Agrobacterium type IV secretion system delivers VirE2 into host cells via conserved endocytosis mediated by clathrin adaptor AP2
Keywords: Agrobacterium, host cells, VirE2, Type IV secretion system, Protein delivery, protein trafficking, endocytosis, clathrin, AP2, cargo motif
Abstract
Agrobacterium tumefaciens can cause crown gall tumors on a wide range of host plants. As a natural genetic engineer, the bacterium can transfer both single-stranded DNA (ssDNA) [transferred DNA (T-DNA)] molecules and bacterial virulence proteins into various recipient cells. Among Agrobacterium-delivered proteins, VirE2 is an ssDNA binding protein that is involved in various steps of the transformation process. However, it is not clear how plant cells receive the T-DNA or protein molecules. Using a split–green fluorescent protein approach, we monitored the VirE2 delivery process inside plant cells in real time. We observed that A. tumefaciens delivered VirE2 from the bacterial lateral sides that were in close contact with plant membranes. VirE2 initially accumulated on plant cytoplasmic membranes at the entry points. VirE2-containing membranes were internalized through clathrin-mediated endocytosis to form endomembrane compartments. VirE2 colocalized with the early endosome marker SYP61 but not with the late endosome marker ARA6, suggesting that VirE2 escaped from early endosomes for subsequent trafficking inside the cells. Dual endocytic motifs at the carboxyl-terminal tail of VirE2 were involved in VirE2 internalization and could interact with the μ subunit of the plant clathrin-associated adaptor AP2 complex (AP2M). Both the VirE2 cargo motifs and AP2M were important for the transformation process. Because AP2-mediated endocytosis is well conserved, our data suggest that the A. tumefaciens pathogen hijacks conserved endocytic pathways to facilitate the delivery of virulence factors. This might be important for Agrobacterium to achieve both a wide host range and a high transformation efficiency.
INTRODUCTION
Agrobacterium tumefaciens causes crown gall tumors on various plants by transferring oncogenic transferred DNA (T-DNA) into plant cells (1–3). Under laboratory conditions, the bacterium can transfer T-DNA into various eukaryotic species, including yeast (4, 5), fungi (6), algae (7), and cultured human cells (8). It has been developed as a DNA delivery vector and is widely used as the workhorse for the genetic engineering of plants (9) and nonplant organisms (10).
During the Agrobacterium-mediated transformation (AMT) process, A. tumefaciens delivers both T-DNA and bacterial virulence proteins into host cells via a bacterial type IV secretion system (T4SS) composed of VirB/VirD4 (11, 12). The Agrobacterium VirB/VirD4 T4SS is the archetype for the T4SS family that is widely used by bacteria to translocate DNA (13, 14) and protein macromolecules (15) to a diverse range of bacterial and eukaryotic cells (16). The Agrobacterium T4SS apparatus comprises 12 bacterial virulence proteins, including VirB1 to VirB11 and VirD4, which form a multisubunit membrane-spanning channel that delivers macromolecules into host cells (17).
Agrobacterium delivers at least five protein substrates into host cells through the T4SS; these include VirD2, VirD5, VirE2, VirE3, and VirF (18–20). The bacterial effectors depend upon their C-terminal positively charged amino acid residues for export into host cells (19). Upon delivery, these effectors may work cooperatively inside host cells and facilitate the transformation process. Host factors have been shown to interact with these effectors and are important for successful transformations (21).
T-DNA is generated inside the bacteria by VirD2 endonuclease (22–24), which remains covalently associated with the 5′ end of the T-DNA and leads the way into host cells through the T4SS (23). Inside the host cytoplasm, the naked T-DNA is hypothesized to be coated with T4SS-delivered VirE2, which is a single-stranded DNA (ssDNA) binding protein, to form the putative “T-complex” (25–27). Both VirD2 and VirE2 contain functional nuclear localization signal (NLS) motifs that interact with host importin α proteins (28). They may work cooperatively for the nuclear import of the T-complex inside host cells.
As an abundant effector protein secreted into recipient cells, VirE2 is also crucial for a variety of other processes during the transformation. In vitro studies have shown that VirE2 forms a voltage-gated and an ssDNA-specific channel on artificial membranes, suggesting that it might facilitate the entry of T-DNA into host cells (29). VirE2 binds to the T-strand in vitro in a cooperative manner, and it has been hypothesized to protect T-DNA from nucleolytic degradation (30, 31). Because VirE2 is the major component of the T-complex, VirE2 trafficking potentially affects the fate of the T-strand.
VirE2 has been shown to participate in nuclear targeting of T-DNA in two different ways. First, two separate NLS motifs that are involved in direct interaction with Arabidopsis importin α isoforms have been identified in the VirE2 molecule (28). Second, VirE2 can also interact with the plant transcription factor VIP1 (32). VIP1 undergoes mitogen-activated protein kinase 3–mediated phosphorylation and nuclear translocation induced by Agrobacterium infection, which might result in the nuclear import of VirE2 and thus the T-strand (33, 34). Inside host nuclei, VIP1 may interact with host histones, and this interaction might help the T-complex to target the host chromatin (34). Moreover, VirE2 may interact with another host protein, VIP2, a putative transcriptional repressor localized to plant nuclei (35). The interaction between VirE2 and VIP2 might facilitate T-strand integration into the host genome (35).
However, it is not clear how plant cells receive the T-DNA or protein molecules. Recently, a split–green fluorescent protein (GFP)–based method (36) was adopted to directly detect the Agrobacterium-delivered VirE2 inside plant cells (37, 38). This split-GFP approach allowed the visualization of VirE2 trafficking in the recipient cells in real time in a natural setting, which indicated that Agrobacterium can deliver VirE2 and transform plant cells at a high efficiency (up to 100%) (37). Here, we report the use of the split-GFP approach to elucidate the VirE2 delivery and trafficking processes.
RESULTS
VirE2 is associated with the host plasma membrane upon delivery
To visualize VirE2 delivery, we expressed the VirE2-GFP11 fusion in A. tumefaciens and GFP1–10 in plant cells. VirE2 delivery into tobacco cells at the early stage was observed using confocal microscopy. The T-DNA free strain EHA105 was used to avoid any potential complication due to T-DNA trafficking. A. tumefaciens EHA105virE2::GFP11–producing VirE2-GFP11 was infiltrated into transgenic Nicotiana benthamiana (Nb308A) leaves expressing both GFP1–10 and DsRed. The VirE2 delivery into the tobacco cells was examined at different time points. As shown in Fig. 1A, a small amount of VirE2 appeared at the tobacco cell borders at 32 hours after agroinfiltration (upper panel). As time passed, more VirE2 was observed at the cell borders. The VirE2 signals became filamentous. Most of the tobacco cells exhibited VirE2 accumulation in the nucleus at 48 hours after agroinfiltration (Fig. 1A, lower panel). VirE2 nuclear localization was confirmed by the three-dimensional visualization method described previously (37). These results indicate that VirE2 first appears at tobacco cell borders and then moves into the nucleus.
Fig. 1. Association of Agrobacterium-delivered VirE2 with the host plasma membrane and endomembrane compartments.
