164], and have afforded significant insight into the role of oxidant stress in toxicology [137,165–169]. Using these sensors involves introduction of an expression vector (wherein the coding sequence is typically placed downstream of a viral promoter) into the cell using either transfection or by transduction with a viral promoter (e.g., adenovirus or lentivirus). An added advantage of live-cell microscopy is that this approach is tolerant of the low expression efficiencies that, since the cells that express the sensor are readily identified visually and can be selected for study. A variety of xenobiotics of interest have been tested in a range of relevant cell culture models refining our understanding of the localization and effect of xenobiotic-induced oxidant stress in toxicologically relevant contexts. These studies are cited in the following section, which reviews genetically-encoded sensors available to monitor oxidative events.
8.1. Glutathione redox potential (EGSH) sensors
In terms of absolute concentration, the GSSG/GSH couple is the dominant redox pair in the cell, since it is present in millimolar concentrations in most cells [101,170]. The relative concentration of members of a redox pair can be calculated with the Nernst equation
using with the known standard redox potential (E°), and the measured redox potential (E) for the pair, in millivolts, and the Faraday and universal gas constants. While the standard redox potential for glutathione (E°GSH) is −280 mV [170], EGSH values in the cytosol of resting cells are considerably more negative, reading −320 mV [101,162], implying that cytosolic glutathione exists largely in the reduced form. Indeed, biochemical studies confirm that healthy cells maintain a large ratio of GSH/GSSG that is supported primarily through the activity of glutathione reductase at the expense of NADPH derived from the pentose phosphate shunt. A practical advantage of the very high ratio of GSH/GSSG in cells is that a small increase in the concentration of GSSG can be sensed as a large change in the EGSH.
EGSH sensors were originally developed to respond to thiol-disulfide equilibria. The original families of genetically-encoded sensors that are used to monitor EGSH were derived independently from YFP and GFP, and are named rxYFP [171] and roGFP [172], respectively. rxYFP and roGFP were subsequently demonstrated to respond to EGSH by inserting into a redox relay through which they ultimately equilibrate with the concentration of GSSG (Fig. 1). The physical readout for these sensors is based on a relative change in the 510 nm emission intensity of the fluorogenic center evoked by excitation with 488 nm and, in the case of roGFP, 405 nm light. Beyond their innovative utility, the introduction of these sensors has revolutionized the approach to study oxidant processes by enabling the quantitative monitoring of EGSH, as a pivotal and well-defined redox event with high spatiotemporal resolution [173–175].
Fig. 1.
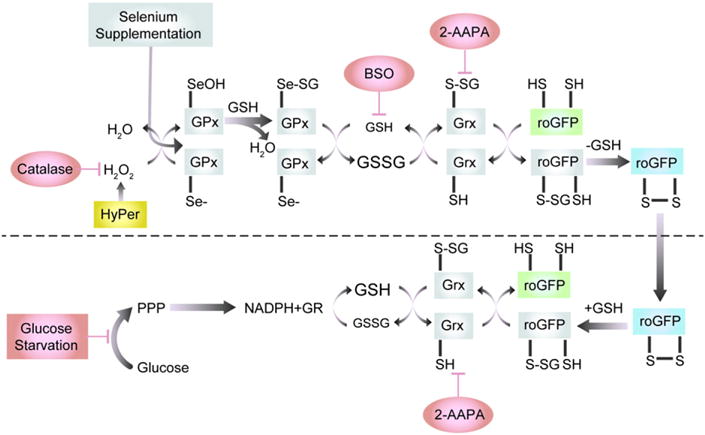
roGFP2 interactions with the glutathione system (adapted from Meyer and Dick 2010). Glutathione peroxidases (GPx) oxidize GSH to GSSG in response to peroxides, including H2O2 and lipid hydroperoxides (LOOHs), thus increasing the glutathione redox potential (EGSH). Abbreviations: LOH, reduced lipid oxide; Se−, reduced selenocyteine; SeOH, oxidized selenocyteine; SeSG, glutathionylated selenocysteine. In response to the increase in GSSG, one of the engineered vicinal cysteines of roGFP2 becomes S-glutathionylated by glutaredoxin (Grx). Glutathionylation in turn causes disulfide bond formation and alteration of the spectral properties of the GFP fluorophore. In the reductive pathway, Grx catalyzes the reduction of roGFP2 disulfide bonds through deglutathionylation as GSSG levels decrease and normal levels of GSH are reestablished by glutathione reductase (GR), at the expense of NADPH, causing a renormalization of EGSH. Glucose and the pentose-phosphate pathway (PPP) create NADPH, which is used by GR to reduce GSSG to GSH.
8.1.1. The yellow fluorescent protein-based redox sensor rxYFP
rxYFP was constructed by introducing substitutions at amino acids 149 and 202 in YFP to cysteines [171]. rxYFP was demonstrated to monitor EGSH by equilibrating with the GSSG/GSH redox pair through its interaction of glutaredoxin-1 (Grx1)-dependent interaction, information that was subsequently utilized to construct a chimeric protein linking rxYFP to Grx1. SinceGrx1-rxYFP is not dependent on interaction with endogenous Grx1, the resulting Grx1-rxYFP fusion protein is a sensor with improved EGSH-sensing kinetics [176]). The utility of rxYFP has been expanded by modified versions that target its expression to a variety of subcellular compartments [177,178]. However, all types of rxYFP are sensitive to pH changes that can be expected to occur within a physiological range, which imposes limitations and validation requirements on their use [101]. The rxYFP sensors also suffer from an additional disadvantage, namely the fact that they are not ratiometric, which renders them susceptible to artifacts caused by photobleaching, variations in intracellular distribution of the sensors and differences in cell morphology (e.g., cell thickness) [101]. On the other hand, oxidized and reduced forms of rxYFP, can be resolved successfully by specialized immunoblot-ting, which is useful to confirm the results obtained from live-cell imaging experiments [179]. The expansion of the color palette available to analyze oxidant events with genetically-encoded sensors is yet another contribution to the field, as in the case of rxRFP, wherein a cpRFP scaffold was utilized to develop a red fluorescent redox sensor. rxRFP is useful for monitoring the general redox health of the cell, since it has been used to detect GSSG, , and ONOO− in live-cell studies [180]. Sugiura et al. recently reported the development of a group of proteins similar to rxYFP, known as oxidation balance sensed quenching (Oba-Q) proteins [181]. Although these sensors are only excitable at one wavelength (and therefore not ratiometric), they are all based on variant chromophores of GFP including Sirius (Oba-Qs), CFP’ (Oba-Qc), and BFP (Oba-Qb), allowing for co-expression of multiple sensors in the same cell without overlap of fluorescence emission spectra.
8.1.2. The green fluorescent protein-based sensor roGFP
The roGFPs are arguably the most important family of sensors to be developed for redox research and, as such, also hold the greatest potential for redox toxicology. roGFP1 was engineered by substituting cysteines at glutamine204 and serine147 to allow the formation of a disulfide bond between the beta strands near the fluorogenic center of GFP under oxidizing conditions [182]. It was subsequently discovered that roGFP interacts only slowly with oxidant species such as H2O2, and that this sensor preferentially equilibrates with the GSH/GSSG redox pair through the intervention of Grx1 [183]. The family of roGFPs has been expanded to generate sensors with midpoint potentials that are useful in the range of glutathione redox potentials that exist in various cellular compartments, including the endoplasmic reticulum ([184, 185] Fig. 2). Organelle targeting versions of roGFP have also been developed to direct the sensor to the cytosol, mitochondrial matrix, mitochondrial intermembrane space, nucleus, peroxisome, endosomes and lysosomes [114,162,164,186–189]. In addition, expression of roGFPs in transgenic animals has successfully resulted in the capability to specifically assess the EGSH of tissues and even specific cells in organ systems [173–175]. (See Fig. 3.)
Fig. 2.
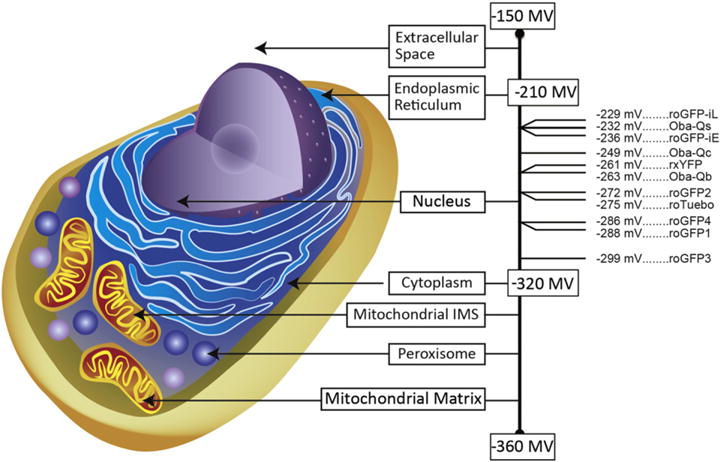
Midpoint Potentials for EGSH Sensors. The average redox potentials for subcellular compartment are shown with the mitochondrial matrix as the most reducing compartment (−360 mV) and the extracellular space as the most oxidizing compartment (−150 mV). Of the available EGSH sensors, only a few have a midpoint potential sufficiently high to be functional in the endoplasmic reticulum (roGFP-iL, roGFP-iE [172], Oba-Qs [181]. A number of sensors can be specifically targeted to the nucleus, cytoplasm, mitochondrial intermembrane space (IMS), peroxisome, or mitochondrial matrix (Oba-Qc, Oba-Qb [181]), rxYFP [171], roGFP1–4 [182], roTurbo [215]. roGFP3 [182] functions optimally in highly reducing compartments such as the mitochondrial matrix.
Fig. 3.
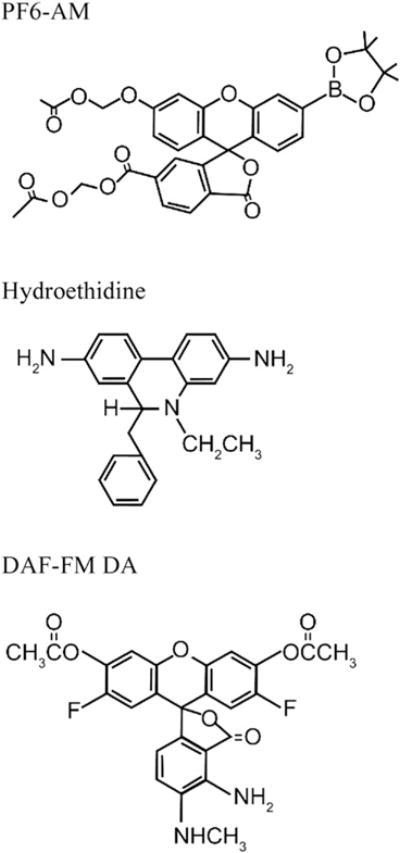
Structures of selected small molecule sensors used for the detection of reactive species. PF6-AM has high specificity for H2O2. Hydroethidine is commonly used for the detection of . DAF-FM DA is designed for the detection of NO.
roGFP1 and roGFP2 (S147C, Q204C, S65T) are currently the most widely used members of the family of roGFPs. While roGFP1 has the advantage of being less sensitive to pH relative to roGFP2, roGFP2 has a wider dynamic range [182]. Additionally, one group has utilized six of the seven mutations of the super-folder GFP with an additional two directed sites on roGFP2 to produce roTurbo-GFP, a sensor that is notably brighter than other roGFPs, which may increase its usefulness in certain applications [190]. Unlike the rxYFPs, all roGFP congeners show two excitation maxima, excitable with the widely accessible 405 and 488 nm lasers found in many imaging systems. As mentioned earlier, probes with fluorescence emission that can be excited with multiple wavelengths are said to be ratiometric, because the intensity of the fluorescence induced by each excitation wavelength can be expressed as a ratio. As also mentioned above, ratiometry offers several important advantages.