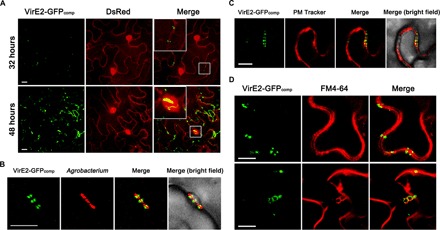
(A) VirE2 detection at different time intervals inside N. benthamiana cells. A. tumefaciens EHA105virE2::GFP11 was infiltrated into transgenic N. benthamiana (Nb308A) leaves expressing both GFP1–10 and DsRed. Projected Z-series images were obtained 32 and 48 hours after agroinfiltration. (B) Accumulation of VirE2 at the cytoplasmic sides of tobacco cells contacting with A. tumefaciens cells. Wild-type N. benthamiana leaves were infiltrated with the DsRed-labeled A. tumefaciens strain EHA105virE2::GFP11(pGFP1–10 and pVBA-RFP), which is also capable of delivering VirE2-GFP11 and the T-DNA expressing GFP1–10. Images were obtained 2 days after agroinfiltration. (C) Accumulation of Agrobacterium-delivered VirE2 at the host plasma membrane. Wild-type N. benthamiana leaves were infiltrated with evenly mixed A. tumefaciens strains EHA105virE2::GFP11(pGFP1–10), which is capable of delivering VirE2-GFP11 and the T-DNA expressing GFP1–10, and EHA105virE2::GFP11(pm-rb), which is capable of delivering VirE2-GFP11 and the T-DNA expressing a plasma membrane (PM) tracker. Images were obtained 2 days after agroinfiltration. (D) Colocalization of Agrobacterium-delivered VirE2 with FM4-64–labeled plasma membranes (upper panel) or endomembrane compartments (lower panel) inside tobacco epidermal cells. Wild-type N. benthamiana leaves were infiltrated with the A. tumefaciens strain EHA105virE2::GFP11(pGFP1–10) and then stained with FM4-64. Images were obtained 2 days after agroinfiltration. Scale bars, 10 μm.
A more detailed time-course study was then conducted to characterize subcellular localizations of Agrobacterium-delivered VirE2 inside plant cells (fig. S1). When VirE2 signals first appeared at 30 hours after agroinfiltration in the transgenic tobacco cells expressing both GFP1–10 and DsRed, 51% of the signals were associated with the plasma membrane (fig. S1B). The percentage of VirE2 associated with the membrane then decreased slightly, and the percentage of VirE2 associated with the cytoplasm increased, as more VirE2 was delivered into the cells. Transient expression of GFP1–10 and DsRed on T-DNA revealed a roughly even distribution of VirE2 among subcellular locations, although the signals appeared 6 hours later (fig. S1D) than the stable expression (fig. S1B), presumably because of the lag time required for the T-DNA delivery and expression. These results are consistent with the observation that VirE2 first appears at the plasma membrane and then moves into the nucleus.
Next, we determined the positioning of A. tumefaciens cells inside plant tissues. The bacterial cells were constructed to express GFP under the control of the virB promoter, and thus, they became fluorescently labeled naturally during agroinfiltration. After GFP-labeled A. tumefaciens cells EHA105(pAT-GFP) were infiltrated into the N. benthamiana leaves, most of the bacterial cells were observed to line up at the intercellular spaces of the tobacco cells (fig. S2A). When GFP-labeled bacterial cells were evenly mixed with DsRed-labeled bacterial cells and infiltrated into the N. benthamiana leaves, we observed that the bacterial cells tightly lined up at the intercellular spaces as single cells (fig. S2B). These results suggest that the limited intercellular spaces of N. benthamiana epidermal cells can only accommodate single bacterial cells, and this space limitation may allow only the lateral side of the bacterium to closely contact with the host cell.
To determine the relative positioning of A. tumefaciens cells and VirE2 delivered into plant cells, we infiltrated wild-type N. benthamiana leaves with the DsRed-labeled A. tumefaciens strain EHA105virE2::GFP11(pGFP1–10 and pVBA-RFP), which is also capable of delivering VirE2-GFP11 and the T-DNA expressing GFP1–10. At 48 hours after agroinfiltration, VirE2 accumulated at the cytoplasmic sides of tobacco cells that were in close contact with A. tumefaciens cells (Fig. 1B). VirE2 was delivered into plant cells from both sides of the bacterial cells. This suggests that a single bacterium can deliver VirE2 into two neighboring host cells simultaneously.
To determine the subcellular location of Agrobacterium-delivered VirE2 inside host cells, a specific plant plasma membrane tracker (39) was transiently expressed inside plant cells by T-DNA delivered by the same bacterial cells delivering VirE2-GFP11. Agrobacterium-delivered VirE2 appeared to colocalize with the transiently expressed plasma membrane tracker (Fig. 1C). VirE2 colocalization with the plasma membrane was confirmed by confocal microscopy using a 100× oil objective (fig. S3). These results suggest that VirE2 is associated with plant cytoplasmic membranes upon delivery.
Agrobacterium-delivered VirE2 is associated with endomembrane compartments
To investigate how the membrane-bound VirE2 moved into the cytoplasm, the fluorescent styryl dye FM4-64 was used to label the membranes and monitor the cellular dynamics (40). This lipophilic dye can label membranes where it is applied, but it cannot penetrate the membranes by itself. This property allowed us to monitor the trafficking process of VirE2-bound membranes. A. tumefaciens EHA105virE2::GFP11 cells were infiltrated into N. benthamiana leaves to start the VirE2 delivery; 48 hours later, the FM4-64 dye was then infiltrated into the same areas. As shown in Fig. 1D, VirE2 colocalized with FM4-64–labeled plasma membranes (upper panel), in a manner similar to that observed when using the plasma membrane tracker (Fig. 1C). VirE2 also colocalized with FM4-64–labeled endomembrane compartments that ranged from 0.8 to 4.5 μm in diameter with an average of 2.2 μm (Fig. 1D, lower panel).
A time-course study was then conducted to characterize the VirE2 colocalization with endomembrane compartments. As shown in fig. S4A, VirE2 appeared at 36 hours after agroinfiltration under a transient expression condition (as observed in fig. S1, C and D); 8.6% of the FM4-64–labeled endomembrane compartments colocalized with the VirE2 signals. The percentage of colocalization increased to 15.3% at 42 hours after agroinfiltration and to 16.5% at 48 hours after agroinfiltration, as the amount of delivered VirE2 increased. These results suggest that VirE2 may use endomembrane compartments for subcellular trafficking.
Moreover, the colocalization of VirE2 with FM4-64–labeled endomembrane compartments continued as the FM4-64–labeled vesicles moved inside the cytoplasm (fig. S4B and movie S1). Their speed of movement ranged from 0.4 to 2.1 μm/s, which is consistent with the endosome dynamics reported in a previous study (41). The data consistently suggest that VirE2 delivered onto host plasma membranes may use host endocytosis for cellular internalization and cytoplasmic movement.
Endocytosis is required for efficient VirE2 trafficking and AMT process
Subsequently, we examined whether the host endocytosis process was required for the internalization of VirE2 protein. It has been reported that plant endocytosis is mediated by clathrin triskelions (42). Overexpression of a C-terminal part of the clathrin heavy chain (Hub) that can bind to and deplete the clathrin light chain would lead to strong dominant-negative effects on clathrin-mediated endocytosis (CME) (43–45).
The effect of Hub overexpression in N. benthamiana leaves was then tested, and the FM4-64 dye was used to monitor the endocytosis process. Transient expression of Hub under a cauliflower mosaic virus (CaMV) 35S promoter markedly decreased the internalization of the FM4-64 dye (fig. S5). This suggests that a dominant negative strategy using Hub could affect endocytosis in N. benthamiana epidermal cells. In addition, Hub overexpression increased VirE2 accumulation at cell borders (Fig. 2, A and B). A time-course experiment demonstrated that VirE2 stayed much longer at the cell borders in the tobacco cells overexpressing Hub compared to the control (fig. S6), indicating that functional clathrin and an active CME process were required for VirE2 departure from the plant cellular membrane.
Fig. 2. Interference with host endocytosis impaired VirE2 trafficking and attenuated virulence.
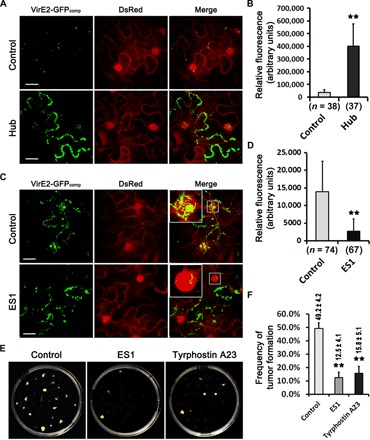
(A) Expression of a defective clathrin Hub impaired VirE2 departure from plasma membranes. N. benthamiana (Nb308A) leaves were infiltrated with the A. tumefaciens strain EHA105virE2::GFP11(pXY01) or EHA105virE2::GFP11(pXY01-Hub), which is capable of delivering VirE2-GFP11 and the T-DNA expressing Hub. Projected Z-series images were obtained 4 days after agroinfiltration. Scale bars, 20 μm. (B) The intensity of VirE2-GFPcomp signals remaining at host cell borders was measured in each image. n, number of images measured. (C) ES1 treatment decreased nuclear accumulation of VirE2 in plant epidermal cells. N. benthamiana (Nb308A) leaves were infiltrated with EHA105virE2::GFP11 alone or together with ES1. Boxed areas are enlarged to highlight the nucleus. Projected Z-series images were obtained 2 days after agroinfiltration. Scale bars, 20 μm. (D) The intensity of VirE2-GFPcomp signals was measured within each host nucleus. n, number of nuclei measured. (E) ES1 or tyrphostin A23 treatment attenuated tumorigenesis on Arabidopsis roots. Chemical-treated Arabidopsis roots were cocultivated with Agrobacterium strain A348 for tumorigenesis. (F) Quantification of the frequency of tumor formation, which is given as a percentage above each column (with ±SE). The quantification was based on the number of tumors formed on 120 root segments. **P < 0.01 (unpaired Student’s t test).
To confirm that host endocytosis is important for VirE2 trafficking, the chemical inhibitor endosidin1 (ES1) was used to interfere with the endocytosis process, because ES1 affects the endocytosis pathway and causes aggregation of early endosomes in Arabidopsis thaliana (46). The marker SYP61-mCherry was transiently expressed to label the highly dynamic round-shaped early endosomes (46, 47) in N. benthamiana epidermal cells. ES1 treatment caused abnormal aggregation of SYP61-mCherry (fig. S7A), indicating aggregation of early endosomes in leaf epidermal cells. Moreover, ES1 treatment caused abnormal VirE2 trafficking in the host cytoplasm; VirE2 accumulated inside the ES1-induced endosome aggregates (fig. S7A and movie S2). Estimation of the colocalization through Pearson’s correlation coefficient suggested VirE2 colocalization with SYP61-mCherry (fig. S7C), indicating VirE2 colocalization with the early endosomes. These results indicate that ES1 can interfere with host endocytosis and thus restrict VirE2 movement.
Early endosomes mainly function as the sorting hub for endocytic trafficking processes in plants; cargoes internalized from the plasma membrane are usually transported to late endosomes and vacuoles for degradation (48). To test whether VirE2 is trafficked to late endosomes, ARA6-DsRed (49, 50) was transiently expressed to label the late endosomes in N. benthamiana epidermal cells. We did not observe any obvious association of VirE2 with the ARA6-DsRed–labeled late endosome structures (fig. S7, B and C), suggesting that VirE2 may escape from early endosomes and move to other locations to avoid degradation in the vacuoles.
The effect of ES1 on nuclear targeting of VirE2 was then tested, as it was shown previously that Agrobacterium-delivered VirE2 was efficiently targeted to the plant nucleus in an NLS-dependent manner (37). As shown in Fig. 2 (C and D), ES1 treatment markedly decreased the nuclear accumulation of VirE2 inside tobacco cells, whereas VirE2 accumulated at the cell borders or inside the cytoplasm. This indicates that ES1 affects VirE2 trafficking rather than the delivery or oligomerization of VirE2. These findings suggest that host endocytosis plays an important role in the cytoplasmic trafficking and subsequent nuclear targeting of VirE2 inside plant cells. To confirm the importance of endocytosis, we studied the effects of chemical inhibitors on AMT, because functional VirE2 is required for the transformation process. Tumorigenesis assays were conducted using A. thaliana roots treated with either ES1 or tyrphostin A23, which is also a CME inhibitor for A. thaliana (51). As shown in Fig. 2 (E and F), treatment with ES1 or tyrphostin A23 significantly attenuated tumorigenesis. These results suggest that interference with host endocytosis can attenuate the stable transformation of plant cells, presumably because blocking endocytosis affects VirE2 movement and thus its role in AMT.
Dual endocytic motifs at the C-terminal tail of VirE2 are required for VirE2 trafficking
Subsequently, we investigated how VirE2 was selected as a cargo for internalization. In general, selection of plasma membrane–associated cargo proteins for internalization depends on the recognition of endocytic signals at the cytosolic side of cargo proteins by a variety of host adaptors (52, 53). Upon delivery into host plant cells through the T4SS, VirE2 might interact with a host adaptor protein at the plasma membrane.
Sequence analysis indicated that VirE2 (accession no. AAZ50538) contained five putative endocytic sorting motifs (Fig. 3A). To test the importance of these putative motifs for VirE2 trafficking, we mutated the potential critical leucine or tyrosine residue for each of the dileucine- or tyrosine-based motifs (54), respectively, to alanine. In addition, a double mutant was constructed for the two tyrosine-based motifs in the C terminus (Fig. 3A). Then, we examined the cellular localization and distribution of Agrobacterium-delivered VirE2 for each of the mutants. Neither single mutation nor double mutation of the dual C-terminal tyrosine-based motifs affected VirE2 delivery to the host cellular membrane (Fig. 3, B and C). However, the double mutation caused a significantly higher level of VirE2 accumulation at the membrane sites (Fig. 3C). Mutations at other putative endocytic motifs of VirE2 did not affect the VirE2 delivery or internalization (fig. S8). These results suggest that the putative dual C-terminal tyrosine-based motifs are important for VirE2 trafficking.
Fig. 3. Mutations at VirE2 endocytic motifs affected VirE2 internalization and impaired AMT.
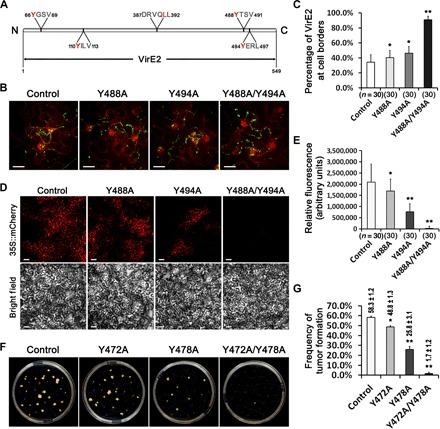
(A) Schematic locations of VirE2 putative endocytic motifs identified by the Eukaryotic Linear Motif resource for functional sites in proteins (www.elm.eu.org). The invariant amino acids of endocytic motifs are labeled in red. (B) Mutations at the dual endocytic motifs at the VirE2 C terminus affected VirE2 internalization into host cells. N. benthamiana (Nb308A) leaves were infiltrated with A. tumefaciens EHA105virE2::GFP11 or mutant strains containing the corresponding tyrosine substitutions with alanine. Projected Z-series images were obtained 2 days after agroinfiltration. Scale bars, 20 μm. (C) The percentage of VirE2 staying at the cell borders is represented as the intensity of VirE2-GFPcomp associated with host cell borders divided by the total intensity in each image. n, number of images measured. (D) Mutations at the dual endocytic motifs at VirE2 C terminus decreased transient transformation efficiency. Wild-type N. benthamiana leaves were infiltrated with A. tumefaciens EHA105 or mutant strains containing the binary vector pQH121-mC (with free mCherry under the control of CaMV 35S promoter in T-DNA). Projected Z-series images were obtained 2 days after agroinfiltration under confocal microscopy with an Olympus UPL SAPO 10× numerical aperture (NA) 0.40 objective. Single optical sections of bright field indicate the cell shapes (lower panel). Scale bars, 50 μm. (E) The intensity of transiently expressed mCherry was measured in each image. n, number of images measured. (F) Mutations at the dual endocytic motifs at the VirE2 C terminus decreased stable transformation efficiency. A. tumefaciens A348 and mutant strains were used for tumor formation assay. (G) Quantification of the frequency of tumor formation, which is given as a percentage above each column (with ±SE). The quantification was based on the number of tumors formed on 120 root segments. *P < 0.05, **P < 0.01 (unpaired Student’s t test).