Although roGFP has proved to be a revolutionary sensor, its response time to changes in EGSH is rather slow because it is limited by the rate of its interaction with endogenous Grx1. In order to improve the response kinetics of roGFP, the group led by Tobias Dick in Heidelberg developed chimeric proteins linking roGFP2 to Grx1 (roGFP2-Grx1 and Grx1-roGFP), the idea being to increase the effective concentration of Grx1 experienced by the sensor molecule [183]. Both iterations of the fusion of roGFP2 to Grx1 succeeded in increasing the response time kinetics. However, only Grx1-roGFP2 targets well to subcellular compartments such as the mitochondrial matrix in mammalian cells, Drosophila and Arabidopsis [191,192]. The construction of another chimeric protein that linked roGFP2 to mycoredoxin-1 (Mrx1), the analog of Grx1 in mycobacterium, is another interesting application further expanding the uses of roGFP [193]. This chimeric protein (roGFP2-Mrx1) monitors the mycothiol redox potential (EMSH) of mycobacterium, useful as a readout of the pathogen/host interactions and the efficacy of antibiotics. The introduction of this sensor expanded the use of roGFP to monitor redox metabolism in prokaryotic models.
8.2. Genetically-encoded sensors of hydrogen peroxide
8.2.1. HyPer
Belousov and colleagues developed a genetically-encoded fluorogenic sensor, which they named HyPer, to specifically detect intracellular levels of H2O2. Structurally, HyPer was formed through circular permutation of YFP (cpYFP) inserted into the bacterial H2O2-sensing protein OxyR1 [194]. The experimental utility of this sensor has been established in a series of studies in which HyPer was targeted to a variety of subcellular organelles, including the mitochondria [195]. HyPer has also been shown to be successfully expressed in zebrafish, revealing the role of H2O2 in developmental growth and wound healing [196, 197]. HyPer has been chimerically linked to the growth factor receptors EGFR and PDGFR in order to create proteins positioned to sense localized increases in H2O2 production [198]. Studies using HyPer have demonstrated that xenobiotic induction of H2O2 is specific to subcellular compartments [114,195,199]. The group that developed HyPer recently reported an improved version, called HyPer-3, that contains point mutations that increase the sensor dynamic range [197].
HyPer is highly sensitive to pH within a physiological range, with a change in pH as small as 0.2 units sufficient to effect a deflection in the excitation ratio reflecting full reduction or excitation of the sensor [194]. Thus it is imperative to monitor pH when using HyPer to monitor H2O2. This can be done using pH specific sensors such as such as pHluorin or pHluorin2 [200], albeit these sensors also emit in the green channel, requiring separate experiments or spectral unmixing [137]. pHRed is an alternative pH sensor that emits in the red channel and can therefore be co-expressed and monitored concurrently with HyPer in the same cells [199,201]. However, the ideal pH sensor to use as a control for HyPer is SypHer, which is a version of HyPer that contains a single point mutation (C199S) that renders it unable to sense H2O2 [202]. Recently a SypHer-2 sensor has been developed which is reported to be a significant improvement upon the original SypHer sensor [203]. It has been argued [204] that cpYFP molecules, including HyPer, may also detect which, if verified, would limit its use as an H2O2 sensor. However, not only is there sufficient evidence to suggest that HyPer is significantly more sensitive to H2O2 than [161], but a recent study demonstrated that cpYFP itself could not be utilized as a sensor [205,206], which should alleviate this concern.
8.2.1.1. Orp-roGFP
For situations where there is a persistent concern regarding the influence of pH or , an alternative to HyPer exists in the form of Orp1-roGFP2. However, Orp1-roGFP2 has not been fully characterized yet (i.e., the reductive pathway has not been identified), and it has yet to be broadly used in mechanistic studies [101,191].
Beyond H2O2, there are other protein ROS sensors under development, including those that sense H2S and ONOO−, two reactive species implicated in signaling as well as oxidant stress [207,208]. Once these sensors are fully characterized, they will be instrumental in developing an integrated understanding of the role of oxidant stress and oxidant signaling in toxicology.
9. Limitations and caveats of using protein sensors in redox toxicology
At least one group has demonstrated that laser excitation can produce ROS through physiological cellular processes and chemical reactions with cell media, potentially creating artifacts and modulating sensor responses [209]. Among the ways to optimize laser-induced fluorescence, such as lowering power and dwell time, the most effective may be to limit the frequency of successive laser excitations to 60 s or longer, a frequency that is well suited for monitoring changes associated with intracellular redox events.
Another aspect that needs to be considered when utilizing genetically encoded sensors is the potential for cellular stress induced by the expression of the sensor. The vector encoding the sensor is introduced into cell cultures through transfection or viral vector transduction and the expression is typically driven by a cytomegalovirus (CMV)-promoter placed upstream of the open reading frame. The use of CMV, SV40 and similar viral promoters is an efficient means of effecting a high level of transcriptional expression in mammalian cells, which can in turn result in the synthesis of a high concentration of the encoded protein, in some cases reaching micromolar levels (Cheng, W-Y et al. unpublished data). However, co-opting cells to express high levels of recombinant proteins, including GFP has been shown to induce cytotoxic effects [210]. Therefore, it is possible that cells that express high levels of the reporter protein have differential sensitivity to xenobiotic exposures that affect their survival and responses to xenobiotic challenge. This survival disadvantage may explain the observation of a decreasing fraction of cells that express GFP in a stably transduced cell line over time [211]. Reducing the heterogeneity in the level of sensor expression in the cells (e.g., through cell sorting selection) or titrating sensor expression may decrease the variability in the response to the xenobiotic challenge.
As identified in this review, some of the sensors available for oxidant stress studies rely on fluorescence that is sensitive to pH, and the possibility that the readout reported by the sensor is an artifact of a change in pH should be investigated with appropriate controls. Although minimally invasive ways to measure intracellular pH exist in the form of dyes or other genetically encoded sensors, the aim should be to maintain constant pH during the experiment by using appropriate environmental conditions (i.e., proper buffers in the exposure media). It should also be noted that some xenobiotic exposures lead to cellular acidification by disrupting the mitochondrial respiratory chain, which could in turn result in driving a redox sensor to its reduced form. This could lead to an impairment or failure to detect an oxidative event by the inactivated sensor.
Another potential problem with expressing redox sensors is that the sensor molecules represents an additional electron sink that could alter the redox relay of interest. In practice, whether sensor expression reaches sufficiently high levels to interfere measurably depends on its concentration relative to cellular relay components. For example, the concentration of roGFP achievable in the cell is expected to be too low relative to those of GSH and other cellular thiols to have much of an impact. On the other hand, it would be difficult to rule out a scenario where the expression of HyPer consumes a significant fraction of the local concentration of H2O2. Experimentally, the approach is to monitor the influence of probe expression on general cellular physiology such as a change in cell growth rate, as well as specific endpoints relevant to the responses being studied (e.g., a change in basal GSH levels, HO-1 gene expression).
10. Specific challenges using oxidant stress sensors in redox toxicology studies
The use of redox sensors in toxicological applications creates a number of uncertainties that require special consideration and experimental validation. With few exceptions (e.g., the ozone-specific sensors) fluorophores appropriate for oxidant stress measurements are developed for applications in cell biology. These sensors are, therefore, designed to report on specific conditions (e.g., EGSH) or concentrations of reactive species (e.g., H2O2) within a range expected for cells at rest or responding to a physiological stimulus. However, the magnitude of redox effects that result from xenobiotic exposures can exceed well beyond those observed in pathophysiology. Similarly, xenobiotics may generate a sensor readout through mechanisms that depart significantly from the established pathway through which the sensor was designed to report. This section will consider issues of concern and illustrate experimental approaches that can be used to validate live-cell imaging data in toxicology studies.
10.1. Dynamic range
As mentioned, many xenobiotics of interest in toxicology have chemical reactivities (e.g., electrophilicity) that may exceed grossly the redox potential of a physiologically relevant oxidant species such as H2O2 that a sensor such as HyPer or PG-1 was designed to detect. Similarly, highly reactive environmental compounds may induce EGSH changes that are outside of the dynamic range within which roGFP is designed to report. In practical terms, this means that exposure to a strong electrophile could result in a shift in the cytosolic EGSH that is too far from the midpoint potential of roGFP to be measured accurately (typically a 40 mV window centered on the sensor midpoint potential [101]. Similarly, localized accumulation of H2O2 generated by a redox-active agent could in theory reach levels that produce a rate of probe oxidization in which essentially every available HyPer molecule is oxidized, thus exhausting the dynamic range of the probe and the signal that can be generated, a point beyond which further increases in the concentration of H2O2 would become unresolvable.
10.2. Artifacts
Beyond dynamic range limitations, the use of oxidant stress sensors in toxicological studies also incurs the potential for generating multiple types of artifacts. For instance, it is theoretically plausible for a xenobiotic to react directly with cellular molecules targeted by the sensor (e.g., cysteinyl thiols in GSH, or Grx) by oxidizing or coordinating them, making them unavailable to participate in redox relays with which the fluorophore equilibrates. Additionally, a toxicant may also attack the sensor molecule itself, damaging the fluorogenic center, decreasing its sensitivity, or rendering it susceptible to other species (e.g., H+, Ca2+). Alternatively, the sensor could possibly be oxidatively altered in a manner that mimics the modification that induces the change in fluorescence under normal conditions, thereby short-circuiting the pathway and creating a potential false positive signal.
In interpreting results from toxicological studies using fluorogenic sensors, it is important to distinguish between frank artifacts and off-target reactivity of a xenobiotic compound that is properly reported by the sensor. As an example, we may consider the case of a hypothetical dithiocarbamate derivative that inhibits Grx, the proximal enzyme in the relay through which roGFP senses the oxidant effect of H2O2 on EGSH (Fig. 1, [101,212]). Since the effect of Grx inhibition would be to sever the ability of roGFP to sense EGSH, any oxidant effect that this dithiocarbamate derivative may have on EGSH, either through the production of H2O2 or by direct reactivity with GSH, could not be reported by the sensor, resulting in a false negative result. In contrast, however, if dithiocarbamate exposure interferes with the pentose phosphate pathway, the effect would be to decrease the production of NADPH. Reduced availability of NADPH, in turn, effectively lowers the reductant tone in the redox relay by decreasing the rate of GSSG reduction, which would be reported by roGFP as an increase in EGSH. In this last scenario, the roGFP sensor responds correctly, notwithstanding the fact that the oxidant stress that induces the change in EGSH is secondary to a metabolic impairment rather than, for example, increased production of H2O2 working through the relay.