Dual endocytic motifs at the C-terminal tail of VirE2 are important for AMT
To determine whether the dual C-terminal tyrosine-based motifs are required for VirE2 function, we conducted assays for transient expression of mCherry under the control of a CaMV 35S promoter on T-DNA. The mCherry expression was analyzed on the basis of the fluorescence intensity. As shown in Fig. 3 (D and E), both single and double mutations at the dual C-terminal endocytic signals significantly decreased the transient AMT efficiency, although the double mutation (Y488A/Y494A) had a more marked effect than the single mutation Y494A, which affected the function more than Y488A. These results suggest that dual C-terminal endocytic signals are required for VirE2 function, and the last endocytic signal at the VirE2 C terminus is more important.
Sequence alignment analysis indicated that the dual C-terminal tyrosine-based endocytic motifs were conserved on VirE2 proteins from different Ti plasmids, suggesting their conserved roles in different Agrobacterium strains (fig. S9). Moreover, mutations at these conserved motifs on VirE2 from the virulent strain A348 also attenuated tumor formation on Arabidopsis root fragments (Fig. 3, F and G). Our results demonstrate that the dual tyrosine–based endocytic signals located at the VirE2 C terminus are important for VirE2 function for both transient and stable AMT processes.
Endocytic motifs at VirE2 C terminus interact with plant AP2M
As CME is facilitated by a group of host adaptors known as “clathrin-associated sorting proteins,” which are responsible for endocytic signal recognition and cargo binding (52, 53), the above results led us to hypothesize that the dual C-terminal tyrosine-based endocytic motifs of VirE2 might be recognized by such a protein. Among them, the adaptor protein 2 (AP-2) complex recognizes the tyrosine-based endocytic signal and binds to it through the C-terminal domain of the μ subunit (AP2M) (55).
To test a potential interaction of VirE2 with the AP-2 complex, we conducted an in vitro pull-down assay with their fusion proteins. As shown in Fig. 4A, when the C-terminal tail was fused to glutathione S-transferase (GST) (GST-VirE2C), VirE2 interacted with the cargo-binding domain of AP2M that was fused to maltose binding protein (MBP) (MBP-AP2MC). However, a double mutation at the dual tyrosine–based endocytic signals eliminated the interaction (Fig. 4B). These results suggest that AP2M recognizes and binds to the VirE2 C-terminal tail through the dual tyrosine–based sorting motifs.
Fig. 4. Endocytic motifs at VirE2 C terminus interacted with AP2M, and ap2m mutations attenuated tumorigenesis.
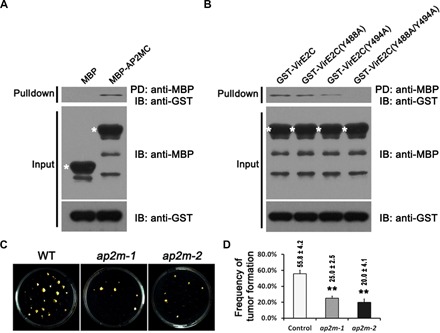
(A) VirE2 C-terminal tail interacted with AP2M. VirE2 C-terminal tail fused onto GST (GST-VirE2C) was used to conduct an in vitro pull-down assay, with AP2M cargo-binding domain fused onto MBP (MBP-AP2MC). The pull-down (upper panel) and 20% of input (lower panels) fractions were analyzed by Western blot. Free MBP and MBP-AP2MC fusion protein are asterisked. (B) Double mutation at the dual endocytic motifs eliminated the interaction between the VirE2 C-terminal tail and AP2M cargo-binding domain. The pull-down (upper panel) and 20% of input (lower panels) fractions were analyzed by Western blot. MBP-AP2MC fusion is asterisked. (C) A. thaliana ap2m-1 and ap2m-2 mutations attenuated tumorigenesis in the root transformation assay. (D) Quantification of the frequency of tumor formation, which is given as a percentage above each column (with ±SE). The quantification was based on the number of tumors formed on 120 root segments. **P < 0.01 (unpaired Student’s t test). PD, pulled down; IB, immunoblot.
To further confirm the importance of host AP-2 complex in the AMT process, we tested tumorigenesis for two AP2M insertional mutants of A. thaliana that have been previously characterized and shown to display defects in endocytosis (56). As shown in Fig. 4 (C and D), both mutants displayed significantly attenuated tumor formation compared to the wild-type control. These results demonstrate that the host AP-2 complex is required for AMT of plant cells.
DISCUSSION
As a natural genetic engineer, A. tumefaciens can cause crown gall disease on an exceptionally wide range of host plants in nature (57). The bacterium can achieve a high efficiency of transformation of up to 100% (37). Here, we show that Agrobacterium uses host endocytic processes to facilitate the delivery and trafficking of one of its virulence factors, VirE2, into host cells. Endocytosis is a well-conserved fundamental process in all eukaryotic cells independent of cell type or differentiation. This may help A. tumefaciens to target a wide range of host cells and achieve a high efficiency of transformation.
We have adopted a split-GFP strategy to monitor Agrobacterium-delivered VirE2 protein in plant cells in real time (37). We observed a close association of Agrobacterium-delivered VirE2 with the host plasma membrane. The VirE2-membrane associations usually appeared at the sites of the bacterial attachment to the plant, suggesting that the associations occurred immediately after VirE2 translocation into plant cells. Previous studies have demonstrated that VirE2 and T-DNA might be translocated into recipient cells separately and then a VirE2-T-DNA coating process likely occurred inside the host cytoplasm (18, 58). Although it is not clear how this occurs, T-DNA and effector proteins presumably share the same T4SS channel for translocation to ensure efficient T-strand coating and protection. This is supported by our observation that VirE2 was initially associated with the plant membrane at the entry point.
The VirE2-membrane association occurred for both T-DNA–containing and T-DNA–free cells, suggesting that it might be due to the affinity of VirE2 with membranes, as demonstrated by previous studies (29). Moreover, in vitro studies have demonstrated that VirE2 may form pores on the plasma membrane (29). It is not clear whether VirE2 formed pores on the host cellular membrane upon its delivery into host cells.
The associations of VirE2 on host plasma membranes usually appeared in pairs. By using red fluorescent protein (RFP)–labeled Agrobacterium cells, we showed that single bacterial cells can laterally translocate VirE2 into two neighboring plant cells (Fig. 1B). Presumably, because of the space limitation, only the lateral sides of a bacterial cell could attach to the two adjacent tobacco cells. This is consistent with the previous observation that the T4SS might form helical arrays around the induced bacterial cells (59).
We hypothesize that VirE2 forms self-aggregated filamentous structures on host plasma membranes upon its delivery into host cells. This may occur independently of the presence or absence of T-DNA. At this stage, it is not clear whether a VirE2 aggregate containing T-DNA is structurally different from a VirE2 aggregate without any T-DNA. Nevertheless, we assume that both types of VirE2 aggregates can depart from the membranes via endocytosis. If T-DNA is present inside a VirE2 aggregate, then the VirE2 molecules in the aggregate should be able to protect the T-DNA from nucleolytic degradation during subsequent trafficking toward the nucleus.
Endocytosis is a widely conserved process in eukaryotic cells that is responsible for the uptake of a variety of molecules from the outside environment. It participates in a great number of cellular functions, including nutrient uptake, signaling transduction, antigen detection, and cell differentiation (60). Although it is known that endocytosis is involved in viral entry into host cells (61), it is not clear whether endocytosis is needed for cellular entry of a virulence protein injected by a bacterial secretion apparatus. Our data demonstrate that Agrobacterium hijacked the host CME pathway for VirE2 internalization into host cells. CME has been shown to be the major pathway in endocytosis (42), and it also plays important roles in antigen perception and initiation of plant defense responses upon pathogen infection (62). It would be of significance to determine whether Agrobacterium might hijack CME-related processes in plant defenses to facilitate infection.
As a highly selective process, recognition and binding of cargo proteins in CME are initiated by clathrin adaptors on the cytoplasmic side of a cell. It is not clear how VirE2 molecules reach the cytoplasmic side of the host cells so that VirE2 can interact with clathrin adaptors. One possibility is that the A. tumefaciens T4SS can deliver VirE2 directly to the cytoplasmic side of host cells. Alternatively, VirE2 may be translocated via additional bacterial or host factor(s) to the cytoplasmic side of host cells. It would be of interest to address this in future studies.
With the help of subsequently recruited accessory proteins, the membrane-associated protein cargoes are internalized to form clathrin-coated vesicles and are then transported to other parts of the cell (52, 53). The AP-2 adaptor complex is composed of two large subunits (α and β2), one medium-sized subunit (μ2), and one small subunit (σ2); it specifically recognizes and binds to the tyrosine-based (YXXØ) and dileucine-based ([DE]XXXL[LI]) sorting motifs on cargo proteins (54). Our results demonstrate that two closely located tyrosine-based motifs on the VirE2 C terminus were responsible for VirE2 interaction with the cargo-binding domain of the μ subunit of the AP-2 adaptor complex. Mutation of the signal sequences decreased VirE2 internalization and impaired the transformation, suggesting that the dual tyrosine–based motifs are important for VirE2 trafficking and function during the AMT process. Mutation of the second signal sequence exhibited a more severe effect on VirE2 function compared with mutation of the first one, suggesting that the second motif might play a more important role in the transformation process. These two motifs might differ in their binding affinity and accessibility to the AP-2 complex.
Previous studies have shown that the AP-2 complex is responsible for cargo transportation between the plasma membrane and early endosomes in plant cells (42, 63). Our data suggest that interaction between VirE2 and the AP-2 complex can facilitate VirE2 internalization from the host plasma membrane. However, the root transformation assays demonstrated that mutations at the critical dual motifs decreased the efficiency of transformation by only 30-fold (Fig. 3G), whereas VirE2 deletion mutants generated virtually no transformation (37). This suggests that VirE2 may be trafficked via an alternative pathway for its role in the transformation process.
We observed that the speed of VirE2 movement together with endomembrane compartments ranged from 0.4 to 2.1 μm/s (fig. S4B and movie S1), which is consistent with our previous studies (37). After VirE2 departs the host membranes via endomembrane compartments, it will be trafficked to subsequent locations. Early endosomes mainly function as the sorting hub for both secretory and endocytic trafficking processes in plants. Cargoes internalized from the plasma membrane are usually transported to late endosomes and vacuoles for degradation or recycling back to the cellular membrane (48). It has been reported that the host endocytic pathway is abused by different pathogens and viruses to facilitate infection (64, 65). Pathogen effectors and viruses have evolved a variety of approaches to escape from host endosomes after internalization (66, 67). To reach the host nucleus, VirE2 should also escape from endosomes to avoid degradation in the host cytoplasm. This is supported by our observation that VirE2 was not associated with late endosomes (fig. S7). Thus, further studies are needed to investigate how VirE2 escapes from endosomes and moves to other parts of host cells.
Our results show that the VirE2 protein efficiently targets the host nucleus in a time-dependent manner. However, a large portion of VirE2 remains in the plant cytoplasm. Presumably, VirE2 is delivered in excess to ensure that the T-strand is well protected from degradation. Alternatively, VirE2 may have the ability to traffic to different locations for other purposes. This remains to be elucidated in future experiments.
MATERIALS AND METHODS
Experimental design
The objective of this report is to study how Agrobacterium delivers the virulence factor VirE2 into plant cells and how Agrobacterium-delivered VirE2 is trafficked inside plant cells.
We adopted a split-GFP–based method that can directly detect the Agrobacterium-delivered VirE2 inside plant cells. This enabled us to monitor the VirE2 delivery process inside plant cells in real time and in a natural setting.
To visualize VirE2 delivery, the VirE2-GFP11 fusion was expressed in A. tumefaciens and GFP1–10 was expressed in plant cells. Upon delivery of VirE2-GFP11 into the plant cells, VirE2-GFPcomp fluorescence signals resulting from the complementation of VirE2-GFP11 and GFP1–10 were visualized. Biochemical, genetic, and bioimaging approaches were used to study the factors and processes that played a role in the delivery and trafficking process of Agrobacterium-delivered VirE2 inside plant cells.
Strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are listed in table S1. A. tumefaciens strains were grown at 28°C in Luria-Bertani (LB) medium supplemented with carbenicillin (100 μg ml−1) or kanamycin (50 μg ml−1) as necessary.
Plant materials
A. thaliana (ecotype Columbia-0) wild-type and mutant plants were used in the root transformation assay. The AP2M insertional mutants ap2m-1 (SALK_083693) and ap2m-2 (CS807972) were obtained from the Arabidopsis Biological Resource Center at Ohio State University. N. benthamiana wild-type and transgenic line Nb308A (expressing GFP1–10 and DsRed) were used in agroinfiltration assays (37).
Constructs
GFP1–10 construct
To construct a binary vector (pXY01) for target gene expression in plant cells, a primer set of 5′-CTAGTCTAGACCCGGGCTCGAGCCATGGGGATCCGAGCTCGAATTTCCCCGATCGTTCAAACATTTGGCAATAAAGTTT-3′ and 5′-CTAGTCTAGAGCTAGCTCCGGACTTAAGA-3′ was used to amplify the binary vector backbone from the plasmid er-gb (39) to generate a DNA fragment, in which the endoplasmic reticulum (ER) marker cassette was replaced with a multiple cloning site sequence; the polymerase chain reaction (PCR) product was then digested with Xba I and self-ligated to generate the binary vector pXY01. The GFP1–10 coding sequence was amplified from pQH308A (37) with primers 5′-CTAGTCTAGAATGGTTTCGAAAGGCGAGGA-3′ and 5′-CGCGGATCCTTATTTCTCGTTTGGGTCTTTGC-3′ and inserted into pXY01 to generate pGFP1–10.
Agrobacterium-labeling constructs
A primer set of 5′-ACGCGTCGACCTCGAGGGGGG-3′ and 5′-ACGCGTCGACTCTCAGTAAAGCGCTGGCTG-3′ was used to amplify the backbone from pCB301 (68); the PCR product was then digested with Sal I and self-ligated to generate pXY301, which lacks the T-DNA right border sequence. A virB promoter region was amplified from the plasmid pTiA6 with primers 5′-ACGCGTCGACATGGGTTTACAGACAGCGTAATCTC-3′ and 5′-ACCTTATCTCCTTAGCTCGCAAC-3′ and cloned into pXY301 to generate pVB. The DsRed coding sequence was amplified with primers 5′-CGGGGTACCATGGCCTCCTCCGAGGACG-3′ and 5′-CGGGGTACCTTACAGGAACAGGTGGTGGCG-3′ and cloned into pVB to generate pVB-RFP. The GFP coding sequence was amplified with primers 5′-CGGGGTACCATGTCTAAAGGTGAAGAATTATTCACTG-3′ and 5′-CGGGGTACCTTATTTGTACAATTCATCCATACCATG-3′ and cloned into pVB to generate pVB-GFP. An ampicillin resistance cassette was then amplified from pACT2 (Clontech) with primers 5′-ATGCAATCATGATTCAAATATGTATCCGCTCAAGAGA-3′ and 5′-ATGCAATCATGACTCACGTTAAGGGATTTTGGACAT-3′ and inserted into pVB-RFP to replace the kanamycin resistance cassette to generate pVBA-RFP.
Hub construct
The 1860–base pair DNA fragment encoding the C-terminal part of CHC1 (At3g11130) was amplified from a total Arabidopsis complementary DNA (cDNA) preparation with primers 5′-TCCCCCCGGGATGAAGAAGTTTAACTTAAATGTTCAGGCTG-3′ and 5′-CGCGGATCCTTAGTAGCCGCCCATCGGT-3′. The PCR fragment was cloned into the vector pXY01 to generate pXY01-Hub, in which the Hub was under the control of a CaMV 35S promoter.
SYP61 constructs
To label the early endosomes, the full-length genomic sequence of A. thaliana SYP61 (At1g28490) was amplified from A. thaliana genomic DNA with primers 5′-CTAGTCTAGAATGTCTTCAGCTCAAGATCCATTCT-3′ and 5′-CCGCTCGAGGGTCAAGAAGACAAGAACGAATAGG-3′ and cloned into the binary vector pXY01 so that SYP61 was under the control of a CaMV 35S promoter. The mCherry coding sequence was subsequently amplified with primers 5′-CCGCTCGAGGGAGGTGGCTCTGGCGGGGGATCAATGGTGAGCAAGGGCGAGGA-3′ and 5′-CGCGGATCCTTACTTGTACAGCTCGTCCATGCCG-3′; the PCR fragment was cloned at the C terminus to generate pXY01-SYP61-mC.