Thus, interpretation of effects observed in cells exposed to xenobiotics requires detailed consideration of alternative mechanisms that can explain the sensor readout, including alternative reactions that would not be considered plausible in a physiological context. On the bright side, experiments conducted to validate the readout of a sensor responding to a xenobiotic exposure often yield data that inform the mechanism of action of the agent of interest, sometimes beyond what the sensor data alone could provide.
10.3. Validating sensor readouts
Experimentally, the genetically encoded sensor readouts can be validated by verifying the role of each enzyme in the redox relay leading to the terminal response sensed by the fluorophore. In the case of stimuli that induce changes in roGFP fluorescence, this means testing the involvement of Gpx and Grx (Fig. 1). Our laboratory used this approach in a recent study [212] that investigated the basis for roGFP2-reported increases in EGSH induced in human airway epithelial cells undergoing real time exposure to O3, a highly reactive environmental electrophile. The requirement for Grx in the O3-induced EGSH change was tested two different ways. First, cells were pretreated with 2-AAPA, an inhibitor of Grx activity, which resulted in a complete suppression of the O3-induced increase in EGSH reported by roGFP. This was followed by comparing cells expressing roGFP2 and Grx1-roGFP2, which confirmed that the rate of O3-induced EGSH sensed by roGFP is limited by Grx availability. The requirement for Gpx in the roGFP redox relay can be conveniently tested in most cultured cell types by exploiting the inducibility of Gpx with selenium supplementation of the culture medium. Overnight preincubation with 100 μM sodium selenate increased intracellular levels of Gpx1 and significantly accelerated the rate of the EGSH increase induced by exposure to O3, demonstrating that Gpx1 has a role in the redox relay that leads to changes in roGFP fluorescence in cells exposed to O3.
On the reductive side of the relay, glutathione reductase (GR) activity to reduce GSSG is dependent on the concentration of NADPH, which can be restricted metabolically by the withdrawal of glucose from the culture medium for 2–4 h prior to exposure. Our experiments showed that glucose deprivation effectively sensitized the cells to increases in EGSH induced by a subsequent exposure to O3 [212], without a detectable alteration in basal EGSH. Depletion of intracellular levels of GSH using an inhibitor of glutathione synthesis, such as buthionine sulfoximine (BSO), is also an effective tool that can be brought to bear in studies aimed at scrutinizing the integrity of the roGFP redox relay in cells exposed to xenobiotics [199].
As an adjunct to the use of species-specific sensors, the involvement of H2O2 in a xenobiotic-induced response may be demonstrated by overexpressing catalase in cells. This approach has been used to support data obtained using HyPer in human respiratory cells exposed to the ambient air electrophile 1.2-naphthoquinone [114,213]. A subsequent study investigated the role of H2O2 in Zn2+-induced oxidative responses using cytosolic as well as ectopic mitochondrial expression of catalase in airway epithelial cells co-expressing HyPer in either compartment, thus providing independent spatial information to confirm the imaging data [199]. This approach could also be used to validate responses reported by small molecule sensors.
11. Future directions in live-cell imaging of toxicological oxidant stress
The growing popularity of the new generation of small molecule and genetically-encoded redox and ROS sensors discussed in this review has generated interest in the development of versions that can be used in specific applications. An example of this is the introduction of roGFP sensors that have midpoint potentials that make them usable in compartments such as the ER that are more oxidizing than the cytosol [101] (Fig. 2). Similarly, the ability to target protein or small molecule sensors to membranes or subcellular compartments is a useful property for a sensor as it leverages the spatial resolution of microscopy, and it can therefore be anticipated that new targetable variants of existing sensors will become available in the future.
Various aspects of the dynamic relationship between the concentration of ROS species and any specific redox status is of considerable mechanistic interest in toxicology research. The ideal approach to investigating these issues is to monitor the behavior of multiple sensors introduced in the same cell. However, for historical and practical reasons, most of the redox sensors available currently are variants of either green fluorescent protein or fluorescein, two green fluorophores with overlapping spectra that are not well resolved using the typical bandpass filters with which microscopy systems are typically equipped. Faced with this challenge, our laboratory developed dynamic spectral unmixing microscopy (DynSUM), specifically to resolve fluorescence signals related to changes in EGSH and H2O2 concentrations reported by roGFP and HyPer, respectively, in human lung cells co-expressing both sensors [137]. Spectral unmixing requires a specialized detector capable of capturing a wide segment of the combined fluorescence spectra emitted by the sample, which is then “unmixed” into its constituent spectra using a statistical algorithm. Future introduction of new sensors based on fluorogenic centers with a broader range of emission spectra will facilitate simultaneous monitoring of oxidative endpoints of interest. New sensors will likely be restricted to red-shifted fluorophores in order to make use of longer excitation wavelengths that are less likely to cause photobleaching. Undoubtedly, redox toxicology research will benefit as the repertoire of sensors to monitor oxidative endpoints in living cells is expanded to include targets of ROS such as lipids, nucleic acids and proteins. A small molecule fluorogenic sensor for the detection of cysteinyl sulfenylation, a pivotal protein modification in redox biology, was described recently [214].
The inherent non-invasiveness of live-cell microscopy permits the behavior of reporters to be monitored with spatiotemporal resolution unrivaled by other experimental techniques. An important advantage of the high temporal resolution afforded by reporter-based live-cell imaging is that it allows access to the mechanistic information encoded in the kinetic relationships between exposure and response. Metrics such as the lag time in the response to an exposure, the rate of the response, the magnitude of the signal achieved, the duration of the peak response, the rate of resolution, etc., all carry quantitative information that can be modeled mathematically to generate an integrated picture of the pathways through which xenobiotic exposure causes adverse effects. Such computational uses of laboratory data represent a growing sector of toxicology research and live-cell imaging data are particularly amenable to this application.
Given the pervasiveness of oxidative effects as toxicological mechanisms and the emergence of cellular redox processes as toxicological targets, the demands placed on methodologies to assess redox toxicology can be expected to grow. The sensor-based live-cell imaging approach described in this review should make it a powerful tool with broad application in the investigation of the multifaceted aspects of xenobiotic-induced oxidant stress.
Footnotes
Disclaimer: The research described in this article has been reviewed by the National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency, and approved for publication. The contents of this article should not be construed to represent agency policy, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
This article is part of a Special Issue entitled Air Pollution, edited by Wenjun Ding, Andrew J. Ghio and Weidong Wu.
Transparency document
The Transparency document associated with this article can be found, in online version.
References
- 1.Cross CE, Valacchi G, Schock B, Wilson M, Weber S, Eiserich J, van der Vliet A. Environmental oxidant pollutant effects on biologic systems: a focus on micronutrient antioxidant-oxidant interactions. Am J Respir Crit Care Med. 2002;166:S44–S50. doi: 10.1164/rccm.2206015. [DOI] [PubMed] [Google Scholar]
- 2.Stringer B, Kobzik L. Environmental particulate-mediated cytokine production in lung epithelial cells (A549): role of preexisting inflammation and oxidant stress. J Toxicol Environ Health A. 1998;55:31–44. doi: 10.1080/009841098158601. [DOI] [PubMed] [Google Scholar]
- 3.Jaeschke H, McGill MR, Ramachandran A. Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: lessons learned from acetaminophen hepatotoxicity. Drug Metab Rev. 2012;44:88–106. doi: 10.3109/03602532.2011.602688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolf MB, Baynes JW. The anti-cancer drug, doxorubicin, causes oxidant stress-induced endothelial dysfunction. Biochim Biophys Acta. 2006;1760:267–271. doi: 10.1016/j.bbagen.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Chakraborti S, Das S, Chakraborti T. Oxidant-mediated activation of cytosolic phospholipase a(2) in pulmonary endothelium: role of protein kinase C alpha and a pertussis toxin-sensitive protein. Endothelium. 2005;12:121–131. doi: 10.1080/10623320500189830. [DOI] [PubMed] [Google Scholar]
- 6.Zengin S, Al B, Yarbil P, Taysi S, Bilinc H, Yildirim C, Aksoy N. Oxidant/antioxidant status in cases of snake bite. J Emerg Med. 2013;45:39–45. doi: 10.1016/j.jemermed.2012.11.097. [DOI] [PubMed] [Google Scholar]
- 7.Pham HT, Maccarone AT, Campbell JL, Mitchell TW, Blanksby SJ. Ozone-induced dissociation of conjugated lipids reveals significant reaction rate enhancements and characteristic odd-electron product ions. J Am Soc Mass Spectrom. 2013;24:286–296. doi: 10.1007/s13361-012-0521-9. [DOI] [PubMed] [Google Scholar]
- 8.Wisthaler A, Weschler CJ. Reactions of ozone with human skin lipids: sources of carbonyls, dicarbonyls, and hydroxycarbonyls in indoor air. Proc Natl Acad Sci U S A. 2010;107:6568–6575. doi: 10.1073/pnas.0904498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumagai Y, Shinkai Y, Miura T, Cho AK. The chemical biology of naphthoquinones and its environmental implications. Annu Rev Pharmacol Toxicol. 2012;52:221–247. doi: 10.1146/annurev-pharmtox-010611-134517. [DOI] [PubMed] [Google Scholar]