ARA6 constructs
To label late endosomes, the coding sequence of A. thaliana ARA6 (At3g54840) was amplified from a total Arabidopsis cDNA preparation with primers 5′-CTAGTCTAGAATGGGATGTGCTTCTTCTCTTCC-3′ and 5′-CTAGTCTAGATGACGAAGGAGCAGGACGAG-3′. The DNA fragment was cloned into the binary vector pXY01 so that ARA6 was under the control of a CaMV 35S promoter. The DsRed coding sequence was subsequently amplified with primers 5′-CCGCTCGAGGGAGGTTCAGGCGGAAGTGCCTCCTCCGAGGACGTCAT-3′ and 5′-CGCGGATCCTTACAGGAACAGGTGGTGGCG-3′. The PCR fragment was added to the C terminus of ARA6 to generate pXY01-ARA6-DsRed.
Transient mCherry expression constructs
The mCherry coding sequence was amplified with primers 5′-CCGCTCGAGATGGTGAGCAAGGGCGAGGA-3′ and 5′-CGGGGTACCTTACTTGTACAGCTCGTCCATGCCG-3′ and cloned into the binary vector pQH121 to generate pQH121-mC, in which the mCherry was under the control of a CaMV 35S promoter.
Pull-down constructs
The coding sequence of the 295–amino acid C-terminal cargo-binding domain of μ2 subunit (AP2M) was amplified from A. thaliana cDNA with primers 5′-CGCGGATCCTCACCATTTTCATCGAAGCCA-3′ and 5′-CTAGTCTAGATCAGCATCTGATCTCGTAAGATCCC-3′; the PCR fragment was cloned into the vector pMAL-c2x (New England Biolabs) to generate pMBP-AP2MC. The coding sequence of the 76–amino acid C-terminal tail of VirE2 was amplified from pTibo542(EHA105) with primers 5′-CGCGGATCCATCGTCGCCGATCGCAA-3′ and 5′-CCGCTCGAGTCAAAAGCTGTTGACGCTTTG-3′; the PCR fragment was cloned into the vector pGEX-4T-1 (GE Healthcare) to generate pGST-VirE2C.
Agroinfiltration
A. tumefaciens cells were grown overnight in LB medium; the cultures were then diluted 50 times in LB medium and grown for a further 5 to 6 hours. The bacteria were collected and resuspended in H2O to OD600 (optical density at 600 nm) = 1.0 unless otherwise specified. The bacterial suspension was infiltrated to the underside of fully expanded N. benthamiana leaves using a syringe. The infiltrated plants were then placed at 22°C under a 16-hour light/8-hour dark photoperiod.
Detection of Agrobacterium-delivered VirE2 in N. benthamiana
Agrobacterium-delivered VirE2 was detected using a split-GFP system, as previously described (37). The VirE2-GFP11 fusion was expressed inside bacteria using the tagged A. tumefaciens strain EHA105virE2::GFP11. GFP1–10 was expressed in plant cells either using a transgenic line Nb308A or by transient expression using A. tumefaciens strains containing the binary plasmid pGFP1–10.
Detection of plant plasma membranes and endosomes
Plasma membranes and endosomes were detected by transient expression with A. tumefaciens strains containing the binary plasmid pm-rb (39), which harbors T-DNA encoding a plasma membrane marker, or pXY01-SYP61-mC or pXY01-ARA6-DsRed, which harbors T-DNA encoding the endocytic marker SYP61 or ARA6, respectively.
FM4-64 staining
FM4-64 (Invitrogen) was infiltrated into the underside of N. benthamiana leaves at a 25 μM concentration in distilled water. Images were taken 1 hour after infiltration.
Transient transformation assay
A. tumefaciens EHA105 or mutant strains containing pQH121-mC, which harbors T-DNA encoding the mCherry, were infiltrated into wild-type N. benthamiana leaves in a low concentration (OD600 = 0.005). Images were obtained 2 days after agroinfiltration and used for intensity calculation.
Stable transformation assay
A. thaliana wild-type or mutant seeds (Col-0) were surface-sterilized using 15% bleach solution and incubated at 4°C for 2 days. The seeds were then placed onto solidified 1/2× Murashige and Skoog (MS) medium [supplemented with 1% sucrose and MES (0.5 g liter−1), pH 5.8] and incubated under a 16-hour light/8-hour dark photoperiod at 22°C for 10 to 12 days. Roots from individual seedlings were cut into 3- to 5-mm segments and mixed with 1 ml of A. tumefaciens cells (A348 or mutant) at a concentration of 1 × 108 cells/ml and spread onto a solidified 1/2× MS plate. The plates were subsequently incubated at 22°C for 36 hours. The root segments were aligned onto 1/2× MS medium plates (40 root segments for each plate; three plates for each treatment) containing cefotaxime (100 μg ml−1) and kept at 22°C for 5 to 6 weeks.
Chemical treatment
A. tumefaciens cells were grown, and the cell concentrations were adjusted to OD600 = 0.5 in H2O; ES1 was added into the cell suspensions at a final concentration of 25 μM. The mixtures were then infiltrated into N. benthamiana leaves. As the control, A. tumefaciens cell suspensions in H2O were infiltrated into N. benthamiana leaves. The infiltrated plants were then placed at 22°C under a 16-hour light/8-hour dark photoperiod.
During a stable transformation assay, A. thaliana roots from individual seedlings were cut into 3- to 5-mm segments and mixed with ES1 or tyrphostin A23 (Sigma) at a final concentration of 60 or 50 μM, respectively, in H2O, and the mixtures were then kept in the dark for 3 hours. As the control, the root fragments were treated with H2O alone. The root fragments were then mixed with A. tumefaciens for root transformation assays, as described above.
In vitro pull-down assay
Fusion proteins were produced using BL21(DE3) Escherichia coli strain. Single colonies of cells were inoculated into LB broth to grow overnight at 37°C. The cell cultures were then diluted into fresh LB broth at OD600 = 0.1 and grown at 37°C for another 1.5 hours until OD600 = 0.6. Expression of the fusion proteins was then induced at 28°C for 6 hours with isopropyl-β-d-thiogalactopyranoside at a final concentration of 1 mM.
Bacterial cells were harvested by centrifugation at 5000g at 4°C for 5 min and washed once with pull-down lysis buffer (50 mM tris-HCl, 50 mM NaCl, pH 7.5). The cells were then resuspended in pull-down lysis buffer containing protease inhibitor cocktail (Nacalai Tesque) and were subjected to brief sonication (12 bursts of 20 s with 40% power). Cell debris was removed by centrifugation at 12,000g at 4°C for 15 min.
The supernatant of bait proteins (MBP or MBP-tagged proteins) was incubated with 150 μl of amylose resin (New England Biolabs) on a rotator at 4°C for 3 hours. The column was then washed five times with the pull-down wash buffer (50 mM tris-HCl, 50 mM NaCl, 0.5% Triton X-100, pH 7.5).
The supernatant of the prey proteins (GST-tagged proteins) was added to the column containing the immobilized MBP-tagged bait protein and incubated on a rotator at 4°C overnight. The column was then washed five times with the pull-down wash buffer, and captured proteins were eluted with the pull-down lysis buffer containing 10 mM maltose. MBP-tagged proteins were detected by immunoblotting with anti-MBP antibody (sc-809, Santa Cruz Biotechnology), and GST-tagged proteins were detected by immunoblotting with anti-GST antibody (sc-459, Santa Cruz Biotechnology).