- 10.Iwamoto N, Sumi D, Ishii T, Uchida K, Cho AK, Froines JR, Kumagai Y. Chemical knockdown of protein-tyrosine phosphatase 1B by 1.2-naphthoquinone through covalent modification causes persistent transactivation of epidermal growth factor receptor. J Biol Chem. 2007;282:33396–33404. doi: 10.1074/jbc.M705224200. [DOI] [PubMed] [Google Scholar]
- 11.Abdel-Malek ZA, Kadekaro AL, Swope VB. Stepping up melanocytes to the challenge of UV exposure. Pigment Cell Melanoma Res. 2010;23:171–186. doi: 10.1111/j.1755-148X.2010.00679.x. [DOI] [PubMed] [Google Scholar]
- 12.Skillrud DM, Martin WJ., 2nd Paraquat-induced injury of type II alveolar cells. An in vitro model of oxidant injury. Am Rev Respir Dis. 1984;129:995–999. doi: 10.1164/arrd.1984.129.6.995. [DOI] [PubMed] [Google Scholar]
- 13.Berisha H, Pakbaz H, Absood A, Foda HD, Said SI. Nitric oxide mediates oxidant tissue injury caused by paraquat and xanthine oxidase. Ann N Y Acad Sci. 1994;723:422–425. [PubMed] [Google Scholar]
- 14.Kang SA, Jang YJ, Park H. In vivo dual effects of vitamin C on paraquat-induced lung damage: dependence on released metals from the damaged tissue. Free Radic Res. 1998;28:93–107. doi: 10.3109/10715769809097880. [DOI] [PubMed] [Google Scholar]
- 15.Aroun A, Zhong JL, Tyrrell RM, Pourzand C. Iron, oxidative stress and the example of solar ultraviolet A radiation. Photochem Photobiol Sci. 2012;11:118–134. doi: 10.1039/c1pp05204g. [DOI] [PubMed] [Google Scholar]
- 16.Letelier ME, Sanchez-Jofre S, Peredo-Silva L, Cortes-Troncoso J, Aracena-Parks P. Mechanisms underlying iron and copper ions toxicity in biological systems: pro-oxidant activity and protein-binding effects. Chem Biol Interact. 2010;188:220–227. doi: 10.1016/j.cbi.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Gao X, Qian M, Campian JL, Marshall J, Zhou Z, Roberts AM, Kang YJ, Prabhu SD, Sun XF, Eaton JW. Mitochondrial dysfunction may explain the cardiomyopathy of chronic iron overload. Free Radic Biol Med. 2010;49:401–407. doi: 10.1016/j.freeradbiomed.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Z, Liu S, Wang X, Khaidakov M, Dai Y, Mehta JL. Oxidant stress in mitochondrial DNA damage, autophagy and inflammation in atherosclerosis. Sci Rep. 2013;3:1077. doi: 10.1038/srep01077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qu B, Li QT, Wong KP, Ong CN, Halliwell B. Mitochondrial damage by the “pro-oxidant” peroxisomal proliferator clofibrate. Free Radic Biol Med. 1999;27:1095–1102. doi: 10.1016/s0891-5849(99)00143-4. [DOI] [PubMed] [Google Scholar]
- 20.Shukla A, Jung M, Stern M, Fukagawa NK, Taatjes DJ, Sawyer D, Van Houten B, Mossman BT. Asbestos induces mitochondrial DNA damage and dysfunction linked to the development of apoptosis. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1018–L1025. doi: 10.1152/ajplung.00038.2003. [DOI] [PubMed] [Google Scholar]
- 21.Gladyshev VN. The free radical theory of aging is dead. Long live the damage theory! Antioxid Redox Signal. 2014;20:727–731. doi: 10.1089/ars.2013.5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terman A. Garbage catastrophe theory of aging: imperfect removal of oxidative damage? Redox Rep. 2001;6:15–26. doi: 10.1179/135100001101535996. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Mori A. Stress, aging, and brain oxidative damage. Neurochem Res. 1999;24:1479–1497. doi: 10.1023/a:1022597010078. [DOI] [PubMed] [Google Scholar]
- 24.Bajt ML, Ramachandran A, Yan HM, Lebofsky M, Farhood A, Lemasters JJ, Jaeschke H. Apoptosis-inducing factor modulates mitochondrial oxidant stress in acetaminophen hepatotoxicity. Toxicol Sci. 2011;122:598–605. doi: 10.1093/toxsci/kfr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajt ML, Knight TR, Lemasters JJ, Jaeschke H. Acetaminophen-induced oxidant stress and cell injury in cultured mouse hepatocytes: protection by N-acetyl cysteine. Toxicol Sci. 2004;80:343–349. doi: 10.1093/toxsci/kfh151. [DOI] [PubMed] [Google Scholar]
- 26.Jaeschke H, Knight TR, Bajt ML. The role of oxidant stress and reactive nitrogen species in acetaminophen hepatotoxicity. Toxicol Lett. 2003;144:279–288. doi: 10.1016/s0378-4274(03)00239-x. [DOI] [PubMed] [Google Scholar]
- 27.Shi J, Feng M, Zhang X, Wei Z, Wang Z. Acute oral toxicity and liver oxidant/antioxidant stress of halogenated benzene, phenol, and diphenyl ether in mice: a comparative and mechanism exploration. Environ Sci Pollut Res Int. 2013;20:6138–6149. doi: 10.1007/s11356-013-1622-0. [DOI] [PubMed] [Google Scholar]
- 28.Cho HY, Gladwell W, Yamamoto M, Kleeberger SR. Exacerbated airway toxicity of environmental oxidant ozone in mice deficient in Nrf2. Oxidative Med Cell Longev. 2013;2013:254069. doi: 10.1155/2013/254069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aksoy L. Evaluation of oxidant-antioxidant status in oral toxicity of fish oil methyl esters and diesel fuel in male rats. Toxicol Ind Health. 2013 doi: 10.1177/0748233713475513. [DOI] [PubMed] [Google Scholar]
- 30.Wessels A, Birmili W, Albrecht C, Hellack B, Jermann E, Wick G, Harrison RM, Schins RP. Oxidant generation and toxicity of size-fractionated ambient particles in human lung epithelial cells. Environ Sci Technol. 2010;44:3539–3545. doi: 10.1021/es9036226. [DOI] [PubMed] [Google Scholar]
- 31.Arafa HM, Aly HA, Abd-Ellah MF, El-Refaey HM. Hesperidin attenuates benzo[alpha] pyrene-induced testicular toxicity in rats via regulation of oxidant/antioxidant balance. Toxicol Ind Health. 2009;25:417–427. doi: 10.1177/0748233709106624. [DOI] [PubMed] [Google Scholar]
- 32.Armutcu F, Gun BD, Altin R, Gurel A. Examination of lung toxicity, oxidant/antioxidant status and effect of erdosteine in rats kept in coal mine ambience. Environ Toxicol Pharmacol. 2007;24:106–113. doi: 10.1016/j.etap.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 33.Turci F, Tomatis M, Gazzano E, Riganti C, Martra G, Bosia A, Ghigo D, Fubini B. Potential toxicity of nonregulated asbestiform minerals: balangeroite from the western Alps. Part 2: oxidant activity of the fibers. J Toxicol Environ Health A. 2005;68:21–39. doi: 10.1080/15287390590523911. [DOI] [PubMed] [Google Scholar]
- 34.Ghio AJ, Peterseim DS, Roggli VL, Piantadosi CA. Pulmonary oxalate deposition associated with Aspergillus niger infection. An oxidant hypothesis of toxicity. Am Rev Respir Dis. 1992;145:1499–1502. doi: 10.1164/ajrccm/145.6.1499. [DOI] [PubMed] [Google Scholar]
- 35.Bus JS, Gibson JE. Paraquat: model for oxidant-initiated toxicity. Environ Health Perspect. 1984;55:37–46. doi: 10.1289/ehp.845537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacic P, Cooksy AL. Unifying mechanism for toxicity and addiction by abused drugs: electron transfer and reactive oxygen species. Med Hypotheses. 2005;64:357–366. doi: 10.1016/j.mehy.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 37.Aguilo JI, Iturralde M, Monleon I, Inarrea P, Pardo J, Martinez-Lorenzo MJ, Anel A, Alava MA. Cytotoxicity of quinone drugs on highly proliferative human leukemia T cells: reactive oxygen species generation and inactive shortened SOD1 iso-form implications. Chem Biol Interact. 2012;198:18–28. doi: 10.1016/j.cbi.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Lemaire P, Livingstone DR. Aromatic hydrocarbon quinone-mediated reactive oxygen species production on hepatic microsomes of the flounder (Platichthys flesus L.) Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1997;117:131–139. doi: 10.1016/s0742-8413(97)00060-1. [DOI] [PubMed] [Google Scholar]
- 39.Wu Z, Du Y, Xue H, Wu Y, Zhou B. Aluminum induces neurodegeneration and its toxicity arises from increased iron accumulation and reactive oxygen species (ROS) production. Neurobiol Aging. 2012;33:199 e191–112. doi: 10.1016/j.neurobiolaging.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 40.Liu R, Liu W, Doctrow SR, Baudry M. Iron toxicity in organotypic cultures of hippocampal slices: role of reactive oxygen species. J Neurochem. 2003;85:492–502. doi: 10.1046/j.1471-4159.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 41.Jia L, Liu Z, Sun L, Miller SS, Ames BN, Cotman CW, Liu J. Acrolein, a toxicant in cigarette smoke, causes oxidative damage and mitochondrial dysfunction in RPE cells: protection by (R)-alpha-lipoic acid. Invest Ophthalmol Vis Sci. 2007;48:339–348. doi: 10.1167/iovs.06-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gobe G, Crane D. Mitochondria, reactive oxygen species and cadmium toxicity in the kidney. Toxicol Lett. 2010;198:49–55. doi: 10.1016/j.toxlet.2010.04.013. [DOI] [PubMed] [Google Scholar]
- 43.McDermott C, O’Donoghue MH, Heffron JJ. n-Hexane toxicity in Jurkat T-cells is mediated by reactive oxygen species. Arch Toxicol. 2008;82:165–171. doi: 10.1007/s00204-008-0286-x. [DOI] [PubMed] [Google Scholar]
- 44.Nicolescu AC, Ji Y, Comeau JL, Hill BC, Takahashi T, Brien JF, Racz WJ, Massey TE. Direct mitochondrial dysfunction precedes reactive oxygen species production in amiodarone-induced toxicity in human peripheral lung epithelial HPL1A cells. Toxicol Appl Pharmacol. 2008;227:370–379. doi: 10.1016/j.taap.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 45.Fennell DA, Corbo M, Pallaska A, Cotter FE. Bcl-2 resistant mitochondrial toxicity mediated by the isoquinoline carboxamide PK11195 involves de novo generation of reactive oxygen species. Br J Cancer. 2001;84:1397–1404. doi: 10.1054/bjoc.2001.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ludewig G, Srinivasan A, Robertson LW. Mechanisms of toxicity of PCB metabolites: generation of reactive oxygen species and glutathione depletion. Cent Eur J Public Health. 2000;8(Suppl):15–17. [PubMed] [Google Scholar]
- 47.Stanislawski L, Lefeuvre M, Bourd K, Soheili-Majd E, Goldberg M, Perianin A. TEGDMA-induced toxicity in human fibroblasts is associated with early and drastic glutathione depletion with subsequent production of oxygen reactive species. J Biomed Mater Res A. 2003;66:476–482. doi: 10.1002/jbm.a.10600. [DOI] [PubMed] [Google Scholar]
- 48.Burkitt MJ, Bishop HS, Milne L, Tsang SY, Provan GJ, Nobel CS, Orrenius S, Slater AF. Dithiocarbamate toxicity toward thymocytes involves their copper-catalyzed conversion to thiuram disulfides, which oxidize glutathione in a redox cycle without the release of reactive oxygen species. Arch Biochem Biophys. 1998;353:73–84. doi: 10.1006/abbi.1998.0618. [DOI] [PubMed] [Google Scholar]