Confocal microscopy
A PerkinElmer UltraView Vox Spinning Disk system with electron-multiplying charge-coupled device cameras was used for confocal microscopy. Agroinfiltrated N. benthamiana leaves were observed 2 days after agroinfiltration unless otherwise specified. To observe the leaf epidermis, agroinfiltrated leaf tissues were detached from N. benthamiana plants and immersed in H2O on a glass slide with a coverslip. All images were processed by Volocity 3D Image Analysis Software 6.2.1. All images were obtained under a confocal microscope with an Olympus UPLSAPO 60× NA 1.20 water-immersion objective unless otherwise specified.
Quantification of fluorescence intensity
Fluorescence intensity was measured using ImageJ (http://rsbweb.nih.gov/ij/).
Statistical analysis
Quantitative data are presented as means ± SE from at least three independent experiments. When appropriate, statistical differences between groups were analyzed using an unpaired Student’s t test. Differences were considered significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank G. Hicks (University of California, Riverside) for providing the chemical inhibitor ES1 and acknowledge the technical assistance of Y. Tong, T. Yan, and S. Xu. Funding: This work was supported by grants from the Singapore Ministry of Education (R-154-000-588-112 and R-154-000-685-112) and the Singapore Millennium Foundation (R-154-000-586-592). Author contributions: X.L. designed and conducted the experiments and wrote the paper. S.Q.P. designed the experiments and wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/3/e1601528/DC1
fig. S1. Time-course study of VirE2 subcellular localizations inside N. benthamiana cells.
fig. S2. Accumulation of Agrobacterium at the intercellular space of infiltrated leaf epidermal cells.
fig. S3. Accumulation of Agrobacterium-delivered VirE2 at the host plasma membrane.
fig. S4. Colocalization of VirE2 with FM4-64–labeled endomembrane compartments in N. benthamiana epidermal cells.
fig. S5. Expression of Hub impaired FM4-64 uptake in N. benthamiana epidermal cells.
fig. S6. Expression of dominant-negative clathrin Hub impaired VirE2 departure from the plasma membrane in N. benthamiana epidermal cells.
fig. S7. Agrobacterium-delivered VirE2 was associated with early endosomes rather than late endosomes.
fig. S8. Mutations at other putative VirE2 endocytic sorting motifs did not affect VirE2 internalization in host cells.
fig. S9. Sequence alignment analysis of VirE2 from different types of Ti plasmids.
table S1. Strains and plasmids used in the studies.
movie S1. Comovement of VirE2 with FM4-64–labeled vesicles.
movie S2. Restriction of VirE2 trafficking in ES1-induced aggregation of SYP61-containing endosomes.
REFERENCES AND NOTES
- 1.Chilton M.-D., Drummond M. H., Merio D. J., Sciaky D., Montoya A. L., Gordon M. P., Nester E. W., Stable incorporation of plasmid DNA into higher plant cells: The molecular basis of crown gall tumorigenesis. Cell 11, 263–271 (1977). [DOI] [PubMed] [Google Scholar]
- 2.Zambryski P., Holsters M., Kruger K., Depicker A., Schell J., Van Montagu M., Goodman H. M., Tumor DNA structure in plant cells transformed by A. tumefaciens. Science 209, 1385–1391 (1980). [DOI] [PubMed] [Google Scholar]
- 3.Albright L. M., Yanofsky M. F., Leroux B., Ma D. Q., Nester E. W., Processing of the T-DNA of Agrobacterium tumefaciens generates border nicks and linear, single-stranded T-DNA. J. Bacteriol. 169, 1046–1055 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bundock P., den Dulk-Ras A., Beijersbergen A., Hooykaas P. J., Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens to Saccharomyces cerevisiae. EMBO J. 14, 3206–3214 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piers K. L., Heath J. D., Liang X., Stephens K. M., Nester E. W., Agrobacterium tumefaciens-mediated transformation of yeast. Proc. Natl. Acad. Sci. U.S.A. 93, 1613–1618 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Groot M. J. A., Bundock P., Hooykaas P. J. J., Beijersbergen A. G. M., Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat. Biotechnol. 16, 839–842 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Kathiresan S., Chandrashekar A., Ravishankar G. A., Sarada R., Agrobacterium-mediated transformation in the green alga Haematococcus pluvialis (Chlorophyceae, Volvocales). J. Phycol. 45, 642–649 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Kunik T., Tzfira T., Kapulnik Y., Gafni Y., Dingwall C., Citovsky V., Genetic transformation of HeLa cells by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 98, 1871–1876 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tzfira T., Citovsky V., Agrobacterium-mediated genetic transformation of plants: Biology and biotechnology. Curr. Opin. Biotechnol. 17, 147–154 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Michielse C. B., Hooykaas P. J. J., van den Hondel C. A. M. J. J, Ram A. F., Agrobacterium-mediated transformation as a tool for functional genomics in fungi. Curr. Genet. 48, 1–17 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Fullner K. J., Lara J. C., Nester E. W., Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273, 1107–1109 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Cascales E., Christie P. J., Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304, 1170–1173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beijersbergen A., Dulk-Ras A. D., Schilperoort R. A., Hooykaas P. J. J., Conjugative transfer by the virulence system of Agrobacterium tumefaciens. Science 256, 1324–1327 (1992). [DOI] [PubMed] [Google Scholar]
- 14.Low H. H., Gubellini F., Rivera-Calzada A., Braun N., Connery S., Dujeancourt A., Lu F., Redzej A., Fronzes R., Orlova E. V., Waksman G., Structure of a type IV secretion system. Nature 508, 550–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cascales E., Christie P. J., The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1, 137–149 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chandran Darbari V., Waksman G., Structural biology of bacterial type IV secretion systems. Annu. Rev. Biochem. 84, 603–629 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Christie P. J., Atmakuri K., Krishnamoorthy V., Jakubowski S., Cascales E., Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59, 451–485 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vergunst A. C., Schrammeijer B., den Dulk-Ras A., de Vlaam C. M. T., Regensburg-Tuink T. J. G., Hooykaas P. J. J., VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290, 979–982 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Vergunst A. C., van Lier M. C. M., den Dulk-Ras A., Stuve T. A. G., Ouwehand A., Hooykaas P. J. J., Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 832–837 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schrammeijer B., den Dulk-Ras A., Vergunst A. C., Jurado Jácome E., Hooykaas P. J. J., Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: Evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 31, 860–868 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelvin S. B., Plant proteins involved in Agrobacterium-mediated genetic transformation. Annu. Rev. Phytopathol. 48, 45–68 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Wang K., Herrera-Estrella L., Van Montagu M., Zambryski P., Right 25 bp terminus sequence of the nopaline T-DNA is essential for and determines direction of DNA transfer from Agrobacterium to the plant genome. Cell 38, 455–462 (1984). [DOI] [PubMed] [Google Scholar]
- 23.Yanofsky M. F., Porter S. G., Young C., Albright L. M., Gordon M. P., Nester E. W., The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell 47, 471–477 (1986). [DOI] [PubMed] [Google Scholar]
- 24.Scheiffele P., Pansegrau W., Lanka E., Initiation of Agrobacterium tumefaciens T-DNA processing. Purified proteins VirD1 and VirD2 catalyze site- and strand-specific cleavage of superhelical T-border DNA in vitro. J. Biol. Chem. 270, 1269–1276 (1995). [DOI] [PubMed] [Google Scholar]