- 49.Kolossov VL, Beaudoin JN, Prabhu Ponnuraj N, DiLiberto SJ, Hanafin WP, Kenis PJ, Gaskins HR. Thiol-based antioxidants elicit mitochondrial oxidation via respiratory complex III. Am J Phys Cell Physiol. 2015 doi: 10.1152/ajpcell.00006.2015. ajpcell 00006 02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winterbourn CC. The challenges of using fluorescent probes to detect and quantify specific reactive oxygen species in living cells. Biochim Biophys Acta. 2014;1840:730–738. doi: 10.1016/j.bbagen.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 51.Kalyanaraman B, Dranka BP, Hardy M, Michalski R, Zielonka J. HPLC-based monitoring of products formed from hydroethidine-based fluorogenic probes — the ultimate approach for intra- and extracellular superoxide detection. Biochim Biophys Acta. 2014;1840:739–744. doi: 10.1016/j.bbagen.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nauseef WM. Detection of superoxide anion and hydrogen peroxide production by cellular NADPH oxidases. Biochim Biophys Acta. 2014;1840:757–767. doi: 10.1016/j.bbagen.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kettle AJ, Albrett AM, Chapman AL, Dickerhof N, Forbes LV, Khalilova I, Turner R. Measuring chlorine bleach in biology and medicine. Biochim Biophys Acta. 2014;1840:781–793. doi: 10.1016/j.bbagen.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 54.Hawkins CL, Davies MJ. Detection and characterisation of radicals in biological materials using EPR methodology. Biochim Biophys Acta. 2014;1840:708–721. doi: 10.1016/j.bbagen.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 55.Carballal S, Bartesaghi S, Radi R. Kinetic and mechanistic considerations to assess the biological fate of peroxynitrite. Biochim Biophys Acta. 2014;1840:768–780. doi: 10.1016/j.bbagen.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thornalley PJ, Rabbani N. Detection of oxidized and glycated proteins in clinical samples using mass spectrometry — a user’s perspective. Biochim Biophys Acta. 2014;1840:818–829. doi: 10.1016/j.bbagen.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 57.Charles R, Jayawardhana T, Eaton P. Gel-based methods in redox proteomics. Biochim Biophys Acta. 2014;1840:830–837. doi: 10.1016/j.bbagen.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 58.Wall SB, Smith MR, Ricart K, Zhou F, Vayalil PK, Oh JY, Landar A. Detection of electrophile-sensitive proteins. Biochim Biophys Acta. 2014;1840:913–922. doi: 10.1016/j.bbagen.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim G, Weiss SJ, Levine RL. Methionine oxidation and reduction in proteins. Biochim Biophys Acta. 2014;1840:901–905. doi: 10.1016/j.bbagen.2013.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weissbach H, Resnick L, Brot N. Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim Biophys Acta. 2005;1703:203–212. doi: 10.1016/j.bbapap.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 61.Chao MW, Po IP, Laumbach RJ, Koslosky J, Cooper K, Gordon MK. DEP induction of ROS in capillary-like endothelial tubes leads to VEGF-A expression. Toxicology. 2012;297:34–46. doi: 10.1016/j.tox.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao D, Willard B, Podrez EA. Analysis of covalent modifications of proteins by oxidized phospholipids using a novel method of peptide enrichment. Anal Chem. 2014;86:1254–1262. doi: 10.1021/ac4035949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Santamarina S, Boronat S, Espadas G, Ayte J, Molina H, Hidalgo E. The oxidized thiol proteome in fission yeast — optimization of an ICAT-based method to identify H2O2-oxidized proteins. J Proteome. 2011;74:2476–2486. doi: 10.1016/j.jprot.2011.05.030. [DOI] [PubMed] [Google Scholar]
- 64.Hidalgo FJ, Alaiz M, Zamora R. A spectrophotometric method for the determination of proteins damaged by oxidized lipids. Anal Biochem. 1998;262:129–136. doi: 10.1006/abio.1998.2758. [DOI] [PubMed] [Google Scholar]
- 65.Gitler C, Zarmi B, Kalef E. General method to identify and enrich vicinal thiol proteins present in intact cells in the oxidized, disulfide state. Anal Biochem. 1997;252:48–55. doi: 10.1006/abio.1997.2294. [DOI] [PubMed] [Google Scholar]
- 66.Niki E. Biomarkers of lipid peroxidation in clinical material. Biochim Biophys Acta. 2014;1840:809–817. doi: 10.1016/j.bbagen.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 67.Arato S, Ito H, Miyashita K, Hayakawa K, Itabashi Y. A facile method for the detection of aldehydes in oxidized lipids using solid-phase microextraction fiber and gas chromatograph equipped with a septum-free injector. J Oleo Sci. 2009;58:17–22. doi: 10.5650/jos.58.17. [DOI] [PubMed] [Google Scholar]
- 68.Kikugawa K, Kato T, Iwata A. Determination of malonaldehyde in oxidized lipids by the Hantzsch fluorometric method. Anal Biochem. 1988;174:512–521. doi: 10.1016/0003-2697(88)90051-6. [DOI] [PubMed] [Google Scholar]
- 69.Collins AR. Measuring oxidative damage to DNA and its repair with the comet assay. Biochim Biophys Acta. 2014;1840:794–800. doi: 10.1016/j.bbagen.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 70.Poulsen HE, Nadal LL, Broedbaek K, Nielsen PE, Weimann A. Detection and interpretation of 8-oxodG and 8-oxoGua in urine, plasma and cerebrospinal fluid. Biochim Biophys Acta. 2014;1840:801–808. doi: 10.1016/j.bbagen.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 71.Ollikainen TR, Linnainmaa KI, Raivio KO, Kinnula VL. DNA single strand breaks and adenine nucleotide depletion as indices of oxidant effects on human lung cells. Free Radic Biol Med. 1998;24:1088–1096. doi: 10.1016/s0891-5849(97)00394-8. [DOI] [PubMed] [Google Scholar]
- 72.Churg A, Keeling B, Gilks B, Porter S, Olive P. Rat mesothelial and tracheal epithelial cells show equal DNA sensitivity to hydrogen peroxide-induced oxidant injury. Am J Phys. 1995;268:L832–L838. doi: 10.1152/ajplung.1995.268.5.L832. [DOI] [PubMed] [Google Scholar]
- 73.Fraga CG, Oteiza PI, Galleano M. In vitro measurements and interpretation of total antioxidant capacity. Biochim Biophys Acta. 2014;1840:931–934. doi: 10.1016/j.bbagen.2013.06.030. [DOI] [PubMed] [Google Scholar]
- 74.Griffith OW. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic Biol Med. 1999;27:922–935. doi: 10.1016/s0891-5849(99)00176-8. [DOI] [PubMed] [Google Scholar]
- 75.Ballatori N, Krance SM, Notenboom S, Shi S, Tieu K, Hammond CL. Glutathione dysregulation and the etiology and progression of human diseases. Biol Chem. 2009;390:191–214. doi: 10.1515/BC.2009.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niki E, Noguchi N. Evaluation of antioxidant capacity. What capacity is being measured by which method? IUBMB Life. 2000;50:323–329. doi: 10.1080/713803736. [DOI] [PubMed] [Google Scholar]
- 77.Nasef N, Belcastro R, Nash A, Bishara R, Iaboni D, Kantores C, Tanswell AK, Jankov RP. Role of ascorbate in lung cellular toxicity mediated by light-exposed parenteral nutrition solution. Free Radic Res. 2011;45:359–365. doi: 10.3109/10715762.2010.532791. [DOI] [PubMed] [Google Scholar]
- 78.Pharikal K, Das PC, Dey CD, Dasgupta S. Tissue ascorbate as a metabolic marker in cadmium toxicity. Int J Vitam Nutr Res (Int Z Vitam Ernahrungsforsch) 1988;58:306–311. Journal international de vitaminologie et de nutrition. [PubMed] [Google Scholar]
- 79.Suwannalert P, Boonsiri P, Khampitak T, Khampitak K, Sriboonlue P, Yongvanit P. The levels of lycopene, alpha-tocopherol and a marker of oxidative stress in healthy northeast Thai elderly. Asia Pac J Clin Nutr. 2007;16(Suppl. 1):27–30. [PubMed] [Google Scholar]
- 80.Bates CJ, Mishra GD, Prentice A. Gamma-tocopherol as a possible marker for nutrition-related risk: results from four National Diet and Nutrition Surveys in Britain. Br J Nutr. 2004;92:137–150. doi: 10.1079/BJN20041156. [DOI] [PubMed] [Google Scholar]
- 81.Mackinnon ES, Rao AV, Josse RG, Rao LG. Supplementation with the antioxidant lycopene significantly decreases oxidative stress parameters and the bone resorption marker N-telopeptide of type I collagen in postmenopausal women. Osteoporos Int. 2011;22:1091–1101. doi: 10.1007/s00198-010-1308-0. [DOI] [PubMed] [Google Scholar]
- 82.Majumdar SK, Shaw GK, Thomson AD. Blood beta-carotene status in chronic alcoholics — a good biochemical marker for malnutrition. Drug Alcohol Depend. 1983;12:111–113. doi: 10.1016/0376-8716(83)90059-5. [DOI] [PubMed] [Google Scholar]
- 83.Ben-Hayyim G, Gromet-Elhanan Z, Avron M. A specific and sensitive method for the determination of NADPH. Anal Biochem. 1969;28:6–12. doi: 10.1016/0003-2697(69)90151-1. [DOI] [PubMed] [Google Scholar]
- 84.Tabata M, Totani M, Murachi T. A chemiluminometric method for NADPH and NADH using a two-enzyme bioreactor and its application to the determination of magnesium in serum. Biomed Chromatogr. 1990;4:123–127. doi: 10.1002/bmc.1130040310. [DOI] [PubMed] [Google Scholar]
- 85.Truong TH, Carroll KS. Redox regulation of protein kinases. Crit Rev Biochem Mol Biol. 2013;48:332–356. doi: 10.3109/10409238.2013.790873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gopalakrishna R, Gundimeda U, Schiffman JE, McNeill TH. A direct redox regulation of protein kinase C isoenzymes mediates oxidant-induced neuritogenesis in PC12 cells. J Biol Chem. 2008;283:14430–14444. doi: 10.1074/jbc.M801519200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Brennan JP, Bardswell SC, Burgoyne JR, Fuller W, Schroder E, Wait R, Begum S, Kentish JC, Eaton P. Oxidant-induced activation of type I protein kinase A is mediated by RI subunit interprotein disulfide bond formation. J Biol Chem. 2006;281:21827–21836. doi: 10.1074/jbc.M603952200. [DOI] [PubMed] [Google Scholar]
- 88.Samet JM, Tal TL. Toxicological disruption of signaling homeostasis: tyrosine phosphatases as targets. Annu Rev Pharmacol Toxicol. 2010;50:215–235. doi: 10.1146/annurev.pharmtox.010909.105841. [DOI] [PubMed] [Google Scholar]
- 89.Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47:1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hybertson BM, Gao B, Bose SK, McCord JM. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol Asp Med. 2011;32:234–246. doi: 10.1016/j.mam.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 92.Elmarakby AA, Faulkner J, Baban B, Sullivan JC. Induction of hemeoxygenase-1 reduces renal oxidative stress and inflammation in diabetic spontaneously hypertensive rats. Int J Hypertens. 2012;2012:957235. doi: 10.1155/2012/957235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hung TC, Huang LW, Su SJ, Hsieh BS, Cheng HL, Hu YC, Chen YH, Hwang CC, Chang KL. Hemeoxygenase-1 expression in response to arecoline-induced oxidative stress in human umbilical vein endothelial cells. Int J Cardiol. 2011;151:187–194. doi: 10.1016/j.ijcard.2010.05.015. [DOI] [PubMed] [Google Scholar]
- 94.Chang MC, Chen LI, Chan CP, Lee JJ, Wang TM, Yang TT, Lin PS, Lin HJ, Chang HH, Jeng JH. The role of reactive oxygen species and hemeoxygenase-1 expression in the cytotoxicity, cell cycle alteration and apoptosis of dental pulp cells induced by BisGMA. Biomaterials. 2010;31:8164–8171. doi: 10.1016/j.biomaterials.2010.07.049. [DOI] [PubMed] [Google Scholar]
- 95.Ferenbach DA, Kluth DC, Hughes J. Hemeoxygenase-1 and renal ischaemia–reperfusion injury. Nephron Exp Nephrol. 2010;115:e33–e37. doi: 10.1159/000313828. [DOI] [PubMed] [Google Scholar]
- 96.Liang L, Gao C, Luo M, Wang W, Zhao C, Zu Y, Efferth T, Fu Y. Dihydroquercetin (DHQ) induced HO-1 and NQO1 expression against oxidative stress through the Nrf2-dependent antioxidant pathway. J Agric Food Chem. 2013;61:2755–2761. doi: 10.1021/jf304768p. [DOI] [PubMed] [Google Scholar]
- 97.Liu CL, Chiu YT, Hu ML. Fucoxanthin enhances HO-1 and NQO1 expression in murine hepatic BNL CL.2 cells through activation of the Nrf2/ARE system partially by its pro-oxidant activity. J Agric Food Chem. 2011;59:11344–11351. doi: 10.1021/jf2029785. [DOI] [PubMed] [Google Scholar]
- 98.Zhu L, Pi J, Wachi S, Andersen ME, Wu R, Chen Y. Identification of Nrf2-dependent airway epithelial adaptive response to proinflammatory oxidant-hypochlorous acid challenge by transcription profiling. Am J Physiol Lung Cell Mol Physiol. 2008;294:L469–L477. doi: 10.1152/ajplung.00310.2007. [DOI] [PubMed] [Google Scholar]
- 99.Aldebasi YH, Aly SM, Rahmani AH. Therapeutic implications of curcumin in the prevention of diabetic retinopathy via modulation of anti-oxidant activity and genetic pathways. Int J Physiol Pathophysiol Pharmacol. 2013;5:194–202. [PMC free article] [PubMed] [Google Scholar]
- 100.Assaf N, Shalby AB, Khalil WK, Ahmed HH. Biochemical and genetic alterations of oxidant/antioxidant status of the brain in rats treated with dexamethasone: protective roles of melatonin and acetyl-L-carnitine. J Physiol Biochem. 2012;68:77–90. doi: 10.1007/s13105-011-0121-3. [DOI] [PubMed] [Google Scholar]
- 101.Meyer AJ, Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal. 2010;13:621–650. doi: 10.1089/ars.2009.2948. [DOI] [PubMed] [Google Scholar]
- 102.Mullassery D, Horton CA, Wood CD, White MR. Single live-cell imaging for systems biology. Essays Biochem. 2008;45:121–133. doi: 10.1042/BSE0450121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Waters JC. Live-cell fluorescence imaging. Methods Cell Biol. 2007;81:115–140. doi: 10.1016/S0091-679X(06)81007-1. [DOI] [PubMed] [Google Scholar]
- 104.Gomes A, Fernandes E, Lima JL. Fluorescence probes used for detection of reactive oxygen species. J Biochem Biophys Methods. 2005;65:45–80. doi: 10.1016/j.jbbm.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 105.Chen X, Zhong Z, Xu Z, Chen L, Wang Y. 2′,7′-Dichlorodihydrofluorescein as a fluorescent probe for reactive oxygen species measurement: forty years of application and controversy. Free Radic Res. 2010;44:587–604. doi: 10.3109/10715761003709802. [DOI] [PubMed] [Google Scholar]
- 106.Brandt R, Keston AS. Synthesis of diacetyldichlorofluorescin: a stable reagent for fluorometric analysis. Anal Biochem. 1965;11:6–9. doi: 10.1016/0003-2697(65)90035-7. [DOI] [PubMed] [Google Scholar]
- 107.Guo H, Aleyasin H, Dickinson BC, Haskew-Layton RE, Ratan RR. Recent advances in hydrogen peroxide imaging for biological applications. Cell Biosci. 2014;4:64. doi: 10.1186/2045-3701-4-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grisham MB. Methods to detect hydrogen peroxide in living cells: possibilities and pitfalls. Comp Biochem Physiol A Mol Integr Physiol. 2013;165:429–438. doi: 10.1016/j.cbpa.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 109.Woolley JF, Stanicka J, Cotter TG. Recent advances in reactive oxygen species measurement in biological systems. Trends Biochem Sci. 2013;38:556–565. doi: 10.1016/j.tibs.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 110.Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic byproduct or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 111.Sies H. Role of metabolic H2O2 generation: redox signaling and oxidative stress. J Biol Chem. 2014;289:8735–8741. doi: 10.1074/jbc.R113.544635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miller EW, Tulyathan O, Isacoff EY, Chang CJ. Molecular imaging of hydrogen peroxide produced for cell signaling. Nat Chem Biol. 2007;3:263–267. doi: 10.1038/nchembio871. [DOI] [PubMed] [Google Scholar]
- 113.Rhee SG. Measuring H2O2 produced in response to cell surface receptor activation. Nat Chem Biol. 2007;3:244–246. doi: 10.1038/nchembio0507-244. [DOI] [PubMed] [Google Scholar]
- 114.Cheng WY, Currier J, Bromberg PA, Silbajoris R, Simmons SO, Samet JM. Linking oxidative events to inflammatory and adaptive gene expression induced by exposure to an organic particulate matter component. Environ Health Perspect. 2012;120:267–274. doi: 10.1289/ehp.1104055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dickinson BC, Huynh C, Chang CJ. A palette of fluorescent probes with varying emission colors for imaging hydrogen peroxide signaling in living cells. J Am Chem Soc. 2010;132:5906–5915. doi: 10.1021/ja1014103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dickinson BC, Tang Y, Chang Z, Chang CJ. A nuclear-localized fluorescent hydrogen peroxide probe for monitoring sirtuin-mediated oxidative stress responses in vivo. Chem Biol. 2011;18:943–948. doi: 10.1016/j.chembiol.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guo B, Xu D, Duan H, Du J, Zhang Z, Lee SM, Wang Y. Therapeutic effects of multifunctional tetramethylpyrazine nitrone on models of Parkinson’s disease in vitro and in vivo. Biol Pharm Bull. 2014;37:274–285. doi: 10.1248/bpb.b13-00743. [DOI] [PubMed] [Google Scholar]
- 118.Duregotti E, Negro S, Scorzeto M, Zornetta I, Dickinson BC, Chang CJ, Montecucco C, Rigoni M. Mitochondrial alarmins released by degenerating motor axon terminals activate perisynaptic Schwann cells. Proc Natl Acad Sci U S A. 2015;112:E497–E505. doi: 10.1073/pnas.1417108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Basu S, Rajakaruna S, Dickinson BC, Chang CJ, Menko AS. Endogenous hydrogen peroxide production in the epithelium of the developing embryonic lens. Mol Vis. 2014;20:458–467. [PMC free article] [PubMed] [Google Scholar]
- 120.Banerjee J, Ghatak P Das, Roy S, Khanna S, Sequin EK, Bellman K, Dickinson BC, Suri P, Subramaniam VV, Chang CJ, Sen CK. Improvement of human keratinocyte migration by a redox active bioelectric dressing. PLoS One. 2014;9:e89239. doi: 10.1371/journal.pone.0089239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ohsaki Y, O’Connor P, Mori T, Ryan RP, Dickinson BC, Chang CJ, Lu Y, Ito S, Cowley AW., Jr Increase of sodium delivery stimulates the mitochondrial respiratory chain H2O2 production in rat renal medullary thick ascending limb. Am J Physiol Ren Physiol. 2012;302:F95–F102. doi: 10.1152/ajprenal.00469.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sakai J, Li J, Subramanian KK, Mondal S, Bajrami B, Hattori H, Jia Y, Dickinson BC, Zhong J, Ye K, Chang CJ, Ho YS, Zhou J, Luo HR. Reactive oxygen species-induced actin glutathionylation controls actin dynamics in neutrophils. Immunity. 2012;37:1037–1049. doi: 10.1016/j.immuni.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang X, Gao F. Imaging mitochondrial reactive oxygen species with fluorescent probes: current applications and challenges. Free Radic Res. 2015;49:374–382. doi: 10.3109/10715762.2015.1014813. [DOI] [PubMed] [Google Scholar]
- 124.Dickinson BC, Lin VS, Chang CJ. Preparation and use of MitoPY1 for imaging hydrogen peroxide in mitochondria of live cells. Nat Protoc. 2013;8:1249–1259. doi: 10.1038/nprot.2013.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dickinson BC, Srikun D, Chang CJ. Mitochondrial-targeted fluorescent probes for reactive oxygen species. Curr Opin Chem Biol. 2010;14:50–56. doi: 10.1016/j.cbpa.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dickinson BC, Chang CJ. A targetable fluorescent probe for imaging hydrogen peroxide in the mitochondria of living cells. J Am Chem Soc. 2008;130:9638–9639. doi: 10.1021/ja802355u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lippert AR, Dickinson BC, New EJ. Imaging mitochondrial hydrogen peroxide in living cells. Methods Mol Biol. 2015;1264:231–243. doi: 10.1007/978-1-4939-2257-4_21. [DOI] [PubMed] [Google Scholar]
- 128.Beyer AM, Durand MJ, Hockenberry J, Gamblin TC, Phillips SA, Gutterman DD. An acute rise in intraluminal pressure shifts the mediator of flow-mediated dilation from nitric oxide to hydrogen peroxide in human arterioles. Am J Physiol Heart Circ Physiol. 2014;307:H1587–H1593. doi: 10.1152/ajpheart.00557.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li B, Iglesias-Pedraz JM, Chen LY, Yin F, Cadenas E, Reddy S, Comai L. Downregulation of the Werner syndrome protein induces a metabolic shift that compromises redox homeostasis and limits proliferation of cancer cells. Aging Cell. 2014;13:367–378. doi: 10.1111/acel.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tao L, Forester SC, Lambert JD. The role of the mitochondrial oxidative stress in the cytotoxic effects of the green tea catechin, (−)-epigallocatechin-3-gallate, in oral cells. Mol Nutr Food Res. 2014;58:665–676. doi: 10.1002/mnfr.201300427. [DOI] [PubMed] [Google Scholar]