- 25.Christie P. J., Ward J. E., Winans S. C., Nester E. W., The Agrobacterium tumefaciens virE2 gene product is a single-stranded-DNA-binding protein that associates with T-DNA. J. Bacteriol. 170, 2659–2667 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Citovsky V., Wong M. L., Zambryski P., Cooperative interaction of Agrobacterium VirE2 protein with single-stranded DNA: Implications for the T-DNA transfer process. Proc. Natl. Acad. Sci. U.S.A. 86, 1193–1197 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen P., Pazour G. J., Anderson D., Das A., Cooperative binding of Agrobacterium tumefaciens VirE2 protein to single-stranded DNA. J. Bacteriol. 171, 2573–2580 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharjee S., Lee L.-Y., Oltmanns H., Cao H., Veena, Cuperus J., Gelvin S. B., IMPa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell 20, 2661–2680 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dumas F., Duckely M., Pelczar P., Van Gelder P., Hohn B., An Agrobacterium VirE2 channel for transferred-DNA transport into plant cells. Proc. Natl. Acad. Sci. U.S.A. 98, 485–490 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rossi L., Hohn B., Tinland B., Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U.S.A. 93, 126–130 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yusibov V. M., Steck T. R., Gupta V., Gelvin S. B., Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. U.S.A. 91, 2994–2998 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tzfira T., Vaidya M., Citovsky V., VIP1, an Arabidopsis protein that interacts with Agrobacterium VirE2, is involved in VirE2 nuclear import and Agrobacterium infectivity. EMBO J. 20, 3596–3607 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Djamei A., Pitzschke A., Nakagami H., Rajh I., Hirt H., Trojan horse strategy in Agrobacterium transformation: Abusing MAPK defense signaling. Science 318, 453–456 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Li J., Krichevsky A., Vaidya M., Tzfira T., Citovsky V., Uncoupling of the functions of the Arabidopsis VIP1 protein in transient and stable plant genetic transformation by Agrobacterium. Proc. Natl. Acad. Sci. U.S.A. 102, 5733–5738 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand A., Krichevsky A., Schornack S., Lahaye T., Tzfira T., Tang Y., Citovsky V., Mysore K. S., Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants. Plant Cell 19, 1695–1708 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cabantous S., Terwilliger T. C., Waldo G. S., Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Li X., Yang Q., Tu H., Lim Z., Pan S. Q., Direct visualization of Agrobacterium-delivered VirE2 in recipient cells. Plant J. 77, 487–495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakalis P. A., van Heusden G. P. H., Hooykaas P. J. J., Visualization of VirE2 protein translocation by the Agrobacterium type IV secretion system into host cells. Microbiologyopen 3, 104–117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson B. K., Cai X., Nebenführ A., A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51, 1126–1136 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G., The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219–230 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Maizel A., von Wangenheim D., Federici F., Haseloff J., Stelzer E. H. K., High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J. 68, 377–385 (2011). [DOI] [PubMed] [Google Scholar]
- 42.McMahon H. T., Boucrot E., Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 12, 517–533 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Liu S.-H., Wong M. L., Craik C. S., Brodsky F. M., Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell 83, 257–267 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J., Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23, 1920–1931 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dhonukshe P., Aniento F., Hwang I., Robinson D. G., Mravec J., Stierhof Y.-D., Friml J., Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr. Biol. 17, 520–527 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Robert S., Chary S. N., Drakakaki G., Li S., Yang Z., Raikhel N. V., Hicks G. R., Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. U.S.A. 105, 8464–8469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foresti O., Denecke J., Intermediate organelles of the plant secretory pathway: Identity and function. Traffic 9, 1599–1612 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Contento A. L., Bassham D. C., Structure and function of endosomes in plant cells. J. Cell Sci. 125, 3511–3518 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Ueda T., Uemura T., Sato M. H., Nakano A., Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40, 783–789 (2004). [DOI] [PubMed] [Google Scholar]
- 50.Ebine K., Fujimoto M., Okatani Y., Nishiyama T., Goh T., Ito E., Dainobu T., Nishitani A., Uemura T., Sato M. H., Thordal-Christensen H., Tsutsumi N., Nakano A., Ueda T., A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat. Cell Biol. 13, 853–859 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Banbury D. N., Oakley J. D., Sessions R. B., Banting G., Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 278, 12022–12028 (2003). [DOI] [PubMed] [Google Scholar]
- 52.Bonifacino J. S., Traub L. M., Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Traub L. M., Tickets to ride: Selecting cargo for clathrin-regulated internalization. Nat. Rev. Mol. Cell Biol. 10, 583–596 (2009). [DOI] [PubMed] [Google Scholar]
- 54.Traub L. M., Bonifacino J. S., Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 5, a016790 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jackson L. P., Kelly B. T., McCoy A. J., Gaffry T., James L. C., Collins B. M., Höning S., Evans P. R., Owen D. J., A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 141, 1220–1229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S. Y., Xu Z.-Y., Song K., Kim D. H., Kang H., Reichardt I., Sohn E. J., Friml J., Juergens G., Hwang I., Adaptor protein complex 2–mediated endocytosis is crucial for male reproductive organ development in Arabidopsis. Plant Cell 25, 2970–2985 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Cleene M., De Ley J., The host range of crown gall. Bot. Rev. 42, 389–466 (1976). [Google Scholar]
- 58.Otten L., De Greve H., Leemans J., Hain R., Hooykaas P., Schell J., Restoration of virulence of Vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol. Gen. Genet. 195, 159–163 (1984). [Google Scholar]
- 59.Aguilar J., Zupan J., Cameron T. A., Zambryski P. C., Agrobacterium type IV secretion system and its substrates form helical arrays around the circumference of virulence-induced cells. Proc. Natl. Acad. Sci. U.S.A. 107, 3758–3763 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu L.-G., Hamid E., Shin W., Chiang H.-C., Exocytosis and endocytosis: Modes, functions, and coupling mechanisms. Annu. Rev. Physiol. 76, 301–331 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mercer J., Schelhaas M., Helenius A., Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833 (2010). [DOI] [PubMed] [Google Scholar]
- 62.Bar M., Avni A., Endosomal trafficking and signaling in plant defense responses. Curr. Opin. Plant Biol. 22, 86–92 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Chen X., Irani N. G., Friml J., Clathrin-mediated endocytosis: The gateway into plant cells. Curr. Opin. Plant Biol. 14, 674–682 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Gruenberg J., van der Goot F. G., Mechanisms of pathogen entry through the endosomal compartments. Nat. Rev. Mol. Cell Biol. 7, 495–504 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Cossart P., Helenius A., Endocytosis of viruses and bacteria. Cold Spring Harb. Perspect. Biol. 6, a016972 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spooner R. A., Smith D. C., Easton A. J., Roberts L. M., Lord J. M., Retrograde transport pathways utilised by viruses and protein toxins. Virol. J. 3, 26 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Personnic N., Bärlocher K., Finsel I., Hilbi H., Subversion of retrograde trafficking by translocated pathogen effectors. Trends Microbiol. 24, 450–462 (2016). [DOI] [PubMed] [Google Scholar]
- 68.Xiang C., Han P., Lutziger I., Wang K., Oliver D. J., A mini binary vector series for plant transformation. Plant Mol. Biol. 40, 711–717 (1999). [DOI] [PubMed] [Google Scholar]
- 69.Hood E. E., Gelvin S. B., Melchers L. S., Hoekema A., New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218 (1993). [Google Scholar]
- 70.Knauf V. C., Nester E. W., Wide host range cloning vectors: A cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8, 45–54 (1982). [DOI] [PubMed] [Google Scholar]
- 71.Chen P.-Y., Wang C.-K., Soong S.-C., To K.-Y., Complete sequence of the binary vector pBI121 and its application in cloning T-DNA insertion from transgenic plants. Mol. Breeding 11, 287–293 (2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/3/e1601528/DC1
fig. S1. Time-course study of VirE2 subcellular localizations inside N. benthamiana cells.
fig. S2. Accumulation of Agrobacterium at the intercellular space of infiltrated leaf epidermal cells.
fig. S3. Accumulation of Agrobacterium-delivered VirE2 at the host plasma membrane.
fig. S4. Colocalization of VirE2 with FM4-64–labeled endomembrane compartments in N. benthamiana epidermal cells.
fig. S5. Expression of Hub impaired FM4-64 uptake in N. benthamiana epidermal cells.
fig. S6. Expression of dominant-negative clathrin Hub impaired VirE2 departure from the plasma membrane in N. benthamiana epidermal cells.
fig. S7. Agrobacterium-delivered VirE2 was associated with early endosomes rather than late endosomes.
fig. S8. Mutations at other putative VirE2 endocytic sorting motifs did not affect VirE2 internalization in host cells.
fig. S9. Sequence alignment analysis of VirE2 from different types of Ti plasmids.
table S1. Strains and plasmids used in the studies.
movie S1. Comovement of VirE2 with FM4-64–labeled vesicles.
movie S2. Restriction of VirE2 trafficking in ES1-induced aggregation of SYP61-containing endosomes.