- 131.Sanders LH, McCoy J, Hu X, Mastroberardino PG, Dickinson BC, Chang CJ, Chu CT, Van Houten B, Greenamyre JT. Mitochondrial DNA damage: molecular marker of vulnerable nigral neurons in Parkinson’s disease. Neurobiol Dis. 2014;70:214–223. doi: 10.1016/j.nbd.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Forman HJ, Maiorino M, Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Robinson KM, Janes MS, Pehar M, Monette JS, Ross MF, Hagen TM, Murphy MP, Beckman JS. Selective fluorescent imaging of superoxide in vivo using ethidium-based probes. Proc Natl Acad Sci U S A. 2006;103:15038–15043. doi: 10.1073/pnas.0601945103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zielonka J, Kalyanaraman B. Hydroethidine- and MitoSOX-derived red fluorescence is not a reliable indicator of intracellular superoxide formation: another inconvenient truth. Free Radic Biol Med. 2010;48:983–1001. doi: 10.1016/j.freeradbiomed.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br J Pharmacol. 2004;142:231–255. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhao H, Kalivendi S, Zhang H, Joseph J, Nithipatikom K, Vasquez-Vivar J, Kalyanaraman B. Superoxide reacts with hydroethidine but forms a fluorescent product that is distinctly different from ethidium: potential implications in intracellular fluorescence detection of superoxide. Free Radic Biol Med. 2003;34:1359–1368. doi: 10.1016/s0891-5849(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 137.Cheng WY, Larson JM, Samet JM. Monitoring intracellular oxidative events using dynamic spectral unmixing microscopy. Methods. 2014;66:345–352. doi: 10.1016/j.ymeth.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 138.Michalski R, Zielonka J, Hardy M, Joseph J, Kalyanaraman B. Hydropropidine: a novel, cell-impermeant fluorogenic probe for detecting extracellular superoxide. Free Radic Biol Med. 2013;54:135–147. doi: 10.1016/j.freeradbiomed.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Patel RP, McAndrew J, Sellak H, White CR, Jo H, Freeman BA, Darley-Usmar VM. Biological aspects of reactive nitrogen species. Biochim Biophys Acta. 1999;1411:385–400. doi: 10.1016/s0005-2728(99)00028-6. [DOI] [PubMed] [Google Scholar]
- 140.Li H, Forstermann U. Nitric oxide in the pathogenesis of vascular disease. J Pathol. 2000;190:244–254. doi: 10.1002/(SICI)1096-9896(200002)190:3<244::AID-PATH575>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 141.Pluth MD, Chan MR, McQuade LE, Lippard SJ. Seminaphthofluorescein-based fluorescent probes for imaging nitric oxide in live cells. Inorg Chem. 2011;50:9385–9392. doi: 10.1021/ic200986v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kojima H, Nakatsubo N, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Detection and imaging of nitric oxide with novel fluorescent indicators: diaminofluoresceins. Anal Chem. 1998;70:2446–2453. doi: 10.1021/ac9801723. [DOI] [PubMed] [Google Scholar]
- 143.Kojima H, Sakurai K, Kikuchi K, Kawahara S, Kirino Y, Nagoshi H, Hirata Y, Nagano T. Development of a fluorescent indicator for nitric oxide based on the fluorescein chromophore. Chem Pharm Bull. 1998;46:373–375. doi: 10.1248/cpb.46.373. [DOI] [PubMed] [Google Scholar]
- 144.Kojima H, Hirotani M, Nakatsubo N, Kikuchi K, Urano Y, Higuchi T, Hirata Y, Nagano T. Bioimaging of nitric oxide with fluorescent indicators based on the rhodamine chromophore. Anal Chem. 2001;73:1967–1973. doi: 10.1021/ac001136i. [DOI] [PubMed] [Google Scholar]
- 145.Gabe Y, Ueno T, Urano Y, Kojima H, Nagano T. Tunable design strategy for fluorescence probes based on 4-substituted BODIPY chromophore: improvement of highly sensitive fluorescence probe for nitric oxide. Anal Bioanal Chem. 2006;386:621–626. doi: 10.1007/s00216-006-0587-y. [DOI] [PubMed] [Google Scholar]
- 146.Vegesna GK, Sripathi SR, Zhang J, Zhu S, He W, Luo FT, Jahng WJ, Frost M, Liu H. Highly water-soluble BODIPY-based fluorescent probe for sensitive and selective detection of nitric oxide in living cells. ACS Appl Mater Interfaces. 2013;5:4107–4112. doi: 10.1021/am303247s. [DOI] [PubMed] [Google Scholar]
- 147.Jourd’heuil D. Increased nitric oxide-dependent nitrosylation of 4.5-diaminofluorescein by oxidants: implications for the measurement of intracellular nitric oxide. Free Radic Biol Med. 2002;33:676–684. doi: 10.1016/s0891-5849(02)00955-3. [DOI] [PubMed] [Google Scholar]
- 148.Lacza Z, Snipes JA, Zhang J, Horvath EM, Figueroa JP, Szabo C, Busija DW. Mitochondrial nitric oxide synthase is not eNOS, nNOS or iNOS. Free Radic Biol Med. 2003;35:1217–1228. doi: 10.1016/s0891-5849(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 149.Roychowdhury S, Luthe A, Keilhoff G, Wolf G, Horn TF. Oxidative stress in glial cultures: detection by DAF-2 fluorescence used as a tool to measure peroxynitrite rather than nitric oxide. Glia. 2002;38:103–114. doi: 10.1002/glia.10024. [DOI] [PubMed] [Google Scholar]
- 150.Nagano T, Takizawa H, Hirobe M. Reaction of nitric oxide with amines in the presence of dioxygen. Tetrahedron Lett. 1995;36:8239–8242. [Google Scholar]
- 151.Garner AL, Croix CMS, Pitt BR, Leikauf GD, Ando S, Koide K. Specific fluorogenic probes for ozone in biological and atmospheric samples. Nat Chem. 2009;1:316–321. doi: 10.1038/nchem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Peng T, Wong NK, Chen X, Chan YK, Ho DH, Sun Z, Hu JJ, Shen J, El-Nezami H, Yang D. Molecular imaging of peroxynitrite with HKGreen-4 in live cells and tissues. J Am Chem Soc. 2014;136:11728–11734. doi: 10.1021/ja504624q. [DOI] [PubMed] [Google Scholar]
- 153.Hou JT, Yang J, Li K, Liao YX, Yu KK, Xie YM, Yu XQ. A highly selective water-soluble optical probe for endogenous peroxynitrite. Chem Commun. 2014;50:9947–9950. doi: 10.1039/c4cc04192e. [DOI] [PubMed] [Google Scholar]
- 154.Sun X, Lacina K, Ramsamy EC, Flower SE, Fossey JS, Qian X, Anslyn EV, Bull SD, James TD. Reaction-based indicator displacement Assay (RIA) for the selective colorimetric and fluorometric detection of peroxynitrite. Chem Sci. 2015;6:2963–2967. doi: 10.1039/c4sc03983a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sun X, Xu Q, Kim G, Flower SE, Lowe JP, Yoon J, Fossey JS, Qian X, Bull SD, James TD. A water-soluble boronate-based fluorescent probe for the selective detection of peroxynitrite and imaging in living cells. Chem Sci. 2014;5:3368–3373. [Google Scholar]
- 156.Clark HA, Barker SLR, Brasuel M, Miller MT, Monson E, Parus S, Shi ZY, Song A, Thorsrud B, Kopelman R, Ade A, Meixner W, Athey B, Hoyer M, Hill D, Lightle R, Philbert MA. Subcellular optochemical nanobiosensors: probes encapsulated by biologically localised embedding (PEBBLEs) Sensors Actuators B Chem. 1998;51:12–16. [Google Scholar]
- 157.Hammond VJ, Aylott JW, Greenway GM, Watts P, Webster A, Wiles C. An optical sensor for reactive oxygen species: encapsulation of functionalised silica nanoparticles into silicate nanoprobes to reduce fluorophore leaching. Analyst. 2008;133:71–75. doi: 10.1039/b711995j. [DOI] [PubMed] [Google Scholar]
- 158.Yang L, Li N, Pan W, Yu Z, Tang B. Real-time imaging of mitochondrial hydrogen peroxide and pH fluctuations in living cells using a fluorescent nanosensor. Anal Chem. 2015;87:3678–3684. doi: 10.1021/ac503975x. [DOI] [PubMed] [Google Scholar]
- 159.Bjornberg O, Ostergaard H, Winther JR. Measuring intracellular redox conditions using GFP-based sensors. Antioxid Redox Signal. 2006;8:354–361. doi: 10.1089/ars.2006.8.354. [DOI] [PubMed] [Google Scholar]
- 160.Ezerina D, Morgan B, Dick TP. Imaging dynamic redox processes with genetically encoded probes. J Mol Cell Cardiol. 2014;73:43–49. doi: 10.1016/j.yjmcc.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 161.Lukyanov KA, Belousov VV. Genetically encoded fluorescent redox sensors. Biochim Biophys Acta. 2014;1840:745–756. doi: 10.1016/j.bbagen.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 162.Schwarzlander M, Dick TP, Meye AJ, Morgan B. Dissecting redox biology using fluorescent protein sensors. Antioxid Redox Signal. 2015 doi: 10.1089/ars.2015.6266. [DOI] [PubMed] [Google Scholar]
- 163.Jiang K, Schwarzer C, Lally E, Zhang S, Ruzin S, Machen T, Remington SJ, Feldman L. Expression and characterization of a redox-sensing green fluorescent protein (reduction-oxidation-sensitive green fluorescent protein) in Arabidopsis. Plant Physiol. 2006;141:397–403. doi: 10.1104/pp.106.078246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Berkelhamer SK, Kim GA, Radder JE, Wedgwood S, Czech L, Steinhorn RH, Schumacker PT. Developmental differences in hyperoxia-induced oxidative stress and cellular responses in the murine lung. Free Radic Biol Med. 2013;61:51–60. doi: 10.1016/j.freeradbiomed.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yu S, Qin W, Zhuang G, Zhang X, Chen G, Liu W. Monitoring oxidative stress and DNA damage induced by heavy metals in yeast expressing a redox-sensitive green fluorescent protein. Curr Microbiol. 2009;58:504–510. doi: 10.1007/s00284-008-9354-y. [DOI] [PubMed] [Google Scholar]
- 166.Loor G, Kondapalli J, Iwase H, Chandel NS, Waypa GB, Guzy RD, Vanden Hoek TL, Schumacker PT. Mitochondrial oxidant stress triggers cell death in simulated ischemia-reperfusion. Biochim Biophys Acta. 2011;1813:1382–1394. doi: 10.1016/j.bbamcr.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Loor G, Kondapalli J, Schriewer JM, Chandel NS, Vanden Hoek TL, Schumacker PT. Menadione triggers cell death through ROS-dependent mechanisms involving PARP activation without requiring apoptosis. Free Radic Biol Med. 2010;49:1925–1936. doi: 10.1016/j.freeradbiomed.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nostbakken OJ, Bredal IL, Olsvik PA, Huang TS, Torstensen BE. Effect of marine omega 3 fatty acids on methylmercury-induced toxicity in fish and mammalian cells in vitro. J Biomed Biotechnol. 2012;2012:417652. doi: 10.1155/2012/417652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ayer A, Sanwald J, Pillay BA, Meyer AJ, Perrone GG, Dawes IW. Distinct redox regulation in sub-cellular compartments in response to various stress conditions in Saccharomyces cerevisiae. PLoS One. 2013;8:e65240. doi: 10.1371/journal.pone.0065240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 171.Ostergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lohman JR, Remington SJ. Development of a family of redox-sensitive green fluorescent protein indicators for use in relatively oxidizing subcellular environments. Biochemistry. 2008;47:8678–8688. doi: 10.1021/bi800498g. [DOI] [PubMed] [Google Scholar]
- 173.Guzman JN, Sanchez-Padilla J, Wokosin D, Kondapalli J, Ilijic E, Schumacker PT, Surmeier DJ. Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature. 2010;468:696–700. doi: 10.1038/nature09536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Breckwoldt MO, Pfister FM, Bradley PM, Marinkovic P, Williams PR, Brill MS, Plomer B, Schmalz A, Clair DKS, Naumann R, Griesbeck O, Schwarzlander M, Godinho L, Bareyre FM, Dick TP, Kerschensteiner M, Misgeld T. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat Med. 2014;20:555–560. doi: 10.1038/nm.3520. [DOI] [PubMed] [Google Scholar]
- 175.Wolf AM, Nishimaki K, Kamimura N, Ohta S. Real-time monitoring of oxidative stress in live mouse skin. J Investig Dermatol. 2014;134:1701–1709. doi: 10.1038/jid.2013.428. [DOI] [PubMed] [Google Scholar]
- 176.Bjornberg O, Ostergaard H, Winther JR. Mechanistic insight provided by glutaredoxin within a fusion to redox-sensitive yellow fluorescent protein. Biochemistry. 2006;45:2362–2371. doi: 10.1021/bi0522495. [DOI] [PubMed] [Google Scholar]
- 177.Hu J, Dong L, Outten CE. The redox environment in the mitochondrial intermem-brane space is maintained separately from the cytosol and matrix. J Biol Chem. 2008;283:29126–29134. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dardalhon M, Kumar C, Iraqui I, Vernis L, Kienda G, Banach-Latapy A, He T, Chanet R, Faye G, Outten CE, Huang ME. Redox-sensitive YFP sensors monitor dynamic nuclear and cytosolic glutathione redox changes. Free Radic Biol Med. 2012;52:2254–2265. doi: 10.1016/j.freeradbiomed.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Banach-Latapy A, He T, Dardalhon M, Vernis L, Chanet R, Huang ME. Redox-sensitive YFP sensors for monitoring dynamic compartment-specific glutathione redox state. Free Radic Biol Med. 2013;65:436–445. doi: 10.1016/j.freeradbiomed.2013.07.033. [DOI] [PubMed] [Google Scholar]
- 180.Fan Y, Chen Z, Ai HW. Monitoring redox dynamics in living cells with a redox-sensitive red fluorescent protein. Anal Chem. 2015;87:2802–2810. doi: 10.1021/ac5041988. [DOI] [PubMed] [Google Scholar]
- 181.Sugiura K, Nagai T, Nakano M, Ichinose H, Nakabayashi T, Ohta N, Hisabori T. Redox sensor proteins for highly sensitive direct imaging of intracellular redox state. Biochem Biophys Res Commun. 2015;457:242–248. doi: 10.1016/j.bbrc.2014.12.095. [DOI] [PubMed] [Google Scholar]
- 182.Hanson GT, Aggeler R, Oglesbee D, Cannon M, Capaldi RA, Tsien RY, Remington SJ. Investigating mitochondrial redox potential with redox-sensitive green fluorescent protein indicators. J Biol Chem. 2004;279:13044–13053. doi: 10.1074/jbc.M312846200. [DOI] [PubMed] [Google Scholar]
- 183.Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. [DOI] [PubMed] [Google Scholar]
- 184.Birk J, Meyer M, Aller I, Hansen HG, Odermatt A, Dick TP, Meyer AJ, Appenzeller-Herzog C. Endoplasmic reticulum: reduced and oxidized glutathione revisited. J Cell Sci. 2013;126:1604–1617. doi: 10.1242/jcs.117218. [DOI] [PubMed] [Google Scholar]
- 185.van Lith M, Tiwari S, Pediani J, Milligan G, Bulleid NJ. Real-time monitoring of redox changes in the mammalian endoplasmic reticulum. J Cell Sci. 2011;124:2349–2356. doi: 10.1242/jcs.085530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Farrow KN, Lee KJ, Perez M, Schriewer JM, Wedgwood S, Lakshminrusimha S, Smith CL, Steinhorn RH, Schumacker PT. Brief hyperoxia increases mitochondrial oxidation and increases phosphodiesterase 5 activity in fetal pulmonary artery smooth muscle cells. Antioxid Redox Signal. 2012;17:460–470. doi: 10.1089/ars.2011.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jiang P, Yamauchi K, Yang M, Tsuji K, Xu M, Maitra A, Bouvet M, Hoffman RM. Tumor cells genetically labeled with GFP in the nucleus and RFP in the cytoplasm for imaging cellular dynamics. Cell Cycle. 2006;5:1198–1201. doi: 10.4161/cc.5.11.2795. [DOI] [PubMed] [Google Scholar]
- 188.Waypa GB, Marks JD, Guzy R, Mungai PT, Schriewer J, Dokic D, Schumacker PT. Hypoxia triggers subcellular compartmental redox signaling in vascular smooth muscle cells. Circ Res. 2010;106:526–535. doi: 10.1161/CIRCRESAHA.109.206334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang H, Limphong P, Pieper J, Liu Q, Rodesch CK, Christians E, Benjamin IJ. Glutathione-dependent reductive stress triggers mitochondrial oxidation and cytotoxicity. FASEB J. 2012;26:1442–1451. doi: 10.1096/fj.11-199869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Dooley CT, Li L, Misler JA, Thompson JH. Toxicity of 6-hydroxydopamine: live cell imaging of cytoplasmic redox flux. Cell Biol Toxicol. 2012;28:89–101. doi: 10.1007/s10565-011-9209-3. [DOI] [PubMed] [Google Scholar]
- 191.Albrecht SC, Barata AG, Grosshans J, Teleman AA, Dick TP. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metab. 2011;14:819–829. doi: 10.1016/j.cmet.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 192.Albrecht SC, Sobotta MC, Bausewein D, Aller I, Hell R, Dick TP, Meyer AJ. Redesign of genetically encoded biosensors for monitoring mitochondrial redox status in a broad range of model eukaryotes. J Biomol Screen. 2014;19:379–386. doi: 10.1177/1087057113499634. [DOI] [PubMed] [Google Scholar]
- 193.Bhaskar A, Chawla M, Mehta M, Parikh P, Chandra P, Bhave D, Kumar D, Carroll KS, Singh A. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 2014;10:e1003902. doi: 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- 195.Malinouski M, Zhou Y, Belousov VV, Hatfield DL, Gladyshev VN. Hydrogen peroxide probes directed to different cellular compartments. PLoS One. 2011;6:e14564. doi: 10.1371/journal.pone.0014564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Niethammer P, Grabher C, Look AT, Mitchison TJ. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bilan DS, Pase L, Joosen L, Gorokhovatsky AY, Ermakova YG, Gadella TW, Grabher C, Schultz C, Lukyanov S, Belousov VV. HyPer-3: a genetically encoded H(2)O(2)probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chem Biol. 2013;8:535–542. doi: 10.1021/cb300625g. [DOI] [PubMed] [Google Scholar]
- 198.Mishina NM, Tyurin-Kuzmin PA, Markvicheva KN, Vorotnikov AV, Tkachuk VA, Laketa V, Schultz C, Lukyanov S, Belousov VV. Does cellular hydrogen peroxide diffuse or act locally? Antioxid Redox Signal. 2011;14:1–7. doi: 10.1089/ars.2010.3539. [DOI] [PubMed] [Google Scholar]
- 199.Wages PA, Silbajoris R, Speen A, Brighton L, Henriquez A, Tong H, Bromberg PA, Simmons SO, Samet JM. Role of H2O2 in the oxidative effects of zinc exposure in human airway epithelial cells. Redox Biol. 2014;3:47–55. doi: 10.1016/j.redox.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mahon MJ. pHluorin2: an enhanced, ratiometric, pH-sensitive green florescent protein. Adv Biosci Biotechnol. 2011;2:132–137. doi: 10.4236/abb.2011.23021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tantama M, Hung YP, Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J Am Chem Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Azarias G, Chatton JY. Selective ion changes during spontaneous mitochondrial transients in intact astrocytes. PLoS One. 2011;6:e28505. doi: 10.1371/journal.pone.0028505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Matlashov ME, Bogdanova YA, Ermakova GV, Mishina NM, Ermakova YG, Nikitin ES, Balaban PM, Okabe S, Lukyanov S, Enikolopov G, Zaraisky AG, Belousov VV. Fluorescent ratiometric pH indicator SypHer2: applications in neuroscience and regenerative biology. Biochim Biophys Acta. 2015;1850:2318–2328. doi: 10.1016/j.bbagen.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wang W, Fang H, Groom L, Cheng A, Zhang W, Liu J, Wang X, Li K, Han P, Zheng M, Yin J, Wang W, Mattson MP, Kao JP, Lakatta EG, Sheu SS, Ouyang K, Chen J, Dirksen RT, Cheng H. Superoxide flashes in single mitochondria. Cell. 2008;134:279–290. doi: 10.1016/j.cell.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schwarzlander M, Murphy MP, Duchen MR, Logan DC, Fricker MD, Halestrap AP, Muller FL, Rizzuto R, Dick TP, Meyer AJ, Sweetlove LJ. Mitochondrial ‘flashes’: a radical concept repHined. Trends Cell Biol. 2012;22:503–508. doi: 10.1016/j.tcb.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 206.Schwarzlander M, Wagner S, Ermakova YG, Belousov VV, Radi R, Beckman JS, Buettner GR, Demaurex N, Duchen MR, Forman HJ, Fricker MD, Gems D, Halestrap AP, Halliwell B, Jakob U, Johnston IG, Jones NS, Logan DC, Morgan B, Muller FL, Nicholls DG, Remington SJ, Schumacker PT, Winterbourn CC, Sweetlove LJ, Meyer AJ, Dick TP, Murphy MP. The ‘mitoflash’ probe cpYFP does not respond to superoxide. Nature. 2014;514:E12–E14. doi: 10.1038/nature13858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chen X, Wu S, Han J, Han S. Rhodamine-propargylic esters for detection of mitochondrial hydrogen sulfide in living cells. Bioorg Med Chem Lett. 2013;23:5295–5299. doi: 10.1016/j.bmcl.2013.07.072. [DOI] [PubMed] [Google Scholar]
- 208.Chen ZJ, Ai HW. A highly responsive and selective fluorescent probe for imaging physiological hydrogen sulfide. Biochemistry. 2014;53:5966–5974. doi: 10.1021/bi500830d. [DOI] [PubMed] [Google Scholar]
- 209.Knight MM, Roberts SR, Lee DA, Bader DL. Live cell imaging using confocal microscopy induces intracellular calcium transients and cell death. Am J Physiol Cell Physiol. 2003;284:C1083–C1089. doi: 10.1152/ajpcell.00276.2002. [DOI] [PubMed] [Google Scholar]
- 210.Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is green fluorescent protein toxic to the living cells? Biochem Biophys Res Commun. 1999;260:712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 211.Villuendas G, Gutierrez-Adan A, Jimenez A, Rojo C, Roldan ER, Pintado B. CMV-driven expression of green fluorescent protein (GFP) in male germ cells of transgenic mice and its effect on fertility. Int J Androl. 2001;24:300–305. doi: 10.1046/j.1365-2605.2001.00302.x. [DOI] [PubMed] [Google Scholar]
- 212.Gibbs-Flournoy EA, Simmons SO, Bromberg PA, Dick TP, Samet JM. Monitoring intracellular redox changes in ozone-exposed airway epithelial cells. Environ Health Perspect. 2013;121:312–317. doi: 10.1289/ehp.1206039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Wages PA, Lavrich KS, Zhang Z, Cheng WY, Corteselli E, Gold A, Bromberg P, Simmons SO, Samet JM. Protein sulfenylation: a novel readout of environmental oxidant stress. Chem Res Toxicol. 2015;28:2411–2418. doi: 10.1021/acs.chemrestox.5b00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yin Q, Huang C, Zhang C, Zhu W, Xu Y, Qian X, Yang Y. In situ visualization and detection of protein sulfenylation responses in living cells through a dimedone-based fluorescent probe. Org Biomol Chem. 2013;11:7566–7573. doi: 10.1039/c3ob41434e. [DOI] [PubMed] [Google Scholar]
- 215.Dooley CT, Dore TM, Hanson GT, Jackson WC, Remington SJ, Tsien RY. Imaging dynamic redox changes in mammalian cells with green fluorescent protein indicators. J Biol Chem. 2004;279:22284–22293. doi: 10.1074/jbc.M312847200. [DOI] [PubMed] [Google Scholar]


